Abstract
For many years Staphylococcus aureus has been recognized as an important human pathogen. In this study, the surfacome and exoproteome of a clinical sample of MRSA was analyzed. The C2355 strain, previously typed as ST398 and spa-t011 and showing a phenotype of multiresistance to antibiotics, has several resistance genes. Using shotgun proteomics and bioinformatics tools, 236 proteins were identified in the surfaceome and 99 proteins in the exoproteome. Although many of these proteins are related to basic cell functions, some are related to virulence and pathogenicity like catalase and isdA, main actors in S. aureus infection, and others are related to antibiotic action or eventually resistance like penicillin binding protein, a cell-wall protein. Studying the proteomes of different subcellular compartments should improve our understanding of this pathogen, a microorganism with several mechanisms of resistance and pathogenicity, and provide valuable data for bioinformatics databases.
Keywords: MRSA, Antibiotic resistance, Proteomics, Surfaceome, Exoproteome
Graphical abstract
Highlights
-
•
We examine the surface proteome and exoproteome of multiresistant strains.
-
•
We identify bacterial infection proteins in the extracellular proteome.
-
•
Confirmation that moonlighting proteins will extend the localization data.
1. Introduction
Staphylococcus aureus is a highly successful opportunistic pathogen that can cause a wide variety of diseases. Infections due to S. aureus were once limited to hospital settings and could be effectively treated with antibiotics. However, S. aureus has an extraordinary capacity to adapt and survive in a great variety of environments which is reflected in its ability to develop antibiotic resistance. In the last decade, a livestock-associated methicillin-resistant clone of S. aureus (LA-MRSA) has emerged that can cause zoonotic disease in humans and appears as a major public health concern. Sequence type (ST) 398 strains were first identified in France in 2005 but came to prominence in the Netherlands as a common component of the skin microbiome of pigs and pig farmers [1].
The bacterial proteome is very dynamic and can change in response to cellular or environmental factors [2]. Molecular and genetic dissection of S. aureus has revealed that a great number of surface adhesins, secreted enzymes, and toxins might be implicated in pathogenesis [3]. The post-genomic era of S. aureus started in 2001 with the publication of the genome sequences of two strains, N315 and Mu50 [4], followed by those of MW2 [5], COL [6], and two clinical isolates [7]. Proteomic tools have been used to characterize proteins involved in the mechanism of antibiotic resistance and pathogenesis of MRSA, mainly focusing on the surface and secreted proteins. The concept of the surfaceome has been described as the surface-exposed proteins including their extracellular epitopes [8]. Surface-exposed integral membrane proteins (IMP) of bacteria are crucial in pathogenesis as they mediate interactions with host cells like adhesion, invasion and reception of signals from the immune response. Some surface IMP sense and respond to the chemical and physical conditions of the external milieu as channels for nutrient acquisition, and others play a role in mounting defenses against host responses by removing antimicrobial or toxic compounds, secreting virulence factors, or sending appropriate signals to the cytoplasmic compartment [9].
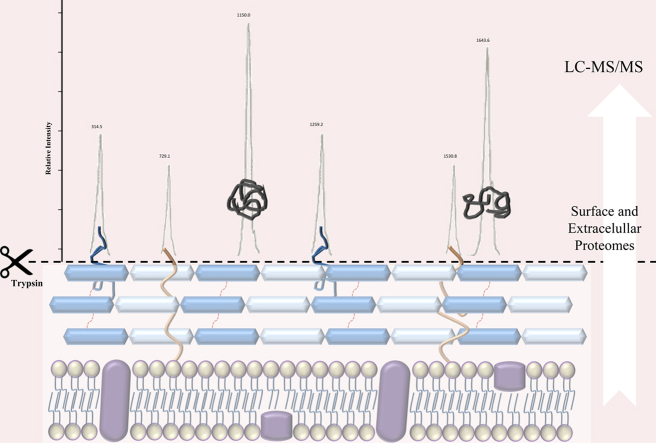
The analysis of hydrophobic surface-exposed IMP remains one of the major challenges for proteomics technologies. Three main approaches are currently taken to identify surface proteins. The first method is based on surface protein prediction by genome analysis using algorithms such as PSORT [10]. The second approach is based on the physical separation of membrane and cell wall fractions from the cytoplasmic fraction followed by identification of the proteins by two-dimensional (2D) polyacrylamide gel electrophoresis (PAGE) or 2D chromatography coupled to mass spectrometry. One such method of defining the surfaceome is cell “shaving”, which involves digesting intact cells with proteases in isotonic solution to release exposed peptides for analysis by LC-MS/MS [11]. Finally, in the third approach, membrane proteins are first defined using one of the two methods described above and then surface localization is confirmed by producing polyclonal antibodies against the recombinant forms of each predicted protein and by assaying antibody binding to whole bacterial cells [12].
Proteomics might provide the most realistic picture of the infective process as it detects the end-products of gene biosynthetic pathways, which essentially determine a biological phenotype. In the present study, we used shotgun proteomics techniques to identify the surface and extracellular proteomes of a clinical MRSA strain of livestock-associated sequence type 398, a strain previously implicated in a human empyema infection, that has the phenotype and genotype of antibiotic resistance [13].
A comprehensive proteomic map of the surfaceome and extracellular proteome was established. Proteins were identified to compare them with those expressed in other MRSA genetic lineages more adapted to the human organism. The protein expression mechanisms of this genetic lineage of MRSA were evaluated in the context of its emergence in animals and later in humans and its very rapid spread in the environment.
2. Material and methods
2.1. Cell surface proteins
2.1.1. Bacterial strains and growth
The MRSA strain C2355 recovered from the pleural liquid of a patient with empyema was used in the present study [13]. C2355 was previously ascribed to the spa-type t011, SCCmec type V, and to ST398. Moreover, this strain shows a phenotype of multiresistance to β-lactams, tetracycline, clindamycin and ciprofloxacin among other antibacterials, and harbors the mecA, tetK, and tetM resistance genes. Cultures were maintained in tryptic soy broth and grown to mid-exponential phase (OD610 nm 0.5) at 37 °C with constant agitation at 200 rpm. For each cell shaving experiment 20 mL of culture was used.
2.1.2. Cell shaving of MRSA ST398
The bacterial cells were harvested at mid-exponential phase by centrifugation at 1000 g for 15 min at 4 °C. Cells were washed three times with wash solution (20 mM Tris–HCl, 150 mM NaCl, pH 7.6) and resuspended in 1.99 mL of hydrolysis buffer (20 mM Tris–HCl, 150 mM NaCl, 10 mM CaCl2, 1 M l-arabinose, pH 7.6). Bacterial suspensions were treated with 2 µg of trypsin for 1 h at 37 °C. Three biological replicates were used.
2.1.3. Collection of peptides from supernatants
The samples were centrifugated at 1000g for 15 min at 4 °C and the supernatants were filtered through a 0.22-µm mesh to completely eliminate cells. Peptides were further digested overnight with 0.6 µg of trypsin at 37 °C, then purified and concentrated with a Sep-Pak C18 Plus Light Cartridge (WATERS, Guyancourt, France) according to the manufacturer's instructions. Peptides were kept in water–acetonitrile (ACN) (98:2, v/v) containing 0.06% trifluoroacetic acid (TFA).
2.2. Extracellular proteins
2.2.1. Bacterial strains and culture conditions
Bacteria were cultured in the chemically defined Staphylococcus siderophores detection (SSD) medium. From stock culture kept at −80 °C, bacterial strains were inoculated in SSD broth and incubated for 16 h at 37 °C. A preculture was set up from one isolated bacterial colony and grown in SSD broth at 37 °C in an orbital shaker (170 rpm). From an inoculum in the exponential growth phase, a 250-mL culture was set up, adjusted to 0.01 (OD600 nm) and incubated as described. For proteomic experiments, sampling was performed in late exponential growth phase (OD600 nm=0.5) after 12 h at 37 °C.
2.2.2. Precipitation of extracellular proteins
The cells in culture were removed by centrifugation (7520g, 15 min, 4 °C) and the supernatants were filtered (0.2-μm mesh) before adding 0.2 mM phenylmethylsulfonyl fluoride to inhibit protease activity. Supernatants were concentrated in a final volume of about 5 mL using a Vivacell-100 centrifugal concentrator (5 kDa cut-off; Sartorius Stedim). Sodium deoxycholate (0.2 mg mL−1) was added to the solution before incubation for 30 min on ice. Sodium deoxycholate supports protein precipitation, which was carried out by adding 10% (w/v) trichloroacetic acid with incubation overnight at 4 °C. After centrifugation (12,000 rpm, 30 min, 4 °C), the precipitate was washed with ice-cold acetone and solubilized in Laemmli buffer. Three protein extracts from three independent cultures were prepared.
2.2.3. SDS PAGE and protein preparation for LC-MS/MS
To eliminate interfering molecules (such as salts) and concentrate proteins in a single band of polyacrylamide gel, samples were submitted to a short SDS-PAGE migration (T=12.5%, C=3.3%) in a BioRad Mini Protean II unit. Protein samples were just allowed to concentrate at the junction between stacking and resolving gel after few minutes migration (25 mA/gel at 100 V). Gel was then stained with Coomassie Brilliant Blue G250 and the single bands in each lane were excised. The proteins in the gel were reduced in 100 mM ammonium bicarbonate with 45 mM dithiothreitol for 45 min at 50 °C, and alkylated with 100 mM iodoacetamide for 20 min at room temperature in the dark. The gel was washed in 25 mM ammonium bicarbonate in 5% ACN for 30 min and twice in 25 mM ammonium bicarbonate in 50% ACN for 30 min each. The gel was dehydrated with 100% acetonitrile and then reswelled in 100 mM ammonium bicarbonate containing 4 µg of trypsin to digest peptides at 37 °C for 5 h. To stop the tryptic digestion and extract the proteins from the gel, 0.1% TFA in 100% ACN was added.
2.3. Protein identification by LC-MS/MS and data analysis
For LC-MS/MS analysis, peptide mixtures were analyzed by nanoflow liquid chromatography using the Ultimate 3000 RSLC (Dionex, Voisins le Bretonneux, France) with nanocapillary columns (15 cm long×75 μm internal diameter; Acclaim Pep Map RSLC, Dionex). The solvent gradient was increased linearly from 4% to 90% ACN in 0.5% formic acid at a flow rate of 300 nL/min for 38 min. The elute was electrosprayed inside an LTQ-VELOS mass spectrometer (Thermo Fisher Scientific, Courtaboeuf, France) through a nanoelectrospray ion source.
Thermo Proteome Discoverer v1.3 was used for raw data file processing. The EBI_Bacteria protein database (containing 25590467 sequences) was used for protein identification. The following parameters were set for the searches: peptide mass tolerance of 1.5 Da, fragment mass tolerance of 0.5 Da, and a maximum of two missed cleavages allowed. Variable modifications considered were methionine oxidation (M) and carbamidomethylation (C) of cysteine. A protein was considered to be valid when a minimum of two unique peptides originating from one protein showed statistically significant (p<0.05) Mascot scores (http://www.matrixscience.com).
2.4. Bioinformatics analysis
Functional annotation of the proteins was determined using the Universal Protein Resource database (UniProt) (http://www.uniprot.org/). Subcellular protein localization was predicted by PSORTb version 3.0 (http://www.psort.org/psortb/).
Prediction of non-classical protein secretion not triggered by signal peptides was detected by the use of the SecretomeP 2.0 server (http://www.cbs.dtu.dk/services /SecretomeP-2.0/).
3. Results and discussion
3.1. Surface proteins
The surface of the bacterial cell forms the interface between pathogen and host, the site of interaction during infection of host tissues. In this study, the surfaceome and exoproteome of a clinical sample of MRSA were analyzed. Strain C2355, which had previously been typed as ST398 and spa-t011, shows a phenotype of multiresistance to antibiotics and carries several resistance genes.
A cell shaving protocol was used to obtain surfaceome fractions. The peptide mass peaks obtained by LC-MS/MS of the fractions were compared with UniProt and NCBI databases. The database identification (ID), molecular weight (MW), isoelectric point value (pI), function, biological process and cellular location of 236 proteins from this subcellular proteome were identified by searching the Uniprot database (Table A1).
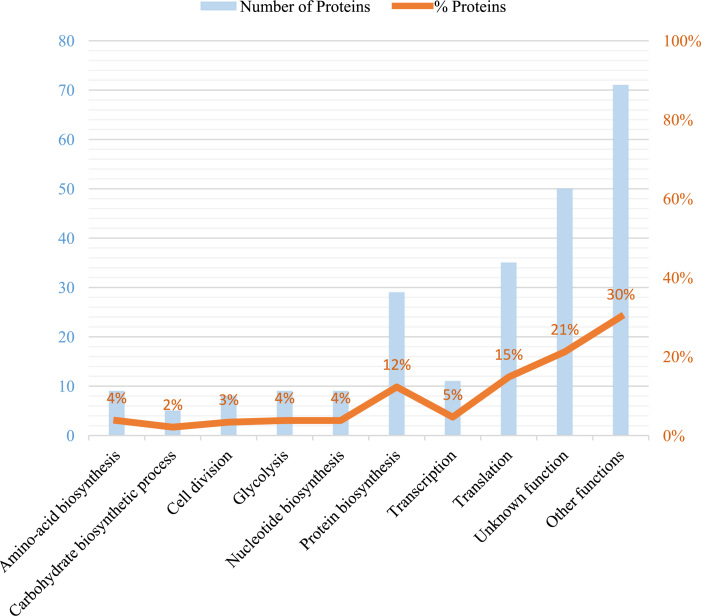
The proteins identified in the cell surface fraction have predicted functional activities in amino acid biosynthesis, carbohydrate processing, cell division, glycolysis, nucleotide biosynthesis, protein biosynthesis, transcription, translation, and 40 other functions (Fig. 1). Translation was the most common function in the proteins identified (15%), although no biological function was associated with about 21% of the identified proteins. The locations of the 236 proteins were predicted using PSORTb software. Overall, 198 proteins (83.9%) were predicted to be cytoplasmic. No location could be predicted for 25 proteins (10.6%). However, 2 proteins (0.8%) are thought to be components of the cell wall and 11 (4.7%) components of the plasma membrane.
Fig. 1.
Biological functions associated with 236 proteins identified in the cell surface fraction. Numbers (blue) and percentages (red) of identified proteins associated with different biological functions. “Other functions” encompasses 40 functions for which 5 or fewer proteins were identified where no functional category contained more than 2% of the total number of proteins.
Gram-positive bacteria such as S. aureus have a cell wall morphology that is strongly resistant to lysis compared to Gram-negative bacteria or animal cells. According to Solis et al. [9], the proteins exposed on the cell surface of S. aureus have a great potential to unravel the molecular basis of pathogenicity and the technique of cell surface shaving is a way to access the exposed peptides. A total of 42 proteins were identified as exposed at the surface including novel ones and new information on the topology of the surface proteins of S. aureus was described [9].
The original cell shaving method [14] used low centrifugal forces for cell washing (3500g) and a brief protein digestion (30 min). In subsequent studies cell washing was more aggressive (12,000g) and tryptic digestion was longer (4 h).
Does the shaving technique access the proteins that are truly exposed on the bacterial surface including those that are theoretically cytoplasmic but routinely identified in surface extracts [9]? Generally, proteins known to be cell wall associated are more abundant early in growth, while proteins known to be secreted into the surrounding medium are more abundant late in growth. Extracellular proteins known to be regulated by the global regulators SarA and Agr were detected at the levels expected for these regulators. Surprisingly, a large percentage of proteins detected in the media during early exponential phase of growth are annotated as or predicted to be cytoplasmic proteins. In the fractionation procedure used here the growth of S. aureus was stopped in the mid-exponential phase, a stage when the bacteria do not secrete much protein and there is less bacterial turnover than during the stationary phase. However, it is possible that phase-dependent growth regulation can make it difficult to identify specific proteins of the stationary phase or proteins expressed in response to extracellular signals, such as cell density or biofilm formation. The shaving technique is still too recent to know how specific the fractionation is [9], so it was necessary to use PSORTb software that predicts the cellular location of the proteins based on the amino-acid sequence (http://psort.hgc.jp/). After the LC-MS/MS analysis and the PSORTb predictions, 13 proteins were identified that can be readily related to a membrane or surface location. Two of them are cell wall proteins, a probable malate:quinone oxidoreductase with catalytic activity (D8HG43, Table A1) and a putative exported protein with unknown biological function (H7G2S8, Table A1). The 11 remaining proteins are predicted to be components of cytoplasmic membranes.
With PSORTb it was possible to identify two important proteins that are exposed on the cell surface. The penicillin binding protein PBP2’ (C5N5L3, Table A1) is essential for cell wall synthesis and are target for inhibiting β-lactam antibiotics. The cell division protein FtsZ (A6U106, Table A1) is essential in the formation of the contractile Z ring at the future site of the septum, thereby regulating the timing and location of cell division. Another function of the ring is to recruit other proteins to produce the new cell wall. FtsZ was originally identified by Copeland in 2007 as a cytoplasmic protein [15]. Finding FtsZ confirms the pertinence of the shaving approach, because it was previously identified in the surfaceome of S. aureus by Dreisbach and coworkers in 2010 [16]. This reinforces the idea, already advocated by researchers in the field of exposed surface proteomics, that it is necessary to revise the models that predict putative protein location in cells.
Actually the major goal of cell shaving protocols is to identify surface proteins without contamination from cytoplasmatic proteins. Cell lysis during the protocol can be one of the explanation for the percentage of cytosolic protein in our shaving extract however it was shown by Dreisbach [16] {Dreisbach, 2010 #14} that the cell lysis due, at least, to the addition of trypsin could be excluded as an explanation for the presence of cytoplasmatic proteins. Our data is not completely conclusive to prove that cytoplasmic proteins are or are not surface-exposed in S. aureus because those proteins can be also exposed by unknown mechanisms or non-classical secretion systems which is a huge problem in the actual bioinformatics models used to predict proteins location {Solis, 2010 #7}. This can be tricky because moonlighting proteins that may perform surface-associated functions in addition to their cytoplasmic role are now well-accepted {Solis, 2010 #7}. Numerous studies have identified proteins that have a moonlighting function on the cell surface. Interestingly, some of these proteins are also involved in pathogenicity. In addition, the surfaceomics approach is particularly amenable to protein expression profiling of small amounts of sample. Only about 107 cells are required for a 2D gel, so one of the limitations of this approach, i.e., the lack of sufficient material for analysis, is overcome [8].
Four ribosomal proteins described in previous studies as being localized on the cell surface of S. aureus were also found in this study. Although ribosomal proteins are theoretically associated with cytoplasm functions, they have recently been identified on the cell surface [16]. PSORTb did not predict that any of the proteins would be at the cell surface. Glyceraldehyde-3-phosphate dehydrogenase 1, clearly a moonlight protein [17], was also present on cell surface.
Finally, about 10% of the proteins had no predicted location but it is possible that some of them are located at the cell surface.
3.2. Extracellular proteins
The mass peaks of the peptides obtained from the medium were compared with protein sequences in the NCBI and UniProt databases. A total of 99 proteins of the extracellular proteome were identified (Table A2). The database ID, MW, pI value, function, biological process and cell location were obtained by interrogation of the Uniprot database (Table A2). The proteins identified outside cells are from several different functional classes, acting in cell division, fatty acid biosynthesis, glycolysis, GMP biosynthesis, pathogenesis, protein biosynthesis, transcription, translation, and 36 other minor functions.
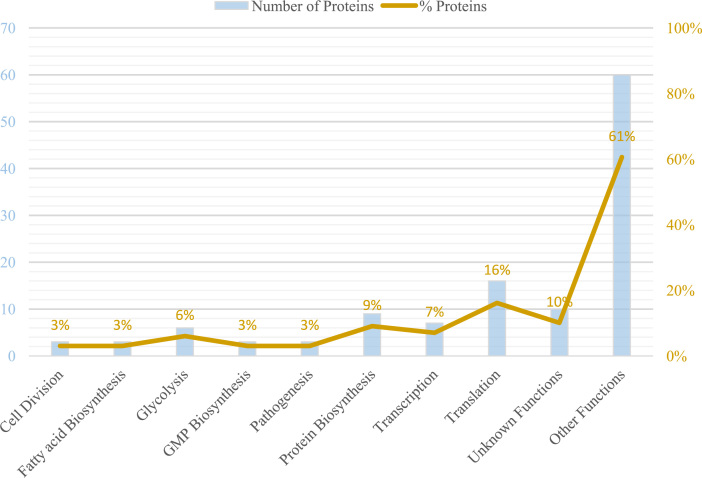
Most proteins (16%) are related to translation activities (Fig. 2). However no associated function was found for as many as 10% of the identified proteins in either of the two databases interrogated. The locations of the 99 identified proteins were predicted by PSORTb. Most proteins, 61 of them, were predicted to be cytoplasmic. For the other proteins, 4 were predicted to be located in plasma membranes, 4 in the cell wall and 13 in the extracellular medium. No location was predicted for 17 proteins.
Fig. 2.
Biological functions associated with 99 proteins identified in extracellular fraction. Number (blue) and percentages (orange) of the identified proteins associated with different biological functions. “Other functions” encompasses 36 functions for which 2 or fewer proteins were identified where no functional category contained more 2% of the total number of proteins.
Like other Gram-positive bacteria, the pathogenicity of S. aureus is largely dependent on the secretion of numerous extracellular virulence factors. Therefore, the clinical performance of an antibiotic in treating S. aureus infection is determined not only by its bactericidal or bacteriostatic activities, but also by how it affects the release of virulence factors.
Some cytoplasmic proteins were detected in the extracellular extracts. This may be because some bacterial cell autolyse or some proteins occur at multiple locations like malate:quinone oxidoreductase and some hypothetical proteins (Table A2).
Biological functions of the identified proteins were diverse though the functions of ~17% were unknown (Table A2). Previous studies showed a similar percentage of proteins with unknown function in S. aureus. Indeed the S. aureus genome codes for around 2600 proteins of which around 1000 proteins are of unknown function [4], [18].
Most well-known virulence factors that are secreted by Gram-positive pathogens have a typical N-terminal signal peptide that directs them into the general secretory pathway (Sec), e.g. exotoxins of S. aureus, the protective antigen of Bacillus anthracis, listeriolysin O of Listeria monocytogenes. Proteins secreted by this pathway are translocated across the cytoplasmic membrane in an unfolded state. Subsequent processing and folding of these proteins take place in the cell wall environment on the trans-side of the membrane [19]. The results of our study are in accordance with those of Wooldridge et al. [19] who showed using SecretomeP software that most S. aureus expo-proteins are exported from the cytoplasm via the Sec pathway. Some studies suggest that MRSA-specific proteins do not follow the signal peptide-dependent secretion pathways, which SecretomeP would predict to be secreted via the classical pathway [20], [21].
Two of the predicted extracellular proteins are known to be related to virulence: the immunoglobulin-binding protein Sbi (A7×659, Table 1) and leukocidin F-subunit (H7G6G2). Sbi is an adhesin that facilitates evasion from the host immune response and colonization of the host by binding to cytoplasmic proteins from host endothelial cells. The leukocidin F-subunit is a component of Panton-Valentine leukocidin (PVL), one of the most important toxins in the infection process of S. aureus. PVL is a two-component leukotoxin that forms pores in cell membranes and is associated with infections of skin and soft tissue [22]. This toxin is encoded by lukF-PV and lukS-PV, two contiguous genes in the genome of various bacteriophages [23].
Table 1.
Proteins identified in the extracellular proteome and their respective predicted locations.
| Accession (Uniprot) | Protein name | Location |
|---|---|---|
| H7G701 | Uncharacterized protein | Extracellular |
| H7G3A4 | Iron-regulated surface protein isdA | Cell wall |
| A7×659 | Immunoglobulin-binding protein sbi | Secreted |
| H0CIZ5 | Triacylglycerol lipase | Secreted |
| H0CJP3 | Sphingomyelin phosphodiesterase | Extracellular |
| G7ZQA4 | Transglycosylase sceD | Extracellular |
| F5WI91 | Heme uptake protein | Cell wall |
| D2FNS2 | Uncharacterized protein | Extracellular |
| H7G6G2 | F subunit of leukocidin | Extracellular |
Two more proteins associated with the pathogenicity of S. aureus were identified but they could not be assigned a location. Both proteins (F9KZ14 and H4BPR3, Table 1) belong to a family of proteins with hemolytic activity. Antibacterial protein 1 (F9KZ14) inhibits the growth of gonococci. Among the extracellular proteins, we identified two putative uncharacterized proteins (H7G701 and D2FNS2).
In the present study, IsdA (H7G3A4, Table 1), a protein that also acts as an adhesin and promotes cell binding to immobilized whole cells proteins, was identified [24]. The IsdA protein is expressed at the S. aureus surface during infection required for human nasal colonization or survival on human skin [24].
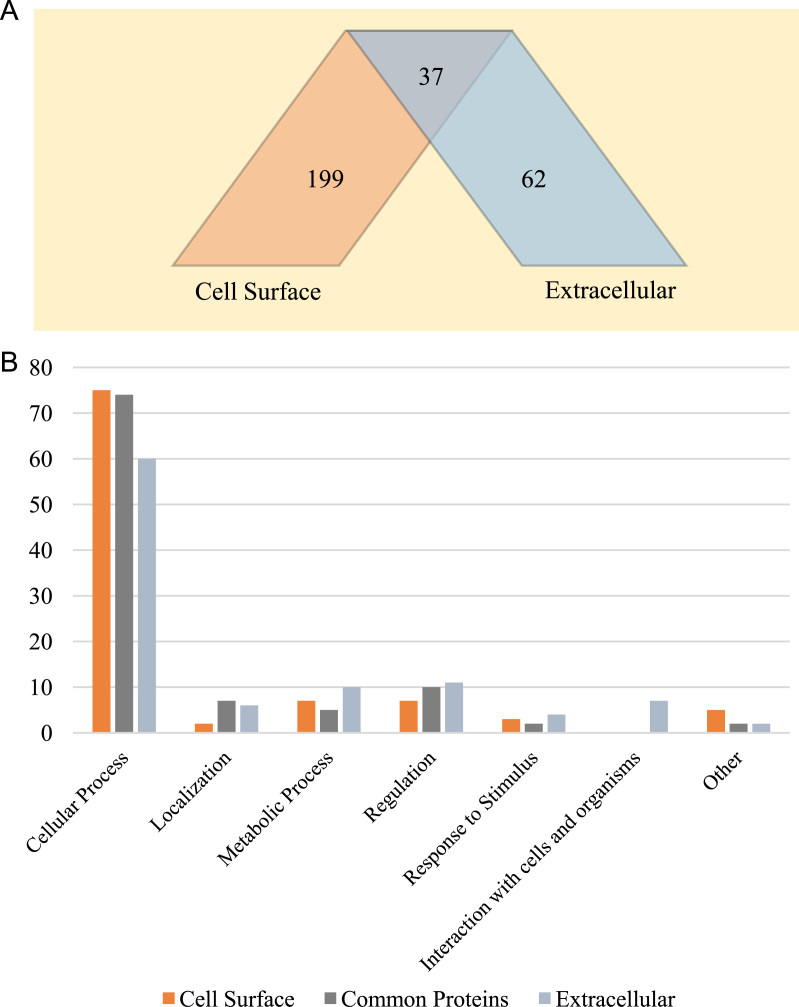
All protein accession numbers, from both proteome fractions, were input into statistical Venn diagram software, to compare the 236 accession identified in the cell surface fraction with the 99 from the extracellular fraction. The output reveals that 37 proteins are common to both proteomes (Fig. 3). These common proteins were analyzed by PSORTb software to predict their location into the cell. STRAP software was used to present the data according to biological processes in which the proteins are implicated (Fig. 3) [25]. As expected, the extracellular proteome is less weighted towards cellular processes but more involved in regulation and metabolic processes and interactions with cells and organisms. The cell surface fraction contained more different kinds of proteins, more than 70% of which are involved in cellular processes. The proteins common to both fractions were mainly related to cellular processes, then to regulation. The presence of cytosolic proteins in both fractions complicates this kind of analysis, but it is still possible to differentiate which processes are most related to different proteomes or subproteomes.
Fig. 3.
(A) Venn diagram generated by STRAP software showing subcellular location of 236 accessions from the cell surface fraction and 99 from the extracellular fraction. The intersection shows the number of proteins common to both fractions. (B) Chart comparing the funtions of cell surface, extracellular and common proteins between these 2 compartments.
All 37 common proteins were predicted to be cytosolic proteins. Even with large quantities of cytosolic proteins in the surfaceome and extracellular proteome, whether contaminants or not, both proteome fractions are distinct and we hypothesize that our methods and protocols are close to obtaining specific subcellular proteomes.
4. Conclusions
The protein profile of MRSA C2355 showed evidence of the microorganism's phenotypic characteristics, consistent our current knowledge of the S. aureus genome with the overall metabolic regulation and cellular physiology expected for MRSA ST398. Identifying bacterial infection proteins like catalase and IsdA in the extracellular proteome is further evidence of the invasiveness of MRSA ST398. Future studies may identify novel determinants for this strain and elucidate the patho-physiological processes that facilitate the survival and replication of MRSA ST398 within the host that can be targeted to find treatments. The moonlight activity of some essential proteins is a challenge for proteomics, then the experimental confirmation of these moonlighting proteins will extend the localization data in databases. Finally, these results show that the cell surface of S. aureus strains is not only highly variable, but also that the proteins displayed are very heterogeneous. Further characterization of environmental antibiotic resistance protein variants will benefit studies of human health and knowledge of the sequence variation of resistance proteins could direct the design of new antibiotics.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi:10.1016/j.bbrep.2015.07.004.
Appendix A. Supplementary material
Supplementary material Appendice Table A1. Identified proteins by cell shaving method of MRSA ST398 based on LC-MS/MS analysis.
Supplementary material Appendice Table A2. Identified proteins by extracellular protein recovery method of MRSA ST398 based on LC-MS/MS analysis.
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 2005;11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilmore J.M., Washburn M.P. Advances in shotgun proteomics and the analysis of membrane proteomes. J. Proteomics. 2010;73:2078–2091. doi: 10.1016/j.jprot.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Novick R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 4.Kuroda M., Ohta T., Uchiyama I., Baba T., Yuzawa H., Kobayashi I. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 5.Baba T., Takeuchi F., Kuroda M., Yuzawa H., Aoki K., Oguchi A. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 6.Gill S.R., Fouts D.E., Archer G.L., Mongodin E.F., Deboy R.T., Ravel J. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 2005;187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holden M.T., Feil E.J., Lindsay J.A., Peacock S.J., Day N.P., Enright M.C. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen P.A., Xu X., Matsunaga J., Sanchez Y., Ko A.I., Haake D.A. Surfaceome of Leptospira spp. Infect. Immun. 2005;73:4853–4863. doi: 10.1128/IAI.73.8.4853-4863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solis N., Larsen M.R., Cordwell S.J. Improved accuracy of cell surface shaving proteomics in Staphylococcus aureus using a false-positive control. Proteomics. 2010;10:2037–2049. doi: 10.1002/pmic.200900564. [DOI] [PubMed] [Google Scholar]
- 10.Nakai K. Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 2000;54:277–344. doi: 10.1016/s0065-3233(00)54009-1. [DOI] [PubMed] [Google Scholar]
- 11.Tjalsma H., Lambooy L., Hermans P.W., Swinkels D.W. Shedding & shaving: disclosure of proteomic expressions on a bacterial face. Proteomics. 2008;8:1415–1428. doi: 10.1002/pmic.200700550. [DOI] [PubMed] [Google Scholar]
- 12.Maione D., Margarit I., Rinaudo C.D., Masignani V., Mora M., Scarselli M. Identification of a universal Group B streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozano C., Aspiroz C., Ezpeleta A.I., Gomez-Sanz E., Zarazaga M., Torres C. Empyema caused by MRSA ST398 with atypical resistance profile, Spain. Emerg. Infect. Dis. 2011;17:138–140. doi: 10.3201/eid1701.100307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Ortega M.J., Norais N., Bensi G., Liberatori S., Capo S., Mora M. Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 2006;24:191–197. doi: 10.1038/nbt1179. [DOI] [PubMed] [Google Scholar]
- 15.A. Copeland, Complete Sequence of Chromosome of Staphylococcus aureus subsp. aureus JH9. The EMBL/GenBank/DDBJ databases 2007. (submitted for publication).
- 16.Dreisbach A., Hempel K., Buist G., Hecker M., Becher D., van Dijl J.M. Profiling the surfacome of Staphylococcus aureus. Proteomics. 2010;10:3082–3096. doi: 10.1002/pmic.201000062. [DOI] [PubMed] [Google Scholar]
- 17.Renier S., Chambon C., Viala D., Chagnot C., Hebraud M., Desvaux M. Exoproteomic analysis of the SecA2-dependent secretion in Listeria monocytogenes EGD-e. J. Proteomics. 2013;80:183–195. doi: 10.1016/j.jprot.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 18.Burlak C., Hammer C.H., Robinson M.A., Whitney A.R., McGavin M.J., Kreiswirth B.N. Global analysis of community-associated methicillin-resistant Staphylococcus aureus exoproteins reveals molecules produced in vitro and during infection. Cell Microbiol. 2007;9:1172–1190. doi: 10.1111/j.1462-5822.2006.00858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wooldridge K. Caister Academic Press; Wymondham: 2009. Bacterial Secreted Proteins: Secretory Mechanisms and Role in Pathogenesis. [Google Scholar]
- 20.Bendtsen J.D., Kiemer L., Fausboll A., Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enany S, Yoshida Y, Yamamoto T. Exploring extra-cellular proteins in methicillin susceptible and methicillin resistant Staphylococcus aureus by liquid chromatography-tandem mass spectrometry. World J. Microbiol. Biotechnol. 2014;30(4):1269–1283. doi: 10.1007/s11274-013-1550-7. [DOI] [PubMed] [Google Scholar]
- 22.Boyle-Vavra S., Daum R.S. Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton–Valentine leukocidin. Lab. Investig.: J. Tech. Methods Pathol. 2007;87:3–9. doi: 10.1038/labinvest.3700501. [DOI] [PubMed] [Google Scholar]
- 23.Boakes E., Kearns A.M., Ganner M., Perry C., Hill R.L., Ellington M.J. Distinct bacteriophages encoding Panton–Valentine leukocidin (PVL) among international methicillin-resistant Staphylococcus aureus clones harboring PVL. J. Clin. Microbiol. 2011;49:684–692. doi: 10.1128/JCM.01917-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke S.R., Andre G., Walsh E.J., Dufrene Y.F., Foster T.J., Foster S.J. Iron-regulated surface determinant protein A mediates adhesion of Staphylococcus aureus to human corneocyte envelope proteins. Infect. Immun. 2009;77:2408–2416. doi: 10.1128/IAI.01304-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhatia V.N., Perlman D.H., Costello C.E., McComb M.E. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal. Chem. 2009;81:9819–9823. doi: 10.1021/ac901335x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Appendice Table A1. Identified proteins by cell shaving method of MRSA ST398 based on LC-MS/MS analysis.
Supplementary material Appendice Table A2. Identified proteins by extracellular protein recovery method of MRSA ST398 based on LC-MS/MS analysis.
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material