Abstract
An imbalance between energy uptake and energy expenditure can lead to obesity and increase the risk of coronary heart disease, high blood pressure, stroke, type II diabetes and some cancers. Given that key elements of the energy pathway are evolutionary conserved, invertebrate research is an attractive alternative that overcomes the many legislative, financial and experimental hurdles typical of research with higher metazoan animals. Recent studies have suggested that some members of the cytochrome P450 superfamily are involved in lipid metabolism in addition to the traditional xenobiotic activity. To investigate this notion in more detail, the present study aimed to pinpoint phenotypic, genetic and genomic-level responses of Caenorhabditis elegans using selected deletion mutants including fat-5 (a member of the Δ9 desaturases) and cyp-35A2 (a member of the cytochrome P450 family). The creation of a fat-5(tm420);cyp-35A2(gk317) mutant uncovered that the deletion of both genes resulted in a strain which is marked by an extended lifespan. Furthermore, it diminished the overall level of Nile Red positive compartments, which is indicative of a change in lipid metabolism. Comprehensive transcriptomics revealed that several genes involved in aging and lipid transport/homeostasis were modulated following the double deletion of fat-5 and cyp-35A2. Taken together, the results suggest the presence of a putative correlation between longevity and lipid regulation and given that both genes have human homologs, this finding may offer a new lead to investigate in higher organisms.
Keywords: C. elegans, Nematode, Cytochrome P450, Fatty acid desaturase, Life-span
Highlights
-
•
C. elegans is a premier invertebrate model in biomedical research.
-
•
Some cytochrome P450s are involved in maintaining a healthy fat composition.
-
•
The desaturase fat-5 is involved in the nutrient sensing pathway.
-
•
We study fat-5, cyp-35A2 and fat-5;cyp-35A2 double mutants.
-
•
We uncover that the double KO has reduced fat content and an extended lifespan.
1. Introduction
The nematode Caenorhabditis elegans is arguably an ideal candidate for lipid and aging research as some 60% of pathways and metabolic activities are conserved [1] and occupies a niche between in vitro and cell culture studies (though cost efficient and fast, lack the whole animal aspect) and mammalian models (which are time consuming and expensive, but clearly of value due to their evolutionary closeness to humans) [2].
Previous work has demonstrated that C. elegans fat-5 (a Δ9 fatty acid desaturase), fat-7 and acs-2 expression is activated by nhr-49 through its interaction with MDT-15 and SBP-1 [30] a member of the sterol-regulatory-element-binding protein (SREBP) family of basic helix-zipper transcription factors. SREBPs are essential for the regulation of cholesterol levels and fatty acid homeostasis in mammals [3]. NHR-49 and NHR-80 are important for the expression of fat-5 and fat-7. When nhr-49 is deleted, the expression of fat-5 and fat-7 decreases dramatically [3]. Moreover, NHR-49 is involved in the regulation of β-oxidation as well as the expression of several essential genes linked to the dietary input. The C. elegans NHR-49 is structurally similar to the mammalian hepatocyte nuclear factor 4 (HNF-4), however, functionally NHR-49 resembles the mammalian peroxisome proliferator-activated receptors (PPARs) [3]. The C. elegans homolog of FOXO, namely DAF-16, is a transcription factor, which is tied into the insulin/IGF-1 signaling-mediated pathway (IIS). It has been reported that DAF-16 extends the lifespan of nematodes by regulating the expression of several genes that are involved in anti-oxidant, anti-microbial and metabolic activities [4], [5], [6], [7], but the precise mechanisms that link DAF-16 and its regulatory effects on the lipid composition remain elusive [8].
Moreover, cytochrome P450s (CYPs), a large family of diverse NADPH-dependent mono-oxygenases, are mainly involved in the metabolism of endogenous compounds, including fatty acids and lipid signaling molecules, as well as oxidation. Gene silencing by RNAi has revealed that CYP-35A, for example, targets the lipid metabolism of the worms [9]. Mammalian studies have revealed that cytochrome P450s play a vital role when relatively long-chain fatty acids, including linoleic and arachidonic acids, are converted into bioactive forms [10].
The C. elegans genome encodes 77 members of the cytochrome P450 superfamily. There is experimental evidence that a number of the cytochrome family members are involved in maintaining a healthy fat composition in worms [9], [12]. It has been suggested that CYPs clustered together are typically involved in xenobiotic activities [10], [13]. Human CYP-1 and CYP-2 are the closest to the members of the C. elegans cyp-35 family, which are involved in metabolizing both exogenous and endogenous compounds such as omega-3 and 6 fatty acids [10], [14], [15], [16]. Furthermore, it has been reconfirmed that inactivation of cyp-35 members results in a reduced accumulation of intestinal fat, therefore suggesting that CYPs are involved in lipid storage [10]. In addition, signal transduction enzymes, transporters, receptors and energy metabolism enzymes were linked to a major fat storage regulator, namely NHR-49 [17], [18]. The authors state that the function of several energy metabolism genes, including fat-5, fat-6 and fat-7 overlap with other regulators, including cyp-35 members. Finally, it was concluded that CYPs and NHRs may share common energy regulatory pathways associated with lipid storage [10].
The CYP-35A protein family is widely expressed within the gut cells, which are also the main sites of fat storage [10]. Indeed, mutations in cyp-35A2, cyp-35A3, cyp-35A4 and cyp-35A5 result in a deficient fatty acid metabolism. Additional members of the CYP protein family have also been reported to be involved in lipid pathways, such as CYP-29A3 and CYP-33E2 which metabolize eicosapentaenic acid, and CYP-31A2 and CYP-31A3, both essential for metabolizing lipids throughout developmental stages [9], [10], [19], [20]. These findings emphasize the complexity of metabolic pathways even in a relatively simple organism such as the nematode C. elegans.
This study utilized the genetically tractable model organism C. elegans to explore the putative link between fat-5 and cyp-35A2 and their interconnected involvement in the aging process and lipid regulation.
2. Materials and methods
2.1. Worm strains and culture
C. elegans strains were maintained at 20 °C on Nematode Growth Media (NGM) plates supplemented with Escherichia coli OP50 as the food source. Wild-type (N2, Bristol) and mutant strains were obtained from the Caenorhabditis Genetics Center, University of Minnesota, MN and included BX107 fat-5(tm420) and VC710 cyp-35A2(gk317). For each assay, nematodes were age-matched using an alkaline hypochlorite treatment to isolate the eggs. Thereafter, eggs were allowed to hatch overnight in M9 solution and arrested at L1 stage. On the following day, age-synchronous L1 worms were transferred to NGM plates and utilized for the assays.
2.2. Crossing single mutants
All single mutants were outcrossed at least 4 times. A fat-5(tm420);cyp-35A2(gk317) double-knockout was created by transferring five young fat-5(tm420) males and one cyp-35A2(gk317) L4 hermaphrodite to a multi-well plate (three replicates). The parent population was removed from each well after the hermaphrodite had laid 20–30 eggs and five males from the F1 generation were transferred to a new multi-well plate and crossed with one L4 fat-5(tm420) hermaphrodite. Once the hermaphrodites laid eggs the F1 generation was removed and at least 24 hermaphrodite trans-heterozygous F2 generation worms were isolated into separate wells of 12-well plates and allowed to reproduce. Genomic DNA (gDNA) was isolated from single F2 parents and the knockout status screened by genomic nested PCR. Homozygous fat-5(tm420);cyp-35A2(gk317) double mutants were cryopreserved for down-stream assays.
2.3. Life history traits (growth and lifespan)
Developmental changes of nematodes were assessed by observing the body size (flat volumetric surface area and length) of individual worms for a period of six days starting from 24 h post-hatch. Digital photographs of 20 age-matched nematodes per condition/strain were taken (at intervals of 24, 48, 72, 96, 120 and 144 h post-hatch) using a digital camera attached to a microscope (Nikon SMZ 800 with DS-2Mv, Nikon UK Ltd.). Thereafter, size of each nematode was quantified via the Image-Pro® Express v5.1 software (Media Cybernetic, Wokingham, UK) by manually tracing to determine flat volumetric surface area and the length. Three biological repeats were performed.
Survival assays were conducted on age-synchronized worm populations. For each condition/strain 4×100 L4 stage C. elegans were transferred onto seeded NGM plates followed by daily transfers to new plates during the reproductive period. Once reproduction had completed, nematodes were transferred every 2–3 days. Survival was assessed by touching the nematodes with a platinum wire and death scored upon failure to respond or move by the prod. Nematodes that had crawled below the agar were censored from further assessment. Statistical analysis was performed using the Log-rank (Mantel–Cox) test and percentage survival of each experimental condition was compared to the wild-type nematodes under control conditions. All lifespan assays were repeated three times as biologically independent replicate groups of worms.
2.4. Nile Red staining
In order to allow the preliminary evaluation of the lipid droplets in C. elegans, fixative and vital staining methods can be applied of which the easiest and most rapid is the vital dye Nile Red staining [21], [22]. A stock solution of 0.5 mg/mL of Nile Red in acetone was prepared (6.2 µM) and stored at −20 °C until further use. NGM plates were inoculated with Nile Red (diluted 1:250 in E. coli OP50) and left at room temperature overnight (in the dark). The following day, age-matched L1 nematodes were transferred to Nile Red/OP50 seeded plates, allowed to grow for 72 h and then individual nematodes were mounted on agarose pads and imaged using an inverted microscope (Nikon Eclipse TE2000-S, Japan) with attached camera. Exposure time and the magnification were consistent throughout imaging (20X magnification). Data were analyzed using the Image Pro-Express software (Media Cybernetics, Buckinghamshire UK) or ImageJ (ImageJ 1.46r, NIH, USA).
2.5. Quantitative real-time PCR
A minimum of 5000 age-matched nematodes at L1 stage were transferred to standard NGM plates inoculated with OP50. Once worms reached the L4 stage, total RNA was extracted using Tri-reagent (Sigma-Aldrich, Dorset, UK) modified to include a homogenization of nematodes by vortex-mixing the samples in Tri-reagent with an equal amount of acid-washed glass beads (Sigma, Dorset, UK). Thereafter, the concentration of extracted total RNA was quantified utilizing a NanoDrop ND-1000 spectrophotometer (Thermo Scientific) followed by the qualitative assessment of RNA quality by 1% agarose gel electrophoresis.
Following the total RNA extraction, complementary DNA strand (cDNA) was synthesized from 1000 ng of RNA by means of an oligo-dT primer (5′-(T)20VN-3). Quantitative real-time PCR was performed using the ABI Prism 7000 platform (Applied BioSystems, Warrington, UK) to measure the relative fold change in gene expression by applying the 2−ΔΔCT method, normalizing to the ribosomal rla-1, an invariant housekeeping gene [1], [23], [24]. Transcriptional responses of fat-5, cyp-35A2, daf-16, sbp-1, nhr-49, cyp-35A5 and cyp-35C1 were measured in different strains of nematodes. Quantification of each mRNA expression level was performed on a minimum of three technical repeats carried out on at least two independent biologically sample groups.
2.6. Data analyses
Throughout the study all statistical analyses were performed using the GraphPad Prism software (GraphPad Software Inc., USA). Error bars present mean±the standard error of the mean (SEM). Statistical significances were quantified by a two-way ANOVA followed by Bonferroni multiple comparison post hoc tests.
3. Results and discussion
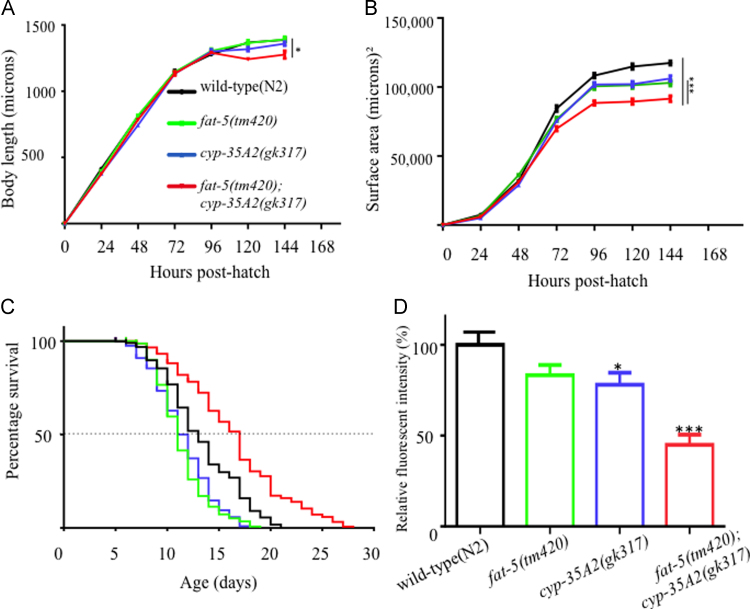
Single fat-5(tm420) and cyp-35A2(gk317) mutants were crossed to create a double mutant to study the presence of putative synergistic/antagonistic effects upon the deletion of fat-5 and cyp-35A2. First, growth parameters (body length and flat volumetric surface area over time) were measured in wild-type, the parental single mutant strains and the double mutant. Irrespective of the mutant background, no marked difference in growth was observed throughout the larval stages L1-L4 (the first 48 h post hatch), but statistically significant changes were observed in older animals. Whilst the final body length was similar in wild-type and the single mutants, fat-5(tm420);cyp-35A2(gk317) was significantly shorter (Fig. 1A). Likewise, the flat volumetric surface area of the double mutant was significantly smaller than the single mutants, which notably were also smaller than the wild-type worms (Fig. 1B). This provided first tantalizing evidence that the double mutation of fat-5 and cyp-35A2 exerts a synergistic effect on the size of adult nematodes. Although the final body size of the double mutant was reduced, percentage survival was significantly extended when compared to wild-type nematodes or the short-lived fat-5(tm420) and cyp-35A2(gk317) mutants (Fig. 1C). Statistical analyses confirmed that the difference in the lifespan of mutants was significant (Supplementary Table S1).
Fig. 1.
Effect of fat-5 and/or cyp-35A2 mutation on life history traits and Nile Red positive compartments. (A and B) Developmental changes were followed from 24 h post-hatch until 144 h. Length (A) and body surface area (B) were measured every 24 h, in wild-type, fat-5(tm420), cyp-35A2-(gk317) and fat-5(tm420);cyp-35A2(gk317) nematodes. The overall length was significantly reduced upon double deletion of fat-5 and cyp-35A2 whereas the overall length of fat-5(tm420) and cyp-35A2(gk317) was not statistically different from wild-type nematodes. The surface area was significantly (*p<0.05) affected in fat-5(tm420) mutants or cyp-35A2(gk317) mutants and resulted in a considerable reduction. The overall decrease in volumetric growth of fat-5(tm420);cyp-35A2 was highly significant (***p<0.001) compared to wild-type and single mutants. Each data represents the average surface area of 20 individual nematodes taken from two biological repeats±the standard error of mean (SEM). (C) Survival rate of wild-type, fat-5(tm420), cyp-35A2(gk317) and the double mutant were compared. Median lifespan and the maximum survival of each strain were determined, and graphs were plotted by the Kaplan–Meier method followed by a Log-Rank (Mantel–Cox) test with 95% significance (GraphPad Prism v5). The median lifespan (50% alive) and maximum lifespan in the double knockout nematodes was significantly extended in comparison to the wild-type as well as the corresponding single knockout strains. Each experimental group of the nematodes was started with a population of 400 age-matched worms and if the nematode was lost/died before day eight of adulthood, the individual nematode was censored from further analysis. (D) The Nile Red staining profile was analyzed in all four age-matched strains at 72 h post-hatch. Quantitative analyses using ImageJ software revealed that the red fluorescent intensity was lower in fat-5(tm420) in comparison to the wild-type, yet this reduction was insignificant. In cyp-35A2(gk317) the relative fluorescent intensity was decreased with a statistical significance (*p<0.05). The fat-5(tm420);cyp-35A2(gk317) double knockout exhibited an analogous, statistically even more significant (***p<0.001) reduction in the relative fluorescent intensity. Each bar graph represents the mean Nile Red intensity measured from 10 worms±the standard error of the mean (SEM). A one-way ANOVA was applied to define the statistical significance.
Others previously reported that cyp-35A2(gk317) mutants are characterized by a decrease in Nile Red staining [12], a finding we were able to confirm (Fig. 1D).
The Nile Red staining pattern of fat-5(tm420) resembled cyp-35A2(gk317) but was statistically not distinguishable from wild-type animals. Noticeable, and analogous to the results obtained from the growth and life-span measurements, a synergistic effect in the Nile Red staining capacity was observed in the fat-5(tm420);cyp-35A2(gk317) double mutant, namely staining was reduced by 57% when compared to wild-type worms (Fig. 1D). To validate whether the staining pattern is limited to the lysosomal compartments (the site of Nile Red staining) further experimentation may include the monitoring of lipid content by other stains (e.g. Bodipy, Oil Red and Sudan Black) or indeed dye-free methodologies such as Coherent Anti-Stokes Raman Scattering (CARS) microscopy.
3.1. The transcription of fat-5 and cyp-35A2
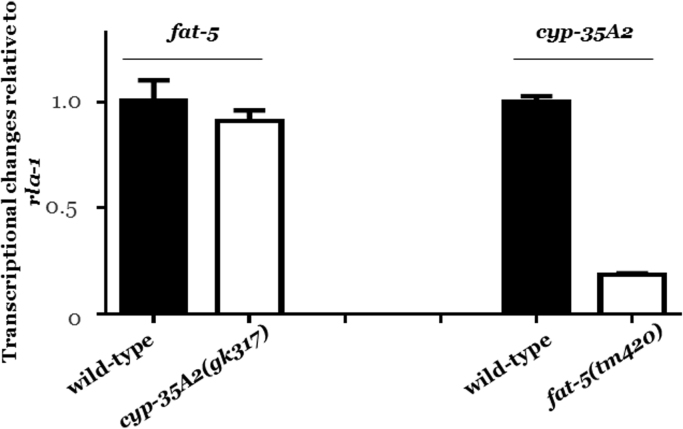
The expression profiles of fat-5 and cyp-35A2 were quantified in wild-type nematodes and the respective single mutants. The expression level of fat-5 was comparable in wild-type and the cyp-35A2 mutant, in contrast, cyp-35A2 expression was suppressed in a fat-5(tm420) background (Fig. 2), tentatively suggesting the presence of a putative interaction between fat-5 and cyp-35A2. It is conceivable that in the absence of fat-5 other members of cytochrome P450 family may compensate and cyp-35A2 may not be directly involved.
Fig. 2.
The transcription levels of fat-5 and cyp-35A2 in wild-type and mutant strains. The basal expression of the respective transcript was normalized to the invariant housekeeping gene rla-1 and then set to 1 in wild-type worms. The expression of fat-5 remained unchanged regardless of the presence (wild-type) or absence of cyp-35A2. However, in the absence of fat-5, cyp-35A2 was significantly down-regulated. Each bar depicts the average from two biologically independent experiments with at least three technical repeats per condition±the standard error of the mean (SEM).
3.2. Transcription factors linked to the deletion of fat-5 and cyp-35A2
In an attempt to explore how the double mutation of fat-5 and cyp-35A2 influences metabolic pathways and leads to physiologically healthier nematodes, the transcriptional response of genes involved in aging and lipid-related pathways were determined by quantitative real-time PCR. Similar to mammalian species, where the ratio between High Density Lipoprotein (HDL) and Low Density Lipoprotein (LDL) accounts for healthy versus unhealthy lipid components, nematodes may alter lipid metabolism by switching into healthier lipid compartments that eventually lead to longevity.
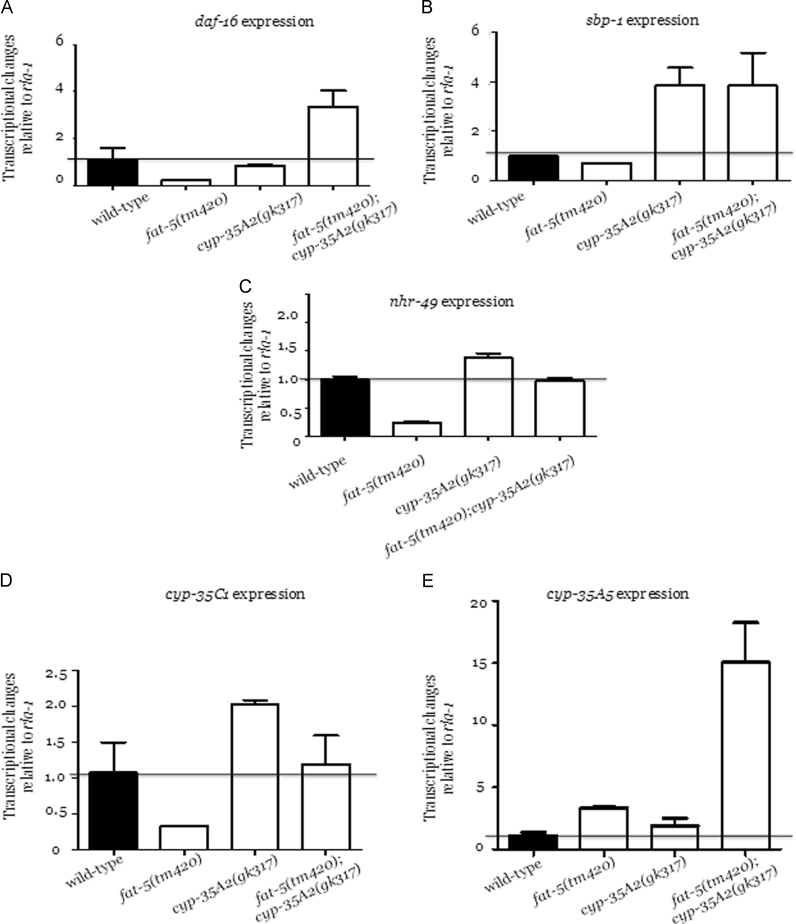
In the absence of fat-5, the desaturation of palmitic acid is impaired, perhaps resulting in a higher concentration of saturated fatty acids. Consequently, elevated levels of saturated fatty acids can influence the cellular membrane fluidity because changes in the fatty acid composition affect the structure of the lipid bilayer. A tougher cell membrane might also protect the cellular compartments from oxidative stress and free radical formation. Therefore, less toxic material can be transported between cells, delaying the aging process [25]. Additionally the C. elegans FOXO homolog, daf-16, is involved in several metabolic pathways including longevity. The expression of this gene was quantified in wild-type, fat-5(tm420), cyp-35A2(gk317) and the fat-5(tm420);cyp-35A2(gk317) double mutant. The expression of daf-16 was, compared to wild-type and the cyp-35A2 mutant, suppressed in the fat-5 mutant background but notably up-regulated in the double mutant (Fig. 3A). As daf-16 is linked to longevity and in some fat regulatory responses, the delayed aging phenotype observed in fat-5(tm420);cyp-35A2(gk317) may suggest that daf-16 induction is causatively linked to the extended lifespan of the double mutant. This hypothesis could be tested further by investigating whether daf-16 RNAi abrogates the enhanced longevity of the double mutant or indeed the nuclear localization of a daf-16:GFP reporter.
Fig. 3.
The transcription levels of daf-16, sbp-1, nhr-49, cyp-35C1 and cyp-35A5 in wild-type, fat-5(tm420), cyp-35A2(gk317) and fat-5(tm420);cyp-35A2(gk317). Genes involved in aging- and lipid-regulatory pathways were selected to evaluate, by qPCR, changes in expression associated with the fat-5 and/or cyp-35A2 genotypes. The expression of daf-16 (Panel A), sbp-1 (Panel B), nhr-49 (Panel C), cyp-35C1 (Panel D) and cyp-35A5 (Panel E) were quantified in all four strains and the transcription level within wild-type was set to 1. Each bar presents the average fold-change of the transcript (normalized to rla-1) determined from two independent experiments with at least three technical repeats in each condition±the standard error of the mean (SEM).
Although DAF-16 has been reported to play a major role in the survival rate of the nematodes, it is also modulated by other factors. Similarly, fat synthesis/breakdown is influenced in C. elegans via DAF-16 activation through a kinase cascade upstream of Δ9 desaturases [3]. This current study links fat-5 expression with the transcription factor daf-16, suggesting the possible existence of a regulatory mechanism between fat-5 and daf-16. Furthermore, an alteration in the fatty acid composition pool leading to the accumulation of more saturated forms potentially renders the membrane more resistant to peroxidation [25]. The mutation of fat-5 and cyp-35A2 together perhaps modulates the fatty acid profile of the animals whereas the single mutation in fat-5 may not necessarily change the fatty acid compositions suggesting that other desaturases such as FAT-6 and FAT-7 may compensate for the loss of function of FAT-5 [3]. Nonetheless, mutation of cyp-35A2 enforces a significant alteration in the physiological state of the animals resulting in a notably extended survival rate.
Several transcription factors, enzymes and regulatory elements work together, which in concert trigger new phenotypic features. Calorie restriction or calorie restriction mimetic conditions and IIS mutants that exhibit higher lipid content are also longer lived [25].
In addition autophagy and longevity have been linked, which is regulated by the DAF-16/FoxO pathway. In addition, once the germline is removed from nematodes or flies, they tend to live longer through DAF-16/FoxO (which is analogous to the insulin/IGF-1 signaling pathway) and lipid metabolism plays an essential role in these animals. Studies have shown that, upon removal of the germline, the expression of LIPL-4 is up-regulated, which is a lipase that is activated through DAF-16/FoxO transcription factor. It has also been shown that lipl-4 is essential for lifespan extension in worms. Interestingly, when lipl-4 is up-regulated it increases the survival rate of the nematodes significantly, which connects lipid metabolism and longevity in nematodes, similar to the lipophagy phenomenon which links autophagy in hydrolyzing lipids and the effect on extended lifespan [26]. This emphasizes not only the complexity of lipid regulation and aging but also the fact that many abundant pathways are involved in a healthy and extended survival rate in nematodes.
The C. elegans homolog of the sterol-regulatory-element-binding protein (SREBP), SBP-1, is an essential component for a healthy lipid metabolism as its deletion has been shown to affect the fat accumulation of the animals [9], [27]. Also, sbp-1 is upstream of fat-5 and regulates fat-5 expression [3]. Although the expression of sbp-1 was not affected in fat-5(tm420), a significant up-regulation of sbp-1 was observed in cyp-35A2(gk317) and fat-5(tm420);cyp-35A2(gk317) (Fig. 3B). This implies that cyp-35A2, rather than fat-5, is linked to sbp-1 expression. Therefore, sbp-1 over-expression might be part of a regulatory mechanism to activate transcription factors and enzymes involved in fatty acid synthesis to maintain the lipid homeostasis. As demonstrated by others, fat-5 is not only regulated by sbp-1 but also by further transcription factors and regulators of lipid metabolism, such as nuclear hormone receptors nhr-49. Furthermore, sbp-1 and nhr-49 share a common co-activator, namely mdt-15 [28], [29], [30].
NHR-49 is responsible for the mitochondrial β-oxidation in addition to fatty acid desaturation, and in the absence of nhr-49 animals exhibit a higher fat content due to the reduced expression of β-oxidation genes [3]. The transcriptional response of nhr-49 was suppressed in fat-5(tm420) but not in cyp-35A2(gk317) nor the double mutant (Fig. 3C), which suggests that the expression of nhr-49 and cyp-35A2 are not correlated. In contrast, the suppression of nhr-49 in fat-5(tm420) implies that these two genes might be functionally linked.
The expression profile of cyp-35C1, which is reportedly co-expressed with cyp-35A2 (information from Wormbase (2014)) was similar to nhr-49, namely suppressed in fat-5(tm420), significantly induced in cyp-35A2(gk317) but not modulated in the fat-5(tm420);cyp-35A2(gk317) double knockout (Fig. 3D). The expression of cyp-35A5, a further transcript co-expressed with cyp-35A2, revealed a modest, yet statistically significant, up-regulation within the fat-5(tm420) and cyp-35A2(gk317) mutant backgrounds, and a striking induction in the fat-5(tm420);cyp-35A2(gk317) double mutant (Fig. 3E). As the cyp-35A/C gene members share protein domains [11], [31], their interaction network is inter-woven. Previous reports have demonstrated that the RNAi of cyp-35A3 and cyp-35A5 affects the normal lipid regulatory pathways in C. elegans [9]. In contrast, Aarnio et al. (2011) stated that only the RNAi of cyp-35A1 and cyp-35A5 (and not cyp-35A2 and cyp-35A4) results in a lower fat content phenotype [10]. Likewise, mutations in cyp-35A1 were shown to result in the over-expression of certain fatty acid synthesizing genes, such as fat-2. Upon the addition of oleic or elaidic triglycerides to the diet of wild-type animals, the cyp-35A1 expression returned to basal levels, but cyp-35A2, cyp-35A4 and cyp-35A5 were significantly down-regulated [10]. The expression of cyp- genes is localized in the intestine of the animals where most fat-droplets are also stored. Lipids are mainly accumulated in the form of triacylglycerides in intestinal lysosome-related organelles and in lipid droplets in the gut, in addition to the epidermal skin-like cells [10].
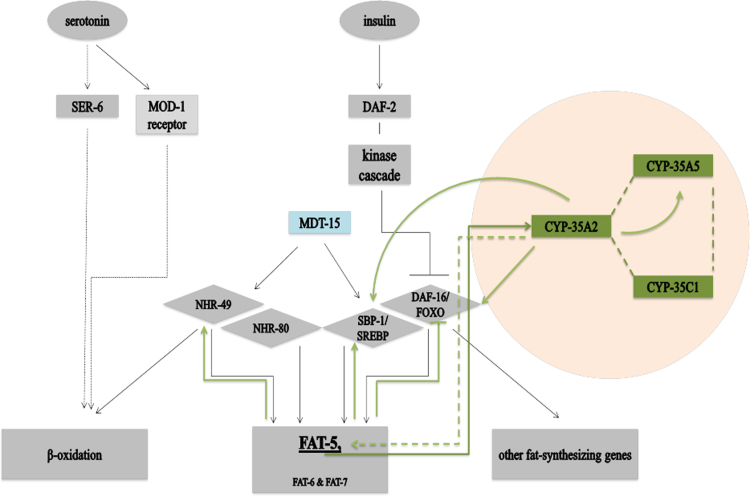
The data obtained from this current study provided circumstantial evidence of a putative correlation between selected genes within the fat-5 and cyp-35A2 pathways, which extends the lipid regulatory network firstly reported by Watts in 2009. The network consists of several transcriptional factors that reside upstream of FAT-5, FAT-6 and FAT-7. By combining the transcriptional responses of fat-5 and cyp-35A2, and corresponding transcription factors and/or co-expressers, it was possible to re-affirm the complexity of the lipid regulatory pathway and provide possible additional insights into the previously proposed longevity and survival mechanisms (Fig. 4).
Fig. 4.
The lipid regulatory pathway based on the nutritional sensing mechanisms. The original pathway, published by Watts in 2009 [3], was expanded to include the findings uncovered by this study (shown in green). Note that only the Δ9 desaturase fat-5 (not fat-6 or fat-7) was investigated. Abbreviations: TGF-β, transforming growth factor-β; SREBP, sterol regulatory element-Binding protein; NHR, nuclear hormone receptor.
4. Conclusions
The fat-5(tm420);cyp-35A2(gk317) is (compared to wild-type and the respective single mutants) long-lived and characterized by a significantly reduced body size. Consistent with this finding is that daf-16 and sbp-1, transcription factors involved in lifespan and lipid pathways [3], [8], are significantly inducted in the double mutant. Interestingly, the double mutant was also marked by a significant up-regulation of cyp-35A5, but not nhr-49 which is reported to be required for a healthy lifespan [3]. These results highlight the complexity of metabolic pathways, where different transcription factors can regulate lifespan, development, lipid and nutrient-sensing pathways, and not all are necessarily involved in phenotypic and/or physiological responses at the same time. Nevertheless, the data supports the notion that the nutrition-sensing pathway and the lipid regulatory feature of cytochrome P450 family members are interconnected (Fig. 4). In conclusion, as many fundamental pathways are conserved between nematodes and humans, the proposed extension of the aging and longevity network may contribute towards uncovering cues that will aid the study of hereditary obesity in mammals and in particular humans.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.06.007.
Appendix A. Supplementary materials
Supplementary material Table S1. Statistical analysis on survival percentages. Kaplan–Meier curves were plotted for each nematode strain (wild-type, fat-5(tm420), cyp-35A2(gk317) and the double knockout fat-5(tm420);cyp-35A2(gk317)). The data were analyzed using the Log-rank (Mantel-Cox) method to define the statistical significance, mean life span as well as median survival.
Supplementary material
Supplementary material
Supplementary material
References
- 1.Qabazard B., Ahmed S., Li L., Arlt V.M., Moore P.K., Sturzenbaum S.R. C. elegans aging is modulated by hydrogen sulfide and the sulfhydrylase/cysteine synthase cysl-2. PloS One. 2013;8:e80135. doi: 10.1371/journal.pone.0080135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng J., Greenway F.L. Caenorhabditis elegans as a model for obesity research. Int. J. Obes. 2005;36(2012):186–194. doi: 10.1038/ijo.2011.93. [DOI] [PubMed] [Google Scholar]
- 3.Watts J.L. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol. Metab.: TEM. 2009;20:58–65. doi: 10.1016/j.tem.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S.S., Kennedy S., Tolonen A.C., Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 5.Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 6.Oh S.W., Mukhopadhyay A., Dixit B.L., Raha T., Green M.R., Tissenbaum H.A. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat. Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 7.Boulias K., Horvitz H.R. The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–450. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shmookler Reis R.J., Xu L., Lee H., Chae M., Thaden J.J., Bharill P., Tazearslan C., Siegel E., Alla R., Zimniak P., Ayyadevara S. Modulation of lipid biosynthesis contributes to stress resistance and longevity of C. elegans mutants. Aging. 2011;3:125–147. doi: 10.18632/aging.100275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashrafi K., Chang F.Y., Watts J.L., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- 10.Aarnio V., Lehtonen M., Storvik M., Callaway J.C., Lakso M., Wong G. Caenorhabditis elegans mutants predict regulation of fatty acids and endocannabinoids by the CYP-35A gene family. Front. Pharmacol. 2011;2:12. doi: 10.3389/fphar.2011.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menzel R., Rodel M., Kulas J., Steinberg C.E. CYP35: xenobiotically induced gene expression in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2005;438:93–102. doi: 10.1016/j.abb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Menzel R., Yeo H.L., Rienau S., Li S., Steinberg C.E., Sturzenbaum S.R. Cytochrome P450s and short-chain dehydrogenases mediate the toxicogenomic response of PCB52 in the nematode Caenorhabditis elegans. J. Mol. Biol. 2007;370:1–13. doi: 10.1016/j.jmb.2007.04.058. [DOI] [PubMed] [Google Scholar]
- 13.Thomas J.H. Rapid birth–death evolution specific to xenobiotic cytochrome P450 genes in vertebrates. PLoS Genet. 2007;3:e67. doi: 10.1371/journal.pgen.0030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz D., Kisselev P., Ericksen S.S., Szklarz G.D., Chernogolov A., Honeck H., Schunck W.H., Roots I. Arachidonic and eicosapentaenoic acid metabolism by human CYP1A1: highly stereoselective formation of 17(R),18(S)-epoxyeicosatetraenoic acid. Biochem. Pharmacol. 2004;67:1445–1457. doi: 10.1016/j.bcp.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 15.Fer M., Dreano Y., Lucas D., Corcos L., Salaun J.P., Berthou F., Amet Y. Metabolism of eicosapentaenoic and docosahexaenoic acids by recombinant human cytochromes P450. Arch. Biochem. Biophys. 2008;471:116–125. doi: 10.1016/j.abb.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Arnold C., Konkel A., Fischer R., Schunck W.H. Cytochrome P450-dependent metabolism of omega-6 and omega-3 long-chain polyunsaturated fatty acids. Pharmacol. Rep.: PR. 2010;62:536–547. doi: 10.1016/s1734-1140(10)70311-x. [DOI] [PubMed] [Google Scholar]
- 17.Van Gilst M.R., Hadjivassiliou H., Jolly A., Yamamoto K.R. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS biology. 2005;3:e53. doi: 10.1371/journal.pbio.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Gilst M.R., Hadjivassiliou H., Yamamoto K.R. A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. USA. 2005;102:13496–13501. doi: 10.1073/pnas.0506234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulas J., Schmidt C., Rothe M., Schunck W.H., Menzel R. Cytochrome P450-dependent metabolism of eicosapentaenoic acid in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2008;472:65–75. doi: 10.1016/j.abb.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Benenati G., Penkov S., Muller-Reichert T., Entchev E.V., Kurzchalia T.V. Two cytochrome P450s in Caenorhabditis elegans are essential for the organization of eggshell, correct execution of meiosis and the polarization of embryo. Mech. Dev. 2009;126:382–393. doi: 10.1016/j.mod.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 21.K. Ashrafi, Obesity and the regulation of fat metabolism (March 9, 2007), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.130.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- 22.Lemieux G.A., Ashrafi K. Insights and challenges in using C. elegans for investigation of fat metabolism. Crit. Rev. Biochem. Mol. Biol. 2015;50:69–84. doi: 10.3109/10409238.2014.959890. [DOI] [PubMed] [Google Scholar]
- 23.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Ackerman D., Gems D. The mystery of C. elegans aging: an emerging role for fat. Distant parallels between C. elegans aging and metabolic syndrome? BioEssays: News Rev. Mol. Cell. Dev. Biol. 2012;34:466–471. doi: 10.1002/bies.201100189. [DOI] [PubMed] [Google Scholar]
- 26.Lapierre L.R., Melendez A., Hansen M. Autophagy links lipid metabolism to longevity in C. elegans. Autophagy. 2012;8:144–146. doi: 10.4161/auto.8.1.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonczy P., Echeverri C., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., Hannak E., Kirkham M., Pichler S., Flohrs K., Goessen A., Leidel S., Alleaume A.M., Martin C., Ozlu N., Bork P., Hyman A.A. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 28.Yang F., Vought B.W., Satterlee J.S., Walker A.K., Jim Sun Z.Y., Watts J.L., DeBeaumont R., Saito R.M., Hyberts S.G., Yang S., Macol C., Iyer L., Tjian R., van den Heuvel S., Hart A.C., Wagner G., Naar A.M. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 29.Taubert S., Van Gilst M.R., Hansen M., Yamamoto K.R., Mediator subunit A. MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomura T., Horikawa M., Shimamura S., Hashimoto T., Sakamoto K. Fat accumulation in Caenorhabditis elegans is mediated by SREBP homolog SBP-1. Genes Nutr. 2010;5:17–27. doi: 10.1007/s12263-009-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakrapani B.P., Kumar S., Subramaniam J.R. Development and evaluation of an in vivo assay in Caenorhabditis elegans for screening of compounds for their effect on cytochrome P450 expression. J. Biosci. 2008;33:269–277. doi: 10.1007/s12038-008-0044-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Table S1. Statistical analysis on survival percentages. Kaplan–Meier curves were plotted for each nematode strain (wild-type, fat-5(tm420), cyp-35A2(gk317) and the double knockout fat-5(tm420);cyp-35A2(gk317)). The data were analyzed using the Log-rank (Mantel-Cox) method to define the statistical significance, mean life span as well as median survival.
Supplementary material
Supplementary material
Supplementary material