Abstract
CK2 is a constitutively active Ser/Thr protein kinase deregulated in cancer and other pathologies, responsible for about the 20% of the human phosphoproteome. The holoenzyme is a complex composed of two catalytic (α or α´) and two regulatory (β) subunits, with individual subunits also coexisting in the cell. In the holoenzyme, CK2β is a substrate-dependent modulator of kinase activity. Therefore, a comprehensive characterization of CK2 cellular function should firstly address which substrates are phosphorylated exclusively when CK2β is present (class-III or beta-dependent substrates). However, current experimental constrains limit this classification to a few substrates. Here, we took advantage of motif-based prediction and designed four linear patterns for predicting class-III behavior in sets of experimentally determined CK2 substrates. Integrating high-throughput substrate prediction, functional classification and network analysis, our results suggest that beta-dependent phosphorylation might exert particular regulatory roles in viral infection and biological processes/pathways like apoptosis, DNA repair and RNA metabolism. It also pointed, that human beta-dependent substrates are mainly nuclear, a few of them shuttling between nuclear and cytoplasmic compartments.
Keywords: CK2, Beta-dependent substrates, Class-III substrates, Holoenzyme-dependent phosphorylation, Bioinformatics, Text mining
Highlights
-
•
The designed linear patterns assist CK2 beta-dependent substrates prediction.
-
•
A high-throughput prediction of CK2 beta-dependent substrates was performed in several organisms including human, mouse and rat.
-
•
The functional classification indicated a role of CK2 beta-dependent regulation in viral infection, apoptosis, DNA repair and RNA metabolism.
-
•
The functional classification indicated that human CK2 beta-dependent substrates are mainly nuclear with a number of them also found in cytoplasm.
1. Introduction
CK2 is a constitutive and ubiquitous Ser/Thr protein kinase that functions as a global regulator of cell survival often found deregulated in cancer and other complex diseases [1], [2], [3], [4]. The CK2 consensus sequence, [pS/pT]-{P}-x-[E/D] or [pS/pT]-{P}-x-pS, constitutes a small motif found so far in more than 300 experimentally determined CK2 substrates among different organisms and viral proteins [3]. Such promiscuity connects CK2-dependent phosphorylation with key cellular biological processes and pathways involved in DNA repair [5], apoptosis [6], survival/proliferation [7], and viral infection [8].
The mammalian CK2 holoenzyme is a heterotetramer composed of two catalytic (CK2α and/or CK2α′) and two regulatory subunits (CK2β) with a α2-β2 stoichiometry (α2-β2, α′2-β2 or α1α′1-β2) [9], [10]. The heterotetramer can dissociate under certain conditions with evidences indicating the presence of individual subunits [11]. The free catalytic monomers maintain its activity against a range of substrates while the regulatory subunits might display CK2-independent functions targeting DNA repair, cell cycle and protein kinases as A-Raf, c-Mos and Chk1 [12]. In the holoenzyme, CK2β is a substrate-dependent modulator of kinase activity. In accordance, Pinna grouped the CK2 substrates in three classes based on the subunit composition of active enzyme [4]. Class-I includes CK2 substrates modified either by CK2α or the holoenzyme, poorly influenced by the CK2β subunit [13]. Class-II, substrates such as calmodulin (CALM), phosphorylated by CK2α alone with holoenzyme formation inhibiting kinase activity [14]; this regulatory effect is mitigated with the addition of polycationic molecules (e.g., histones, polylysine and polyamines) [15]. Class-III substrates are beta-dependent and their phosphorylation relies on the integrity of the N terminal acidic loop of this subunit (sequence D55LEPDEELED64). This is true for Rev [16], [17] and eIF2β [18] where electrostatic contacts between basic residues on the substrates and the acidic loop enhance kinase-substrate binding.
The classification of CK2 substrates based on active enzyme subunit composition provides a framework for understanding the regulatory function of CK2β subunit. However, few experiments exist that functionally explore such classification [17], [18], [19], [20], [21], [22], [23]; this reflects practical limitations of elucidating kinase specificity. In this regard, high-throughput methods provide an alternative to traditional techniques for phosphosites identification (e.g., ELISA and phospho-specific antibodies). For instance, mass spectrometry-driven approaches figure at the most promising high-throughput techniques with a subsequent increase in the use of in silico high-throughput methods for data interpretation. Currently, in silico methodologies enclose motif-based identification of phosphorylation sites, structural information integration, integration of phosphorylation site structural context, phospho-clusters modeling, integration of Protein–Protein Interaction Network (PPIN) information, and multi-organisms prediction [24], [25]. Here, we propose four linear patterns for identifying class-III CK2 sites. These patterns were constructed manually based on literature and database information describing well known class-III substrates.
2. Materials and methods
2.1. CK2 substrates
The information of class-III CK2 substrates for motif design was obtained by mining PubMed for functional relationships using RLIMS-P [26] and Chilibot [27] text mining tools (Fig. S1). The experimentally determined CK2 substrates were obtained from PhosphoSitePlus resource [28] and the literature. The PhosphoSitePlus information was extracted from the “Substrates of” CK2 protein, selecting both “CK2A1” and “CK2A2” entries (S1 File). Literature information was gathered from the works of Meggio and Pinna, 2003 [3] and Bian et al., 2013 [29] (S1 File) in the form of phosphorylated peptides and UniProt entries, respectively. The phosphorylated peptides reported by Pinna et al., were mapped to UniProt identifiers using BLAST program, set to default values.
2.2. Class-III substrate prediction
The class-III substrates prediction (S2 File) was performed with the ScanProsite tool [30] using the four designed linear patterns and the UniProt identifiers obtained from PhosphoSitePlus resource and the literature (S1 File). The parameters selected for the run were Option 3 (i.e., submit protein sequences and motifs to scan them against each other) and match mode “greedy, overlap, includes” as pattern options.
2.3. CK2 substrates three-dimensional structures
The PDB IDs corresponding to the predicted class-III substrates were retrieved from UniProt (S3 File). The three-dimensional structures of the experimentally determined and predicted class-III substrates were retrieved from PDB database [31] (S3 File) for interactive visualization and analysis using Chimera program [32]. The predicted class-III phosphopeptides were mapped to the PDB sequences in FASTA format using PeptideMatch from PIR [33].
2.4. Sequence alignment
The sequence multiple alignment of Vpu sequences from HIV1 was performed using Clustal Omega program [34].
2.5. Functional classification of predicted class-III substrates
The functional classification of human predicted class-III substrates was performed with DAVID [35], GeneCoDis [36] and NetVenn [37] enrichment analysis tools based on GO biological process, cellular component, cancer genes and/or KEGG pathway annotations. The default parameters were selected and the significance level was set at 0.05 (P<0.05 considered significant). For DAVID analysis, the background list was selected as the CK2 substrates identified in Bian et al. study. The protein function and the sequence related annotations were extracted from UniProtKB [38] and from the literature using Chilibot [27], RLIMS-P [26] and GoPubmed [39] information retrieval and text mining tools.
2.6. Network analysis
Network representation and analysis were performed with NetworkAnalyst tool [40] using the protein list functionality and the predicted class-III substrates from Bian et al. dataset as the input list. The network, 3509 nodes (193 seeds) and 8032 edges obtained from InnateDB interaction dataset was trimmed to keep only the seed nodes (class-III substrates predicted from Bian et al. dataset) and their first neighbors. The obtained network, 607 nodes (193 seeds) and 2801 edges, fitted the recommended size (200–2000 nodes) to avoid dense network and facilitate subsequent analyses. Network nodes were functionally explored based on KEGG pathway annotation and on their degree.
3. Results and discussion
3.1. Linear pattern design for class-III substrate prediction
Pattern-matching is a useful approach for deciphering kinase–substrates relationships at phosphosite level. Here, we designed four linear patterns for matching CK2 class-III sites based on available information of known class-III substrates. First, we used text mining for extracting information on class-III substrates. As a result, we found significant mentions on: Rev protein from HIV1 virus [16] and eIF2β human translation factor [18], two well-studied CK2 substrates (Table 1). We also detected the yeast transcriptional regulator Nrg1 [41], the bovine ferredoxin Fdx1 [42], the human transporter CFTR [43], the human homeoprotein SIX1 [19], the human transcription factor Olig2 [20], the human pancreas/duodenum homeobox protein 1 [21], [22], and the chick homeobox protein En2 [23] as beta-dependent substrates (Table 1).
Table 1.
In vitro CK2 class-III substrates retrieved from the literature.
| Protein name (UniProt ID) | Class-III sites/basic cluster | Maximum distancea |
|---|---|---|
| Rev | S5GDSDEDLLKAVRLIKFLYQSNPPPNPEGTRQARRNRRRR44 | 29 |
| (REV_HV1H3, | ||
| REV_HV1B1) | ||
| eIF2β (IF2B_HUMAN) | S2GDEMIFDPTMSKKKKKKKK20 | 8 |
| Nrg1 | R152KQRTDPRNTLSDEE166 | 7 |
| (NRG1_YEAST) | ||
| Fdx1b | H56LIFEQHIFEKLEAITDEENDMLDLAYGLTDRSRLGCQICLTK98 | 12 |
| (ADX_HUMAN) | ||
| CFTR | R1446VKLFPHRNSSKCKSKPQIAALKEETEEE1474 | 13 |
| (CFTR_HUMAN) | ||
| Olig2 (OLIG2_HUMAN) | S84STSSAAASSTKKDKKQMTEPELQQLRLKINSRERKR120 | 27 |
| SIX1 (SIX1_HUMAN) | R110VRRKFPLPRTIWDGEETSYCFKEKSRGVLREWYAHNPYPSPREKR155 | 5/28 |
| Pdx1 (PDX1_HUMAN) | R197RMKWKKEEDKKRGGGTAVGGGGVAEPEQDCAVTSGEE234 | 30 |
| En2 (HME2_CHICK) | S150SDSDSSQAGSNAGNQPMLWPAWVYCTRYSDRPSSGPRSRKPKKK194 | 33 |
Maximum number of amino acids in the substrate from CK2 consensus sequence ([pS/pT]-{P}-x-[E/D] or [pS/pT]-{P}-x-pS, underlined text) to a mapped basic cluster. For Nrg1, Olig2, SIX1, Pdx1, and En2 substrates the basic clusters were selected considering Rev's basic cluster position as reference.
The position of the phosphosite Thr-71 refers to the mature protein and differs from the position reported in UniProt where the residue corresponds to Thr-131.
Second, we found relevant for holoenzyme-mediated phosphorylation the interaction between a cluster of basic residues in Rev and eIF2β substrates (Rev: R38RNRRRR44 and eIF2β: K14KKKKKK20) and the acidic loop of CK2β (Table 1). The basic cluster in both substrates is located relatively far from the phosphorylatable residue (Table 1) and the acidic loop has been described as the binding region for polybasic substances [14]. The physical association of these determinants mitigates the intrinsic down-regulatory function adjudicated to the acidic loop, allowing the phosphorylation of the substrates [16]. For example, Rev and CFTR-derived peptides lacking the basic cluster lose class-III behavior [16], [17], [43]. In addition, polycationic effectors inhibits holoenzyme-dependent Rev phosphorylation indicating a competitive mechanism in which binding of the acidic loop to a basic cluster in Rev is imperative for substrate phosphorylation [16]. Similarly, polylysine has a negative effect on eIF2β phosphorylation. For instance, the addition of polylysine to the eIF2β 1-22 peptide, which contains the basic cluster, prevents peptide binding to the CK2β acidic loop decreasing its affinity for the holoenzyme [18].
CK2 phosphorylates eIF2β protein at Ser-2 residue and Rev protein at Ser-5 and Ser-8. In these substrates, the basic cluster is positioned down-stream the consensus sequence to a maximum distance of 8 and 29 amino acids, respectively (Table 1). Interestingly, we found that the other selected class-III substrates also contain a basic cluster relatively far from the phosphoacceptor site at a maximum distance of 33 residues (Table 1). This basic cluster primarily locates down-stream the modified residue(s). However, for Fdx1, CFTR, SIX1 and Pdx1 proteins a basic cluster was found up-stream the modified residue (Table 1) allowing us to speculate that this determinant might act equally up-stream or down-stream to regulate beta-dependent phosphorylation.
Since little information is available regarding CK2 substrates structure and the phosphoacceptor sites spatial coordinates, we considered only elements from the primary structure. In correspondence, we designed two linear phosphorylation patterns starting with the CK2 consensus sequence (positions 0–+3). Next, we included the minimum and the maximum number of amino acids (x) allowed between CK2 consensus sequence and the acidic cluster, five and 33, respectively. The lower limit is in agreement with the minimum number of residues that in an extended conformation fit the distance between CK2α active site and the acidic loop in CK2β (Fig. 1). In this case we considered Cα-Cα distance as ~3.8 Å [44] and the hydrogen bond distance between catalytic Asp-156 and phosphorylable residue of ~3.0 Å. The upper limit was fixed based on the maximum number of amino acids from consensus sequence to the basic cluster in the validated class-III substrates (Table 1). Finally, we included at least three basic residues to match the basic cluster (Table 1). This element fits the basic cluster of mentioned substrates except for Fdx1 and CFTR proteins. The basic residues of these substrates are spaced irregularly compared with the others from which the former pattern can be deducted (Table 1).
Fig. 1.
Surface representation of the three-dimensional structure of the human CK2 holoenzyme (PDB ID: 1JWH). Catalytic (CK2α) and regulatory (CK2β) subunits are colored in yellow and purple, respectively. In one of the catalytic subunits, the active site is occupied by the ATP analog adenylyl imidodiphosphate (AMPPNP). In a close view, residues of two important functional regions, the basic cluster in CK2α and the acid loop in CK2β, are denoted as balls-and-sticks in colors blue and red, respectively. The minimal distance between the catalytic residue Asp-156 and the acidic loop in CK2β (Asp-55 residue) is indicated with a dashed line (39.5 Å).
The defined linear phosphorylation patterns are:
-
1.
[ST]-{P}-x-[ED]-x(5,33)-[KR]-x-[KR](2) or [ST]-{P}-x-[ED]-x(5,33)-[KR](2)-x-[KR]
-
2.
[ST]-{P}-x-S-x(5,33)-[KR]-x-[KR](2) or [ST]-{P}-x-S-x(5,33)-[KR](2)-x-[KR]
We also considered pattern variants where the basic cluster is located up-stream the CK2 consensus sequence. The up-stream linear patterns are:
-
3.
[KR]-x-[KR](2)-x(5,33)-[ST]-{P}-x-[ED] or [KR](2)-x-[KR]-x(5,33)-[ST]-{P}-x-[ED]
-
4.
[KR]-x-[KR](2)-x(5,33)-[ST]-{P}-x-S or [KR](2)-x-[KR]-x(5,33)-[ST]-{P}-x-S
3.2. CK2 class-III substrates prediction
A global classification of CK2 substrates based on the subunit composition of active enzyme is an ambitious and demanding experimental task that will further increase our knowledge on this kinase function. As previously reported, the use of in silico tools is widely-accepted for kinase motif prediction and mapping of phosphorylation sites [30], [45]. Here, we applied phosphorylation-patterns to predict class-III phosphorylation sites from sets of experimentally determined CK2 substrates. As a result, those proteins containing at least one match were considered tentative CK2 class-III substrates (S2 File). Next, we functionally analyzed the obtained results based on GO biological process, cellular component and KEGG pathway annotations.
The class-III pattern-driven prediction was performed on 913 experimentally determined CK2 substrates, 1745 sites, retrieved from PhosphoSitePlus database and the literature. This set includes data on CK2 phosphoacceptor sites for several organisms such as human, mouse and rat (Table 2). The work reported by Meggio and Pinna summarizes the information of phosphoacceptor sites of 33 different organisms and virus [3]. Here, we selected 13 of them to match those represented in the PhosphoSitePlus resource also considering the information available in the literature relative to CK2 phosphorylation.
Table 2.
CK2 in vitro substrates analyzed for class-III site prediction.
| Source | Organism | No. proteins | No. phosphosites |
|---|---|---|---|
| PhosphoSitePlus | Chicken | 4 | 9 |
| Cow | 17 | 40 | |
| Human | 199 | 406 | |
| Mouse | 48 | 94 | |
| Rabbit | 12 | 17 | |
| Rat | 37 | 86 | |
| Dog | 3 | 6 | |
| Meggio and Pinna[2] | Bovine | 9 | 14 |
| Fruit fly | 7 | 15 | |
| Epstein-Barr virus | 2 | 4 | |
| Chicken | 2 | 4 | |
| Hepatitis delta virus | 1 | 1 | |
| Human | 78 | 135 | |
| HIV1 | 3 | 5 | |
| Mouse | 9 | 12 | |
| Rabbit | 7 | 13 | |
| Rat | 11 | 22 | |
| Baker’s yeast | 4 | 7 | |
| SV40 | 1 | 3 | |
| African clawed frog | 2 | 3 | |
| Bian et al. [29] | Human | 575 | 986 |
The prediction shows that 327 substrates, 467 sites, out of the total matched the described phosphorylation patterns (Table 3). Here, the number of substrates analyzed for prediction was 883 instead of 913 given that the ScanProSite tool eliminated 30 proteins from which the corresponding UniProt entries were either merged or deleted (Table 3). The rabbit, chicken, and African clawed frog (Xenopus laevis) sets stood out with more than 50% of the proteins having at least one class-III site; contrastingly, in other organisms such as human, mouse and fruit fly the matches represent around one third of the corresponding proteins (Table 3). Nevertheless, a more systematic annotation of CK2 phosphoacceptor sites that provide comparable data among organisms is needed.
Table 3.
Class-III substrates and phosphosites predicted based on the designed linear patterns across several organisms and viruses.
| Organism | No. proteins | No. Class III proteins (%) | No. phosphosites | No. class III sites (%) |
|---|---|---|---|---|
| Human | 734 | 274 (37) | 1380 | 392 (28) |
| Mouse | 50 | 16 (32) | 98 | 25 (26) |
| Rat | 39 | 11 (28) | 90 | 16 (18) |
| Bovine | 20 | 7 (35) | 51 | 10 (20) |
| Rabbit | 14 | 7 (50) | 28 | 7 (25) |
| Fruit fly | 7 | 2 (29) | 15 | 3 (20) |
| Chicken | 4 | 2 (50) | 11 | 4 (36) |
| Baker's yeast | 4 | 1 (25) | 7 | 1 (14) |
| HIV-1 | 3 | 2 (67) | 5 | 3 (60) |
| Epstein-Barr virus | 2 | 2 (100) | 4 | 2 (50) |
| African clawed frog | 2 | 1 (50) | 3 | 1 (33) |
| Hepatitis delta virus | 1 | 1 (100) | 1 | 1 (100) |
| SV40 | 1 | 1 (100) | 3 | 2 (67) |
| Dog | 2 | – | 6 | – |
| Eliminated a | 30 | – | 45 | – |
| Total | 913 | 327 | 1745 | 467 |
CK2 substrates eliminated from the analysis by ScanProsite tool since the corresponding UniProt entry was either merged or deleted from the database.
3.3. Structural considerations
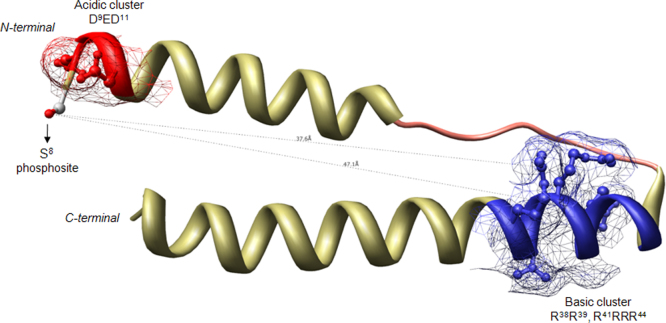
The sequence between the phosphosite(s) and the basic cluster forms an extended region in eIF2β and a structural HLH (helix–loop–helix) motif in Rev [16], [17], [18] (Fig. 2). Rev basic cluster localizes at a greater distance compared with eIF2β (Table 1) but the existence of a HLH motif might bring closer these determinants due to loop flexibility (Fig. 2) [16], [17]. The deletion of the loop in Rev protein reduces holoenzyme phosphorylation efficiency [16]. This illustrates the importance of considering information of protein secondary structure when analyzing beta-dependent phosphorylation. However, experimental information is missing on known class-III substrates that allow us to comprehensively describe the vicinity of the phosphosites in a structural basis. For example, among the other experimentally determined class-III substrates only Fdx1 and SIX1 have structural information available; in both cases the sequence between the phosphosite and the basic cluster forms an extended region (Fig. 3).
Fig. 2.
Ribbon representation of CK2 holoenzyme substrate Rev helix–loop–helix motif (PDB ID: 3LPH). The phosphosite Ser-8 is shown as balls-and-sticks. The acidic residues in the vicinity of Ser-8 and those within the basic cluster are colored in red and blue, respectively. The helixes and the loop of the HLH motif are depicted in yellow and pink, respectively. The distances between Ser-8 phosphosite and the first (Arg-38, 47.1 Å) and last amino acid (Arg-44, 37.6 Å) of the basic cluster in the substrate are represented in dashed lines.
Fig. 3.
Ribbon representation of class-III CK2 substrates A) Fdx1 (PDB ID: 3N9Y) and B) SIX1 (PDB ID: 4EGC). The phosphosite and the basic residues within the basic cluster are shown as balls-and-sticks. The acidic residues in the vicinity of the phosphosite and those within the basic cluster are colored in red and blue, respectively. The sequence that contains the loop and the basic cluster is shown in yellow. The distances between the phosphosite and the first amino acid of the basic cluster for each of the substrates are represented in dashed lines.
Based on Rev and eIF2β information, we searched for the occurrence of phosphopeptides located in a HLH motif or in an extended region. First, we retrieved the PDB sequences in FASTA format of the predicted substrates with associated PDB entries (146 substrates out of 327) (S3 File). Then, we mapped the predicted phosphopeptides to the sequences identifying 29 substrates (48 phosphopeptides) with at least one hit (in a total of 187 structures) (S3 File). Further sequence visualization and analysis discarded 15 of the substrates since they had important missing residues inside regions that matched the phosphopeptides (S3 File). From the 14 remaining substrates, two matched the HLH motif (S2 Fig.), two the extended region (S2 Fig.) and the remaining ones showed other motifs (S3 File). Correspondingly, this analysis pointed that in the 40% of the studied substrates; the class-III phosphopeptides locate in a region of the protein where the basic cluster may be closer to the phosphorylation site compared with what is expected in primary sequence observation.
3.4. Intrinsic complexity of CK2 substrate classification
CK2 substrate classification is a simplistic view of a complex regulatory process. Here, we provide two relevant examples on calmodulin (CALM) and B23 CK2 substrates that illustrate the complexity of class-III site prediction and that how a cautious interpretation is needed when analyzing the prediction results.
CALM is a class-II substrate [46] for which phosphorylation is enhanced by polybasic peptides such as polylysines and protamines [15], [47]. CK2 phosphorylates CALM in vitro with the catalytic subunit modifying the Thr-79, Ser-81, Ser-101, and Thr-117 residues [15]. In the present study, these sites were used for class-III prediction and no matches were expected. Conversely, we identified the Ser-101 residue, located in the Ca-binding loop III of the protein [48], as a putative class-III site. To explain this result we searched the literature and found a work by Arrigoni et al. 2004 that explored the CK2-dependent phosphorylation of these sites using large synthetic peptides of the protein [15]. They concluded that CK2 phosphorylation of different CALM peptides lacking the C-terminal domain (essential for conferring class-II substrate behavior and polylysine dependency) occurs in the presence of either the catalytic subunit or the holoenzyme. Noticeably, two of these peptides, residues 93-106 and 54–106, included the Ser-101 phosphosite [15]. In this example, is made evident that the full-length protein dictates the behavior of the different phosphosites and that the classification of the protein based on in vitro experiments using peptides or in silico prediction could be misleading.
Other example that showcases classification complexity is the CK2-mediated phosphorylation of B23. This protein has been previously regarded as a class-I CK2 substrate [13]. However, in a recent work by Zanin et al. the authors tested the effect of the beta-dependent phosphorylation inhibitor CK2-MCP peptide on B23. Conversely, they observed a certain inhibitory effect indicating a possible dependence on the regulatory subunit. In agreement, our analysis predicted the Ser-125 residue of B23 as a putative class-III site (Table 2). Although this finding supports the use of the designed patterns for capturing beta-dependent regulatory effect further experiments are mandatory to comprehensively describe B23 substrate behavior in vivo.
3.5. Class-III substrates functional classification
3.5.1. Role of CK2 holoenzyme phosphorylation in viral infection
The relevance of CK2 in viral infection is accepted with the enzyme interacting and/or phosphorylating both viral and infectivity-related host proteins. CK2β subunit binds EBNA1 [49] and IE63 [50] viral proteins while CK2α associates to NSI [51], pUL84 [52] and RIG-I (cellular viral infection defense) [53]. An example of functional association between this kinase and viral proteins is the induction of parvovirus capsid phosphorylation by NS1/CK2α complex [51].
The Table 4 summarizes the identified class-III proteins/sites relative to the four viruses selected for prediction and experimental information of the kinase assay. It includes the Ser-5 and Ser-8 phosphoacceptorsites of HIV-1 Rev protein used in pattern design. Literature search shows that the identification of CK2 phosphoacceptor sites among these proteins was carried out using the holoenzyme and that no analysis of subunit contribution was performed as it is the case for Rev protein (Table 4, S4 File). Thus, in the absence of complementary information, pattern-matching becomes a tool of choice guiding the detection of potential class-III sites. For example, in our analysis Vpu, BZLF1, LT-AG, Nef, LT-Ag and EBNA2 matched the designed patterns and were classified as beta-dependent substrates. However, for Nef, LT-Ag and EBNA2 substrates, in particular, literature analysis does suggest that the relevance of the basic cluster in the phosphorylation reaction might be only evident in the full-length protein since derived peptides lacking the basic determinant are phosphorylated by the holoenzyme. This probably resembles the class-I behavior observed in Ser-5 and Ser-8 phosphoacceptor sites in the context of the Rev peptide [16].
Table 4.
CK2 phosphorylated viral proteins and their tentative class-III sites.
| Virus | Protein Name | CK2 PhosphoSitePlus | Kinase assay substrate type | CK2 kinase assay | References |
|---|---|---|---|---|---|
| (Class-III prediction) | |||||
| HIV1 | Rev | Ser-5, Ser-8 | protein, Rev 1-26 peptide | Recombinant α and β subunits of human CK2. | [16], [17] |
| Vpu | Ser-52, Ser-56 | protein, Vpu 32-81 peptide | Recombinant human CK2. | [36], [37] | |
| HIV2 | Nef | Ser-92 | Nef87-96 (MDDVDSDDDD) peptide | Purified CK2 (bovine liver, lung, or brain) | [56] |
| Epstein-barr virus | EBNA2 | Ser-469, Ser-470 | protein, EBNA2 462-474 (FETTESPSSDEDY) peptide | Recombinant CK2. | [57] |
| BZLF1 | Ser-167, Ser-173 | protein | Recombinant CK2. | [58], [59], [60] | |
| SV40 | LT-AG | Ser-106, Ser-111, Ser-112 | LT-AG 106-119 (SEEMPSSDDEATAD) peptide | Purified CK2 (bovine liver, lung, or brain) | [56] |
| Hepatitis delta virus | HDAg | Ser-2 | protein | No kinase assay. In vivo labeling and kinase inhibitor experiments | [61] |
Overall, our results for Vpu and BZLF1 proteins might serve to speculate a functional relationship between these viral proteins and CK2 beta-dependent phosphorylation. Hence, based on this assumption, we searched the literature and found interesting evidences connecting Vpu with CK2 beta-dependent phosphorylation. For BZLF1 protein, literature information was insufficient for a further analysis (S4 File).
Vpu is a HIV1 viral protein involved in CD4 proteasomal degradation and in the inhibition of protein transport from ER-Golgi complex to the plasma membrane; functions regulated by CK2-dependent phosphorylation of the Ser-52 and Ser-56 residues [62]. These phosphosites were characterized in vitro using CK2 holoenzyme and Vpu recombinant protein and synthetic peptides (Table 4) [55]. The studied peptides constitute variants of the hydrophilic C-terminal domain (residues 32–81) which besides the CK2 consensus sequence contains a basic cluster (R34QRK37) at an appropriate distance up-stream the phosphosites [54]. In correspondence both sites were predicted as class-III.
The E47RAEDSGNESEG58 dodecapeptide is considered the major determinant of Vpu CK2-mediated phosphorylation [63]. This is a highly conserved sequence found in Vpu protein of different HIV1 strains [63]. To investigate the significance of the basic cluster we ran a BLAST search against UniProt database and identified 10 Vpu proteins (reviewed status) sharing 100% of sequence identity to the dodecapeptide (Fig. S3). The alignment of these 10 sequences shows that the basic cluster is also well-conserved (Fig. S3). This finding suggests that the acidic cluster might be a relevant regulatory element of CK2-mediated phosphorylation in Vpu. The validation of this viral protein as a class-III substrate might provide a strategy in which targeting of holoenzyme integrity will impair protein function in HIV viral infection.
3.5.2. Functional classification of human class-III substrates
Bian et al. performed a global screening of human CK2 substrates where they identified 575 substrates of the kinase (988 sites) (Table 2, S1 File), a list enriched in spliceosomal proteins. This experiment combined in vitro CK2 holoenzyme reaction and immobilized proteome along with phosphoproteomics. The results were refined by filtering the data based on in vivo phosphosites information extracted from phosphorylation databases [29]. Importantly, the authors used CK2 holoenzyme for kinase reaction most likely marginalizing the phosphorylation of class-II sites [29].
Class-III prediction of Bian et al. substrates pointed out that 219 out of the 575 proteins might correspond to this category (Table 2). The functional classification of these putative class-III substrates might serve to speculate, considering the predictive nature of this approach, which biological processes and pathways linked to CK2 signaling might be targeted by class-III phosphorylation inhibitors. As a result, the functional analysis relative to the background of identified CK2 substrates (S5 File) indicated a significant enrichment of RNA processing biological process (P value=0.003) and of nuclear lumen subcellular localization (P value=0.01) with proteins from both the nucleolus (P value=0.02) and the nucleoplasm part (P value=0.03). This suggests that the holoenzyme might exert a key role regulating the function of nuclear located CK2 substrates that function on RNA processing events like RNA splicing and mRNA processing. Likewise, selecting the human genome as the background list (S5 File) indicated a marked enrichment of processes related to RNA processing (P value=6.31e−10) and of nuclear localization (P value=8.48e−63) (Fig. 4). Other enriched biological processes and cellular components are regulation of transcription (P value=3.73e−4), cell cycle (P value=2.24e−2), apoptosis (P value=0.007), cytoplasm (P value=2.22e−17), and cytoskeleton (P value=0.003) (Fig. 4).
Fig. 4.
Biological processes and cellular components enriched in CK2 class-III substrates predicted from Bian et al. dataset. All annotations are significantly enriched (P value<0.05).
A previous study by Filhol et al. analyzed the intracellular dynamic of CK2 subunits distribution using live-cell imaging [11]. The authors found that the holoenzyme is targeted to the cytoplasm and that binding to certain proteins such as FGF-2 enhances its nuclear accumulation. In addition CK2 independent subunits are imported to nucleus where they may associate to form the holoenzyme or interact with nuclear proteins and mediate independent functions [11]. The subcellular location analysis (S5 File) shows an enrichmentof nuclear (137 proteins) and cytoplasmic (79 proteins) class-III predicted substrates with an overlap of 62 proteins that localize to both compartments (Fig. 4). In correspondence, we also studied the subcellular location of human class-III substrates predicted from PhosphoSitePlus dataset. Similarly, the substrate list was enriched in the nuclear (42 proteins) and cytoplasm (33 proteins) locations with an overlap of 26 proteins. Thus, the results suggest that beta-dependent phosphorylation could modulate primarily the function of nuclear-located CK2 substrates. In addition, protein overlap points to a possible link between cytoplasmic class-III substrates phosphorylation and their redistribution to the nucleus.
The involvement of CK2 in DNA repair and its regulation has been well-documented with the identification for instance of BRCA1, APC and P53 as binding partners and/or substrates of the enzyme [64], [65], [66]. The biological process enrichment analysis (S5 File) supports a possible functional relationship between beta-dependent phosphorylation and the response to DNA damage stimulus and DNA repair. For example, the protein MDC1 (Mediator of DNA damage checkpoint protein 1), here predicted as a class-III substrate, is a scaffold for the recruitment of DNA repair and signal transduction proteins to damaged sites required for cell cycle arrest. Its phosphorylation by CK2 promotes the assembly and retention of the MRN complex to DSBs for the initiation of HR repair [67].
RNA interference and down-regulation of CK2 abundance level result in potent induction of apoptosis [68]. Here a total of 9 proteins associated to apoptosis were predicted as class-III substrates (DIDO1, CDK11A, PDCD4, CLSPN, FXR1, ZC3H8, DPF2, DDX41, and PSMD2) (S5 File) Likewise, some of the predicted class-III substrates were classified as cancer genes based on Futreal et al. 2004 (DEK, NUP98, PSIP1, MYH9, and HSP90AB1) [69] and Gene database (CDC5L, DEK, MTDH, CLSPN, SLC4A1AP, RSF1, and PDCD4) information (S5 File). For example, DEK is an oncogene with chromosomal aberration in AML and the encoded protein is involved in chromatin organization and in splice site selection during mRNA processing. CK2-mediated phosphorylation of the C-terminal domain modifies DNA binding properties of DEK [70].
Furthermore, the mapping of predicted class-III substrates in the human protein–protein interaction network (S5 File) situates them as member of functional modules that correspond to intracellular signaling pathways such as: cell cycle, RNA transport, Wnt signaling pathway, and P53 signaling pathway (Fig. 5). We also mapped the predicted substrates to modules associated to pathways in cancer including prostate, pancreatic and lung cancer and chronic myeloid leukemia. Finally, the network analysis also pointed to the possible modulation of hub proteins including HDAC1, NCL and ESR1 and of proteins essential for connectivity between modules like ATRX, CPSF2 and CHD1 (S5 File).
Fig. 5.
Protein–protein interaction network depicting the functional heterogeneity of class-III substrates predicted from Bian et. al dataset (193 seed nodes) and of their first neighbors (414 nodes). Enriched pathways: cell cycle (pink, P value=1.64e−11), RNA transport (green, P value=2.41e−11), Pathways in cancer (red, P value=9.98e−05), Adherens junction (yellow, P value=2.39e−04), Wnt signaling pathway (blue, P value=0.005), and P53 signaling pathway (violet, P value=0.03). Node size is proportional to node degree.
4. Conclusions
The four designed linear patterns aimed to assist CK2 beta-dependent substrates prediction. The discrimination power of these patterns relies on the recognition of a basic cluster at a suitable distance from the phosphoacceptor site in the substrate. Since the patterns utilized the CK2 consensus sequence for matching substrates we performed the prediction of class-III substrates on experimentally determined CK2 substrates to reduce noise. As a result we obtained a list of 327 predicted class-III substrates, 467 sites. The functional classification of these substrates indicated a role of beta-dependent regulation in viral infection and biological processes and pathways such as apoptosis, DNA repair and RNA metabolism. It also suggested that the human substrates are primarily nuclear located with a number of them also found in cytoplasm. A cautious interpretation of these results is needed since this analysis derives from an in silico predictive approach. Thus, future experiments are required to validate the results of the prediction and the functional analysis.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2015.08.011
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Guerra B., Issinger O.-G. Protein kinase CK2 in human diseases. Curr. Med. Chem. 2008;15:1870–1886. doi: 10.2174/092986708785132933. [DOI] [PubMed] [Google Scholar]
- 2.Salvi M., Sarno S., Cesaro L., Nakamura H., Pinna L.A. Extraordinary pleiotropy of protein kinase CK2 revealed by weblogo phosphoproteome analysis. Biochim. Biophys. Acta. 2009;1793:847–859. doi: 10.1016/j.bbamcr.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 4.Pinna L.A. Protein kinase CK2: a challenge to canons. J. Cell Sci. 2002;115:3873–3878. doi: 10.1242/jcs.00074. [DOI] [PubMed] [Google Scholar]
- 5.Olsen B.B., Wang S.-Y., Svenstrup T.H., Chen B.P.C., Guerra B. Protein kinase CK2 localizes to sites of DNA double-strand break regulating the cellular response to DNA damage. BMC Mol. Biol. 2012;13:7. doi: 10.1186/1471-2199-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Maira G., Brustolon F., Bertacchini J., Tosoni K., Marmiroli S., Pinna L.A., Ruzzene M. Pharmacological inhibition of protein kinase CK2 reverts the multidrug resistance phenotype of a CEM cell line characterized by high CK2 level. Oncogene. 2007;26:6915–6926. doi: 10.1038/sj.onc.1210495. [DOI] [PubMed] [Google Scholar]
- 7.St-Denis N.A., Litchfield D.W. Protein kinase CK2 in health and disease: From birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell. Mol. Life Sci. 2009;66:1817–1829. doi: 10.1007/s00018-009-9150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perea S.E., Reyes O., Baladron I., Perera Y., Farina H., Gil J., Rodriguez A., Bacardi D., Marcelo J.L., Cosme K., Cruz M., Valenzuela C., López-Saura P.A., Puchades Y., Serrano J.M., Mendoza O., Castellanos L., Sanchez A., Betancourt L., Besada V. CIGB-300, a novel proapoptotic peptide that impairs the CK2 phosphorylation and exhibits anticancer properties both in vitro and in vivo. Mol. Cell. Biochem. 2008;316:163–167. doi: 10.1007/s11010-008-9814-5. [DOI] [PubMed] [Google Scholar]
- 9.Litchfield D.W. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niefind K., Guerra B., Ermakowa I., Issinger O.G. Crystal structure of human protein kinase CK2: insights into basic properties of the CK2 holoenzyme. EMBO J. 2001;20:5320–5331. doi: 10.1093/emboj/20.19.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filhol O., Nueda A., Martel V., Gerber-Scokaert D., Benitez M.J., Souchier C., Saoudi Y., Cochet C. Live-cell fluorescence imaging reveals the dynamics of protein kinase CK2 individual subunits. Mol. Cell. Biol. 2003;23:975–987. doi: 10.1128/MCB.23.3.975-987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibby A.C., Litchfield D.W. The multiple personalities of the regulatory subunit of protein kinase CK2: CK2 dependent and CK2 independent roles reveal a secret identity for CK2beta. Int. J. Biol. Sci. 2005;1:67–79. doi: 10.7150/ijbs.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanin S., Sandre M., Cozza G., Ottaviani D., Marin O., Pinna L.A., Ruzzene M. Chimeric peptides as modulators of CK2-dependent signaling: mechanism of action and off-target effects. Biochim. Biophys. Acta. 2015 doi: 10.1016/j.bbapap.2015.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Meggio F., Boldyreff B., Issinger O.G., Pińna L.A. Casein kinase 2 down-regulation and activation by polybasic peptides are mediated by acidic residues in the 55-64 region of the beta-subunit A study with calmodulin as phosphorylatable substrate. Biochemistry. 1994;33:4336–4342. doi: 10.1021/bi00180a030. [DOI] [PubMed] [Google Scholar]
- 15.Arrigoni G., Marin O., Pagano M.A., Settimo L., Paolin B., Meggio F., Pinna L.A. Phosphorylation of calmodulin fragments by protein kinase CK2 Mechanistic aspects and structural consequences. Biochemistry. 2004;43:12788–12798. doi: 10.1021/bi049365c. [DOI] [PubMed] [Google Scholar]
- 16.Marin O., Sarno S., Boschetti M., Pagano M.A., Meggio F., Ciminale V., D’Agostino D.M., Pinna L.A. Unique features of HIV-1 Rev protein phosphorylation by protein kinase CK2 (’casein kinase-2’) FEBS Lett. 2000;481:63–67. doi: 10.1016/s0014-5793(00)01971-2. [DOI] [PubMed] [Google Scholar]
- 17.Meggio F., Marin O., Boschetti M., Sarno S., Pinna L.A. HIV-1 Rev transactivator: a beta-subunit directed substrate and effector of protein kinase CK2. Mol. Cell. Biochem. 2001;227:145–151. [PubMed] [Google Scholar]
- 18.Poletto G., Vilardell J., Marin O., Pagano M.A., Cozza G., Sarno S., Falqués A., Itarte E., Pinna L.A., Meggio F. The regulatory beta subunit of protein kinase CK2 contributes to the recognition of the substrate consensus sequence A study with an eIF2 beta-derived peptide. Biochemistry. 2008;47:8317–8325. doi: 10.1021/bi800216d. [DOI] [PubMed] [Google Scholar]
- 19.Ford H.L., Landesman-Bollag E., Dacwag C.S., Stukenberg P.T., Pardee A.B., Seldin D.C. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J. Biol. Chem. 2000;275:22245–22254. doi: 10.1074/jbc.M002446200. [DOI] [PubMed] [Google Scholar]
- 20.Huillard E., Ziercher L., Blond O., Wong M., Deloulme J.-C., Souchelnytskyi S., Baudier J., Cochet C., Buchou T. Disruption of CK2beta in embryonic neural stem cells compromises proliferation and oligodendrogenesis in the mouse telencephalon. Mol. Cell. Biol. 2010;30:2737–2749. doi: 10.1128/MCB.01566-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng R., Al-Quobaili F., Müller I., Götz C., Thiel G., Montenarh M. CK2 phosphorylation of Pdx-1 regulates its transcription factor activity. Cell. Mol. Life Sci. 2010;67:2481–2489. doi: 10.1007/s00018-010-0348-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochscherf J., Lindenblatt D., Steinkrüger M., Yoo E., Ulucan O., Herzig S., Issinger O.-G., Helms V., Götz C., Neundorf I., Niefind K., Pietsch M. Development of a high-throughput screening-compatible assay to identify inhibitors of the CK2α/CK2β interaction. Anal. Biochem. 2014;468C:4–14. doi: 10.1016/j.ab.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Maizel A., Tassetto M., Filhol O., Cochet C., Prochiantz A., Joliot A. Engrailed homeoprotein secretion is a regulated process. Development. 2002;129:3545–3553. doi: 10.1242/dev.129.15.3545. [DOI] [PubMed] [Google Scholar]
- 24.Palmeri A., Ferrè F., Helmer-Citterich M. Exploiting holistic approaches to model specificity in protein phosphorylation. Front. Genet. 2014;5:315. doi: 10.3389/fgene.2014.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman R.H., Zhang J., Zhu H. Toward a systems-level view of dynamic phosphorylation networks. Front. Genet. 2014;5:263. doi: 10.3389/fgene.2014.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M.Torii, G.Li, Z.Li, R.Oughtred, F.Diella, I.Celen, C.N.Arighi, H.Huang, K.Vijay-Shanker, C.H.Wu, RLIMS-P: an online text-mining tool for literature-based extraction of protein phosphorylation information, Database (Oxford)., 2014 (2014). [DOI] [PMC free article] [PubMed]
- 27.Chen H., Sharp B.M. Content-rich biological network constructed by mining PubMed abstracts. BMC Bioinformatics. 2004;5:147. doi: 10.1186/1471-2105-5-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian Y., Ye M., Wang C., Cheng K., Song C., Dong M., Pan Y., Qin H., Zou H. Global screening of CK2 kinase substrates by an integrated phosphoproteomics workflow. Sci. Rep. 2013;3:3460. doi: 10.1038/srep03460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Castro E., Sigrist C.J.A., Gattiker A., Bulliard V., Langendijk-Genevaux P.S., Gasteiger E., Bairoch A., Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berman H.M., Battistuz T., Bhat T.N., Bluhm W.F., Bourne P.E., Burkhardt K., Feng Z., Gilliland G.L., Iype L., Jain S., Fagan P., Marvin J., Padilla D., Ravichandran V., Schneider B., Thanki N., Weissig H., Westbrook J.D., Zardecki C. The Protein Data Bank. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 32.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 33.Wu C.H., Yeh L.-S.L., Huang H., Arminski L., Castro-Alvear J., Chen Y., Hu Z., Kourtesis P., Ledley R.S., Suzek B.E., Vinayaka C.R., Zhang J., Barker W.C. The Protein Information Resource. Nucleic Acids Res. 2003;31:345–347. doi: 10.1093/nar/gkg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J.D., Higgins D.G. Vol. 7. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega; p. 539. (Mol. Syst. Biol.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Tabas-Madrid D., Nogales-Cadenas R., Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40:W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Thilmony R., Gu Y.Q. NetVenn: an integrated network analysis web platform for gene lists. Nucleic Acids Res. 2014;42:W161–W166. doi: 10.1093/nar/gku331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M.Magrane, U.Consortium, UniProt Knowledgebase: a hub of integrated protein data, Database (Oxford)., 2011 (2011) bar009. [DOI] [PMC free article] [PubMed]
- 39.Doms A., Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33:W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia J., Benner M.J., Hancock R.E.W. NetworkAnalyst--integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014;42:W167–W174. doi: 10.1093/nar/gku443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berkey C.D., Carlson M. A specific catalytic subunit isoform of protein kinase CK2 is required for phosphorylation of the repressor Nrg1 in Saccharomyces cerevisiae. Curr. Genet. 2006;50:1–10. doi: 10.1007/s00294-006-0070-5. [DOI] [PubMed] [Google Scholar]
- 42.Bureik M., Zöllner A., Schuster N., Montenarh M., Bernhardt R. Phosphorylation of bovine adrenodoxin by protein kinase CK2 affects the interaction with its redox partner cytochrome P450scc (CYP11A1) Biochemistry. 2005;44:3821–3830. doi: 10.1021/bi047697b. [DOI] [PubMed] [Google Scholar]
- 43.Venerando A., Franchin C., Cant N., Cozza G., Pagano M.A., Tosoni K., Al-Zahrani A., Arrigoni G., Ford R.C., Mehta A., Pinna L.A. Detection of phospho-sites generated by protein kinase CK2 in CFTR: mechanistic aspects of Thr1471 phosphorylation. PLoS One. 2013;8:e74232. doi: 10.1371/journal.pone.0074232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews B.W. Stoichiometry versus hydrophobicity in protein folding. J. Biomol. Struct. Dyn. 2011;28(589–91):669–674. doi: 10.1080/073911011010524956. discussion. [DOI] [PubMed] [Google Scholar]
- 45.Diella F., Cameron S., Gemünd C., Linding R., Via A., Kuster B., Sicheritz-Pontén T., Blom N., Gibson T.J. PhosphoELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bidwai A.P., Reed J.C., Glover C.V. Phosphorylation of calmodulin by the catalytic subunit of casein kinase II is inhibited by the regulatory subunit. Arch. Biochem. Biophys. 1993;300:265–270. doi: 10.1006/abbi.1993.1037. [DOI] [PubMed] [Google Scholar]
- 47.Meggio F., Boldyreff B., Marin O., Marchiori F., Perich J.W., Issinger O.G., Pinna L.A. The effect of polylysine on casein-kinase-2 activity is influenced by both the structure of the protein/peptide substrates and the subunit composition of the enzyme. Eur. J. Biochem. 1992;205:939–945. doi: 10.1111/j.1432-1033.1992.tb16860.x. [DOI] [PubMed] [Google Scholar]
- 48.Nelson M.R., Thulin E., Fagan P.A., Forsén S., Chazin W.J. The EF-hand domain: a globally cooperative structural unit. Protein Sci. 2002;11:198–205. doi: 10.1110/ps.33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivachandran N., Cao J.Y., Frappier L. Epstein-Barr virus nuclear antigen 1 Hijacks the host kinase CK2 to disrupt PML nuclear bodies. J. Virol. 2010;84:11113–11123. doi: 10.1128/JVI.01183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wadd S., Bryant H., Filhol O., Scott J.E., Hsieh T.Y., Everett R.D., Clements J.B. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J. Biol. Chem. 1999;274:28991–28998. doi: 10.1074/jbc.274.41.28991. [DOI] [PubMed] [Google Scholar]
- 51.Nüesch J.P.F., Rommelaere J. NS1 interaction with CKII alpha: novel protein complex mediating parvovirus-induced cytotoxicity. J. Virol. 2006;80:4729–4739. doi: 10.1128/JVI.80.10.4729-4739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Y., Pari G.S. Interaction of human cytomegalovirus pUL84 with casein kinase 2 is required for oriLyt-dependent DNA replication. J. Virol. 2009;83:2393–2396. doi: 10.1128/JVI.02339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Z., Ren H., Liu Y., Teeling J.L., Gu J. Phosphorylation of RIG-I by casein kinase II inhibits its antiviral response. J. Virol. 2011;85:1036–1047. doi: 10.1128/JVI.01734-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.P.Henklein, U.Schubert, O.Kunert, S.Klabunde, V.Wray, K.D.Klöppel, M.Kiess, T.Portsmann, D.Schomburg, Synthesis and characterization of the hydrophilic C-terminal domain of the human immunodeficiency virus type 1-encoded virus protein U (Vpu), Pept. Res., 6 79–87. [PubMed]
- 55.Schubert U., Henklein P., Boldyreff B., Wingender E., Strebel K., Porstmann T. The human immunodeficiency virus type 1 encoded Vpu protein is phosphorylated by casein kinase-2 (CK-2) at positions Ser52 and Ser56 within a predicted alpha-helix-turn-alpha-helix-motif. J. Mol. Biol. 1994;236:16–25. doi: 10.1006/jmbi.1994.1114. [DOI] [PubMed] [Google Scholar]
- 56.Marshak D.R., Carroll D. Synthetic peptide substrates for casein kinase II. Methods Enzymol. 1991;200:134–156. doi: 10.1016/0076-6879(91)00135-j. [DOI] [PubMed] [Google Scholar]
- 57.Grässer F.A., Göttel S., Haiss P., Boldyreff B., Issinger O.G., Mueller-Lantzsch N. Phosphorylation of the Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 1992;186:1694–1701. doi: 10.1016/s0006-291x(05)81604-3. [DOI] [PubMed] [Google Scholar]
- 58.El-Guindy A.S., Paek S.Y., Countryman J., Miller G. Identification of constitutive phosphorylation sites on the Epstein-Barr virus ZEBRA protein. J. Biol. Chem. 2006;281:3085–3095. doi: 10.1074/jbc.M506076200. [DOI] [PubMed] [Google Scholar]
- 59.Kolman J.L., Taylor N., Marshak D.R., Miller G. Serine-173 of the Epstein-Barr virus ZEBRA protein is required for DNA binding and is a target for casein kinase II phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10115–10119. doi: 10.1073/pnas.90.21.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Guindy A.S., Miller G. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 2004;78:7634–7644. doi: 10.1128/JVI.78.14.7634-7644.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeh T.S., Lo S.J., Chen P.J., Lee Y.H. Casein kinase II and protein kinase C modulate hepatitis delta virus RNA replication but not empty viral particle assembly. J. Virol. 1996;70:6190–6198. doi: 10.1128/jvi.70.9.6190-6198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vincent M.J., Abdul Jabbar M. The human immunodeficiency virus type 1 Vpu protein: a potential regulator of proteolysis and protein transport in the mammalian secretory pathway. Virology. 1995;213:639–649. doi: 10.1006/viro.1995.0035. [DOI] [PubMed] [Google Scholar]
- 63.Schubert U., Schneider T., Henklein P., Hoffmann K., Berthold E., Hauser H., Pauli G., Porstmann T. Human-immunodeficiency-virus-type-1-encoded Vpu protein is phosphorylated by casein kinase II. Eur. J. Biochem. 1992;204:875–883. doi: 10.1111/j.1432-1033.1992.tb16707.x. [DOI] [PubMed] [Google Scholar]
- 64.O’Brien K.A., Lemke S.J., Cocke K.S., Rao R.N., Beckmann R.P. Casein Kinase 2 Binds to and Phosphorylates BRCA1. Biochem. Biophys. Res. Commun. 1999;260:658–664. doi: 10.1006/bbrc.1999.0892. [DOI] [PubMed] [Google Scholar]
- 65.Homma M.K., Li D., Krebs E.G., Yuasa Y., Homma Y. Association and regulation of casein kinase 2 activity by adenomatous polyposis coli protein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5959–5964. doi: 10.1073/pnas.092143199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKendrick L., Milne D., Meek D. Protein kinase CK2-dependent regulation of p53 function: evidence that the phosphorylation status of the serine 386 (CK2) site of p53 is constitutive and stable. Mol. Cell. Biochem. 1999;191:187–199. [PubMed] [Google Scholar]
- 67.Becherel O.J., Jakob B., Cherry A.L., Gueven N., Fusser M., Kijas A.W., Peng C., Katyal S., McKinnon P.J., Chen J., Epe B., Smerdon S.J., Taucher-Scholz G., Lavin M.F. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res. 2010;38:1489–1503. doi: 10.1093/nar/gkp1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slaton J.W., Unger G.M., Sloper D.T., Davis A.T., Ahmed K. Induction of apoptosis by antisense CK2 in human prostate cancer xenograft model. Mol. Cancer Res. 2004;2:712–721. [PubMed] [Google Scholar]
- 69.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kappes F., Damoc C., Knippers R., Przybylski M., Pinna L.A., Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol. Cell. Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material