Abstract
gabra2 gene codes for the alpha-2 subunit of the GABAA receptor, one of the ionotropic receptors which has been related to anxiety, depression and other behavioural disorders, including drug dependence and schizophrenia. GABAergic signalling also plays a role during development, by promoting neural stem cell maintenance and renewal. To investigate the role of gabra2 in CNS development, gabra2 deficient zebrafish were generated. The pattern of proliferation during the embryonic development was disrupted in morphant embryos, which also displayed an increase in the number of apoptotic nuclei mainly at the mid- and hindbrain regions. The expression of several genes (notch1, pax2, fgf8 and wnt1) known to contribute to the development of the central nervous system was also affected in gabra2 morpholino-injected embryos, although no changes were found for pax6a and shh a expression. The transcriptional activity of neuroD (a proneural gene involved in early neuronal determination) was down-regulated in gabra2 deficient embryos, and the expression pattern of gad1b (GABA-synthesising enzyme GAD67) was clearly reduced in injected fish. I propose that gabra2 might be interacting with those signalling pathways that regulate proliferation, differentiation and neurogenesis during the embryonic development; thus, gabra2 might be playing a role in the differentiation of the mesencephalon and cerebellum. Given that changes in GABAergic circuits during development have been related to several psychiatric disorders, such as autism and schizophrenia, this work might be helpful to understand the role of neurotransmitter systems during CNS development and to assess the developmental effects of several GABAergic drugs.
Abbreviations: CNS, central nervous system; fgf8, fibroblast growth factor 8; GABA, γ-aminobutyric acid; GABAA, gamma-aminobutyric acid (GABA) A receptor; gabra2, gamma-aminobutyric acid receptor subunit alpha-2; gad1b, glutamate decarboxylase; hpf, hours post-fertilisation; ISH, in situ hybridisation; KCC2, neuron-specific potassium/chloride cotransporter 2; MHB, mid-hindbrain boundary; neuroD, neurogenic differentiation; notch1a, notch homologue 1a; ORF, open reading frame; pax2a, paired box gene 2a; pax6a, paired box gene 2a; shh a, sonic hedgehog; wnt1, wingless-type MMTV integration site family, member 1
Keywords: GABA, Gabra2, Central nervous system, Development, Proliferation, Differentiation
Highlights
-
•
gabra2 might have a role in the regulation of proliferation during CNS development.
-
•
The expression of notch1, pax2a, fgf8 and wnt1 is altered in gabra2 deficient fish.
-
•
neuro D expression, is down-regulated in the absence of a functional Gabra2.
-
•
The generation of GABAergic neurons might be reduced in gabra2 morphants.
-
•
gabra2 may interact with several signalling pathways that harness CNS development.
1. Introduction
The γ-aminobutyric acid (GABA) is the main inhibitory neurotransmitter in the vertebrate central nervous system (CNS), and elicits its actions by binding to three different classes of receptors: the ionotropic receptors GABAA, and GABAC, and the metabotropic receptors GABAB. [1]. The GABAA receptors are ligand-gated chloride channels, which usually reduce the membrane excitability by reducing the membrane potential to chloride potential (approximately −80 mM in neurons) [2], and thus inhibiting neurotransmitter release. The prototypical GABAA receptor is formed by five protein subunits arranged in a ring, shaping a central chloride pore. Each subunit contains four transmembrane α-helices, a C-terminal and a large N-terminal domains, both located on the extracellular face of the plasma membrane. There are different types of subunits that can be combined in several ways to form a functional GABAA receptor, although most of them contain two alpha, two beta and one gamma subunit [3]. Different receptor subtypes display differences in their location, affinity for GABA, the properties of the chloride channel and in their pharmacology. The GABAA receptor has one binding site for GABA, its agonists (e.g. muscimol) and antagonists (e.g. bicuculline) at the α–β subunit interface, and several other binding sites for different modulators, as for example benzodiazepines (at the α–γ interface), neurosteroids and ethanol [4], [5], [6], [7].
Although GABA is an inhibitory neurotransmitter in the adult brain, there is evidence that it also has an excitatory role during the CNS development in mammals [8]. In the immature brain, GABAergic synapses, which are formed prior to the glutamatergic synapses, are mainly excitatory, due to the increased concentration of intracellular chloride ions; in this way, GABA keeps immature neurons in a depolarised state. The transition of GABA signalling takes place after the expression of the neuron-specific potassium/chloride cotransporter 2 (KCC2), which shifts the chloride equilibrium potential, and then GABAergic synapses become inhibitory [9], [10]. Besides, GABAergic signalling also plays a role in neural stem cell maintenance and renewal, as stem cells express GABA receptors and release GABA [11]. It seems that auto/paracrine GABA action upon GABAA receptors reduces proliferation of neural stem cells by blocking them in the S phase, but it does not affect differentiation [12], [13]. Pleiotropic effects of GABA signalling during brain development include modulation of proliferation and neuronal maturation. During cortical development, migrating interneurons release GABA, which activates GABAA receptors in the radial glia, thus reducing progenitor proliferation and the migration of immature pyramidal neurons. GABA signalling is also required for dendritic arborisation and synaptogenesis in cortical neurons. In the subventricular zone, GABA reduces proliferation and migration of young neurons to the rostral migratory stream. In the other stem cell niche of the adult brain, GABA release regulates the synaptic integration and maturation of newborn granule cells in the dentate gyrus of the hippocampus [14].
The pharmacological interest of GABAA receptor is unquestionable, as it is the target of several neuroactive drugs, such as benzodiazepines, barbiturates, picrotoxine and muscimol. Besides, GABAA receptor also plays a role in the ethanol-elicited depressor response [15]. GABAA receptors mediate many behavioural effects of ethanol, including sedation, reduction in anxiety, motor coordination impairment [16], stimulation of the mesolimbic reward circuitry [17] and withdrawal symptoms [16]. It has been reported that the GABAA receptor alpha-2 subunit (gene name, gabra2) is a key protein in the control of spinal pain, as knock-out mice for gabra2 gene displayed reduced analgesia after benzodiazepine administration [18]. This finding represents a further contribution to develop novel antinociceptive agents that can specifically target this receptor subtype, thus eliciting analgesia but avoiding undesirable side-effects. Besides, α2-containing GABAA receptors have also been linked to anxiety, depression and other behavioural disorders, including drug dependence and schizophrenia [19]. Evidence from knock-out studies performed in mice indicates that gabra2 subunit may represent a pharmacological target to develop antidepressant drugs [20]. Also, several haplotypes of gabra2 gene have been associated to ethanol, nicotine and cannabis dependence [21], [22].
The validation of novel therapeutic agents and their potential effects on the desired targets requires a straightforward system to perform in vivo tests. In this line, the zebrafish clearly shows many advantages [23], [24]. It seems that GABA depletion is a common feature in treatment-resistant seizures, and GABAergic neurotransmission could stand for a novel therapeutic target. In this line, zebrafish seizure model has shown similar behavioural responses to mice models when treated with anti epileptic drugs [25] and locomotor assays with zebrafish larvae can be used to screen for novel anticonvulsant drugs [26]. Edge preference is a behavioural assay with zebrafish larvae that measures anxiety, and the anxiolytic effect of benzodiazepines that bind to GABAA receptors is reflected as a reduction in edge preference [27]. In this work I analyse the role of the gabra2 gene in the CNS development, and its potential cross-talk with other morphogen cascades and neurotransmitter systems that control neuronal development. These findings might be helpful to evaluate the developmental effects of several drugs that target the GABAergic system.
2. Materials and methods
2.1. Zebrafish maintenance, breeding and drug treatments
The experiments were performed using the wild type AB zebrafish line. General maintenance and care of fish were carried out according to standard protocols in our own zebrafish facility. Animals were maintained at a constant temperature of 28 °C in a 14 h light cycle and fed three times a day. Embryos were obtained by natural mating and cultured in E3 medium, with or without the addition of 0.003% 1-Phenyl-2-thiourea (PTU, Sigma) to inhibit pigmentation. Embryos were staged according to hours post fertilisation (hpf) [28]. In all experiments, adequate measures were taken to minimise pain or discomfort and animals were handled according to the guidelines of the European Communities Council Directive 2010/63/UE, to the current Spanish legislation for the use and care of animals RD 53/2013 (BOE-A-2013-1337) and to the Guide for the Care and Use of Laboratory animals as adopted and promulgated by the U.S. National Institutes of Health (2011). All experiments were performed at the University of Salamanca with the approval of the University of Salamanca Animal Care Committee.
Zebrafish embryos were exposed to 1 μM GABA, 1 μM muscimol or 100 μM bicuculline in E3 embryo buffer from 5 hpf to 24 hpf. A control group (no drug treatment) was also included for comparison.
2.2. Cloning
The complete open reading frame (ORF) from gabra2 gene was cloned using standard procedures and the oligonucleotides described in Table S1. Briefly, a partial ORF for gabra2 gene was initially cloned and then the amplification of the 5′ unknown sequence was achieved by 5′ RACE (5′/3′ RACE Kit, Roche Applied Science) following the protocol described by the supplier; then two oligonucleotides were designed to amplify the complete ORF and subcloned in pCR®4-TOPO® vector (Invitrogen, Life Technologies). The partial ORF for gad1b was also amplified and subcloned into the same vector. To confirm the sequence of the cloned inserts, DNA sequencing was performed by the Sequencing Facility (Servicio de Secuenciación, Universidad de Salamanca, Spain). These clones were used to generate the corresponding in situ riboprobes.
2.3. Sequence analysis
Amino acid sequences of Gabra2 proteins from zebrafish (dr – Danio rerio), human (hs – Homo sapiens), chimpanzee (pt – Pan troglodytes), cow (bt – Bos taurus), mouse (mm – Mus musculus), rat (rn – Rattus norvegicus), xenopus (xt – Xenopus tropicalis) and two other fish species (tn – Tetraodon nigroviridis and ga – Gasterosteus aculeatus) were obtained from the Treefam record of the family (Database accession number TF315453) and were automatically aligned with MUSCLE [29]. The resulting alignment was analysed using JalView [30] and the ClustalX colouring scheme was used. SMART software (http://smart.embl-heidelberg.de/) was used to perform the protein structure analysis.
2.4. Homology modelling
The 3D structure of Gabra2 subunit from D. rerio was predicted by homology modelling using Pyhre2 server [31] using the crystal structure of the human β3 homopentamer of GABRAA receptor (4COF PDB ID) [32] as template. Phyre2 investigator was run to perform an in-depth analysis of model quality and conservation, and benzodiazepine binding domain was predicted by Fpocket [33]. Binding loops for GABA and for benzodiazepines were determined by homology using clustalW alignment with β3, α1, α2 and α6 subunits of human GABAA receptor. The amino acid sequence of these subunits was retrieved form UNIPROT. The 3D modelled structure was visualised and molecular graphics and analyses were performed with the UCSF Chimera package [34].
2.5. In situ hybridisation
The following riboprobes were used: fgf8 (fibroblast growth factor 8), gad1b (glutamate decarboxylase GAD67), neuroD (neurogenic differentiation), notch1a (notch homologue 1a), pax2a (paired box gene 2a), pax6a (paired box gene 2a), shh a (sonic hedgehog a) and wnt1 (wingless-type MMTV integration site family, member 1a). The probes were transcribed from their templates using the DIG RNA Labelling Kit (SP6/T7) (Roche Applied Science) following the manufacturer's instructions. In situ hybridisations were performed on wholemounts as previously described [35], and probes were detected using BM purple or NBT/BCIP reagents (Roche Applied Science). Control experiments were always performed in parallel. Images were taken with a Leica M165FC Stereomicroscope. 25 embryos were scored for each treatment and for each time point, and each experiment was repeated three times.
2.6. MO injections: knockdown assays at one-cell stage
The translation blocking antisense morpholino GAGAATGACCCCGTCGCAGAATCAT complementary to the region from +1 to +25 of the dre-gabra2 ORF was purchased from GeneTools LLC and was microinjected at the 1-cell stage at a concentration of 0.25 mM. As control, the standard control morpholino (also available from GeneTools) was injected at the same concentration.
2.7. Immunohistochemistry
Immunohistochemical studies were performed either on wholemounts or on cryosections and analysed with Leica M165FC Stereomicroscope or with an Olympus AX70 fluorescence microscope. In some cases, images were taken with a confocal microscope (Leica microsystems). The rabbit anti-histone H3 (phospho Ser10, Abcam Cat# ab5176, RRID:AB_304763) was used as primary antibody (1:100) and Alexa Fluor 594 conjugated anti-rabbit (Molecular Probes (Invitrogen) Cat# A11012, RRID:AB_141359) as secondary antibody (1:500). Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Roche Applied Science). TUNEL assays to determine apoptosis were performed in toto using the ApopTag® Peroxidase In Situ Apoptosis Detection Kit (Millipore) following the manufacturer’s instructions.
2.8. Quantitative real-time PCR analysis
qPCR assays were performed as previously described [36]. Total RNA from zebrafish embryos was isolated using the Trizol reagent (Invitrogen, Life Technologies), treated with DNase I (New England Biolabs) and 1 μg of total RNA was reverse transcribed with the QuantiTect Reverse Transcription Kit (QiaGen). The primers used to specifically amplify the DNA fragments are listed in Table S2. These primers were designed with the Universal ProbeLibrary Assay Design Center webtool from Roche Applied Science (https://www.roche-applied-science.com/sis/rtpcr/upl/acenter.jsp?id=030000), or taken from [37]. RT-PCR experiments were performed following the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines [38]. The amplification reactions were carried out in 25 µL final volume using Power SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies) and the real-time polymerase chain reaction was performed using the ABI Prism 7300 Applied Biosystems (Life Technologies). Ct values were calculated with the SDS v1.3.1 software (Applied Biosystems, Life Technologies) for each gene and the cDNA abundance of each transcript was calculated relative to the expression of the housekeeping gene eef1a1l1 using the REST-384© v2 software [39]. Also, standard curves for each gene and negative controls (NTC: no template control, and RNA: RNA which was not reverse transcribed) were included in each PCR reaction. Experiments were performed in triplicates and repeated four times. Statistical analysis was performed using Graph Pad Prism software.
3. Results
3.1. Characterisation of GABAA receptor alpha 2 subunit (gabra2) in the zebrafish
The complete open reading frame of a gabra2 gene from zebrafish was cloned using standard methodology. Briefly, the sequence of several partial ESTs were obtained from PubMed (National Library of Medicine: http://www.ncbi.nlm.nih.gov/pubmed), EMBL-EBI (http://www.ebi.ac.uk), ENSEMBL (http://www.ensembl.org/index.html) and ZFIN (The Zebrafish Model Organism Database; http://zfin.org). CAP3 programme was used to assemble all these ESTs in a putative cDNA sequence, and oligonucleotides were designed to verify that the predicted cDNA was indeed expressed in the zebrafish. Given that part of the 5′ region was still missing, a 5′RACE was performed, and finally the complete cDNA sequence of a gabra2 gene was cloned (Fig. S1) in the pCR2.1 TOPO vector.
Genomic analysis revealed that this gabra2 gene (ENSEMBL ID: ENSDARG00000070005) is located in chromosome 13 and comprises at least 10 exons which span along 31 Kbp (11998048–12029410). There are another 3 gene predictions in chromosome 13 that match gabra2 gene (ENSDARG00000087129, ENSDARG00000091459 and ENSDARG00000034011) but they only contain partial cDNAs. Up to now, it is not known whether these predictions stand for tandem repeats or if they are assembly errors which have not been manually curated so far.
The following protein domains have been identified in zebrafish Gabra2 using the SMART software (Fig. S1): (a) a signal peptide (residues 1–27); (b) two regions with intrinsic disorder (residues 30–38 and 437–451); (c) a extracellular ligand binding domain “Neur_chan_LBD” (PFAM accession number PF02931), spanning amino acid residues 42 to 250; and (d) a transmembrane domain “Neur_chan_memb” (PFAM accession number PF02932) from residue 257 to 434, which contains four transmembrane α-helices. These two latter domains are characteristic of the ligand-gated ion channel superfamily, which comprises GABAA, nicotinic acetylcholine, glycine, 5-HT3 serotonine and the ionotropic glutamate receptors.
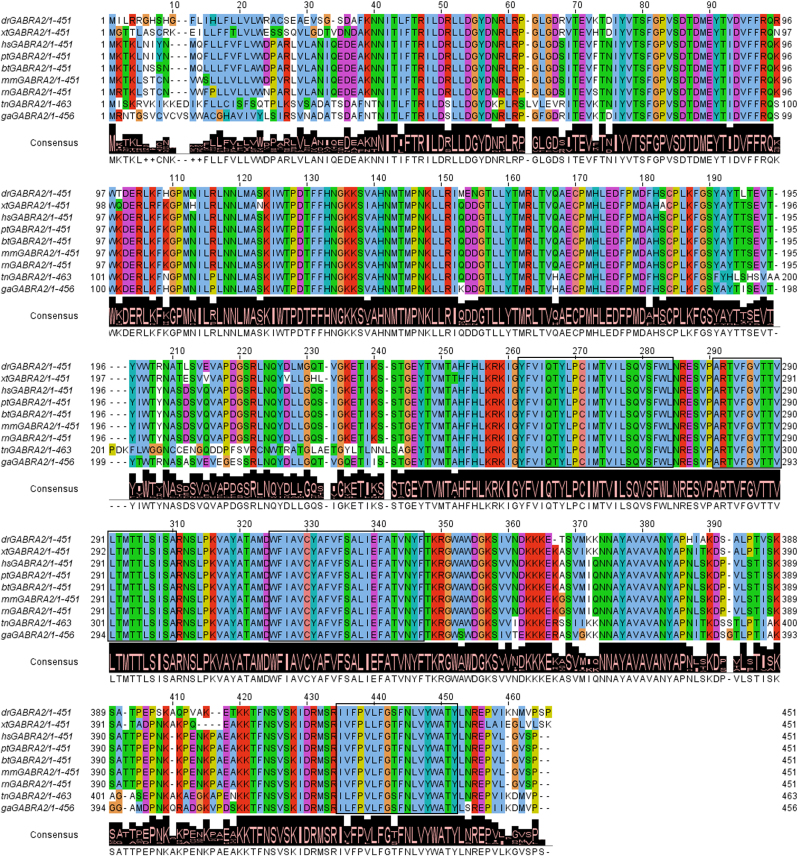
To determine if Gabra2 protein was conserved among the evolutionary scale, the full-length sequences for Gabra2 protein in different vertebrate species (mammals, amphibians and fish) were obtained from TreeFam (Database accession number TF315453) and aligned with MUSCLE from JalView (Fig. 1). The protein sequence is highly conserved (about 81% identity of all aligned sequences when compared to Gabra2 from zebrafish, except for the N-terminal signal peptide and three internal regions (residues 200–230, 350–370 and 390–420). In the case of Tetraodon, there are some amino acid substitutions that could be due to sequencing errors or to a poor sequence quality.
Fig. 1.
Multiple sequence alignment of Gabra2 proteins from zebrafish (dr – Danio rerio), human (hs – Homo sapiens), chimpanzee (pt – Pan troglodytes), cow (bt – Bos taurus), mouse (mm – Mus musculus), rat (rn – Rattus norvegicus), xenopus (xt – Xenopus tropicalis) and two other fish species (tn – Tetraodon nigroviridis and ga – Gasterosteus aculeatus). The initial set of sequences was obtained from the Treefam record of the family (Database accession number TF315453) and were automatically aligned using MUSCLE and manually curated with JalView. The consensus sequence and conservation logo is shown below. The ClustalX colouring scheme is used and the four transmembrane regions are boxed. Note the high degree of sequence similarity for Gabra2 protein among all vertebrate species.
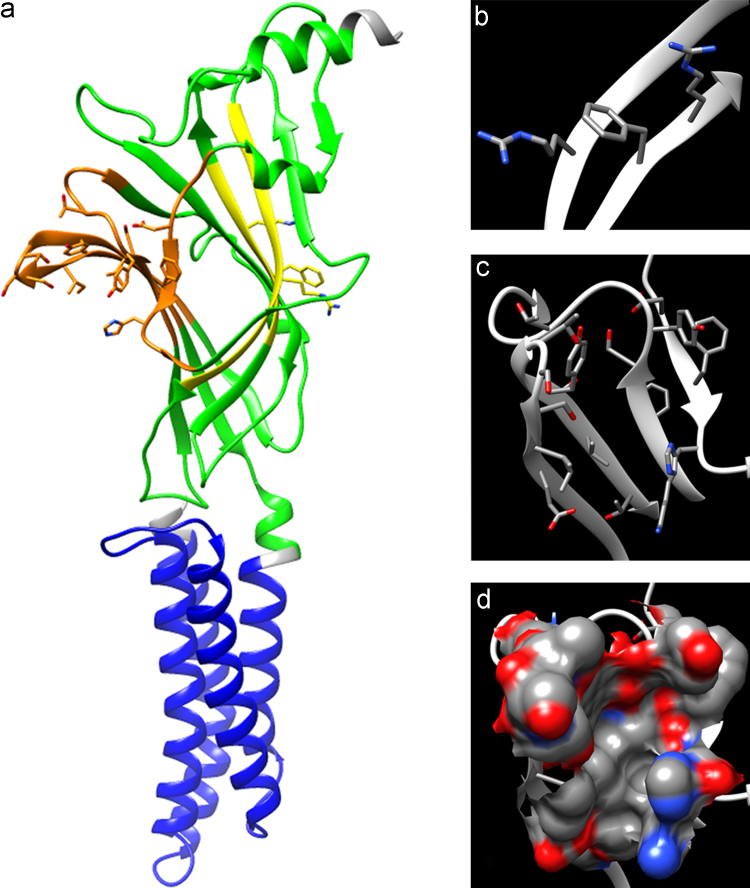
In an attempt to build a homological model, the predicted zebrafish Gabra2 protein sequence was submitted to Phyre2 to generate alignments against proteins with published structures. The human β3 homopentamer of GABAA receptor was the most closely related protein whose crystal structure has been solved (4COF PDB ID) [32], so that it was selected as template for homology-based modelling, as the functional domains are conserved. Residues 36–438 were modelled at 100% confidence (Fig. 2), so that the homological model of Gabra2 is consistent with conservation of the overall three-dimensional structure between these two proteins. Functional domains were annotated and binding pockets were predicted by homology using clustalW alignment with β3, α1, α2 and α6 subunits of human GABAA receptor. The transmembrane region displays the characteristic 4 α-helices and the extracellular domain is mainly formed by β-sheets connected by coiled regions. GABA binding loop D spans amino acid residues Asp90 to Arg 96 and loop E spans Thr153 to Ala 164 (Fig. S1). GABA contacting amino acid residues Phe92, Arg94 (in Loop D) and Arg159 (in loop E) [40], [41] are conserved within this domain. Benzodiazepine binding loop A spans amino acid Trp122 to Lys132, loop B Leu182 to Val194 and loop C Thr224 to Ala242 (Fig. S1); residues known to be important for benzodiazepine binding [42], [43] are fully conserved. Besides, the benzodiazepine binding pocket was also predicted in this domain, which supports the existence of a functional benzodiazepine binding domain.
Fig. 2.
Gabra2 homological model. (a) The 3D structure of zebrafish gabra2 subunit was generated by homology modelling using the crystal structure of human GABRB3 (PDB ID 4COF) as template. The protein structure is displayed as silhouette (rounded ribbon), and the Neur_chan_memb domain is coloured in blue, the Neur_chan_LBD domain in green and the two regions with intrinsic disorder are left in white. In the extracellular domain, GABA binding loops D and E are coloured in yellow and benzodiazepine binding loops A–C in orange. Side chains of important amino acid residues for ligand binding (either for GABA or for benzodiazepines) are shown and coloured by heteroatom. (b) GABA binding loops D (Asp90 – Arg 96) and E (Thr153 – Ala 164). Side chains of Phe92, Arg94 and Arg159 (amino acid residues known to interact with GABA) are shown and coloured by element. (c) Benzodiazepine binding loops A–C (Trp122 – Lys132, Leu182 – Val194 and Thr224 – Ala242). Important amino acid residues for ligand binding are shown and coloured by element. (d) Molecular surface of benzodiazepine binding site in GABRA2 subunit.
3.2. gabra2 developmental expression
The spatio-temporal distribution of gabra2 was assayed by wholemount in situ hybridisation on staged embryos which were fixed at different developmental stages (Fig. 3). To reveal the expression in some internal nuclei, 20 μm-thick sections were obtained from the stained embryos. The onset of expression begins before the end of the CNS segmentation period, as a diffuse expression in the neural tube was already seen at the stage of 11 somites, with stronger labelling at the presumptive mid- and hindbrain areas, and at the tip of the tail. At 24 hpf the staining was more intense and localised in several positive nuclei of the brain and in the floor plate of the spinal cord. From 48 hpf until the end of the embryonic development, gabra2 expression was found in the three brain regions (fore-, mid- and hindbrain), whereas the positive labelling in the spinal cord was significantly reduced.
Fig. 3.
Developmental expression of gabra2 transcript. Analysis of dre-gabra2 expression by in situ hybridisation on wholemount embryos of (a) 11 somites, (b) 17 somites, (c) 20 hpf, (d) 24 hpf, (e) 30 hpf, (f) 36 hpf, (g) 42 hpf, (h) 48 hpf, (i) 3 dpf and (j) 4 dpf. (a–j) lateral views are shown, where embryos are oriented with the anterior towards the left. Inset of (g): ventral view of the head. (k–o) higher magnification images taken from 20 μm-thick sections that were obtained from stained embryos. (k) transverse section (oriented dorsal up) of a 24 hpf embryo at the level of the spinal cord, where intense labelling is seen at the floor plate of the neural tube. (l) coronal section (oriented anterior to the left) of an embryo of 48 hpf; staining is found lateral to the olfactory bulbs. (m and n) coronal sections (oriented anterior to the left) of embryos of 72 hpf, where gabra2 expressing cells are located at surroundings of the olfactory bulbs (m), in the ganglion cell layer of the retina (m) and in some nuclei of the thalamus and of the medulla oblongata (n). (o) coronal section (oriented anterior to the left) of an embryo of 4 dpf, where the olfactory epithelium is labelled with the gabra2 ISH probe.
3.3. Effects of dre-gabra2 knocking down on the development of the CNS
Since it has been shown that the GABAergic system is involved in the development of the CNS and in the maintenance of neural stem cells, I wanted to determine the effects of the absence of a functional Gabra2 protein. To achieve this, I have used a translation blocking morpholino to block the expression of gabra2 during development in zebrafish embryos. This morpholino did not significantly alter the expression levels of gabra3 (0.91±0.02 fold change at 24 hpf), which codes for another alpha subunit (α3) closely related to gabra2. An up-regulation of c-fos mRNA levels was observed in the morphant embryos (6.72±0.75 fold change at 24 hpf and 2.40±0.35 fold change at 48 hpf, as determined by qPCR). Morpholino microinjection produced several macroscopic defects, as smaller size of the embryos, changes in the morphology of the brain and an increase in the size of the ventricles (Fig. S2). These observations led me to think that gabra2 gene might be involved in CNS development.
3.4. gabra2 morphants display changes in the proliferation pattern in the CNS
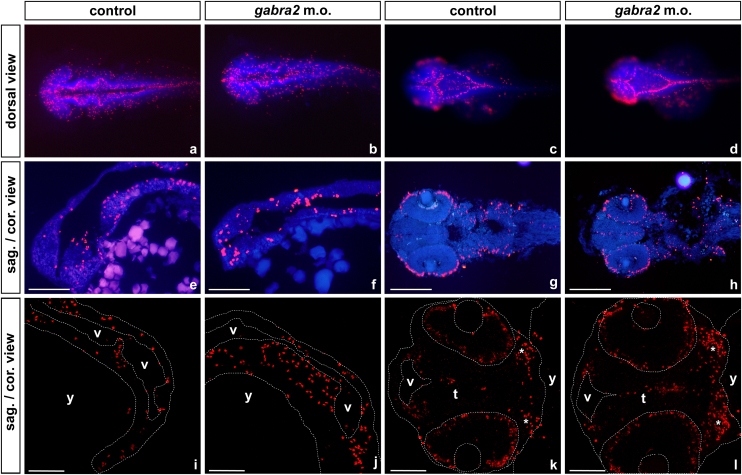
To determine whether knocking-down gabra2 expression affects CNS development by disrupting the proliferation pattern, phospho-histone 3 (Ser10) immunostaining (which specifically labels cells on the M phase) was performed in control and in morphant embryos of 24 and 48 hpf, both on wholemounts and on cryosections. The results are summarised in Fig. 4. At 24 hpf, the proliferation pattern was disorganised in gabra2 deficient embryos: in control embryos, positive cells were found along the periventricular layer of the brain, and mitotic cells were more widespread and dispersed in the morphants. Thus, the characteristic pattern of proliferation in the brain was lost, and many mitotic cells were found scattered outside of the proliferation niches. At 48 hpf, proliferation was more restricted to the periventricular areas of the brain, which will harbour the stem-cell niches in the adult CNS. At this stage, the morphant embryos seemed to recover the wild-type pattern of proliferation, as the walls of the third ventricle were clearly labelled; yet this labelling was more intense in gabra2 deficient embryos than in controls. In fact, experiments performed on 14 μm-thick cryosections showed an increase in the number of mitotically active cells along the midline of the tegmentum, in the retina and in the branchial arches. These results indicate that knocking-down gabra2 expression might be altering the proliferation of stem-cells and/or neural progenitors during the embryonic development.
Fig. 4.
Proliferation studies on gabra2 deficient embryos. Phospho-histone 3 (Ser10) immunostaining [23] on zebrafish control embryos and on embryos that were microinjected with gabra2 morpholino. In all cases embryos are oriented with the anterior towards the left and in (a–h) cell nuclei were counterstained with DAPI (blue). (a–d) Images from wholemount immunostainings that were taken with a Leica M165FC Stereomicroscope a and b) Wholemount embryos of 24 hpf, where it can be seen that proliferation pattern in the periventricular areas of the brain is disrupted in the morphant embryos. (c and d) At 48 hpf, the proliferative pattern is mainly restored, although the labelling for the mitotic marker is increased in the periventricular areas of the mid- and hindbrain from morpholino-injected embryos. (e–h) Images from 14 μm thick cryosections that were taken with an Olympus AX70 fluorescence microscope. (e and f) sagittal views of the fore- and midbrain from embryos of 24 hpf; in control embryos, positive cells are scattered throughout the ventral area of the brain, while mitotic cells show a more anterior and dorsal location in the morphant. (g and h) coronal sections from embryos of 48 hpf; proliferative cells are found along the midline of the tegmentum and in the outermost external cell layer of the retina in the morpholino-injected embryos, whereas control embryos display a more restricted pattern of proliferation in the CNS. (i–l) Higher resolution images of 14 μm thick cryosections taken with a Leica confocal microscope. The periphery of the embryos, the eyes, lens, ventricles and yolk are outlined with dashed lines. (i and j) mid-sagittal section of the hindbrain from embryos of 24 hpf, where it can be observed a dense distribution of positive cells along the wall of the ventricle in the morphant embryo. (k and l) coronal section of the developing brain at 48 hpf. The gabra2 deficient embryos display more labelled cells in the midline of the tegmentum and in the gill arches than controls. Labels: t: tegmentum; v: ventricle; y: yolk: *: gill arches. Scale bars: e and f: 200 μm; g and h: 100 μm; (i–l): 75 μm.
Each new cell can either keep its mitotic potential and remain in an undifferentiated state (as stem or progenitor cell) or can exit the cell cycle and differentiate into a specialized cell type. Cells which fail to acquire a specific fate usually undergo programmed cell death; thus, an increase in apoptosis can compensate for over-proliferation. To test whether this is the case in gabra2 deficient embryos, TUNEL assays have been performed (Fig. 5). Indeed, more apoptotic nuclei were found in several brain areas which displayed an excess of cells undergoing cell division. At 24 hpf, the hindbrain region in the morphant embryos displayed a greater number of TUNEL-positive cells as compared to controls. Apoptosis was clearly reduced at 48 hpf, although some differences were observed between controls and gabra2 deficient fish: while apoptosis is mostly absent in control cases, morpholino-injected embryos showed intense labelling in the walls of the third ventricle, as well as in some regions of the forebrain, namely the prectectum and the tectum opticum.
Fig. 5.
Apoptosis assays on gabra2 morphant embryos. TUNEL staining on gabra2 deficient embryos of 24 hpf (a and b – dorsal views; e and f – lateral views) and 48 hpf (c and d – dorsal views; g and h – 20 μm thick coronal sections). At 24 hpf, there is an increase in the number of apoptotic nuclei (which are positive for peroxidase labelling) in the hindbrain region of morpholino-injected embryos [69.30±15.52 apoptotic nuclei in control embryos as compared to 164.6±36.87 in morphant embryos; * p<0.05 as determined by one sample t-test]. Apoptosis is reduced at 48 hpf when compared to morphants at 24 hpf (* p<0.05 as determined by one sample t-test), although gabra2 deficient fish display apoptotic cells in the walls of the third ventricle and in some regions of the forebrain. [36.87±4.67 apoptotic nuclei in control embryos as compared to 77.06±5.34 in morphant embryos; ** p<0.01 as determined by one sample t-test]. (a–f) Embryos are oriented anterior towards the left and posterior towards the right. (g and h) Orientation of embryos is anterior up; scale bar: 100 μm.
3.5. Expression of several progenitor/differentiation markers in the morphant CNS
To assess the developmental effects of the absence of a functional Gabra2 protein, I have analysed the expression of several genes known to be part of those signalling cascades that are involved in the development of the CNS and neurogenesis. The changes in mRNA levels have been determined by qPCR (Table 1) and the differences in the expression pattern have been assayed by in situ hybridisation (Figs. 6 and S3). The following genes were studied: fgf8 (fibroblast growth factor 8), neuroD (neurogenic differentiation), notch1a (notch homologue 1a), pax2a (paired box gene 2a), pax6a (paired box gene 2a), shh a (sonic hedgehog) and wnt1 (wingless-type MMTV integration site family, member 1a). fgf8 mRNA expression was increased at both stages, whereas the opposite effect was seen for neuroD and pax2a. Interestingly, notch1a and wnt1 mRNA expression was down-regulated at 24 hpf, but no significant changes were observed at 48 hpf.
Table 1.
Changes in the expression levels of several progenitor/differentiation markers in dre-gabra2 deficient fish.
| gene | 24 hpf | 48 hpf |
|---|---|---|
| fgf8 | 1.42±0.10* | 1.51±0.07** |
| neuroD | 0.70±0.08* | 0.49±0.10* |
| notch1a | 0.55±0.03*** | 1.92±0.50 (n.s.) |
| pax2a | 0.65±0.10* | 0.74±0.10 (n.s.) |
| pax6a | 0.92±0.08 (n.s.) | 1.00±0.10 (n.s.) |
| shh a | 0.84±0.10 (n.s.) | 1.15±0.16 (n.s.) |
| wnt1 | 0.42±0.05** | 1.25±0.34 (n.s.) |
Results represent the relative gene expression fold-changes of several genes known to be part of signalling cascades that determine the development of the CNS in gabra2 morpholino-injected embryos of 24 hpf and 48 hpf. Data shown are normalised to dre-eef1a1l1. n.s. denotes p>0.05 (not significant);
denotes p<0.05;
denotes p<0.01;
denotes p<0.001 (unpaired Student t-test, n=4). 50 embryos were used for each treatment and for each time point; the experiment was performed in triplicates and repeated four times.
Fig. 6.
Effect of the knocking down of gabra2 gene on the expression pattern of several developmental markers. ISH studies were conducted on control embryos (a, c, e, g, i, k, m, o, q and s) and on gabra2 morphants (b, d, f, h, j, l, n, p, r and t) at 24 hpf (a, b, e, f, i, j, m, n, q and r) and 48 hpf (c, d, g, h, k, l, o, p, s and t) to establish the expression pattern of several markers known to be involved in CNS development: (a–d) fgf8; (e–h) neuroD; (i–l) notch1a; (m–p) pax2a; and (q–t) wnt1. Note that the expression patterns of all genes are altered in gabra2 deficient fish, and that the mid- and hindbrain regions are more affected than the forebrain. Embryos are oriented with the anterior towards the left and posterior towards the right. (a, b, k, l and o–t) dorsal views; (c–j, m and n) lateral views.
At 24 hpf, the expression pattern of fgf8 was more restricted in gabra2 deficient fish, as staining was localised at the presumptive mid-hindbrain boundary (MHB), but no changes in the expression pattern were observed at 48 hpf. There was a strong a generalised reduction of neuroD pattern of expression at 24 hpf, and at 48 hpf the staining a the cerebellum was lost. notch1a labelling was weaker in gabra2 morphants of 24 hpf, although the pattern of expression remained unaltered; however, al 48 hpf there was a stronger signal in the cephalic region, which might correspond to the tendency observed in the qPCR results (the expression levels were up-regulated, although they did not reach statistical significance). The expression pattern of pax2a became more localised in morphant embryos of 24 hpf, as very strong labelling was found a the anterior part of the forebrain, at the cerebellum and at the hindbrain. At 48 hpf, the expression of pax2a, which is restricted to the cerebellum and medulla oblongata, was weaker in gabra2 deficient fish than in control embryos. At 24 hpf wnt1 labelling in gabra2 morphants was clearly weaker, as all expression areas, namely the dorsal midbrain, MHB, dorsal hindbrain and spinal chord, seemed to be affected. At 48 hpf the staining was stronger at the dorsal lateral hindbrain (that is, the walls of the third ventricle), which might resemble the qPCR results (an up-regulation of the levels of expression, although not statistically significant). The expression pattern of both pax6a and shh a remained unaltered in gabra2 deficient fish (Fig. S3), which is in accordance with the obtained qPCR results for these two genes.
3.6. Expression of gad1b in gabra2 deficient fish
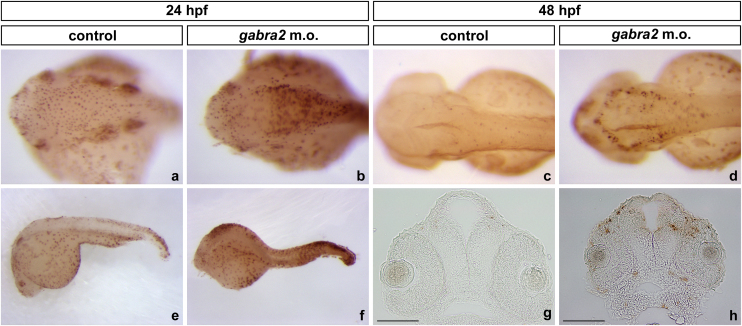
gad1b codes for the GABA-synthesising enzyme GAD67, so that it can be used to cheque if the GABAergic neurotransmitter system is altered in gabra2 injected fish. gad1b expression was reduced at both at 24 hpf and 48 hpf, as determined by ISH (Fig. 7). At 24 hpf, the staining was weaker at the mid- and hindbrain regions, while at 48 hpf a moderate effect is observed: the labelling was parcially lost, and it remained restricted to three paired nuclei: the olfactory bulbs at the telencephalon (pretectum and tectum opticum) and two paired nuclei at the hindbrain.
Fig. 7.
Expression pattern of gad1b on gabra2 deficient fish. The expression pattern of gad1b (GABA-synthesising enzyme GAD67)-GABAergic marker-was assayed by ISH on (a and c) control and b and d) gabra2 morphant embryos at 24 hpf (a and b) and 48 hpf (c and d). Note the reduction of the areas of expression (as determined by a decrease in the stained area) found for this gene at both stages. Dorsal views are shown, in which embryos are oriented with the anterior towards the left and posterior towards the right.
3.7. GABAergic drugs and proliferation during CNS development
To assess whether GABAergic activation can elicit changes in the proliferation pattern during CNS development, zebrafish embryos were exposed to GABAergic drugs from 5 hpf to 24 hpf. The following agents were used: 1 μM GABA; 1 μM muscimol, the prototypic GABAA agonist [44]; and 100 μM bicuculline, a selective GABA antagonist [45]. Proliferation was assessed by phospho-histone 3 (Ser10) immunostaining on wholemounts, but no significant changes in the proliferative pattern were observed between control embryos and drug-treated groups (Fig. S4). TUNEL assays (peroxidase staining) for detection of apoptotic cells were also conducted, and only a slight increase in peroxidase labelling was found in the forebrain of those embryos exposed to GABAergic agents.
4. Discussion
The interest in GABA signalling not only lies on the fact that it is the main inhibitory neurotransmitter in the brain, but also it has been related to several neurological disorders, as for example epilepsy, anxiety, depression and schizophrenia [19], [46]. Thus, the study of GABAergic neurotransmitter system could bring novel therapeutical approaches. Studies from knock-out mice revealed that the GABAA receptor subtype containing α2 subunit could be the target for novel antidepressant drugs [20]. Besides, GABA is one of the mediators of ethanol effects in the CNS, and GABAA receptors play a central role in the acute and chronic effects of ethanol [17], [47]. There is also a close link between the GABAergic and opioidergic systems, as opiates are able to inhibit GABA-mediated synaptic transmission in certain brain regions [48]. Conversely, GABAergic neurons in the ventral tegmental area and in the nucleus accumbens are involved in modulating the reinforcing properties of opiates [49]. In fact, recent evidence shows that selective agents targeting α2 and α3 subtypes of GABAA receptors could be of great use as analgesics to treat chronic pain, as for example neuropathic pain [50], [51].
GABA is a rather ancient molecule, as it is also found in plants and GABAergic signalling is already present in invertebrates [52]. The role of GABA is not only restricted to synaptic transmission, but also it has been related to cell proliferation and differentiation [53]. It has been reported that GABAA activation reduces the proliferation of embryonic stem cells and neural crest stem cells in mouse embryos, as autocrine / paracrine signalling causes cell arrest in the S phase; as a consequence, there is a net reduction in the stem cell progenies [13]. In fact, GABAA signalling helps to regulate the stem cell niche [11]. Cortical development and neurogenesis are also controlled by GABA and glutamate; these two neurotransmitters are localised in the ventricular and subventricular zone, where the newborn cortical neurons arise. GABAA and non-NMDA (mainly AMPA and kainate) receptor signalling are able to decrease DNA synthesis, while antagonists elicit the opposite effect [54]. Therefore, alterations in GABAergic circuits during development have been related to several psychiatric disorders, such as autism and schizophrenia [55]. For all these reasons, I aimed to characterize the developmental effects of gabra2 deficiency in the zebrafish.
The molecular characterisation of Gabra2 has revealed that the amino acid sequence of Gabra2 protein is highly conserved throughout the evolutionary scale of vertebrates (80% percentage of identity between mammalian, amphibian and fish homologues), which may serve as evidence for functional conservation. Besides, the crystal structure of Gabra subunits has not been experimentally solved yet, so it was necessary to generate a homological model of Gabra2. The high degree of domain conservation and structural similarity among neurotransmitter-gated ion channels allowed me to build the 3D structure of zebrafish Gabra2, and this prediction is in agreement with the conservation of the function and with the clustalw alignment. Two binding domains are identified in the same regions as in GABRB3, and key amino acids for GABA and benzodiazepine binding are present. These features suggest that the intermolecular interactions of α2 (namely, with zebrafish β and γ subunits) may be largely conserved, so that zebrafish Gabra2 protein may display the same functionality as its human homologue. Therefore, results obtained in zebrafish can be extrapolated to higher vertebrates, thus yielding reliable estimations about the role of GABA in vertebrate development.
gabra2 expression is detected at early stages of embryonic development in the main subdivisions of the CNS, which may suggest that gabra2 is playing a role in development, by controlling proliferation and/or differentiation. As development proceeds, staining becomes more restricted to certain areas of the fore- and midbrain. A morpholino-based approach allowed me to block the expression of gabra2 gene and to analyse its effect in CNS development. Although morpholinos have been extensively used to specifically knock-down gene expression in zebrafish, it has been recently stated that this technique shows certain limitations, and a certain degree of off-target effects cannot be ruled out [56]. The morphant embryos were viable and did not show a strong macroscopic phenotype, as they only display a smaller body size and a modest swelling of the brain ventricles; this weak phenotype could be due to the existence of compensatory mechanisms in the early embryo. However, the pattern of proliferation was clearly disrupted in all brain regions, as dividing cells were found in anomalous locations. An increase in apoptotic cells was observed at 24 hpf mainly in the hindbrain. At 48 hpf stronger labelling was found for both mitotic and apoptotic markers at the walls of the third ventricle. These findings indicate that those cells which are abnormally dividing may be unable to acquire a specific cell fate, and hence they undergo programmed cell death. It is not known whether this aberrant proliferation may result in depletion of stem- or progenitor cells in the mature CNS, as it has been previously reported [57], [58]. Taken together, our results indicate that gabra2 might be involved in shaping the proliferating microenvironment in the early embryo.
The process of differentiation into specialized cell types takes place during the embryonic development, and is tightly regulated by signalling cascades. The absence of a functional Gabra2 seems to affect the development of the mid- and hindbrain regions. It is well known that the isthmic organiser signalling at the MHB is essential for the proper development and regionalisation of the mid- and hindbrain. notch1, pax2, fgf8 and wnt1 are the major players in controlling MHB establishment and positioning [59]. Hence I have analysed if these signalling pathways are altered in gabra2 deficient embryos. notch1 is involved in the regulation of the stem-cell niche, both during the embryonic development as well as in the adult organism, as it controls the balance between the stem-cell self renewal, proliferation and progenitor differentiation [60]. notch1 also plays a role in boundary formation and restricts Ffg8 signalling from the organiser [61]. pax2, together with wnt1 and fgf8, is involved in the maintenance of the MHB isthmic organiser [62]. In fact, pax2 is the inducer of fgf8 [59] and Fgf8 signalling from the isthmic organiser determines the anteroposterior neural patterning in the midbrain and cerebellum [63]. wnt1 regulates mid- and hindbrain (mainly cerebellum) development, and is involved in establishing the MHB [64]. These genes are mis-expressed in gabra2 morphants, as their transcriptional level is altered and their pattern of expression is different from those found in control embryos. In fact, it seems that the staining is more restricted, and stronger labelling is found at the border between mesencephalon and cerebellum. Hence, it is possible that gabra2 might be playing a role in the embryonic development of the mid- and hindbrain. Interestingly, neurodevelopmental abnormalities have also been related to misexpression of the neuron-specific potassium/chloride cotransporter 2 (KCC2), as KCC2 overexpression perturbs neuronal differentiation and development in mouse [65] and zebrafish embryos [66]. In mice, early KCC2 expression impairs proper axon maturation in the hindbrain and spinal cord [65], and in zebrafish KCC2 interacts with the actin cytoskeleton to modulate neuronal differentiation, although the active form of this chloride exporter is not required for this interaction [66].
Phospho-histone 3 immunostaining indicates that there are more proliferating cells in the morphant CNS, so it is possible that fewer cells are undergoing differentiation towards a neuronal lineage. Therefore I have also analysed the expression of neuroD: this proneural gene is implicated in early neuronal determination, and is essential for the differentiation of granule cells in the cerebellum and hippocampus [67], [68]. Indeed, neuroD expression is down-regulated in gabra2 deficient embryos.
pax6, which is a highly conserved transcription factor, plays a key role in CNS development, as for example it is involved in eye development and in the patterning of the neural tube. pax6 also regulates proliferation and differentiation in the neurogenic niches, namely the subgranular zone of the dentate gyrus in the hippocampus and in the subventricular zone [69]. Besides, shh is a diffusible morphogen which is involved in the control of numerous developmental processes, from rather early events (as it is the case of the dorso-ventral patterning of the neural tube) to the proliferation of neural precursors during embryonic development and in the adult organism [70]. shh and pax6 regulate the balance between glutamategic and GABAergic neurogenesis [71]. However, no clear changes in the expression of these two genes were observed in the morphant embryos. This result can be explained by the presence of redundant signalling pathways or of compensatory mechanisms.
On a further step I have evaluated if the GABAergic neurotransmitter system is affected in gabra2 deficient fish. gad1 (GABA-synthesising enzyme GAD67) expression, a marker for GABAergic cells, was significantly reduced in the morphant embryos; this outcome may indicate that the generation of GABAergic cells is reduced in gabra2 morpholino-injected fish. Interestingly, gad1 expression is reduced in the pretectum and tectum opticum, and these two areas are known to harbour a rich population og GABA-positive cells [72].
I also wanted to check whether GABAergic drugs can induce changes in proliferation and in apoptosis. I have tested the effects of GABA, muscimol (a prototypic GABAA agonist [44]) and bicuculline (a selective GABA antagonist [45]). None of them were able to modify the pattern of cell proliferation at 24 hpf. However, repeated exposure to GABA and muscimol can induce tolerance, by promoting down-regulation of the receptors [73], and/or even at the mRNA level [74]. This possibility, as well as receptor desensitization after prolongued agonist exposure, cannot be ruled out in order to explain the apparent lack of effect of GABAergic drugs on CNS development.
5. Conclusion
gabra2 codes for the GABAA receptor alpha-2, and it amino acid sequence is highly conserved among vertebrate evolution, which may serve as evidence for functional conservation. Our results indicate that gabra2 might be involved in shaping the proliferating niches in the early embryo. Besides, gabra2 may also interact with those signalling pathways that control proliferation, differentiation and neurogenesis during the embryonic development, principally in the differentiation of the mesencephalon and cerebellum. However, considering the weak phenotype observed for the morphant embryos, it can be concluded that gabra2 is not a major player in the CNS development, or maybe a compensatory mechanism could overcome the absence of a functional gabra2. These findings will help to understand the role of neurotransmitter systems during CNS development.
Declarations of interest
The author of manuscript declare that the paper contains no conflict of interest that would prejudice its impartially.
Author contributions
Veronica Gonzalez-Nunez: Conception and design, financial support, collection and assembly of data, data analysis and interpretation, manuscript writing and final approval of manuscript.
Funding
This work was supported by grant from Consejería de Sanidad, Junta de Castilla y León (SAN673/SA25/08).
Acknowledgements
VGN wants to thank Prof. Raquel E. Rodriguez for reagents, for providing lab space to perform the experiments, for valuable scientific discussions and for critical reading of the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.08.003.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Watanabe M., Maemura K., Kanbara K., Tamayama T., Hayasaki H. GABA and GABA receptors in the central nervous system and other organs. Int. Rev. Cytol. 2002;213:1–47. doi: 10.1016/s0074-7696(02)13011-7. [DOI] [PubMed] [Google Scholar]
- 2.Olsen R.W. GABA. In: Davis K.L., editor. Neuropsychopharmacology: The Fifth Generation of Progress, American College of Neuropsychopharmacology. American College of Neuropsychopharmacology; Brentwood, Tennesse: 2002. pp. 159–168. [Google Scholar]
- 3.Olsen R.W., Tobin A.J. Molecular biology of GABAA receptors. FASEB J. 1990;4:1469–1480. doi: 10.1096/fasebj.4.5.2155149. [DOI] [PubMed] [Google Scholar]
- 4.Macdonald R.L., Olsen R.W. GABAA receptor channels. Annu. Rev. Neurosci. 1994;17:569–602. doi: 10.1146/annurev.ne.17.030194.003033. [DOI] [PubMed] [Google Scholar]
- 5.Burt D.R., Kamatchi G.L. GABAA receptor subtypes: from pharmacology to molecular biology. FASEB J. 1991;5:2916–2923. doi: 10.1096/fasebj.5.14.1661244. [DOI] [PubMed] [Google Scholar]
- 6.Hevers W., Luddens H. The diversity of GABAA receptors. Pharmacological and electrophysiological properties of GABAA channel subtypes. Mol. Neurobiol. 1998;18:35–86. doi: 10.1007/BF02741459. [DOI] [PubMed] [Google Scholar]
- 7.Barnard E.A., Skolnick P., Olsen R.W., Mohler H., Sieghart W., Biggio G., Braestrup C., Bateson A.N., Langer S.Z. International Union of Pharmacology. XV. Subtypes of gamma-aminobutyric acidA receptors: classification on the basis of subunit structure and receptor function. Pharmacol. Rev. 1998;50:291–313. [PubMed] [Google Scholar]
- 8.Ben-Ari Y., Khazipov R., Leinekugel X., Caillard O., Gaiarsa J.L. GABAA, NMDA and AMPA receptors: a developmentally regulated ‘menage a trois’. Trends Neurosci. 1997;20:523–529. doi: 10.1016/s0166-2236(97)01147-8. [DOI] [PubMed] [Google Scholar]
- 9.Ben-Ari Y. Excitatory actions of gaba during development: the nature of the nurture. Nat. Rev. Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- 10.Yamada J., Okabe A., Toyoda H., Kilb W., Luhmann H.J., Fukuda A. Cl- uptake promoting depolarizing GABA actions in immature rat neocortical neurones is mediated by NKCC1. J. Physiol. 2004;557:829–841. doi: 10.1113/jphysiol.2004.062471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng L., Tang Y.B., Sun F., An S.M., Zhang C., Yang X.J., Lv H.Y., Lu Q., Cui Y.Y., Hu J.J., Zhu L., Chen H.Z. Non-neuronal release of gamma-aminobutyric Acid by embryonic pluripotent stem cells. Stem Cells Dev. 2013;22:2944–2953. doi: 10.1089/scd.2013.0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Ari Y., Crepel V., Represa A. Seizures beget seizures in temporal lobe epilepsies: the boomerang effects of newly formed aberrant kainatergic synapses. Epilepsy Curr. 2008;8:68–72. doi: 10.1111/j.1535-7511.2008.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andang M., Hjerling-Leffler J., Moliner A., Lundgren T.K., Castelo-Branco G., Nanou E., Pozas E., Bryja V., Halliez S., Nishimaru H., Wilbertz J., Arenas E., Koltzenburg M., Charnay P., El Manira A., Ibanez C.F., Ernfors P. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. doi: 10.1038/nature06488. [DOI] [PubMed] [Google Scholar]
- 14.Wang D.D., Kriegstein A.R. Defining the role of GABA in cortical development. J. Physiol. 2009;587:1873–1879. doi: 10.1113/jphysiol.2008.167635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzdak P.D., Schwartz R.D., Skolnick P., Paul S.M. Ethanol stimulates gamma-aminobutyric acid receptor-mediated chloride transport in rat brain synaptoneurosomes. Proc. Natl. Acad. Sci. USA. 1986;83:4071–4075. doi: 10.1073/pnas.83.11.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang J., Olsen R.W. Alcohol use disorders and current pharmacological therapies: the role of GABA(A) receptors. Acta Pharmacol. Sin. 2014;35:981–993. doi: 10.1038/aps.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies M. The role of GABAA receptors in mediating the effects of alcohol in the central nervous system. J. Psychiatry Neurosc. 2003;28:263–274. [PMC free article] [PubMed] [Google Scholar]
- 18.Knabl J., Witschi R., Hosl K., Reinold H., Zeilhofer U.B., Ahmadi S., Brockhaus J., Sergejeva M., Hess A., Brune K., Fritschy J.M., Rudolph U., Mohler H., Zeilhofer H.U. Reversal of pathological pain through specific spinal GABAA receptor subtypes. Nature. 2008;451:330–334. doi: 10.1038/nature06493. [DOI] [PubMed] [Google Scholar]
- 19.Engin E., Liu J., Rudolph U. Alpha2-containing GABA(A) receptors: a target for the development of novel treatment strategies for CNS disorders. Pharmacol. Ther. 2012;136:142–152. doi: 10.1016/j.pharmthera.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vollenweider I., Smith K.S., Keist R., Rudolph U. Antidepressant-like properties of alpha2-containing GABA(A) receptors. Behav. Brain Res. 2011;217:77–80. doi: 10.1016/j.bbr.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhart M., Weerts E.M., McCaul M.E., Guo X., Yan X., Kranzler H.R., Li N., Wand G.S. GABRA2 markers moderate the subjective effects of alcohol. Addict. Biol. 2013 doi: 10.1111/j.1369-1600.2012.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philibert R.A., Gunter T.D., Beach S.R., Brody G.H., Hollenbeck N., Andersen A., Adams W. Role of GABRA2 on risk for alcohol, nicotine, and cannabis dependence in the Iowa Adoption Studies. Psychiatr. Genet. 2009;19:91–98. doi: 10.1097/YPG.0b013e3283208026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ridges S., Heaton W.L., Joshi D., Choi H., Eiring A., Batchelor L., Choudhry P., Manos E.J., Sofla H., Sanati A., Welborn S., Agarwal A., Spangrude G.J., Miles R.R., Cox J.E., Frazer J.K., Deininger M., Balan K., Sigman M., Muschen M., Perova T., Johnson R., Montpellier B., Guidos C.J., Jones D.A., Trede N.S. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119:5621–5631. doi: 10.1182/blood-2011-12-398818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jo D.H., Son D., Na Y., Jang M., Choi J.H., Kim J.H., Yu Y.S., Seok S.H., Kim J.H. Orthotopic transplantation of retinoblastoma cells into vitreous cavity of zebrafish for screening of anticancer drugs. Mol. Cancer. 2013;12:71. doi: 10.1186/1476-4598-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leclercq K., Afrikanova T., Langlois M., De Prins A., Buenafe O.E., Rospo C.C., Van Eeckhaut A., de Witte P.A., Crawford A.D., Smolders I., Esguerra C.V., Kaminski R.M. Cross-species pharmacological characterization of the allylglycine seizure model in mice and larval zebrafish. Epilepsy Behav. 2015;45:53–63. doi: 10.1016/j.yebeh.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Afrikanova T., Serruys A.S., Buenafe O.E., Clinckers R., Smolders I., de Witte P.A., Crawford A.D., Esguerra C.V. Validation of the zebrafish pentylenetetrazol seizure model: locomotor versus electrographic responses to antiepileptic drugs. PloS One. 2013;8:e54166. doi: 10.1371/journal.pone.0054166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richendrfer H., Pelkowski S.D., Colwill R.M., Creton R. On the edge: pharmacological evidence for anxiety-related behavior in zebrafish larvae. Behav. Brain Res. 2012;228:99–106. doi: 10.1016/j.bbr.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 29.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Troshin P.V., Procter J.B., Barton G.J. Java bioinformatics analysis web services for multiple sequence alignment--JABAWS:MSA. Bioinformatics. 2011;27:2001–2002. doi: 10.1093/bioinformatics/btr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley L.A., Sternberg M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 32.Miller P.S., Aricescu A.R. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Guilloux V., Schmidtke P., Tuffery P. Fpocket: an open source platform for ligand pocket detection. BMC Bioinform. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez-Nunez V., Nocco V., Budd A. Characterization of drCol 15a1b: a novel component of the stem cell niche in the zebrafish retina. Stem Cells. 2010;28:1399–1411. doi: 10.1002/stem.461. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez-Nunez V., Jimenez Gonzalez A., Barreto-Valer K., Rodriguez R.E. In vivo regulation of the mu opioid receptor: role of the endogenous opioid agents. Mol. Med. 2013;19:7–17. doi: 10.2119/molmed.2012.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattingly C.J., Hampton T.H., Brothers K.M., Griffin N.E., Planchart A. Perturbation of defense pathways by low-dose arsenic exposure in zebrafish embryos. Environ. Health Perspect. 2009;117:981–987. doi: 10.1289/ehp.0900555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carpenter T.S., Lau E.Y., Lightstone F.C. A role for loop F in modulating GABA binding affinity in the GABA(A) receptor. J. Mol. Biol. 2012;422:310–323. doi: 10.1016/j.jmb.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 41.Wagner D.A., Czajkowski C., Jones M.V. An arginine involved in GABA binding and unbinding but not gating of the GABA(A) receptor. J. Neurosci. 2004;24:2733–2741. doi: 10.1523/JNEUROSCI.4316-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson S.M., Morlock E.V., Satyshur K.A., Czajkowski C. Structural requirements for eszopiclone and zolpidem binding to the gamma-aminobutyric acid type-A (GABAA) receptor are different. J. Med. Chem. 2008;51:7243–7252. doi: 10.1021/jm800889m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morlock E.V., Czajkowski C. Different residues in the GABAA receptor benzodiazepine binding pocket mediate benzodiazepine efficacy and binding. Mol. Pharmacol. 2011;80:14–22. doi: 10.1124/mol.110.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krogsgaard-Larsen P., Hjeds H., Curtis D.R., Lodge D., Johnston G.A. Dihydromuscimol, thiomuscimol and related heterocyclic compounds as GABA analogues. J. Neurochem. 1979;32:1717–1724. doi: 10.1111/j.1471-4159.1979.tb02284.x. [DOI] [PubMed] [Google Scholar]
- 45.Curtis D.R., Duggan A.W., Felix D., Johnston G.A. GABA, bicuculline and central inhibition. Nature. 1970;226:1222–1224. doi: 10.1038/2261222a0. [DOI] [PubMed] [Google Scholar]
- 46.Wong C.G., Bottiglieri T., Snead O.C., 3rd GABA, gamma-hydroxybutyric acid, and neurological disease. Ann. Neurol. 2003;54(Suppl 6):S3–S12. doi: 10.1002/ana.10696. [DOI] [PubMed] [Google Scholar]
- 47.Grobin A.C., Matthews D.B., Devaud L.L., Morrow A.L. The role of GABA(A) receptors in the acute and chronic effects of ethanol. Psychopharmacology. 1998;139:2–19. doi: 10.1007/s002130050685. [DOI] [PubMed] [Google Scholar]
- 48.Vaughan C.W., Ingram S.L., Connor M.A., Christie M.J. How opioids inhibit GABA-mediated neurotransmission. Nature. 1997;390:611–614. doi: 10.1038/37610. [DOI] [PubMed] [Google Scholar]
- 49.Xi Z.X., Stein E.A. GABAergic mechanisms of opiate reinforcement. Alcohol Alcohol. 2002;37:485–494. doi: 10.1093/alcalc/37.5.485. [DOI] [PubMed] [Google Scholar]
- 50.Mirza N.R., Munro G. The role of GABA(A) receptor subtypes as analgesic targets. Drug News Perspect. 2010;23:351–360. doi: 10.1358/dnp.2010.23.6.1489909. [DOI] [PubMed] [Google Scholar]
- 51.Munro G., Ahring P.K., Mirza N.R. Developing analgesics by enhancing spinal inhibition after injury: GABAA receptor subtypes as novel targets. Trends Pharmacol. Sci. 2009;30:453–459. doi: 10.1016/j.tips.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Ben-Ari Y., Gaiarsa J.L., Tyzio R., Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol. Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- 53.Nakamichi N., Takarada T., Yoneda Y. Neurogenesis mediated by gamma-aminobutyric acid and glutamate signaling. J. Pharmacol. Sci. 2009;110:133–149. doi: 10.1254/jphs.08r03cr. [DOI] [PubMed] [Google Scholar]
- 54.LoTurco J.J., Owens D.F., Heath M.J., Davis M.B., Kriegstein A.R. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 1995;15:1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 55.Chattopadhyaya B. Molecular mechanisms underlying activity-dependent GABAergic synapse development and plasticity and its implications for neurodevelopmental disorders. Neural Plast. 2011;2011:734231. doi: 10.1155/2011/734231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bedell V.M., Westcot S.E., Ekker S.C. Lessons from morpholino-based screening in zebrafish. Brief. Funct. Genom. 2011;10:181–188. doi: 10.1093/bfgp/elr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Encinas J.M., Michurina T.V., Peunova N., Park J.H., Tordo J., Peterson D.A., Fishell G., Koulakov A., Enikolopov G. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–579. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imayoshi I., Sakamoto M., Yamaguchi M., Mori K., Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bally-Cuif L., Wassef M. Determination events in the nervous system of the vertebrate embryo. Curr. Opin. Genet. Dev. 1995;5:450–458. doi: 10.1016/0959-437x(95)90048-l. [DOI] [PubMed] [Google Scholar]
- 60.Androutsellis-Theotokis A., Leker R.R., Soldner F., Hoeppner D.J., Ravin R., Poser S.W., Rueger M.A., Bae S.K., Kittappa R., McKay R.D. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- 61.Tossell K., Kiecker C., Wizenmann A., Lang E., Irving C. Notch signalling stabilises boundary formation at the midbrain-hindbrain organiser. Development. 2011;138:3745–3757. doi: 10.1242/dev.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Picker A., Scholpp S., Bohli H., Takeda H., Brand M. A novel positive transcriptional feedback loop in midbrain-hindbrain boundary development is revealed through analysis of the zebrafish pax2.1 promoter in transgenic lines. Development. 2002;129:3227–3239. doi: 10.1242/dev.129.13.3227. [DOI] [PubMed] [Google Scholar]
- 63.Carl M., Wittbrodt J. Graded interference with FGF signalling reveals its dorsoventral asymmetry at the mid-hindbrain boundary. Development. 1999;126:5659–5667. doi: 10.1242/dev.126.24.5659. [DOI] [PubMed] [Google Scholar]
- 64.McMahon A.P., Joyner A.L., Bradley A., McMahon J.A. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell. 1992;69:581–595. doi: 10.1016/0092-8674(92)90222-x. [DOI] [PubMed] [Google Scholar]
- 65.Horn Z., Ringstedt T., Blaesse P., Kaila K., Herlenius E. Premature expression of KCC2 in embryonic mice perturbs neural development by an ion transport-independent mechanism. Eur. J. Neurosci. 2010;31:2142–2155. doi: 10.1111/j.1460-9568.2010.07258.x. [DOI] [PubMed] [Google Scholar]
- 66.Reynolds A., Brustein E., Liao M., Mercado A., Babilonia E., Mount D.B., Drapeau P. Neurogenic role of the depolarizing chloride gradient revealed by global overexpression of KCC2 from the onset of development. J. Neurosci. 2008;28:1588–1597. doi: 10.1523/JNEUROSCI.3791-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyata T., Maeda T., Lee J.E. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wullimann M.F., Mueller T., Distel M., Babaryka A., Grothe B., Koster R.W. The long adventurous journey of rhombic lip cells in jawed vertebrates: a comparative developmental analysis. Front. Neuroanat. 2011;5:27. doi: 10.3389/fnana.2011.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osumi N., Shinohara H., Numayama-Tsuruta K., Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 70.Marti E., Bovolenta P. Sonic hedgehog in CNS development: one signal, multiple outputs. Trends Neurosci. 2002;25:89–96. doi: 10.1016/s0166-2236(02)02062-3. [DOI] [PubMed] [Google Scholar]
- 71.Robertshaw E., Matsumoto K., Lumsden A., Kiecker C. Irx3 and Pax6 establish differential competence for Shh-mediated induction of GABAergic and glutamatergic neurons of the thalamus. Proc. Natl. Acad. Sci. USA. 2013;110:E3919–E3926. doi: 10.1073/pnas.1304311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mueller T., Vernier P., Wullimann M.F. A phylotypic stage in vertebrate brain development: GABA cell patterns in zebrafish compared with mouse. J. Comp. Neurol. 2006;494:620–634. doi: 10.1002/cne.20824. [DOI] [PubMed] [Google Scholar]
- 73.Mehta A.K., Ticku M.K. Chronic GABA exposure down-regulates GABA-benzodiazepine receptor-ionophore complex in cultured cerebral cortical neurons. Brain Res. Mol. Brain Res. 1992;16:29–36. doi: 10.1016/0169-328x(92)90190-m. [DOI] [PubMed] [Google Scholar]
- 74.Hirouchi M., Ohkuma S., Kuriyama K. Muscimol-induced reduction of GABAA receptor alpha 1-subunit mRNA in primary cultured cerebral cortical neurons. Brain Res. Mol. Brain Res. 1992;15:327–331. doi: 10.1016/0169-328x(92)90125-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material