Abstract
Glutathione peroxidase (GPX) plays a pivotal role in the protection of cells against oxidative damage. The green alga Chlamydomonas reinhardtii expresses both selenocysteine-containing GPX and the non-selenium GPX homolog (GPXH). We previously reported that supplementation of selenium to algal culture induces GPXH to exhibit GPX activity. Here we investigated the incorporation of selenium into GPXH and its causal relationship with the upregulation of the enzymatic activity. GPXH was purified from algal cells grown with selenium and proteolytically digested into four fragments. Selenium content analysis for these proteolytic fragments confirmed that GPXH-incorporated selenium is predominantly enriched in a fragment that carries the putative catalytic residue Cys-38. We next constructed three kinds of engineered GPXH proteins by substituting Ser for one of three Cys residues in native GPXH, Cys-38, -66, and -84, using a bacterial overexpression system, resulting in Cys38Ser, Cys66Ser, and Cys84Ser derivatives, respectively. Of these, the Cys66Ser and Cys84Ser derivatives exhibited the same level of selenium-dependent GPX activity as the normal recombinant GPXH, whereas the Cys38Ser mutant GPXH not only lost its activity completely but also demonstrated severely impaired incorporation of selenium. These findings strongly suggest that selenium is post-translationally assimilated into the Cys-38 of the GPXH protein, thereby enhancing its enzymatic activity.
Abbreviations: GPX, glutathione peroxidase; GPXH, glutathione peroxidase homolog
Keywords: Chlamydomonas reinhardtii, Glutathione peroxidase, Green alga, Selenium, Selenoenzyme
Highlights
-
•
Non-Se algal GPX was characterized in terms of Se-associated structure–function.
-
•
Se was found to be specifically bound to the catalytic Cys of the GPX.
-
•
Se-binding targeted to the active site was required for GPX up-regulation.
-
•
This is the first evidence for Se-mediated post-translational activation of plant GPX.
1. Introduction
GPX catalyzes the reduction of H2O2 and organic hydroperoxides with reduced glutathione. Since the discovery of GPX [1], it has been intensively studied and demonstrated to be essential in the organism's defensive response to oxidative stress [2], [3]. Mammalian GPX isozymes, except for epididymal secretory GPX, contain selenocysteine in their primary structure; the selenocysteine residue is found in the active site and is crucial for their biological activity [2], [4]. In all organisms where selenocysteine-containing proteins (selenoproteins) have been found, this rare amino acid is encoded by an in-frame UGA codon that is usually a translational stop codon; however, the presence of a unique stem-loop structure in the mRNA designated as the selenocysteine insertion sequence element precludes translational termination and promotes selenocysteine insertion into the growing polypeptide chain [5], [6], [7]. So far, known selenoproteins are of animal and bacterial origin; the presence of selenocysteine has not been clearly identified in any plant or yeast proteins [8]. As for GPX, numerous groups have cloned GPX-like homologs from plants and yeast, although all these genes encode Cys rather than selenocysteine in their putative catalytic site [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19]. However, the green alga Chlamydomonas reinhardtii has exceptionally been demonstrated to express selenocysteine-containing GPX and possess the selenocysteine insertion system [20], [21].
We have previously found that GPX-like activity was induced by growing C. reinhardtii in the presence of selenite [22]. This GPX-like protein is strictly compartmentalized to the cytosol in contrast to the predicted localization of the above mentioned selenoprotein-type of GPX to the mitochondria [20] and is closely related to classical GPXs from mammals with respect to its enzymological, physicochemical, and immunological properties [23], [24]. Thereafter, a cDNA clone encoding the algal GPX-like protein, tentatively called the GPX homolog (GPXH), was isolated and characterized [25]. The deduced amino acid sequence of GPXH was highly homologous to GPX-like proteins from higher plants and yeast. Moreover, GPXH was found to contain a Cys residue at position 38 from the N-terminal (Cys-38) in place of the catalytic selenocysteine in GPX as with other non-animal GPX homologs. As such, the genome of C. reinhardtii encodes both GPX with co-translationally inserted selenocysteine and its paralogous variant, GPXH, which is independent of selenocysteine. Whereas GPXH requires inorganic selenium for enzymatic activity despite the incompatibility of its translation with selenocysteine insertion machineries, the underlying process of such a non-canonical mode of selenium dependency remains to be clarified. In this study, we addressed the molecular characterization of GPXH in terms of selenium incorporation and the functional link between selenium-mediated modification of GPXH and its GPX activity.
2. Materials and methods
2.1. Organisms and growth conditions
Chlamydomonas reinhardtii Dangeard C9 was cultured on Allen's medium [26] supplemented with 17 µM sodium selenate to induce GPX activity at 26 °C for 7 days under illumination at 240 µmol/m2/s and air bubbling at 4 L/min.
Escherichia coli BL21(DE3)pLysS transformants were grown in LB medium at 37 °C. For protein overexpression, when bacterial growth became equivalent to an A600 of 0.6, IPTG and sodium selenite were added at 0.4 mM and 67 µM, respectively that was followed by additional cultivation for 6 h.
2.2. Preparation of cell extracts
C. reinhardtii cells were collected by centrifugation at 3000×g for 5 min, suspended in buffer A (0.1 M Tris–HCl, pH 8.2, and containing 0.3 M sucrose and 5 mM glutathione), and then disintegrated by sonication at 10 kHz for a total of 5 min with 10 intervals of 30 s each. E. coli cells were harvested by centrifugation at 6,000×g for 10 min, suspended in buffer A, and then sonicated at 10 kHz for a total of 1 min with three intervals of 15 s each. For both cell types, the cell extract was centrifuged at 12,000×g for 20 min, and the resulting supernatant was used for crude enzyme preparation.
2.3. Enzyme assay
GPX activity was spectrophotometrically assayed at 30 °C in the presence of glutathione reductase, which reverses oxidized glutathione formed by GPXH catalysis, according to Takeda et al. [24]. The reaction mixture contained 0.1 M Tris–HCl at pH 8.2, 1 mM glutathione, 0.4 mM NADPH, 0.2 mM H2O2, 1 unit of glutathione reductase, and GPXH preparation in a total volume of 1 mL.
2.4. Purification of GPXH from algal and bacterial cells
Crude enzyme solution was loaded onto a HiPrep Q XL 16/10 column (GE Healthcare) pre-equilibrated with the above mentioned buffer A. After washing the column with 60 mL buffer A, the proteins were eluted with a linear NaCl gradient (0–300 mM, 216 mL) in buffer A. To the active fractions combined, ammonium sulfate was added to 30% saturation, and the resulting precipitate was removed by centrifugation at 100,000×g for 30 min. The supernatant was charged onto a HiPrep Phenyl FF 16/10 column (GE Healthcare) that was pre-equilibrated with buffer A containing 30%-saturated ammonium sulfate, followed by washing with 60 mL of the same buffer. Elution was performed with a linear gradient of 30–0% ammonium sulfate saturation in buffer A (216 mL). The active fractions were precipitated with buffer A containing 80%-saturated ammonium sulfate and then dissolved in 2 mL of buffer A. The concentrated sample was applied on a Superdex 200 Increase 10/300 GL column (GE Healthcare) that was pre-equilibrated with buffer A containing 0.15 M NaCl. The column was developed with the same buffer, and the active fractions were saved as the purified enzyme preparation.
2.5. Proteolysis and sequence analysis
Proteolysis of GPXH was performed with endoproteinase Arg-C at an enzyme:substrate molar ratio of 1:100 for 18 h at 37 °C in 50 mM potassium phosphate buffer, pH 8.0. The degradation products were separated by Tricine-SDS-PAGE and then transferred onto a PVDF membrane according to the method of Ploug et al. [27]. For amino acid sequencing, each proteolytic fragment was recovered by electroelution and subjected to an Applied Biosystems 492 A Sequencer. The peptide solution thus obtained was also used for selenium content determination.
2.6. Experiments of molecular biology
For expression of recombinant GPXH protein, a synthetic DNA fragment covering the C. reinhardtii GPXH coding sequence (DDBJ/EMBL/GenBank Acc. no. AF014927) was cloned into pET3a (Takara Bio) and transformed into E. coli BL21(DE3)pLysS. Site-directed mutagenesis of recombinant GPXH was performed using the algal GPXH cDNA sequence cloned into pET3a as a template and the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). The oligonucleotide primers designed to replace Cys-38, Cys-66, and Cys-84 with Ser were as follows: forward Cys38Ser, 5′-GCTAGCAAGAGCGGCTTTACG-3′; reverse Cys38Ser, 5′-CGTAAAGCCGCTCTTGCTAGC-3′; forward Cys66Ser, 5′-GGCTTCCCCAGCAA CCAGTTTG-3′; reverse Cys66Ser, 5′-CAAACTGGTTGCTGGGGAAGCC-3′; forward Cys84Ser, 5′-GGGAGTTCAGCCAGCGCAAT-3′; reverse Cys84Ser, 5′-ATTGCGCTGGCTGAACTCCC-3′ (The underlined bases indicate the mutated ones). Introduction of the directed mutations was confirmed by DNA sequencing over the entire cDNA region.
2.7. Quantification of selenium and protein
Selenium content in the protein was fluorometrically assayed using 2,3-diaminonaphthalene after digestion with HNO3 and HClO4 according to the method of Bayfield and Romalis [28]. Protein levels were determined by Bradford's method using bovine serum albumin as the standard [29].
3. Results and discussion
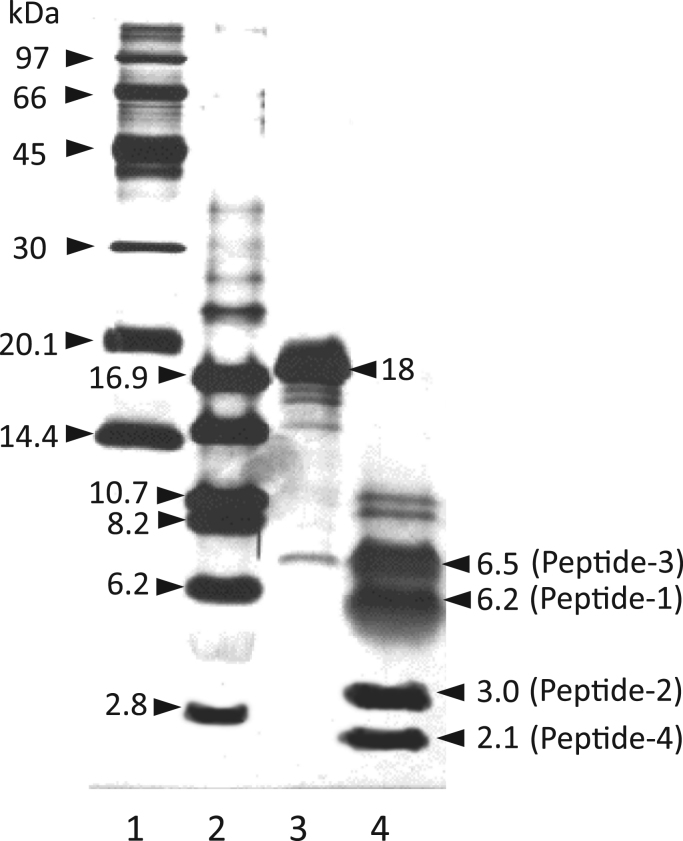
Our previous data revealed that the GPXH obtained from C. reinhardtii cells cultured with selenite contains a stoichiometric amount of selenium [23]. For the purpose of defining the binding site for selenium within the GPXH molecule, GPXH was purified to homogeneity from selenium-supplemented algal cells and was subjected to endoproteinase digestion (Fig. 1). As a result, four species of peptide fragments, Peptides 1–4 (6.2 kDa, 3.0 kDa, 6.5 kDa, and 2.1 kDa in size), were generated, and each peptide was identified by N-terminal sequencing (Fig. 2). We then determined the selenium content in these peptides. Although quantitative estimation of the selenium content was not possible because of unintended release of the protein-bound selenium during the sample preparation under rather harsh conditions, a significant amount of selenium was detected only in the 6.2 kDa fragment, i.e., residues 1–57, that encompasses the putative GPXH active site surrounding Cys-38, whereas only negligible amounts of selenium were found in the other peptides (data not shown). This result indicates that selenium is not inserted into GPXH in a non-specific manner but is regionally targeted to the catalytically essential moiety of the enzyme.
Fig. 1.
Tricine-SDS-PAGE analysis of proteolytically digested GPXH. GPXH purified from C. reinhardtii cells cultured with selenite was digested with endoproteinase Arg-C, separated by Tricine-SDS-PAGE, and detected by silver staining. Lanes: 1 and 2, size markers; 3, purified intact GPXH; and 4, proteolytic fragments of GPXH. The molecular weights of the size markers and GPXH peptides are given to the left and the right, respectively, of each lane.
Fig. 2.
Amino acid sequence of GPXH proteolytic products.After separation by Tricine-SDS-PAGE (Fig. 1), four species of proteolytic products, Peptides 1–4, were transferred to a PVDF membrane and separately recovered for identification and selenium quantification. The numbers indicate the position of the amino acid residues relative to the N-terminal. Amino acid sequences experimentally determined are underlined.
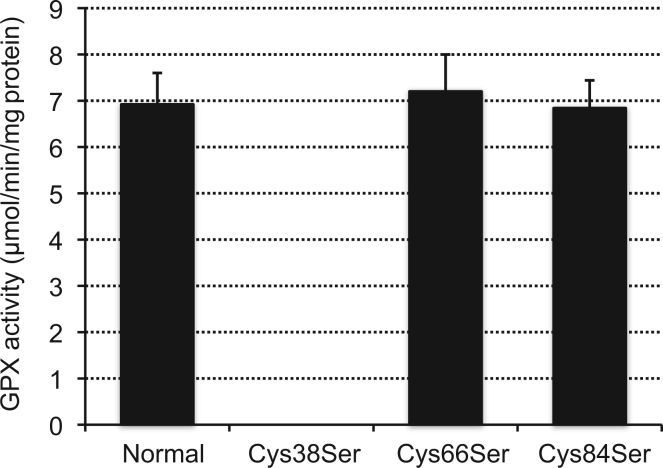
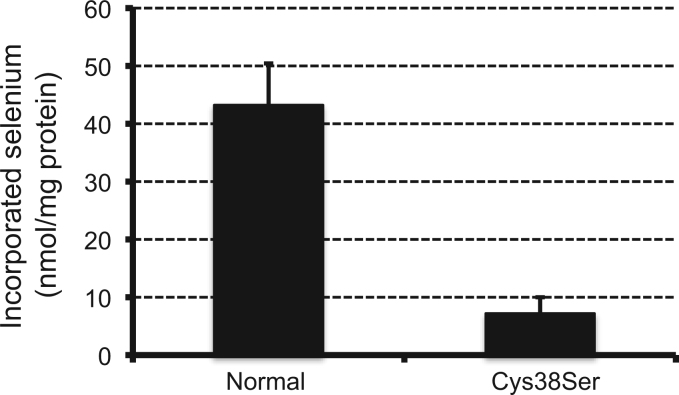
Based on the above finding, using recombinant GPXH, we examined the binding of selenium to GPXH and its influence on the enzymatic activity. We created a series of mutant GPXHs by replacing Cys-38, -66, or -84 with Ser, which were termed as Cys38Ser, Cys66Ser, and Cys84Ser, respectively, as well as a non-mutated normal GPXH recombinant. These constructs were expressed in bacterial transformants cultured with selenite to induce GPX activity, and then crude enzyme solution was prepared for each type of recombinant GPXH. Enzyme assay for GPX revealed that Cys66Ser and Cys84Ser exhibit selenium-induced GPX activity comparable with that of the normal recombinant, whereas the activity in Cys38Ser is completely eliminated (Fig. 3). We subsequently purified the normal and Cys38Ser GPXH recombinants and compared their capacity to incorporate selenium. As shown in Fig. 4, substitution of Cys-38 to Ser resulted in an 83% decrease in selenium incorporation into the GPXH molecule. The residual incorporation observed in the mutant GPXH could be attributed to Cys-66 and Cys-84 that are considered to be much less reactive to selenium modification than Cys-38.
Fig. 3.
GPX assay of recombinant GPXH. Cell extracts were individually prepared from selenium-supplemented E. coli expressing non-mutated normal GXPH and three forms of mutant GPXH derivatives, Cys38Ser, Cys66Ser, and Cys84Ser, and assayed for GPX activity. The mean values of five independent experiments±S.D. are shown.
Fig. 4.
Selenium incorporation into recombinant GPXH protein. Non-mutated normal GPXH and its Cys38Ser mutant derivative were individually purified to homogeneity from selenium-supplemented E. coli expressing each construct, and the protein-bound selenium was quantified. The mean values of four independent measurements±S.D. are shown.
The present study indicates that selenium is almost exclusively bound to Cys-38 of the GPXH protein at the post-translational level, which most likely triggers activation of this enzyme. This conclusion can be explained by the established fact that selenium possesses a more powerful redox potential; hence, reacts much faster with its substrate compared with sulfur. Moreover, this finding is consistent with previous reports describing that replacement of the catalytic selenocysteine with Cys in mammalian GPX led to a reduction in enzymatic activity by more than 99% [30]; on the contrary, an amino acid substitution of native Cys for selenocysteine at the active site of citrus GPX conferred a four-fold higher peroxidase activity to this mutated enzyme [31]. To our knowledge, our finding presents the first evidence for the close association between the enzymatic activity of plant GPX and its Cys-targeted post-translational modification with selenium, and given the crucial importance of GPX in plant physiology, a similar functional correlation is likely the case for other non-selenium peroxidases.
Our additional interest is the chemical form of selenium bound to Cys-38 of the GPXH protein. Stadtman and co-workers proposed for several species of enzyme the formation of protein perselenide (protein-S-Se-) as a product generated from post-translational insertion of selenium into a reactive Cys residue of these enzymes [32], [33]. They postulated that the protein-bound selenium is then released in the presence of reducing agents and transferred to an as yet unidentified delivery pathway for further selenium metabolism. Another laboratory presented a model to explain the selenium-catalyzed oxidation of carbon monoxide (CO) at the active domain of CO dehydrogenase [34], [35]. In this case, an S-selanylcysteine residue of this protein resulting from post-translational selenium incorporation, which is equivalent to the above protein perselenide, is predicted to serve as the central component in the catalytic reaction. On the basis of these reports, it is likely that the selenium atom bound to catalytically essential Cys forms such a perselenide derivative in the GPXH protein as well. Although the biological significance of the existence of GPXH apart from the selenoprotein-type GPX in algae is unknown, post-translational modulation of GPX activity would permit plants to elicit a more flexible and quick response in the face of environmental perturbations, such as oxidative stress, for which exogenous selenium is a key factor.
4. Conclusions
This study highlighted the functional correlation between the activity of algal non-selenium GPX and its selenium incorporation. Here, externally added selenium was specifically bound to the catalytic Cys of the GPX, and also this selenium-binding targeted to the active site was found to be essential for GPX up-regulation. Our finding will provide novel insight in the molecular understanding of plant defense mechanisms against oxidative damage.
Acknowledgments
We thank Dr. Shigeru Shigeoka for his kind advice and encouragement. We also thank Kumiko Yamamoto and Masato Yoshii for their assistance in conducting the experiments. This work was supported by the Grant-in-Aid for Scientific Research (C) (T.T.: 20580066) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.08.018.
Appendix A. Supplementary material
Supplementary material
References
- 1.Mills G.C. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J. Biol. Chem. 1957;229:189. [PubMed] [Google Scholar]
- 2.Bela K., Horvath E., Galle A., Szabados L., Tari I., Csiszar J. Plant glutathione peroxidases: emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015;176:192–201. doi: 10.1016/j.jplph.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Brigelius-Flohe R., Banning A., Schnurr K. Selenium-dependent enzymes in endothelial cell function. Antioxid. Redox Signal. 2003;5:205–215. doi: 10.1089/152308603764816569. [DOI] [PubMed] [Google Scholar]
- 4.Margis R., Dunand C., Teixeira F.K., Margis-Pinheiro M. Glutathione peroxidase family – an evolutionary overview. FEBS J. 2008;275:3959–3970. doi: 10.1111/j.1742-4658.2008.06542.x. [DOI] [PubMed] [Google Scholar]
- 5.Stadtman T.C. Selenocysteine. Annu. Rev. Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 6.Burk R.F., Hill K.E. Orphan selenoproteins. Bioessays. 1999;21:231–237. doi: 10.1002/(SICI)1521-1878(199903)21:3<231::AID-BIES7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 7.Allmang C., Wurth L., Krol A. The selenium to selenoprotein pathway in eukaryotes: more molecular partners than anticipated. Biochim. Biophys. Acta. 1790;2009:1415–1423. doi: 10.1016/j.bbagen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Gladyshe V.N., Kryukov G.V. Evolution of selenocysteine-containing proteins: significance of identification and functional characterization of selenoproteins. Biofactors. 2001;14:87–92. doi: 10.1002/biof.5520140112. [DOI] [PubMed] [Google Scholar]
- 9.Criqui M.C., Jamet E., Parmentier Y., Marbach J., Durr A., Fleck J. Isolation and characterization of a plant cDNA showing homology to animal glutathione peroxidases. Plant Mol. Biol. 1992;18:623–627. doi: 10.1007/BF00040684. [DOI] [PubMed] [Google Scholar]
- 10.Depege N., Drevet J., Boyer N. Molecular cloning and characterization of tomato cDNAs encoding glutathione peroxidase-like proteins. Eur. J. Biochem. 1998;253:445–451. doi: 10.1046/j.1432-1327.1998.2530445.x. [DOI] [PubMed] [Google Scholar]
- 11.Holland D., Ben-Hayyim G., Faltin Z., Camoin L., Strosberg A.D., Eshdat Y. Molecular characterization of salt-stress-associated protein in citrus: protein and cDNA sequence homology to mammalian glutathione peroxidases. Plant Mol. Biol. 1993;21:923–927. doi: 10.1007/BF00027124. [DOI] [PubMed] [Google Scholar]
- 12.Jung B.G., Lee K.O., Lee S.S., Chi Y.H., Jang H.H., Kang S.S., Lee K., Lim D., Yoon S.C., Yun D.J., Inoue Y., Cho M.J., Lee S.Y. A Chinese cabbage cDNA with high sequence identity to phospholipid hydroperoxide glutathione peroxidases encodes a novel isoform of thioredoxin-dependent peroxidase. J. Biol. Chem. 2002;277:12572–12578. doi: 10.1074/jbc.M110791200. [DOI] [PubMed] [Google Scholar]
- 13.Li W.-J., Feng H., Fan J.-H., Zhang R.-Q., Zhao N.-M., Liu J.-Y. Molecular cloning and expression of a phospholipid hydroperoxide glutathione peroxidase homolog in Oryza sativa. Biochim. Biophys. Acta. 2000;1493:225–230. doi: 10.1016/s0167-4781(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 14.Mullineaux P., Karpinski S., Jimenez A., Cleary S., Robinson C., Creissen G. Identification of cDNAs encoding plastid-targeted glutathione peroxidase. Plant J. 1998;13:375–379. doi: 10.1046/j.1365-313x.1998.00052.x. [DOI] [PubMed] [Google Scholar]
- 15.Rodriguez Milla M.A., Maurer A., Rodriguez Huete A., Gustafson J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003;36:602–615. doi: 10.1046/j.1365-313x.2003.01901.x. [DOI] [PubMed] [Google Scholar]
- 16.Roeckel-Drevet P., Gagne G., de Labrouhe D., Dufaure J., Nicolas P., Drevet J. Molecular charcerization, organ distribution and stress-mediated induction of two glutathione peroxidase-encoding mRNAs in sunflower (Helianthus annuus) Physiol. Plant. 1998;103:385–394. [Google Scholar]
- 17.Sugimoto M., Sakamoto W. Putative phospholipid hydroperoxide glutathione peroxidase gene from Arabidopsis thaliana induced by oxidative stress. Genes Genet. Syst. 1997;72:311–316. doi: 10.1266/ggs.72.311. [DOI] [PubMed] [Google Scholar]
- 18.Xiao-Dong Y., Jun L.W., Yuan L.J. Isolation and characterization of a novel PhGPx gene in Raphanus sativus. Biochem. Biophys. Acta. 2005;1728:199–205. doi: 10.1016/j.bbaexp.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Inoue Y., Matsuda T., Sugiyama K., Izawa S., Kimura A. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:27002–27009. doi: 10.1074/jbc.274.38.27002. [DOI] [PubMed] [Google Scholar]
- 20.Fu L.H., Wang X.F., Eyal Y., She Y.M., Donald L.J., Standing K.G., Ben-Hayyim G. A selenoprotein in the plant kingdom. Mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii glutathione peroxidase. J. Biol. Chem. 2002;277:25983–25991. doi: 10.1074/jbc.M202912200. [DOI] [PubMed] [Google Scholar]
- 21.Novoselov S.V., Rao M., Onoshko N.V., Zhi H., Kryukov G.V., Xiang Y., Weeks D.P., Hatfield D.L., Gladyshev V.N. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokota A., Shigeoka S., Onishi T., Kitaoka S. Selenium as inducer of glutathione peroxidase in low-CO2-grown Chlamydomonas reinhardtii. Plant Physiol. 1988;86:649–651. doi: 10.1104/pp.86.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shigeoka S., Takeda T., Hanaoka T. Characterization and immunological properties of selenium-containing glutathione peroxidase induced by selenite in Chlamydomonas reinhardtii. Biochem. J. 1991;275:623–627. doi: 10.1042/bj2750623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda T., Nakano Y., Shigeoka S. Effects of selenite, CO2 and illumination on the induction of selenite-dependent glutathione peroxidase in Chlamydomonas reinhardtii. Plant Sci. 1993;94:81–88. [Google Scholar]
- 25.Leisinger U., Ruefenacht K., Zehnder A.J.B., Eggen R.I.L. Structure of a glutathione peroxidase homologous gene involved in the oxidative stress response in Chlamydomonas reinhardtii. Plant Sci. 1999;149:139–149. [Google Scholar]
- 26.Allen M.M. Simple conditions for the growth of unicellular blue-green algae in plates. J. Phycol. 1968;4:1–4. doi: 10.1111/j.1529-8817.1968.tb04667.x. [DOI] [PubMed] [Google Scholar]
- 27.Ploug M., Jensen A.L., Barkholt V. Determination of amino acid compositions and NH2-terminal sequences of peptides electroblotted onto PVDF membranes from tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis: application to peptide mapping of human complement component C3. Anal. Biochem. 1989;181:33–39. doi: 10.1016/0003-2697(89)90390-4. [DOI] [PubMed] [Google Scholar]
- 28.Bayfield R.F., Romalis L.F. pH control in the fluorometric assay for selenium with 2,3-diaminonaphthalene. Anal. Biochem. 1985;144:569–576. doi: 10.1016/0003-2697(85)90155-1. [DOI] [PubMed] [Google Scholar]
- 29.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 30.Maiorino M., Aumann K.D., Brigelius-Flohe R., Doria D., van den Heuvel J., McCarthy J., Roveri A., Ursini F., Flohe L. Probing the presumed catalytic triad of selenium-containing peroxidases by mutational analysis of phospholipid hydroperoxide glutathione peroxidase (PHGPx) Biol. Chem. Hoppe Seyler. 1995;376:651–660. doi: 10.1515/bchm3.1995.376.11.651. [DOI] [PubMed] [Google Scholar]
- 31.Hazebrouck S., Camoin M., Faltin Z., Strosberg A.D., Eshdat Y. Substituting selenocysteine for catalytic cysteine 41 enhances enzymatic activity of plant phospholipid hydroperoxide glutathione peroxidase expressed in Escherichia coli. J. Biol. Chem. 2000;275:28715–28721. doi: 10.1074/jbc.M004985200. [DOI] [PubMed] [Google Scholar]
- 32.Lacourciere G.M., Levine R.L., Stadtman T.C. Direct detection of potential selenium delivery proteins by using an Escherichia coli strain unable to incorporate selenium from selenate into proteins. Proc. Natl. Acad. Sci. USA. 2002;99:9150–9153. doi: 10.1073/pnas.142291199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogasawara Y., Lacourciere G.M., Ishii K., Stadtman T.C. Characterization of potential selenium-binding proteins in the selenophosphate synthetase system. Proc. Natl. Acad. Sci. USA. 2005;102:1012–1016. doi: 10.1073/pnas.0409042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobbek H., Gremer L., Meyer O., Huber R. Crystal structure and mechanism of CO dehydrogenase, a molybdo iron-sulfur flavoprotein containing S-selanylcysteine. Proc. Natl. Acad. Sci. USA. 1999;96:8848–8889. doi: 10.1073/pnas.96.16.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer O., Gremer L., Ferner R., Ferner M., Dobbek H., Gnida M., Meyer-Klaucke W., Huber R. The role of Se, Mo and Fe in the structure and function of carbon monoxide dehydrogenase. Biol. Chem. 2000;381:865–876. doi: 10.1515/BC.2000.108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material