Abstract
Background
While electrospun materials have been frequently used in tissue engineering no wound dressings exist that significantly improved wound healing effectively.
Methods
We succeeded to fabricate three-dimensional (3D) electrospun poly(D,l-lactide) (PLA) fiber mats into which nanospheres, formed from amorphous calcium polyphosphate (polyP) nanoparticles (NP) and encapsulated retinol (“retinol/aCa-polyP-NS” nanospheres [NS]), had been incorporated.
Results
Experiments with MC3T3-E1 cells revealed that co-incubation of the cells with Ca-polyP together with retinol (or incubation with retinol/aCa-polyP-NS) resulted in a significant synergistic effect on cell growth compared with particle-free polyP complexed with Ca2+ or amorphous Ca-polyP NPs and retinol alone. Incubation of the cells in the presence of the retinol/aCa-polyP NSs also caused a significant increase of the expression levels of the genes encoding for the fatty acid binding protein 4 (FABP4), as well as of the genes encoding for leptin and the leptin receptor. In contrast, the single components, soluble Na-polyP, complexed to Ca2+, or retinol-free aCa-polyP NPs, and retinol, had no significant effect on the expression of these genes.
Conclusions
These results indicate that the PLA fibers, supplemented with aCa-polyP-NP or retinol/aCa-polyP-NS, elicit morphogenetic activity, suggesting that these fiber mats, along with the antibacterial effect of polyP, have a beneficial potential as wound dressings combining antimicrobial and regenerative (wound healing) properties.
General significance
The PLA-based fiber mats, containing retinol and polyP nanoparticles, provide promising bioactive meshes that are urgently needed as dressings for chronic wounds.
Keywords: Polyphosphate, Retinol, Electrospinning, Wound healing, Skin damage
Graphical abstract
Highlights
• Effect of PLA fiber mats with polyphosphate/retinol nanospheres on gene expression.
• Increased expression of FABP4, leptin and leptin receptor in MC3T3-E1 cells.
• Application as wound dressings, combining antimicrobial and regenerative properties.
1. Introduction
Wound healing is a complex and sequential, biological process that involves at least four consecutive but overlapping and highly programmed phases (reviewed in: [1]): Hemostasis, inflammation, proliferation, and remodeling. First, during the hemostasis period that immediately starts after wound setting vascular constriction and fibrin clot formation occurs. Second after bleeding is under control, pro-inflammatory cytokines and growth factors, including platelet-derived growth factor, fibroblast growth factor and epidermal growth factor, are released. Parallel with these events, a sequential infiltration of neutrophils, macrophages and lymphocytes, as well as platelets takes place that contributes to the prevention of blood loss [2]. The function of these cells is the elimination of invading microorganisms as well as the removal of cellular debris within the damaged tissue region. These cells produce and release protease(s) and reactive oxygen species, which might cause bystander damage, but also morphogenetically active polymers, e.g. polyphosphate (polyP) from platelets [3]. Third, during granulation tissue or new stroma formation, especially during the early phase of cutaneous wound repair, new capillaries endowing the neostroma with its granular appearance and macrophages, fibroblasts, and blood vessels move into the wound [4]. Finally, fourth, remodeling occurs that results in scar formation. Especially, during this phase the interaction of the cells, involved in remodeling, requires, for functional regeneration, the synthesis of the extracellular matrix by fibroblasts [5].
Both local and systemic factors are involved in wound healing. Major local factors that directly influence wound healing include (i) oxygenation/superoxide radical formation, involved in energy production and/or oxidative killing of pathogens, (ii) infections via microorganisms which are normally restricted to the skin surface but might invade the underlying tissue strata. While inflammation is a physiological period in the course of the wound-healing process, during which contaminating microorganisms are eliminated, a prolonged inflammation period caused by bacterial endotoxins and mediated by pro-inflammatory cytokines, e.g. interleukin-1 and tumor necrosis factor-α, may lead to an extended or even chronic inflammatory status. The main determining systemic factors affecting wound healing are the age status of the patient which results in a temporal delay in wound healing [6]. The major alterations are delayed T-cell migration into the damaged wound area, reduced macrophage phagocytic activity and delayed reepithelialization as well as collagen synthesis. Likewise, the shift in the balance between female estrogens (estrone and 17β-estradiol) and male androgens (testosterone and 5α-dihydrotestosterone) during aging adversely affect wound healing [7]. In addition, physical and psychological stresses that are associated with cardiovascular disease or cancer compromise wound healing. Furthermore, the impaired healing in patients with diabetes involving both hypoxia and dysfunction in fibroblasts and other epidermal cells impairs angiogenesis and neo-vascularization (reviewed in: [1]).
The well established and described medications of wound healing involve glucocorticoid steroid drugs, non-steroidal anti-inflammatory compounds or even chemotherapeutics. Adversely determining factors during wound healing are distinct, unhealthy lifestyle habits, like obesity, excessive alcohol consumption or smoking [8]. Wound healing supportive nutrients, like polyunsaturated fatty acids, positively influence the production of the essential or pro-inflammatory cytokines or vitamins; e.g., retinol and vitamin C show potent anti-oxidant, anti-inflammatory and, in turn, pro-wound healing effects [9].
As mentioned above, during wound healing cytokines and growth factors are released that initiate and maintain the interaction between the cells and control the regenerative response of the infiltrating cells. Besides of those soluble compounds external factors are efficient as well as regeneration-promoting stimuli. Among those is retinoic acid and its precursor retinol. Retinoic acid causes a differential gene expression by activating the genes controlling the pathways involved in retinoic acid esterification, synthesis from its precursor(s), and metabolism [10]; simultaneously, retinoic acid down-regulates the expression of the genes encoding for lipid metabolism during keratinocyte differentiation. In spite of an increased fibroblastic proliferation potency elicited by retinoic acid, the production of collagen was found to be diminished. In contrast to retinoic acid retinol has been proven to improve significantly wound healing [11].
It is well established that retinol is metabolically transformed into retinoic acid, especially in vivo by an enzymatically mediated two-step conversion via retinal and causes a differential gene expression within the retinoic acid nuclear receptors complex [12]. In response cellular proliferation and differentiation is modulated. However, only a small fraction of topically applied retinol is metabolized to retinoic acid, while it undergoes an alternative conversion into a series of metabolites, e.g. 14-hydroxy-retro-retinol which is involved in regulation of cell growth and death of lymphocytes. Finally, retinol was found to produce reactive oxygen species that activate different protein kinase pathways likewise involved in the control of morphologic and proliferative alterations in some human cells [13].
A new aspect in wound healing via restoration of endothelial progenitor cell functions is the finding that leptin, administered systemically and topically improves re-epithelialization of wounds in mice [14]. In addition, these authors described that keratinocytes, located at the wound margins, express the leptin-receptor subtype ObRb during the repair phase. Furthermore, leptin elicits a mitogenic stimulus to human keratinocyte cells in vitro. Those data are strongly corroborated by the finding that ob/ob (leptin null), and db/db (leptin receptor null), mouse strains show severe deficiencies in cutaneous wound repair [15]. Interestingly enough leptin primarily released from adipocytes but also synthesized in placenta, ovaries or skeletal muscle regulates fat deposition in the body (reviewed in: [16]). The leptin receptor has been originally identified in the hypothalamus. Applying both the techniques of polymerase chain reaction (PCR) and in situ hybridization evidence could be presented in a mouse model that leptin expression is acutely up-regulated in experimental wounds [17], suggesting that leptin is acutely up-regulated in injured skin tissue. Since also the leptin receptor is expressed in those cells autocrine/paracrine regulatory signaling cycle(s) have been proposed [17].
It is the aim of the present study to fabricate three-dimensional (3D) porous mats comprising loosely connected fibers with high porosity and high surface area by electrospinning. This widely used technology utilizes electrical forces to produce polymer fibers with diameters ranging from 2 nm to several micrometers and using polymer solutions of both natural and synthetic origin (reviewed in: [18]). This technology offers the favorable potential to synthesize novel natural nanofibers comprising controllable pore structures. As recently outlined only a few wound dressings have been developed that combine both prevention of microbial infiltration and stimulating wound cell regeneration [9]. In the present study we have developed wound covering meshes/mats that combine the property to allow a gas exchange environment with antibacterial properties as well as with a morphogenetic activity for the cells involved in wound healing. We succeeded to encapsulate retinol into nanospheres [19], formed from polyP nanoparticles, and fabricated them into electrospun fibers made of poly(D,l-lactide) (PLA). It is shown in the present study, that those mats retain the morphogenetic activity, to increase the expression of the fatty acid binding protein 4 (FABP4) as well as of leptin and its receptor, determined to be elicited by the retinol/polyP nanospheres that are not embedded into PLA. In previous studies it has been determined that polyP comprises also an antibacterial activity [20] and hemostatic activity [20].
For the administration of polyP and retinol to the cells had been fabricated into nanospheres; polyP acted as a bioscaffold for the production of nanospheres into which retinol has been encapsulated to form the nanospheres. Those polyP nanoparticles had been fabricated as amorphous particles; in those state, polyP maintained its morphogenetic activity and can be taken up by endocytosis [19]. For the studies here osteoblast-like MC3T3-E1 cells have been used that had been implicated in wound healing as well [21].
2. Material and methods
2.1. Materials
Na-polyphosphate (Na-polyP) with an average chain length of 30 phosphate units (NaPO3)n=30 was obtained from Merck Millipore (#106529; Darmstadt; Germany), all-trans retinol (#95144; ≥97.5%, Mr 286.45) was from Sigma (Taufkirchen; Germany).
2.2. Cultivation of MC3T3-E1 cells
The mouse calvaria cells MC3T3-E1 cells (ATCC-CRL-2593; #99072810; Sigma) were cultivated in a-MEM (Gibco-Invitrogen, Darmstadt; Germany) containing 20% fetal calf serum (FCS; Gibco). In addition, the medium was supplemented with 2 mM l-glutamine, 1 mM Na-pyruvate and 50 μg/ml of gentamycin. The cells were incubated in 25 cm2 flasks or in 24-well plates (Greiner Bio-One, Frickenhausen; Germany) in an incubator at 37 °C and 5% CO2. After reaching 80% confluency, the cells were detached using trypsin/EDTA and then subcultured at a density of 5·103 cells/ml. The cells were seeded at a density of 5·103 cells/well. Medium/serum change was every 3 d.
As indicated, the following polyP preparations were added to the cells; (i) “Na-polyP”, stoichiometrically complexed with Ca2+ (molar ratio of 1:2 [phosphate monomer:Ca2+]; [22]), (ii) “aCa-polyP-NP” nanoparticles (see below), (iii) “retinol/aCa-polyP-NS” nanospheres (see below) or (iv) “retinol”. Retinol was dissolved in ethanol (1 mg/ml) and then diluted in DMSO (dimethyl sulfoxide). Incubation was performed for 3 d.
In the indicated experiments, circular fiber mat samples (diameter of 15 mm) were inserted into each 24-well of the plates and seeded with MC3T3-E1 cells at a density of 5·103 cells/well. Then the assays were incubated for 3 d and subjected to microscopic as well as polymerase chain reaction analyses. In the assays to determine cell viability and growth, MC3T3-E1 cells were seeded onto the 15 mm (diameter) mat samples for 2 or 4 d.
2.3. Preparation of Ca-polyP nanoparticles and Ca-polyP/retinol nanospheres
The amorphous Ca-phosphate nanoparticles, aCa-polyP-NP, were prepared, as described recently [19]. The resulting “aCa-polyP-NP”, with a Ca2+ to phosphate stoichiometric ratio of 2:1, were found to be amorphous, as verified by Fourier transformed infrared spectroscopy and X-ray diffraction analysis.
The amorphous retinol/Ca-polyP nanospheres, retinol/aCa-polyP-NS, were prepared under avoidance of light. A retinol solution (100 mg/50 ml absolute ethanol), containing 2.8 g of CaCl2, was added drop-wise to a Na-polyP solution (1 g in 100 ml water). To avoid phase separation 2 g of poly(ethylene glycol) (PEG) (P5413; Sigma-Aldrich; average mol wt 8000) were added to the Na-polyP solution. After stirring for 6 h, the particles formed were collected by filtration. The nanospheres were found to contain 100 mg retinol/g nanospheres (≈1 µM), applying the SbCl3-based spectroscopic technique [23]. In contrast to the nanoparticles (size of the globular particles between 70 and 200 nm), formed without retinol (aCa-polyP-NP), the retinol-containing nanospheres (average diameter of 200–350 nm) were colored in light yellow.
2.4. Fabrication of poly(lactic acid) fiber mats
Poly(lactic acid)-based nanofibers were prepared as described [24]; a schematic outline is given in Fig. 1. Poly(D,l-lactide) (PLA) was obtained from Sigma (mol wt 75,000–120,000; P1691). PLA was mixed with PEG (in a ratio 80:20 wt%) and then added to chloroform at a final concentration of 20% (w/v). The PLA/PEG-chloroform suspension was stirred at 40 °C for 6 h, until complete dissolution of the polymers was obtained. Retinol was added to the PLA solution to give a final concentration of 20 wt% (with respect to PLA). Where indicated, Ca-polyP nanoparticles, aCa-polyP-NP, prepared according to Müller et al. [19]. were added to the PLA solution, containing already retinol, at a fixed concentration of 10 wt%. The weight ratio between CaCl2 and polyP had been adjusted to 2:1. The suspension was sonicated for 15 min immediately prior to electrospinning to assure a suitable dispersion of the nanospheres in the PLA solution.
Fig. 1.
Preparation of PLA-based electrospun mats. (A) PLA, the basic fiber matrix, is dissolved in chloroform. (B) Retinol (20 wt%) is added to this solution, followed by addition of the nanoparticles, aCa-polyP-NP (10 wt%). After a short stirring period, the suspension is sonicated immediately prior to (C) electrospinning. A spun mat is shown in the insert (a), a SEM image; an enlarged schematic outline of the fibers with their integrated retinol/Ca-polyP nanoparticles (retinol/Ca-polyP-NP) is depicted in (b). Fabricated mats are shown that are formed of (D) PLA alone, (E) PLA and retinol and (F) PLA, retinol and Ca-polyP.
Electrospinning was performed with the electrospinning unit (Spraybase; Profector Life Sciences, Dublin; Ireland) with some modifications, adapted to the PLA-based spinning material. The solutions were poured into the plastic syringe, equipped with a blunt-ended stainless steel needle (18-gauge) which was connected to a high-voltage supply. The solutions were spun at a feed rate of 1 ml per h and at a voltage of 17 kV; during the process the distance between nozzle and the collector was kept at 150 mm. A metallic net was used as the collector in all cases, except for the electrospinning of the mats. The fiber mats were removed from the collectors and dried overnight. During the complete procedures light exposure was avoided as so far as it is possible. The average diameter of the fibers within the mats vary between 0.5 and 2 µm.
2.5. Chemical characterization by Ca-phosphate by FTIR
Fourier transformed infrared (FTIR) spectroscopy in the attenuated total reflectance (ATR) mode was applied, using the Varian 660-IR spectrometer with Golden Gate ATR auxiliary (Agilent, Darmstadt; Germany). Spectra between the wavenumbers 4000 and 600 cm−1 were recorded.
2.6. Cell viability assay
Cell proliferation/cell viability was determined by the colorimetric XTT method ([Na-3′[1-(phenylamino-carbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene sulfonic acid]) using the “Cell Proliferation Kit II” (Roche, Mannheim; Germany). The absorbance was determined at 650 nm and subtracted from the background values (500 nm). Viable cells were determined after 72 h.
In the cell proliferation assays, performed on mats, incubation was terminated after 48 h or 96 h. The MC3T3-E1 cells were seeded onto the 15 mm (diameter) mat samples for 2 or 4 d. Then the serum/medium was aspirated and the XTT assay was performed.
2.7. Staining of the cells
The MC3T3-E1 cells were stained with Nile Blue A (Basic Blue 12, Nile blue sulfate; Sigma N0766) as described [25]. This staining reagent highlights in histological samples neutral lipids (triglycerides, cholesteryl esters, steroids) in pink, while acids (fatty acids, chromolipids, phospholipids) are stained in blue.
2.8. Reverse transcription-quantitative real-time PCR analyses
The gene expression levels were determined by applying the technique of reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR). The cells were incubated in medium/serum for 3 d in the absence or presence of 3 µg/ml of polyP (in the soluble form or in nanoparticles/nanospheres) or of 3 µM retinol. Then the cells were harvested, their RNA was isolated and subjected to RT-qPCR. The following primer pairs, matching with the respective mouse genes, were used: Fatty acid binding protein 4 (Mus musculus; accession number NM_024406) Fwd: 5′-CGATGAAATCACCGCAGACGAC-3′ [nt278 to nt299] and Rev: 5′-ACCACCAGCTTGTCACCATCTC-3′ [nt412 to nt392]; product size 135 bp; leptin (M. musculus; NM_008493) Fwd: 5′-GAAGAGACCGGGAAAGAGTGACAG-3′ [nt2888 to nt2911] and Rev: 5′-TGACCAAGGTGGCATAGCACAG-3′ [nt3040 to nt3019]; size 153 bp; and leptin receptor, transcript variant 2 (M. musculus; NM_010704) Fwd: 5′-GTGTGAGGAGGTACGTGGTGAAG-3′ [nt2570 to nt2592] and Rev: 5′-CCGAGGGAATTGACAGCCAGAAC-3′ [nt2708 to nt2686]; size 139 bp. The GAPDH [glyceraldehyde 3-phosphate dehydrogenase (Mus GAPDH; NM_008084)] was used as reference gene Fwd: 5′-TCACGGCAAATTCAACGGCAC-3′ [nt200 to nt220] and Rev: 5′-AGACTCCACGACATACTCAGCAC-3′ [nt338 to nt316; size 139 bp]. Amplification was performed in an iCycler (Bio-Rad, Hercules, CA; USA) applying the respective iCycler software. After determination of the Ct values the expression of the respective transcripts was calculated [26]. The expression levels of the respective genes were determined and the calculated values were correlated to the expression values for genes in untreated cells; this value was set to 1. The ratios between the levels in the cells, exposed to retinol or polyP alone or together, were calculated and then plotted.
2.9. Electron microscopy and energy dispersive X-ray spectroscopy
For the scanning electron microscopic (SEM) analyses a HITACHI SU 8000 electron microscope (Hitachi High-Technologies Europe GmbH, Krefeld, Germany) was employed. For the visualization of the cells, attached to the spun mats, the samples were remover after incubation and subjected to fixation, dehydration and air drying.
EDX spectroscopy was performed with an EDAX Genesis EDX System attached to a scanning electron microscope (Nova 600 Nanolab; FEI, Eindhoven, The Netherlands) operating at 10 kV with a collection time of 30–45 s. Areas of approximately 10 μm2 were analyzed by EDX.
2.10. Statistical analysis
After finding that the values follow a standard normal Gaussian distribution, the results were statistically evaluated using paired Student's t-test [27].
3. Results
3.1. Effect of polyP and polyP-retinol laden nanoparticles/nanospheres on cell growth
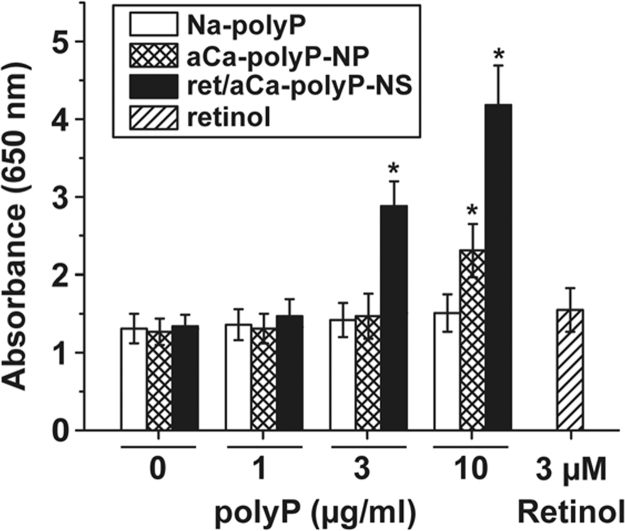
It is the aim of this series of experiments to elucidate if polyP and retinol, administered together to the cells, display an interacting effect on cell growth and metabolism. Both compounds, separately added, display miscellaneous anabolic effects on cells in vitro. As examples, polyP causes an inducing effect on biomineralization/hydroxyapatite formation which is paralleled with an increased induction of the gene encoding for alkaline phosphatase [22]. Likewise, retinol contributes to the anabolic metabolic pathways in a series of processes, e.g. embryonic development or regulation of epithelial and hematopoietic cellular differentiation. In the present study and using MC3T3-E1 cells it is found that both particle-free Na-polyP, complexed with Ca2+, as well as nanoparticles, formed by amorphous Ca-polyP, have no effect on the number of cells during a 72 h incubation period below a concentration of 3 µg/ml (Fig. 2). However, if the nanoparticle preparation, aCa-polyP-NP, is added to the cells at a concentration of 10 µg/ml, a significant increase in the number of viable cells, based on the XTT reduction assay, is measured during the incubation period. Retinol, added as a single component, at a concentration of 3 µM also does not affect cell growth significantly.
Fig. 2.
Influence of polyP, encapsulated in nanoparticles or together with retinol in nanospheres, on cell viability (XTT colorimetric assay). The MC3T3-E1 cells were incubated with different concentrations of polyP (given in µg/ml) or retinol (µM), as indicated, for 72 h. Then the cells were assayed with XTT. The cells were exposed to different concentrations of Na-polyP, complexed with Ca2+, aCa-polyP-NP nanoparticles containing only polyP, retinol/aCa-polyP-NS nanospheres that contain in addition of polyP retinol, as well as with retinol. The results are expressed as means (n=10 experiments each)±standard error of the mean; *p<0.01.
If retinol is encapsulated into Ca-polyP-based nanoparticles and by that is forming the retinol/Ca-polyP nanospheres, a strong amplification of cell growth is measured (Fig. 2). Already at the concentration of 3 µg/ml the retinol/aCa-polyP-NS increase the absorbance in the XTT assay from ≈1.3 A260 units to 2.88 units, a value which increased to 4.18 units in assays containing 10 µg/ml. The concentration of retinol in 3 µg/ml of nanospheres is approximately 0.3 µg/ml (≈1 µM), a retinol level found not to change the growth of the cells (not shown). This is a first indication reflecting that Ca-polyP together with retinol acts synergistically in the system used here.
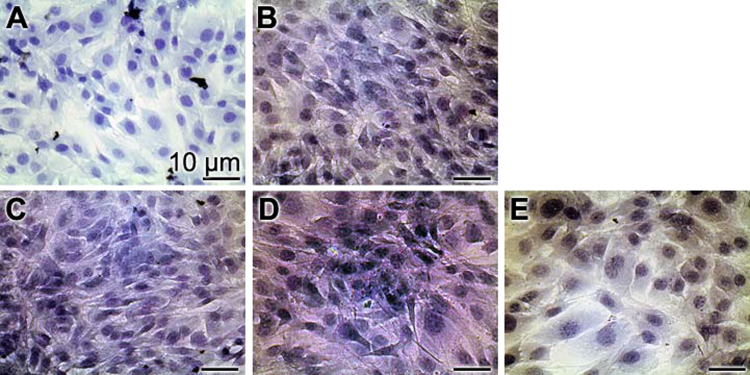
The cells, grown as monolayers, were stained with Nile Blue A (Fig. 3) The cells were incubated with 3 µg/ml of Na-polyP, aCa-polyP-NP or retinol/aCa-polyP-NS, as well as with 3 µM retinol. In the controls (Fig. 3A) the cells are dominantly stained in blue while all cell exposed to soluble Na-polyP (Fig. 3B), to polyP nanoparticles (Fig. 3C), to nanospheres (Fig. 3D), or to retinol (Fig. 3E) highlight in light/bright pink. This result is taken as an indication that the cells incubated in the presence of polyP, as well as of retinol contain more fatty acids, chromolipids and/or phospholipids than the controls. In addition, it is apparent that the densities of the cell layers in the polyP-treated cells as well as in the retinol-treated cells are higher, compared to the controls.
Fig. 3.
Staining of MC3T3-E1 cells with Nile Blue A. The cells were incubated for 72 h (A) without addition of any component, or with 3 µg/ml of (B) Na-polyP, (C) aCa-polyP-NP or (D) retinol/aCa-polyP-NS, as well as with (E) 3 µM retinol. It is seen that the polyP-retinol-treated cells highlight in pink, while the controls appear in blue.
3.2. Effect of polyP and retinol on gene expression (FABP4, leptin and leptin receptor)
The expression levels of the fatty acid binding protein 4 (FABP4) as well as of leptin and the corresponding leptin receptor were assessed by RT-qPCR, using the house-keeping gene GAPDH as a reference. The MC3T3-E1 cells remained either untreated or were exposed to 3 µg/ml of soluble Na-polyP, aCa-polyP nanoparticles or retinol/aCa-polyP nanospheres. In this series also the incubation with free retinol (3 µM) was included. The incubation period of the cells was set to 3 d. Then the RNA was extracted and the expression levels were determined and correlated to the one of GAPDH; this ratio was set to 1.
The expression level of all three genes, FABP4, leptin, and leptin receptor, did not change significantly, with respect to the untreated control, if the cells were incubated with soluble Na-polyP, complexed to Ca2+ (Fig. 4). However, addition of the polyP nanoparticles, aCa-polyP-NP, caused a significant upregulation of the steady-state-expression of both the FABP4 (2.3-fold), the leptin (4.2-fold) and the leptin receptor gene (3.8-fold). Even more pronounced is the increase if the nanospheres, retinol/aCa-polyP-NS, are added to the cells; in those assays the amount of transcripts increases for FABP4 (5.8-fold), the leptin (11.6-fold) and the leptin receptor gene (8.3-fold). If the cells are exposed to retinol alone only a non-significant increase is measured (Fig. 4).
Fig. 4.
Steady-state-expression of the genes encoding the FABP4 receptor, as well as leptin or leptin receptor determined in untreated MC3T3-E1 cells (stippled bars) or in cells exposed for 3 d either to 3 µg/ml of soluble Na-polyP (open bars), aCa-polyP-NP nanoparticles (cross hatched), retinol/aCa-polyP-NS nanospheres (black bars) or to 3 µM retinol (hatched). After incubation RNA was isolated from the cultures and analyzed by RT-qPCR by using GAPDH as reference gene. The expression ratios were correlated to GAPDH in the respective culture series; the ratio obtained was set to 1. SD are shown (5 experiments/time point); *p<0.01.
3.3. Fabrication of the electrospun mats
The effect of both retinol and amorphous Ca-polyP nanoparticles, if embedded into the electrospun fibrous mats, on the expression of FABP4, leptin and leptin receptor was determined. As outlined under “Section 2” the components retinol and Ca-polyP nanoparticles had been incorporated into the PLA-based fiber mesh (Fig. 1). PLA was mixed with PEG in a ratio of 80:20 wt% and then dissolved in chloroform as outlined under “Section 2”. Subsequently retinol was added at 20 wt% (with respect to PLA) to the PLA solution. In addition, the aCa-polyP-NP nanoparticles were added as well, reaching a final concentration of 10 wt%. The suspension formed was sonicated and used for the electrospinning procedure.
Mats with diameters of up to 20 cm were fabricated, woven from ≈2–4 µm fibers of an average mesh size of 10–20 µm. The color of the mats became yellowish if the fibers contained retinol; hence they had to be protected from light. While the pure PLA mats appeared in white (Fig. 1D), the mats spun from both PLA/retinol solution (Fig. 1E) and from PLA/retinol/aCa-polyP-NS suspension appear in yellow (Fig. 1F).
3.4. Characterization of the mats
FTIR analyses were conducted to assess structural changes and/or molecular chain interactions within the PLA:nanospheres fiber mats. The following samples were included: PLA basic matrix, retinol, PLA containing 20% retinol as well as PLA containing 20% retinol and 10% Ca-polyP nanoparticles. As shown in Fig. 5, all 5 samples tested caused a pronounced peak for the PLA polyester with the strong carbonyl band (C=O) at 1746/1749 cm−1, a C–H bending vibrations of CH3 at 2982 and 2930 as well as at 1445 and 1380 cm−1, and the asymmetric C–O stretching vibrations at 1300–1000 cm−1. In contrast to the pure PLA samples, the absorption band of C=O of the carbonyl groups in PLA shifts from 1746 cm−1 to 1749 cm−1 or 1748 cm−1; this absorption intensity shifts also after addition of retinol and Ca-polyP (Fig. 5). Moreover, a similar shift occurred for the stretching vibrations within the wavenumber range between 1300 and 1000 cm−1.
Fig. 5.
FTIR spectra of PLA (PLA), and retinol (Retinol) as well as of PLA, containing 20 wt% retinol (PLA-ret), and finally of PLA, composed of retinol and aCa-polyP-NP nanoparticles (PLA-ret/aCa-polyP-NP), as described under “Section 2”. The spectra were recorded between (A) wavenumbers 4000 to 775 cm−1 and (B) wavenumbers 2000 and 775 cm-1.
The FTIR spectrum for retinol shows the characteristic retinol band at 1549 cm−1 (hydroxyl group of retinol), a signal that disappeared in the spectrum for PLA, containing retinol (Fig. 5). In addition, in retinol a wide range of signals between wavenumbers 1100 and 800 cm−1. These findings indicate that PLA into which the components polyP and retinol had been encapsulated show weak interactions to the PLA matrix.
The EDX analyses of the PLA fiber material (Fig. 6A) as well as the PLA/retinol spinning material (Fig. 6B) show only the expected signals for C and O. In contrast, if the PLA-aCa-polyP-NP polymer is analyzed then the signals for Ca, P and Na appear in addition to C and O (Fig. 6C). The Na peaks reflect the residual Na+ that originate form the original Na-polyP material, used for the conversion to the insoluble Ca-polyP.
Fig. 6.
EDX analysis of the polymer used for electrospinning. The analysis was performed with (A) PLA polymer only, (B) PLA fibrous material that contained in addition retinol, or (C) PLA that has been supplemented with aCa-polyP-NP nanoparticles prior to the spinning process. The signals for C, O, Na, P and Ca are marked.
The diameters of the fibers composing the mats are as follows (number of determinations is 10): PLA fibers 3.8±1.3 µm, PLA fibers with integrated aCa-polyP-NP 4.3±1.6 or with retinol/aCa-polyP-NS 3.1±1.8.
3.5. Functional studies of the polyP-containing fiber mats
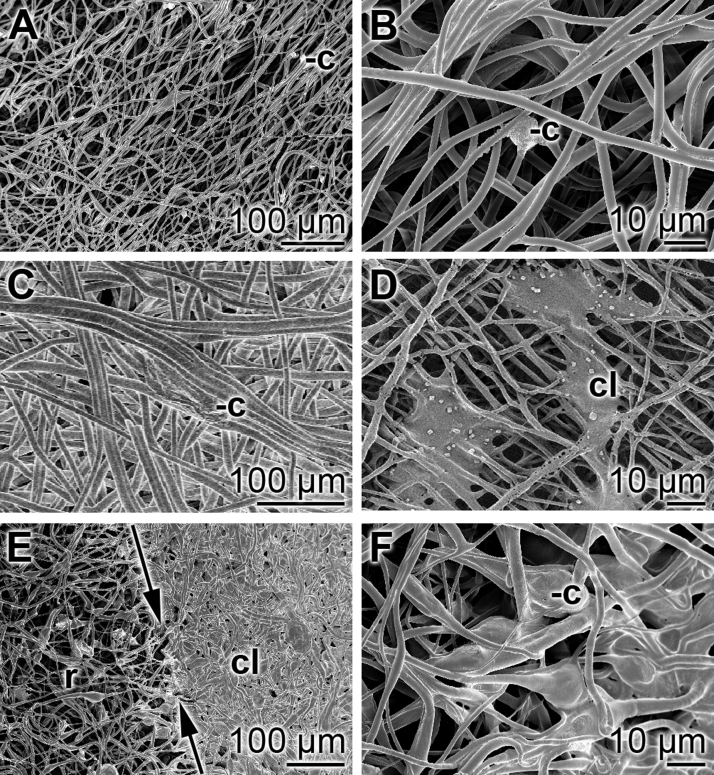
The MC3T3-E1 cells were seeded onto the circularly sliced samples of the spun mats and incubated for 3 d. In the first series of experiments the mats were taken after incubation fixed, dehydrated and then subjected to SEM analysis. The results show that almost no cells could be visualized onto those fibrous mats that had been spun with PLA alone (Fig. 7A and B). A higher density of cells can be resolved on PLA mats, fabricated with aCa-polyP-NP (Fig. 7C and D). In contrast, if the cells had been cultivated onto PLA fibers, supplemented with retinol/aCa-polyP-NS, densely packed cell layer are observed (Fig. 7E and F).
Fig. 7.
SEM analysis of fibrous mats after cultivation of cells. Precisely fitting mat samples were cut and submersed into the 24-well plates; 5·103 cells/well were seeded and incubation was performed for 3 d. Then the mats were taken, fixed and dehydrated and subsequently subjected to SEM analysis. (A and B) Fibrous mats spun with PLA only; very rarely cells/cell clusters could be identified (-c). (C and D) In a scattered pattern cells (c) are arranged onto mats, fabricated with PLA and aCa-polyP-NP; infrequently dense cell layers (cl) can be resolved. (E and F) In contrast, dense cell layers (cl) are seen that have been formed during the incubation period onto fibers formed from PLA/retinol/aCa-polyP-NS. Occasionally, within the rim region (r), the transition from the densely packed cells (in C at the right) to the less populated areas can be resolved (left). The dividing line is marked by two single headed arrows.
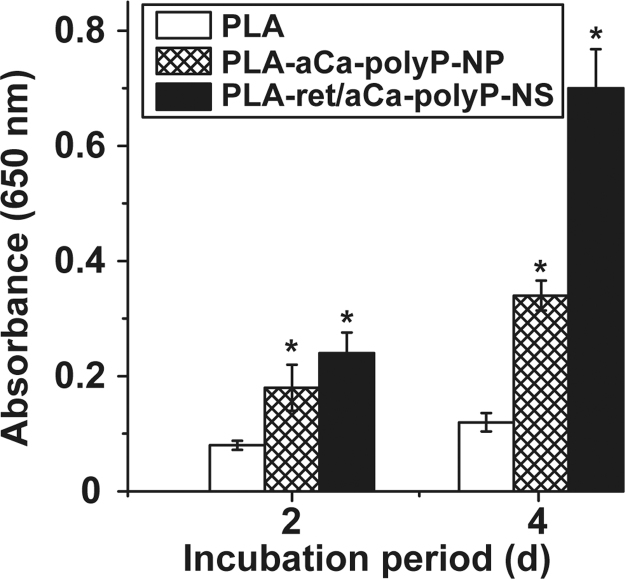
The viability/growth of MC3T3-E1 cells, cultured for 2 or 4 d onto plain PLA fiber mats, or mats supplemented with either aCa-polyP-NP or retinol/aCa-polyP-NS was determined by using the XTT assay. The results revealed (Fig. 8) that only a slight increase in the absorbance values (at 650 nm) is measured after 2 d with (0.084±0.009) and 4 d (0.128±0.024) in cultures growing on PLA fibers only. The respective absorbance values at time zero in all assays had been determined with 0.021±0.003. In contrast, a significant and strong increase in cell number is seen in the assays with mats fabricated of PLA, supplemented with both aCa-polyP-NP and retinol/aCa-polyP-NS. After the incubation period of 4 d an increase in the absorbance values to 0.349±0.026 is measured in assays with mats made with PLA and aCa-polyP-NP, or to 0.705±0.068 in assays with PLA fibers, supplemented with with retinol/aCa-polyP-NS.
Fig. 8.
Determination of cell viability (XTT colorimetric assay) of MC3T3-E1 cells, cultured for 2 or 4 d onto plain PLA fiber mats (open bars), or mats supplemented with aCa-polyP-NP (cross hatched) or with retinol/aCa-polyP-NS (closed). The cell viability/growth of the cells onto mats, supplemented with either aCa-polyP-NP or with retinol/aCa-polyP-NS, is significantly higher compared to cultures with plain PLA fibers. Means (n=10)±standard error of the mean; *p<0.01.
To support quantitatively the functional properties of the different mats for the cellular activity the steady-state-expression levels for the three genes, studied here FABP4, leptin, and leptin receptor, were determined by RT-qPCR analyses. The expression values have been correlated to the expression of the reference gene GAPDH; those values are subsequently proportioned to the ratio of those genes to GAPDH measured in the cells that have been used for seeding (immediately after detachment and a handling period of 2 h). It is seen that no measurable expression values of the genes in cells cultivated onto purely PLA mats can be given, due to the presence of only a few cells that survived onto those fibers (Fig. 9). In contrast, both the steady-state-expression levels for FABP4, leptin and leptin receptor undergo significant elevation onto PLA-aCa-polyP-NP fiber mats with 1.2-fold, 2.3-fold and 2.6-fold, respectively. A likewise strong induction is seen for those genes, if grown for 3 d onto PLA-retinol/aCa-polyP-NS fibers, with 3.7-fold, 5.8-fold and 6.1-fold, respectively (Fig. 9).
Fig. 9.
RT-qPCR analyses to assess the number of transcripts for the three genes FABP4, leptin, and leptin receptor. After seeding, the MC3T3-E1 cells were cultivated for 3 d onto mats fabricated from PLA only (numbers not determinable [n.d.]), onto spun fibers made of PLA and aCa-polyP-NP (cross hatched bars), or of PLA and retinol/aCa-polyP-NS (black bars). The expression values for FABP4, leptin, and leptin receptor were referred to the GAPDH expression. In turn, those numbers were correlated to the expression values of the genes under study and GAPDH, measured in cells that have been used for seeding (after detachment from the culture bottoms and a subsequently handling period for 2 h); those ratios were set to 1. SD are shown (5 experiments/time point); *p<0.01.
Based on these experiments we conclude that those PLA fibers, if supplemented with either aCa-polyP-NP or retinol/aCa-polyP-NS, elicit morphogenetic activity as monitored for the expression of the genes encoding for FABP4, leptin and leptin receptor.
4. Discussion
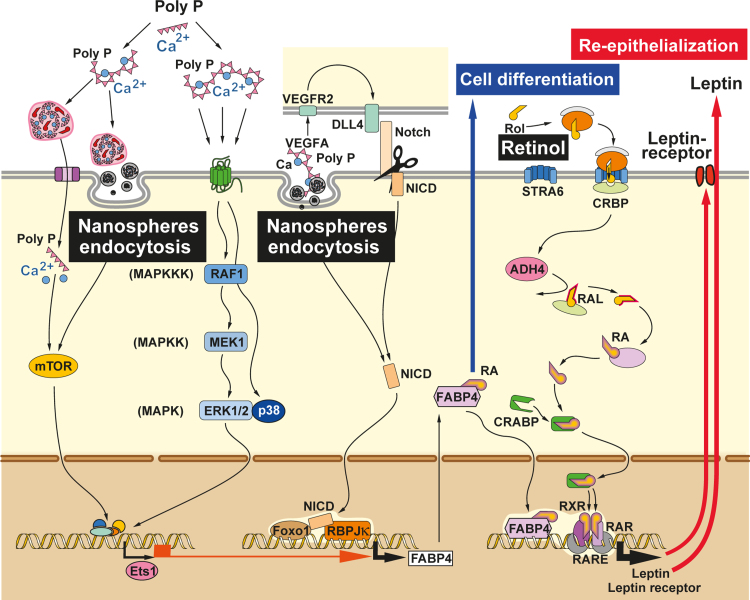
In the present study we report that polyP, fabricated as nanoparticles, if administered together with retinol and packed into fibrous mats elicit a synergistic expression of genes, encoding for leptin/leptin receptor as well as for the FABP4 in MC3T3-E1 cells. Both cascades are involved in the regulation not only of the overall energy metabolism of cells but also in cell growth and differentiation control. Important for the study described here is that both pathways (leptin/leptin receptor and FABP4) are affected by retinoids, more specific by retinoic acid. The functional interaction between leptin and leptin receptor can regulate growth and differentiation of e.g. hematopoietic cells and macrophages as well as re-epithelialization of wounds in mice [14]; FABP4 controls cell differentiation and proliferation in a series of cell types, among them endothelial cells; scheme in Fig. 10.
Fig. 10.
Proposed effect of polyP and retinol on the expression of the fatty acid binding protein 4 (FABP4) and leptin/leptin receptor. It has been proposed that polyP acts via the MAPK signaling route (p38) MAPK pathway and/or the mTOR complexes 1 and 2. The FABP4 expression might include the transcription factor Ets1. This pathway acts positively on the FABP4 expression level that is correlated with the polyP/DLL4-NOTCH signaling pathway as well as with Foxo1. VEGFA (vascular endothelial growth factor) induces, after binding to its receptor (VEGFAR) and activation of DLL4 (delta-like ligand-4), the paracrine activation of NOTCH signaling, resulting in FABP4 expression. Intermediately, the intracellular domain (NICD) of the NOTCH receptor translocates to the nucleus and bind to the transcription factor complex, composed of Foxo1 and the DNA-binding protein RBP-Jκ, through which FABP4 transcription is induced. FABP4 protein migrates into the cytoplasm and binds RA (retinoic acid). It is proposed that RA with FABP4 re-enters the nucleus and forms the RXR–RAR (retinoid X receptor-retinoic acid receptor heterodimer). In this transcription complex, at the RA response element (RARE), the genes encoding for leptin and leptin receptor are increasingly transcribed. RA is synthesized via retinol (Rol), that is taken up by the cells via STRA6 (membrane transporter) where it is bound to CRBP (cellular retinol-binding protein), undergoes oxidation by the ADH4 (alcohol dehydrogenase 4) to RAL (retinal) and finally to RA, which is finally translocated via CRABP (a retinoic acid-binding protein) to the nucleus where it binds at the RARE (retinoic acid response element) of the DNA under formation of the RXR/RAR complex. In conclusion, polyP causes an upregulation of the binding protein FABP4 which accelerates the retinol effect on the expression of leptin and leptin receptor. These two arms, FABP4 increased synthesis and RAR transcriptional promoting activity, positively affect cell differentiation and re-epithelization.
Retinoic acid has been found to suppress leptin synthesis and secretion in adipocytes in vitro. However, if administered for a short period retinoic acid elevates leptin expression in vivo, in rats [28], more specific in syncytiotrophoblasts that are forming the outer layer of the chorionic villi of the human placenta. In general, the effect of retinoic acid appears to function in a tissue-specific manner and apparently elicits no effect on leptin gene expression in osteoblasts. Interestingly, the leptin receptor has been found to be co-expressed in leukemic cells with the leukemia-retinoic acid receptor, a finding that opens a new therapeutic target for leukemia therapy [29]. These data might imply that retinoids, if they display a anabolic morphogenetic effect, act via the retinol–retinal–retinoic acid-retinoid X receptor α (RXR) on leptin/leptin receptor gene expression. These in vitro data had been corroborated by in vivo studies revealing that retinoids modulate anabolically hepatic leptin signaling and insulin sensitivity in mice [30].
The fatty acid binding protein 4 (FABP4) is a member of the family of intracellular FABPs that facilitates the compartmental distribution of fatty acids inside the cell. FABP4 binds with high affinity long-chain fatty acids (Kd≈100 nM) but also retinoic acid with a lower affinity (Kd≈50 µM) as described [31]. Originally FABP4 had been described as an adipocytes-specific protein, and later identified also in other cell types, e.g. macrophages or endothelial cells during their processes of regeneration [32]. The expression of FABP4 is controlled by the vascular endothelial growth factor A/vascular endothelial growth factor 2 (VEGFA/VEGFR2) pathway in endothelial cells [33]. This important signaling protein is involved in both vasculogenesis and in angiogenesis; in addition, VEGFA stimulates endothelial cell mitogenesis and cell migration. A reduced expression of FABP4 reduces endothelial cell proliferation, migration, and sprouting. Finally, the FABPs have been described to be necessary for cell motility within regenerative epidermis during wound healing [34]. In the present study it is shown that the polyP-based nanospheres elicit an increased expression of the FABP4 gene.
Recently it has been established that VEGFA after binding to the VEGFR2 causes an activation of the Notch ligand DLL4 through which the Notch signaling pathway directly regulates FABP4 gene expression [33]. Intermediately, the Notch intracellular domain (NICD) is translocated to the nucleus and, in concert with the insulin-responsive Foxo1 transcription factor as well as other with other transcription factors, e.g. RBPJκ, causes FABP4 gene expression. In turn FABP4 binds in the cytoplasm to both long chain fatty acids and retinoic acid and delivers those specific ligands from the cytosol to the nuclear receptor PPARγ [peroxisome proliferator-activated receptors gamma]. There, this receptor forms a heterodimer with the RXRs and causes induction of transcription; RXR is activated by the retinol metabolite 9-cis retinoic acid not only in fat-related cells but also in keratinocytes [35]. These cross-talks are sketched in Fig. 10.
A second pathway had been identified through which FABP4 expression is controlled; the mitogen-activated protein kinase (MAPK) signaling route, targeting the transcription factors c-jun and c-myc [36]. This finding is interesting since also polyP has been found to use the signal transduction pathway MAPK for inducing of genes, e.g. the heat shock protein 27 [37]. However, the data presented indicate that the p38/MAPK path, initiated by polyP, is used and not the c-jun. In turn, future studies must clarify to which extent a switch between the different three main signaling modules (ERK1/2, JNK/p38 and ERK5) of the MAPK pathways targeting FABP4 can occur. Based on these data we postulate that polyP acts on two levels: on the steady-state-expression of FABP4; first via the MAPK signaling on the expression of FABP4 and, if of polyP and retinol is packed into nanospheres, of both the leptin and its corresponding leptin receptor. Both arms of the signal transduction chain cause an increased proliferation/differentiation, combined with an amplification of the retinol-caused activation of the RXR, resulting in a promoted transcription of those downstream target genes that either are involved in the pleiotropic effects of the retinoids during differentiation or control the energy balance, inflammation, and vascularization [38]. Interesting are the recent data that indicate that polyP activates the mTOR complexes 1 and 2 in endothelial cells [39], a complex that is crucially involved in the cellular energy metabolism [40]. This finding fits with the observation, documented in the present study that polyP induces the lipid reservoirs in the MC3T3-E1 cells used.
In conclusion, the data present here indicate that polyP, packed with retinol into nanospheres, activate/modulate via mTOR and the MAPK pathway the expression of the transcription factor ETS1 [41], which in turn is assumed to interact synergistically with the Foxo1/NICD-RBPJκ complex under expression of FABP4 gene. Comprehensive data on FTIR and EDAX spectra of the particles have been given recently [19], [42].
FABP4 in turn binds to retinoic acid in the cytoplasm and returns back to the nucleus. There an increase expression and synthesis of leptin and leptin receptor takes place, a ligand-receptor loop that positively affects re-epithelialization. FABP4 additionally affects the cell proliferation and differentiation.
It should be stressed that the published effect of polyP to increase the expression of the alkaline phosphatase [22], and the report that this enzyme is an indicator for a beneficial effect on wound healing [43], additionally support our presented findings that this polymer has a beneficial effect on wound healing.
The retinol concentration in the retinol/Ca-polyP-NS nanospheres was determined to be 100 mg/g nanoparticles, applying the SbCl3-based spectroscopic technique [23]. Adding those particles to a basiscreme at a concentration of 2% for cosmetic and/or dermatological applications, a final available retinol concentration would be 0.2%; this concentration meets the recommendation for ingredients in cosmetic products, e.g. body lotions or creams [44]. With respect to wound dressings, fabricated in the present study, the fibers contain 10 wt% of nanospheres, yielding to a final retinol concentration of 1% of retinol.
Future studies are directed to investigate the kinetics of degradation of the retinol/Ca-polyP-NS nanospheres by cutaneous alkaline phosphatase, and the cellular uptake of the liberated retinol, as well as possible stategies to stabilize the polyP in order to prolong the biological activity of the retinol/Ca-polyP-NS based electrospun mats for potential use as dressings in wound repair. In addition, cryosectioning/transmission electron microscopy studies will be performed to assess the distribution of the nanoparticles within the fibers of the mats. Future studies will also include degradation kinetics of the mats as well as of the nanoparticles and the nanospheres. The data presented here indicate that the PLA-based fiber mats, containing retinol and polyP nanoparticles have to be considered not only as advanced, interactive electrospun meshes but also as bioactive meshes that are urgently needed as dressings for chronic wounds.
Acknowledgments
W.E.G. M. is a holder of an ERC Advanced Investigator Grant (No. 268476 BIOSILICA). We thank Dipl. Ing. G. Glaßer (“Elektronenmikroskopie”; Max Planck Institute for Polymer Research, Mainz, Germany) for very expert and helpful SEM analyses. This work was supported by Grants from the Deutsche Forschungsgemeinschaft (Schr 277/10-3), the European Commission (“Bio-Scaffolds”, No. 604036; “CoreShell”: No. 286059; “MarBioTec*EU-CN*”: No. 268476; and “BlueGenics”: No. 311848) and the International Human Frontier Science Program (RG-333/96-M).
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2015.08.007.
Contributor Information
Werner E.G. Müller, Email: wmueller@uni-mainz.de.
Xiaohong Wang, Email: wang013@uni-mainz.de.
Appendix A. Supplementary material
Supplementary material
References
- 1.Guo S., Dipietro L.A. Factors affecting wound healing. J. Dent. Res. 2010;89:219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nurden A.T., Nurden P., Sanchez M., Andia I., Anitua E. Platelets and wound healing. Front. Biosci. 2008;13:3532–3548. doi: 10.2741/2947. [DOI] [PubMed] [Google Scholar]
- 3.Faxälv L., Boknäs N., Ström J.O., Tengvall P., Theodorsson E., Ramström S., Lindahl T.L. Putting polyphosphates to the test: evidence against platelet-induced activation of factor XII. Blood. 2013;122:3818–3824. doi: 10.1182/blood-2013-05-499384. [DOI] [PubMed] [Google Scholar]
- 4.Clark R.A.F. Wound repair. Curr. Opin. Cell Biol. 1989;1:1000–1008. doi: 10.1016/0955-0674(89)90072-0. [DOI] [PubMed] [Google Scholar]
- 5.McDougall S., Dallon J., Sherratt J., Maini P. Fibroblast migration and collagen deposition during dermal wound healing: mathematical modelling and clinical implications. Philos. Trans. A: Math. Phys. Eng. Sci. 2006;364:1385–1405. doi: 10.1098/rsta.2006.1773. [DOI] [PubMed] [Google Scholar]
- 6.Keylock K.T., Vieira V.J., Wallig M.A., DiPietro L.A., Schrementi M., Woods J.A. Exercise accelerates cutaneous wound healing and decreases wound inflammation in aged mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008;294:R179–R184. doi: 10.1152/ajpregu.00177.2007. [DOI] [PubMed] [Google Scholar]
- 7.Gilliver S.C., Ashworth J.J., Ashcroft G.S. The hormonal regulation of cutaneous wound healing. Clin. Dermatol. 2007;25:56–62. doi: 10.1016/j.clindermatol.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Ahn C., Mulligan P., Salcido R.S. Smoking – the bane of wound healing: biomedical interventions and social influences. Adv. Skin Wound Care. 2008;21:227–238. doi: 10.1097/01.ASW.0000305440.62402.43. [DOI] [PubMed] [Google Scholar]
- 9.Abrigo M., McArthur S.L., Kingshott P. Electrospun nanofibers as dressings for chronic wound care: advances, challenges, and future prospects. Macromol. Biosci. 2014;14:772–792. doi: 10.1002/mabi.201300561. [DOI] [PubMed] [Google Scholar]
- 10.Lee D., Stojadinovic O., Krzyzanowska A., Vouthounis C., Blumenberg M., Tomic-Canic M. Retinoid-responsive transcriptional changes in epidermal keratinocytes. J. Cell Physiol. 2009;220:427–439. doi: 10.1002/jcp.21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keleidari B., Mahmoudieh M., Bahrami F., Mortazavi P., Aslani R.S., Toliyat S.A. The effect of vitamin A and vitamin C on postoperative adhesion formation: a rat model study. J. Res. Med. Sci. 2014;19:28–32. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang L., Tankersley L.R., Tang M., Potter J.J., Mezey E. Regulation of alpha 2(I) collagen expression in stellate cells by retinoic acid and retinoid X receptors through interactions with their cofactors. Arch. Biochem. Biophys. 2004;428:92–98. doi: 10.1016/j.abb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Gelain D.P., Pasquali M.A., Caregnato F.F., Castro M.A., Moreira J.C. Retinol induces morphological alterations and proliferative focus formation through free radical-mediated activation of multiple signaling pathways. Acta Pharmacol. Sin. 2012;33:558–567. doi: 10.1038/aps.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frank S., Stallmeyer B., Kämpfer H., Kolb N., Pfeilschifter J. Leptin enhances wound re-epithelialization and constitutes a direct function of leptin in skin repair. J. Clin. Invest. 2000;106:501–509. doi: 10.1172/JCI9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman J.M., Halaas J.L. Leptin and the regulation of body weight in mammals. Nature. 1998;395:763–770. doi: 10.1038/27376. [DOI] [PubMed] [Google Scholar]
- 16.Coppari R., Bjørbæk C. Leptin revisited: its mechanism of action and potential for treating diabetes. Nat. Rev. Drug Discov. 2012;11:692–708. doi: 10.1038/nrd3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murad A., Nath A.K., Cha S.T., Demir E., Flores-Riveros J., Sierra-Honigmann M.R. Leptin is an autocrine/paracrine regulator of wound healing. FASEB J. 2003;17:1895–1897. doi: 10.1096/fj.03-0068fje. [DOI] [PubMed] [Google Scholar]
- 18.Baji A., Mai Y.W., Wong S.C., Abtahi M., Chen P. Electrospinning of polymer nanofibers: effects on oriented morphology, structures and tensile properties. Compos. Sci. Technol. 2010;70:703–718. [Google Scholar]
- 19.Müller W.E.G., Tolba E., Schröder H.C., Wang S., Glaßer G., Muñoz-Espí R., Link T., Wang X.H. A new polyphosphate calcium material with morphogenetic activity. Mater. Lett. 2015;148:163–166. [Google Scholar]
- 20.Lorencová E., Vltavská P., Budinský P., Koutný M. Antibacterial effect of phosphates and polyphosphates with different chain length. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2012;47:2241–2245. doi: 10.1080/10934529.2012.707544. [DOI] [PubMed] [Google Scholar]
- 21.Hatakeyama N., Kojima T., Iba K., Murata M., Thi M.M., Spray D.C., Osanai M., Chiba H., Ishiai S., Yamashita T., Sawada N. IGF-I regulates tight-junction protein claudin-1 during differentiation of osteoblast-like MC3T3-E1 cells via a MAP-kinase pathway. Cell Tissue Res. 2008;334:243–254. doi: 10.1007/s00441-008-0690-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller W.E.G., Wang X.H., Diehl-Seifert B., Kropf K., Schloßmacher U., Lieberwirth I., Glasser G., Wiens M., Schröder H.C. Inorganic polymeric phosphate/polyphosphate as an inducer of alkaline phosphatase and a modulator of intracellular Ca2+ level in osteoblasts (SaOS-2 cells) in vitro. Acta Biomater. 2011;7:2661–2671. doi: 10.1016/j.actbio.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Subramanyam G.B., Parrish D.B. Colorimetric reagents for determining vitamin A in feeds and foods. J. Assoc. Off. Anal. Chem. 1976;59:1125–1130. [PubMed] [Google Scholar]
- 24.Müller W.E.G., Tolba E., Schröder H.C., Diehl-Seifert B., Link T., Wang X.H. Biosilica-loaded poly(Ɛ-caprolactone) nanofibers mats provide a morphogenetically active surface scaffold for the growth and mineralization of the osteoclast-related SaOS-2 cells. Biotechnol. J. 2014;9:1312–1321. doi: 10.1002/biot.201400277. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi T., Kato S. Impact of diabetes mellitus on myocardial lipid deposition: an autopsy study. Pathol. Res. Pract. 2014;210:1018–1025. doi: 10.1016/j.prp.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Wiens M., Wang X.H., Schröder H.C., Kolb U., Schloßmacher U., Ushijima H., Müller W.E.G. The role of biosilica in the osteoprotegerin/RANKL ratio in human osteoblastlike cells. Biomaterials. 2010;31:7716–7725. doi: 10.1016/j.biomaterials.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Sachs L. Springer; Berlin: 1984. Angewandte Statistik; p. 242. [Google Scholar]
- 28.Krskova-Tybitanclova K., Macejova D., Brtko J., Baculikova M., Krizanova O., Zorad S. Short term 13-cis-retinoic acid treatment at therapeutic doses elevates expression of leptin, GLUT4, PPAR gamma and aP2 in rat adipose tissue. J. Physiol. Pharmacol. 2008;59:731–743. [PubMed] [Google Scholar]
- 29.Tabe Y., Konopleva M., Munsell M.F., Marini F.C., Zompetta C., McQueen T., Tsao T., Zhao S., Pierce S., Igari J., Estey E.H., Andreeff M. PML–RAR alpha is associated with leptin-receptor induction: the role of mesenchymal stem cell-derived adipocytes in APL cell survival. Blood. 2004;103:1815–1822. doi: 10.1182/blood-2003-03-0802. [DOI] [PubMed] [Google Scholar]
- 30.Tsuchiya H., Ikeda Y., Ebata Y., Kojima C., Katsuma R., Tsuruyama T., Sakabe T., Shomori K., Komeda N., Oshiro S., Okamoto H., Takubo K., Hama S., Shudo K., Kogure K., Shiota G. Retinoids ameliorate insulin resistance in a leptin-dependent manner in mice. Hepatology. 2012;56:1319–1330. doi: 10.1002/hep.25798. [DOI] [PubMed] [Google Scholar]
- 31.Matarese V., Bernlohr D.A. Purification of murine adipocytes lipid-binding protein. Characterization as a fatty acid- and retinoic acid-binding protein. J. Biol. Chem. 1988;263:14544–14551. [PubMed] [Google Scholar]
- 32.Lee M.Y., Tse H.F., Siu C.W., Zhu S.G., Man R.Y., Vanhoutte P.M. Genomic changes in regenerated porcine coronary arterial endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2007;27:2443–2449. doi: 10.1161/ATVBAHA.107.141705. [DOI] [PubMed] [Google Scholar]
- 33.Harjes U., Bridges E., McIntyre A., Fielding B.A., Harris A.L. Fatty acid-binding protein 4, a point of convergence for angiogenic and metabolic signaling pathways in endothelial cells. J. Biol. Chem. 2014;289:23168–23176. doi: 10.1074/jbc.M114.576512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusakari Y., Ogawa E., Owada Y., Kitanaka N., Watanabe H., Kimura M., Tagami H., Kondo H., Aiba S., Okuyama R. Decreased keratinocyte motility in skin wound on mice lacking the epidermal fatty acid binding protein gene. Mol. Cell. Biochem. 2006;284:183–188. doi: 10.1007/s11010-005-9048-8. [DOI] [PubMed] [Google Scholar]
- 35.Yoshimura K., Uchida G., Okazaki M., Kitano Y., Harii K. Differential expression of heparin-binding EGF-like growth factor (HB-EGF) mRNA in normal human keratinocytes induced by a variety of natural and synthetic retinoids. Exp. Dermatol. 2003;12(Suppl 2):S28–S34. doi: 10.1034/j.1600-0625.12.s2.5.x. [DOI] [PubMed] [Google Scholar]
- 36.Girona J., Rosales R., Plana N., Saavedra P., Masana L., Vallvé J.C. FABP4 induces vascular smooth muscle cell proliferation and migration through a MAPK-dependent pathway. PLoS One. 2013;8(11):e81914. doi: 10.1371/journal.pone.0081914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Segawa S., Fujiya M., Konishi H., Ueno N., Kobayashi N., Shigyo T., Kohgo Y. Probiotic-derived polyphosphate enhances the epithelial barrier function and maintains intestinal homeostasis through integrin-p38 MAPK pathway. PLoS One. 2011;6(8):e23278. doi: 10.1371/journal.pone.0023278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plutzky J. The PPAR-RXR transcriptional complex in the vasculature: energy in the balance. Circ. Res. 2011;108:1002–1016. doi: 10.1161/CIRCRESAHA.110.226860. [DOI] [PubMed] [Google Scholar]
- 39.S.M.H. Mehr, P.Dinarvand, A.R.Rezaie, Inorganic polyphosphate activates mTOR complexes 1 and 2 in vascular endothelial cells. 56th ASH Annual Meeting in San Francisco, CA, Session 302, Abstract 1441, 2014.
- 40.Laplante M., Sabatini D.M. mTOR signaling at a glance. J. Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X.H., Schröder H.C., Feng Q., Diehl-Seifert B., Grebenjuk V.A., Müller W.E.G. Isoquercitrin and polyphosphate co-enhance mineralization of human osteoblast-like SaOS-2 cells via separate activation of two RUNX2 cofactors AFT6 and Ets1. Biochemical. Pharmacol. 2014;89:413–421. doi: 10.1016/j.bcp.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 42.Müller W.E.G., Tolba E., Schröder H.C., Diehl-Seifert B., Wang X.H. Retinol encapsulated into amorphous Ca2+ polyphosphate nanospheres acts synergistically in MC3T3-E1 cells. Eur. J. Pharm. Biopharm. 2015;93:214–223. doi: 10.1016/j.ejpb.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Alpaslan G., Nakajima T., Takano Y. Extracellular alkaline phosphatase activity as a possible marker for wound healing: a preliminary report. J. Oral. Maxillofac. Surg. 1997;55:56–62. doi: 10.1016/s0278-2391(97)90447-x. [DOI] [PubMed] [Google Scholar]
- 44.R.B. Hetland, Risk assessment of vitamin A (retinol and retinyl esters) in cosmetics, in: Opinion of the Panel on Food Additives, Flavourings, Processing Aids, Materials in Contact with Food and Cosmetics of the Norwegian Scientific Committee for Food Safety, Doc. no.: 10-405-3, ISBN: 978-82-8259-059-4, 2012, p. 74
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material