Abstract
Solution structures of nucleosomes containing a human histone variant, H2A.Z.1, were measured by small-angle X-ray and neutron scatterings (SAXS and SANS). SAXS revealed that the outer shape, reflecting the DNA shape, of the H2A.Z.1 nucleosome is almost the same as that of the canonical H2A nucleosome. In contrast, SANS employing a contrast variation technique revealed that the histone octamer of the H2A.Z.1 nucleosome is smaller than that of the canonical nucleosome. The DNA within the H2A.Z.1 nucleosome was more susceptible to micrococcal nuclease than that within the canonical nucleosome. These results suggested that the DNA is loosely wrapped around the histone core in the H2A.Z.1 nucleosome.
Abbreviations: SAXS, small-angle X-ray scattering; SANS, small-angle neutron scattering; CV-SANS, small-angle neutron scattering employing contrast variation technique
Keywords: Nucleosome, H2A.Z.1, Small-angle X-ray scattering, Small-angle neutron scattering, Contrast variation, Stuhrmann plot
Highlights
-
•
Outer shape of H2A.Z.1 nucleosome is similar to that of canonical one.
-
•
Histone octamer of H2A.Z.1 nucleosome is smaller than that of canonical one.
-
•
The change of histone octamer might give H2A.Z.1 nucleosome dynamical properties.
1. Introduction
Chromatin is composed of repeated protein-DNA complexes, referred to as nucleosomes. The protein portion of the nucleosome is a histone octamer, consisting of two copies of the histone H2A, H2B, H3, and H4 molecules, and about 150 base pairs of DNA are wrapped around the disk-shaped histone core [1]. The DNA and the histone octamer are tightly bound in the nucleosome, which functions as a storage form for eukaryotic genomic DNA. However, the functional regions of the genomic DNA must become accessible to the enzymes that mediate DNA metabolism, such as transcription, recombination, repair, and replication [2], [3]. To do so, the nucleosome must relax the bound DNA. For example, on active genes, the genomic DNA must detach from the histone core to allow the access of transcription factors [2]. Therefore, the structural versatility and dynamics of the nucleosome are an important research target to understand the mechanisms of genomic DNA regulation and maintenance in eukaryotes.
Non-allelic histone variants are considered to be genomic DNA regulators, since they induce structural alterations of the nucleosome by replacing the canonical histones [4], [5]. In fact, by solution scattering measurements, we previously reported that in the H2A.B nucleosome, in which the H2A histones are replaced by the H2A.B variants, the DNA was partially peeled off [6], [7]. H2A.B is a distant H2A variant, which shares about 50% amino acid identity with H2A [8]. Among the H2A variants, H2A.Z, the major one (10% of all histone H2A species, while H2A.B is less than 0.2%), is predominantly localized around the promoters of active genes, and might be responsible for producing a nucleosome-free region near the transcription start sites [9], [10], [11]. Therefore, much interest has been focused on the structure of the H2A.Z nucleosome, in which the H2A molecules are replaced by H2A.Z. Two H2A.Z subtypes, H2A.Z.1 and H2A.Z.2, have been identified in higher eukaryotes [12]. Recently, our group reported the crystal structures of the H2A.Z.1 and H2A.Z.2 nucleosomes and their functional analyses [13]. The crystal structures of the H2A.Z.1 and H2A.Z.2 nucleosomes are not significantly different, but fluorescence recovery after photobleaching analyses revealed that H2A.Z.1 is more rapidly exchanged in living cells, as compared to the canonical H2A and H2A.Z.2 nucleosomes [13]. These results suggest that the H2A.Z.1 nucleosome could be structurally unstable in vivo.
To study the solution structure of the H2A.Z.1 nucleosome, we employed small-angle-X-ray scattering (SAXS) and small-angle neutron scattering utilizing a contrast variation method (CV-SANS). SAXS is a very powerful tool to reveal the outer shapes of biomolecules in solution. In the SAXS analysis of nucleosomes, the DNA portion has higher scattering power than that of a histone octamer in nucleosomes. Therefore, it is difficult to observe the structure of a histone octamer without the sugar-contrast variation technique [14]. As compared with X-ray scattering, neutron scattering has an isotope effect, because the neutrons are scattered by the nucleus. The isotope effect is most distinguished between a proton (scattering length: −3.74 fm) and a deuteron (scattering length: +6.64 fm), indicating that the scattering length density (SLD) of the solvent can be controlled by using the proper mixture of H2O and D2O. This technique for small-angle neutron scattering is called the (solvent) contrast variation method (CV-SANS), and it is very useful to observe the inner structures of complexes consisting of domains with different SLDs in an aqueous solution [15], [16], [17], [18], [19], [20], such as nucleosomes [7], [22], [23], [24] (see details in Section 2.3).
Here, we report the results of SAXS and CV-SANS measurements of the H2A.Z.1 nucleosome in an aqueous solution, and discuss its structural differences from the canonical H2A nucleosome and the variant H2A.B nucleosome.
2. Materials and methods
2.1. Preparation of nucleosomes
Histones H2A, H2A.Z.1, H2B, H3.1, and H4 were bacterially expressed and purified, as described previously [13]. To form the histone octamers containing H2A and H2A.Z.1, histones were mixed in 20 mM Tris–HCl (pH 7.5) buffer, containing 7 M guanidine hydrochloride and 20 mM 2-mercaptoethanol, and the samples were dialyzed against 10 mM Tris–HCl (pH 7.5) buffer, containing 2 M NaCl, 1 mM EDTA, and 5 mM 2-mercaptoethanol. The resultant histone octamers were then purified by HiLoad 16/60 Superdex 200 gel filtration column chromatography (GE Healthcare). For the nucleosome reconstitution, the histone octamers and 145 base pairs of the Widom 601 DNA [21] were mixed in 10 mM Tris–HCl (pH 7.5) buffer, containing 2 M KCl, 1 mM EDTA, and 1 mM dithiothreitol, and the KCl concentration was gradually decreased to 0.25 M. The reconstituted nucleosomes were separated from the free histones and unbound DNA by native polyacrylamide gel electrophoresis (PAGE), using a Prep Cell apparatus (Bio-Rad).
2.2. SAXS measurement
The SAXS experiments were performed with an upgraded apparatus at BL10C of the Photon Factory, at the Institute of Materials Structure Science (IMSS), High Energy Accelerator Research Organization (KEK), Tsukuba, Japan. The wavelength of the incident X-ray beam was tuned to 1.488 Å, and the reciprocal space ranging from 0.008 to 0.26 Å−1 was measured with a two-dimensional position-sensitive detector, PILATUS3 2M. By using the software for SAXS image data processing, SAngler, incident beam normalization, transmission correction, solvent subtraction, and circular average were applied, and then the scattering profiles of the nucleosomes were obtained.
The analyzed nucleosomes were the canonical H2A nucleosome and the variant H2A.Z.1 nucleosome. For each SAXS experiment, we prepared for 0.05 mL sample solution of which the nucleosome concentration was 3.0 mg/mL and the buffer solvent was 20 mM Tris–HCl (pH 7.5) containing 50 mM NaCl and 1 mM dithiothreitol. In addition, dilute samples (0.5 mg/mL) were also measured, to confirm that particle interference can be ignored under the 3.0 mg/mL solvent conditions with 50 mM NaCl.
2.3. CV-SANS measurement
CV-SANS is a powerful technique to observe the partial structures of complex materials consisting of domains/molecules with different SLDs [7], [22], [23]. The scattering intensity from the solution is proportional to the square of the scattering contrast between solute and solvent, Δρ2=(ρsolute−ρsolvent)2, where ρsolute and ρsolvent are the SLDs of the solute and solvent, respectively. Nucleosomes consists of different biomolecules which have different SLDs: The SLDs of histones and DNA are 0.221 (fm/Å3) and 0.394 (fm/Å3), respectively. Here, the SLDs of H2O and D2O are −0.056 (fm/Å3) and 0.636 (fm/Å3). Therefore, in a 65% D2O solution, because the SLD of the DNA is matched to that of the 65% D2O solvent, we can erase the scattering due to DNA and selectively observe the structure of the histone octamer in a nucleosome. Moreover, in a 40% D2O solution, we selectively observe the DNA by the same concept of contrast matching with the exchange of the roles.
We used two SANS spectrometers, TAIKAN at J-PARC/MLF, Tokai, Ibaraki, Japan for mainly structural check of nucleosomes and also D11 instrument, installed at the Institut Laue-Langevin (ILL), Grenoble, France for CV-SANS experiment. In the CV-SANS experiments with D11, the SANS intensities were measured in the q-range of 0.0085–0.13 Å−1 at a constant temperature of 25 °C. The observed SANS intensities were corrected for the background, empty cell and buffer scatterings, and the transmission factors, using the standard D11 data processing software, GRASP, and finally converted to one-dimensional scattering profiles.
The samples were the canonical H2A nucleosome and the variant H2A.Z.1 nucleosome, at a concentration of 3.0 mg/mL in a 50 mM NaCl aqueous solution. In consideration of the contrasts between histone/DNA and solvent, four sample solutions with different H2O/D2O ratios were employed: 0% D2O solution with the amount of 0.40 mL (reference), 40% D2O solution with the amount of 0.40 mL (matching point for histone), 65% D2O solution with the amount of 0.80 mL (matching point for DNA), and 100% D2O solution with the amount of 0.80 mL (another reference).
2.4. Micrococcal nuclease assay
The nucleosomes (0.4 μg) containing H2A or H2A.Z.1 were treated with 1.2 units of micrococcal nuclease (MNase, Takara) for 0, 5, 10, 15, and 20 min at 25 °C in 42 mM Tris–HCl (pH 7.5) buffer, containing 10 mM NaCl, 10% glycerol, 1 mM CaCl2, 2.5 ng/μl BSA, and 2.6 mM dithiothreitol. The reactions were then stopped by adding proteinase K (Roche). The resulting DNA fragments were extracted by phenol/chloroform/isoamyl alcohol and purified by ethanol precipitation. The purified DNA fragments were analyzed by 6% non-denaturing PAGE with ethidium bromide staining.
3. Results and discussion
3.1. Overall structures: SAXS measurement
To analyze the solution structures of the nucleosomes by SAXS and SANS, we prepared highly purified nucleosomes containing H2A and H2A.Z.1. To do so, we reconstituted the nucleosomes containing H2A and H2A.Z.1 in vitro. We confirmed that the nucleosomes containing H2A and H2A.Z.1 were highly purified and the four types of histones were present in the same molar ratios in the nucleosomes (Fig. S1).
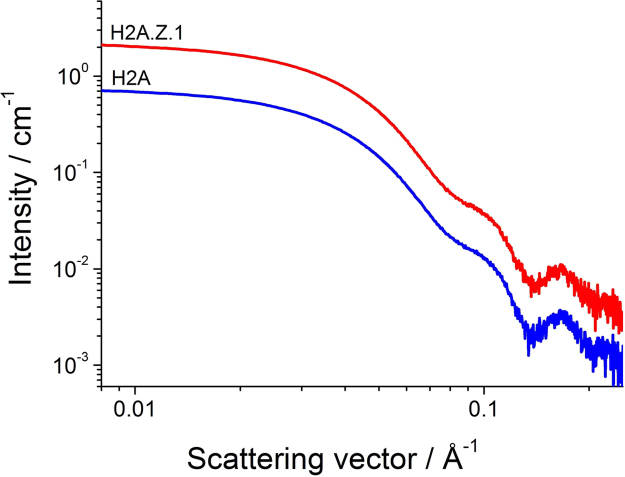
The SAXS profiles of the canonical H2A nucleosome and the variant H2A.Z.1 nucleosome are shown in Fig. 1. The SAXS profile of the H2A.Z.1 nucleosome is almost the same as that of the canonical H2A nucleosome. This feature was quantitatively clarified by an examination of the gyration radii and a comparison of the distance distribution functions. The gyration radii were 44.6 Å and 44.4 Å for the H2A and H2A.Z.1 nucleosomes, respectively (Fig. S2), and the largest distances, Dmax, of both nucleosomes were 128 Å (Fig. S3). In comparison with another variant nucleosome, the H2A.B nucleosome, this structural coincidence between the canonical H2A nucleosome and variant H2A.Z.1 nucleosome is remarkable, as the Rg and Dmax of the H2A.B nucleosome are 49.4±0.4 Å and 195 Å, respectively [6], reflecting the fact that the edges of the DNA in the H2A.B nucleosome are detached from the histone core. In contrast, even though the H2A.Z.1 nucleosome is also a variant nucleosome, its outer shape is similar to that of the canonical nucleosome, as shown in their crystallographic structures [13], [21]. This feature is also seen with dummy atom models of the H2A nucleosome and H2A.Z.1 nucleosome built based on their SAXS profiles (Fig. S4). The both models show a similar disk shape, indicating the all DNA segments of both nucleosomes are wrapped around their histone cores. In conclusion, the SAXS measurement did not reveal any free DNA segments, and thus the DNA ends are stably bound to the histone core, as in the crystal structure.
Fig. 1.
SAXS profiles of H2A nucleosomes (blue) and H2A.Z.1 nucleosomes (red). The scattering intensity of H2A.Z.1 nucleosome was multiplied by three.
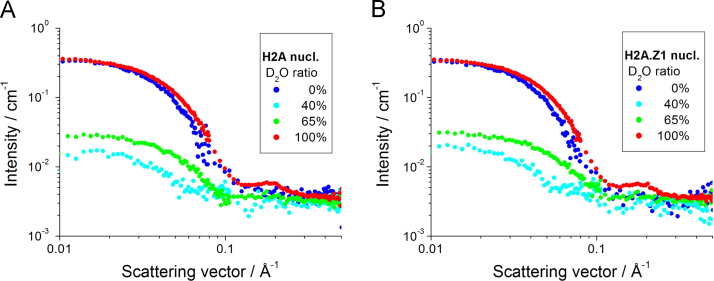
3.2. Partial structure: CV-SANS measurement
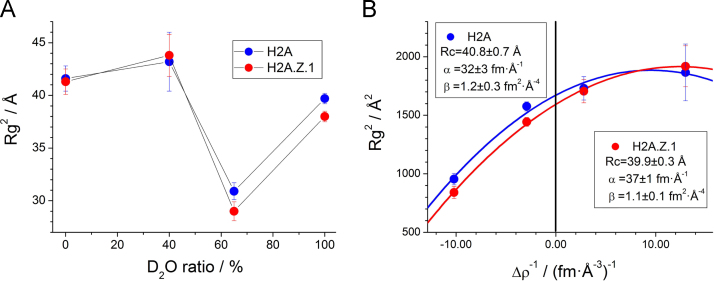
We wanted to determine whether the H2A.Z.1 nucleosome has any structural differences from the canonical H2A nucleosome in solution. Therefore, we employed CV-SANS to analyze the inner structure of the H2A.Z.1 nucleosome in solution. The CV-SANS profiles of the H2A and H2A.Z.1 nucleosomes are shown in Fig. 2 at four contrast points: 0%, 40%, 65%, and 100% D2O. The dependence of the gyration radius on the contrast is shown in Fig. 3A. The gyration radius reasonably exhibits the minimum and maximum values in the 65% and 40% D2O solutions, in which the inner histones and the outer DNA shapes are selectively observed, respectively. Their average values are observed in the 0% and 100% D2O solutions. As shown in Fig. 3A, the gyration radii are slightly different between the canonical H2A and H2A.Z.1 nucleosomes at several contrast points, but it is difficult to clarify the structural differences between them. Therefore, we performed the Stuhrmann's analysis to derive the SLD distribution [14], [15], [16], [17], [18], [19], [20], which reflects the spatial configurations of the DNA (with the higher SLD) and the histones (with the lower SLD) in a nucleosome. In the Stuhrmann's formalization, the square of the gyration radius, Rg2, is expressed as a quadratic function of the inverse of contrast, 1/Δρ:
| (1) |
where Stuhrmann's parameters, Rc, α, and β, are the gyration radius at infinite contrast, the second moment of the internal SLD fluctuations, and a measure of the displacement of the SLD distribution, respectively (see the mathematical formalization of the parameters in “Stuhrmann's parameters” Supplement). As shown in Fig. 3B, the observed Rg2 values for both the H2A and H2A.Z.1 nucleosomes were well reproduced with Eq. (1), and the Stuhrmann's parameters (displayed in the boxes of Fig. 3B) were obtained with least square fitting. Rc corresponds to the only geometrical parameter of the nucleosomes that was unaffected by the inner distribution of the SLD. Here, the Rcs of both the H2A and H2A.Z.1 nucleosomes are almost the same, indicating that there are no large differences in their outer shapes, in accordance with the SAXS results. We then examined the α-values, the second moments of the internal SLD fluctuations, and found a slight but significant difference: the α-values of the H2A and H2A.Z.1 nucleosomes were 32 fm/Å and 37 fm/Å, respectively. In the case of a nucleosome, the meaning of the α-value is simple: a larger value corresponds to the expansion of DNA and/or the compaction of a histone octamer. Considering the SAXS results, which revealed that the DNA is not detached from the histone core and is not expanding, the larger α-value of the H2A.Z.1 nucleosome suggests that the histone octamer of the H2A.Z.1 nucleosome is more compacted than that of the H2A nucleosome. Finally, we checked the β-values. In the case of a nucleosome, the β-value shows the degree of spatial difference in the center of mass between the DNA and the histone octamer. The β-values obtained for both nucleosomes are quite similar (1.2 fm2/Å4 and 1.1 fm2/Å4 for the H2A and H2A.Z.1 nucleosomes, respectively). This means that the DNA configurations in both nucleosomes are the same, and the histone octamer in the H2A.Z.1 nucleosome is symmetrically compacted. The H2A.B nucleosome has a larger α-value (60 fm/Å) and β-value (2.16 fm2/Å4), indicating that the DNA configuration of H2A.B nucleosome is asymmetrically expanded and, therefore, the DNA segments are detached at the ends [7].
Fig. 2.
SANS profiles of the canonical H2A nucleosome (A) and the H2A.Z.1 nucleosome (B). Blue, cyan, green, and red circles depict the scattering at the contrast points of 0%, 40%, 65%, and 100% D2O, respectively.
Fig. 3.
(A) Gyration radii as a function of D2O concentration. Blue and red circles depict Rg values of the canonical H2A and variant H2A.Z.1 nucleosomes, respectively. The straight lines are guides for visualization. (B) Stuhrmann plots (Rg2vs. Δρ−1) of the nucleosomes. Blue and red circles depict Rg2 values of the canonical H2A and variant H2A.Z.1 nucleosomes, respectively. Curved lines represent the results of the least-squares fitting. The inserted boxes show the calculated Stuhrmann's parameters.
To summarize the structural studies with the SAXS and CV-SANS experiments, the outer shape of the H2A.Z.1 nucleosome is similar to that of the H2A nucleosome, in contrast to another variant H2A.B nucleosome. This indicates that the DNA shapes are similar in both the H2A an H2A.Z nucleosomes. In contrast, the size of the histone octamer in the H2A.Z.1 nucleosome is clearly smaller than that in the H2A nucleosome. Notably, this size difference is not apparent in the crystal structures of the H2A.Z.1 and H2A nucleosomes [13]. Therefore, the octamer size difference found in this study may reflect the difference in the histone octamer dynamics in solution.
3.3. The DNA-histone interaction is less tight
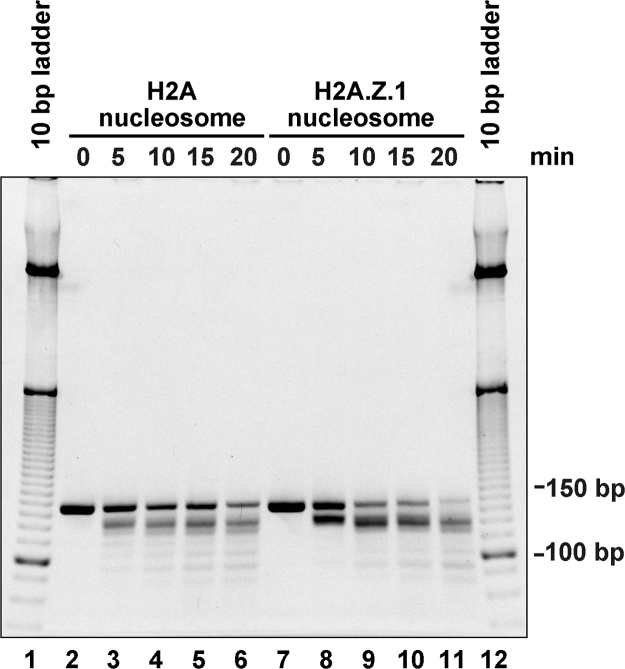
If the histone octamer in the H2A.Z.1 nucleosome is more compact, without affecting the DNA shape, then the DNA should be less tightly bound to the histone core. To test this possibility, we performed a micrococcal nuclease (MNase) assay. In this assay, MNase preferentially degrades looser DNA ends in nucleosomes. As shown in Fig. 4, the DNA in the H2A.Z.1 nucleosome was clearly more susceptible than that in the H2A nucleosome. Since the DNA configuration in the H2A.Z1 nucleosome is almost the same as that in the H2A nucleosome in solution (Figs. 1, Appendix A, Appendix A), the enhanced susceptibility may be responsible for the compacted histone octamer.
Fig. 4.
MNase assay. The nucleosomes containing H2A or H2A.Z.1 were treated with MNase for 0, 5, 10, 15, and 20 min at 25 °C. The resulting DNA fragments were analyzed by 6% non-denaturing PAGE with ethidium bromide staining. Lanes 1 and 12 indicate the DNA markers. Experiments with the H2A nucleosome (lanes 2–6) and the H2A.Z.1 nucleosome (lanes 7–11) are presented.
3.4. Perspective
In conclusion, the H2A.Z.1 nucleosome has a similar structure to that of the canonical H2A nucleosome, and does not exhibit a drastic structural change, such as the large DNA-detachment observed in the variant H2A.B nucleosome in solution. However, H2A.Z.1 undergoes a subtle change, in which the histone octamer is compacted in solution. The crystal structure of the H2A.Z.1 nucleosome revealed that the histone octamer is not clearly compacted, as compared to that of the H2A nucleosome. This discrepancy suggests that the histone dynamics is different between the H2A.Z.1 and H2A nucleosomes in solution. In the H2A.Z.1 nucleosome, the histone octamer may move dynamically, resulting in the smaller average size of the octamer. This dynamic nature of the H2A.Z.1 nucleosome may reflect rapid exchange of H2A.Z.1 in cells [13]. It is also possible that the histone tails of the H2A.Z.1 nucleosome may be less expanded toward the solvent, resulting in the smaller size of the histone core. The histone tails are not visible in the available crystallographic structures of the H2A.Z.1 nucleosomes [13], [25]. The specific physical characteristics of the histone octamer containing H2A.Z.1 in the nucleosome may directly reflect its function, and thus further studies are needed.
Acknowledgments
We would like to thank Dr. Nobutaka Shimizu for his kind assistance with the SAXS experiment at BL10C of KEK-PF. The SAXS measurements were performed with the approval of the Photon Factory Program Advisory Committee (Proposal nos. 2014G127 and 2014G162). We would also like to express our appreciation to Dr. Takashi Oda for his help analysis about the SAXS data. The SANS measurements were performed under proposal No. 8-03-838 at ILL and also under No. 2014A0060 at J-PATC/MLF. This work was partly supported by MEXT KAKENHI Grant nos. 26116509 and 26102524 (to M.S.), and 25116002 (to H.K.) and JSPS KAKENHI Grant nos. 24310068 and 15H02042 (to M.S.). This work was partly supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science), from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and a grant from the Japan Agency for Medical Research and Development (AMED) (to H.K.). N.H. and H.K. are also members of the Waseda Research Institute for Science and Engineering.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.08.019.
Contributor Information
Masaaki Sugiyama, Email: sugiyama@rri.kyoto-u.ac.jp.
Hitoshi Kurumizaka, Email: kurumizaka@waseda.jp.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Venkatesh S., Workman J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 3.Misteli T., Soutoglou E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat. Rev. Mol. Cell Biol. 2009;10:243–254. doi: 10.1038/nrm2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biterge B., Schneider R. Histone variants: key players of chromatin. Cell Tissue Res. 2014;356:457–466. doi: 10.1007/s00441-014-1862-4. [DOI] [PubMed] [Google Scholar]
- 5.Bönisch C., Hake S.B. Histone H2A variants in nucleosomes and chromatin: more or less stable? Nucleic Acids Res. 2012;40:10719–10741. doi: 10.1093/nar/gks865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arimura Y., Kimura H., Oda T., Sato K., Osakabe A., Tachiwana H., Sato Y., Kinugasa Y., Ikura T., Sugiyama M., Sato M., Kurumizaka H. Structural basis of a nucleosome containing histone H2A.B/H2A.Bbd that transiently associates with reorganized chromatin. Sci. Rep. 2013;3 doi: 10.1038/srep03510. 1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugiyama M., Arimura Y., Shirayama K., Fujita R., Oba Y., Sato N., Inoue R., Oda T., Sato M., Heenan R.K., Kurumizaka H. Distinct features of the histone core structure in nucleosomes containing the histone H2A.B variant. Biophys. J. 2014;106:2206–2213. doi: 10.1016/j.bpj.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadwick B.P., Willard H.F. A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J. Cell Biol. 2001;152:375–384. doi: 10.1083/jcb.152.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishibashi T., Li A., Eirín-López J.M., Zhao M., Missiaen K., Abbott D.W., Meistrich M., Hendzel M.J., Ausió J. H2A.Bbd: an X-chromosome-encoded histone involved in mammalian spermiogenesis. Nucleic Acids Res. 2010;38:1780–1789. doi: 10.1093/nar/gkp1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piña B., Suau P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev. Biol. 1987;123:51–58. doi: 10.1016/0012-1606(87)90426-x. [DOI] [PubMed] [Google Scholar]
- 11.Soboleva T.A., Nekrasov M., Pahwa A., Williams R., Huttley G.A., Tremethick D.J. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat. Struct. Mol. Biol. 2012;19:25–30. doi: 10.1038/nsmb.2161. [DOI] [PubMed] [Google Scholar]
- 12.Eirín-López J.M., González-Romero R., Dryhurst D., Ishibashi T., Ausió J. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol. Biol. 2009;9:31. doi: 10.1186/1471-2148-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horikoshi N., Sato K., Shimada K., Arimura Y., Osakabe A., Tachiwana H., Hayashi-Takanaka Y., Iwasaki W., Kagawa W., Harata M., Kimura H., Kurumizaka H. Structural polymorphism in the L1 loop regions of human H2A.Z.1 and H2A.Z.2. Acta Cryst. 2013;D69:2431–2439. doi: 10.1107/S090744491302252X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoko Y., Yamamoto M., Fujiwara S., Ueki T. X-ray scattering study of the shape of the DNA region in nucleosome core particle with synchrotron radiation. J. Biochem. 1992;111:310–316. doi: 10.1093/oxfordjournals.jbchem.a123755. [DOI] [PubMed] [Google Scholar]
- 15.Stuhrmann H.B. Contrast variation application in small-angle neutron scattering experiments. J.Phys.: Conf. Ser. 2012;351:012002. [Google Scholar]
- 16.Stuhrmann H.B. Small-angle scattering and its interplay with crystallography, contrast variation in SAXS and SANS. Acta Cryst. 2008;A64:181–191. doi: 10.1107/S0108767307046569. [DOI] [PubMed] [Google Scholar]
- 17.H.B. Stuhrmann, K.H. Nierhaus, in: R.B. Knott, B.P. Schoenborn (Eds.), Neutrons in Biology, Plenum Press, 1996, pp. 397–413.
- 18.L.A. Feigin, D.I. Svergun, in: G.W. Taylor (Ed.), Structure Analysis by Small-Angle X-Ray and Neutron Scattering, Plenum Press, 1987, pp. 115–131.
- 19.Stuhrmann H.B., Tardieu A., Mateu L., Sardet C., Luzzati V., Aggerbeck I., Scanu A.M. Neutron scattering study of human serum low density lipoprotein. Proc. Nat. Acad. Sci. USA. 1975;72:2270–2273. doi: 10.1073/pnas.72.6.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuhrmann H.B. Neutron small-angle scattering of biological macromolecules in solution. J. Appl. Cryst. 1974;7:173–175. [Google Scholar]
- 21.Lowary P.T., Widom J., New D.N.A. Sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 22.Pardon J.F., Worcester D.L., Wooley J.C., Tatchell K., Van Holde K.E., Richards B.M. Low-angle neutron scattering from chromatin subunit particles. Nucleic Acids Res. 1975;4:2163–2176. doi: 10.1093/nar/2.11.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suau P., Kneale G.G., Braddock G.W., Baldwin J.P., Bradbury E.M. A low resolution model for the chromatin core particle by neutron scattering. Nucleic Acids Res. 1977;4:3769–3786. doi: 10.1093/nar/4.11.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sibbet G.J., Carpenter B.G., Ibel K., May R.P., Kneale G.G. Neutron-scattering studies of accurately reconstituted nucleosome core particles and the effect of ionic strength on core particle structure. Eur. J. Biochem. 1983;133:393–398. doi: 10.1111/j.1432-1033.1983.tb07475.x. [DOI] [PubMed] [Google Scholar]
- 25.Suto R.K., Clarkson M.J., Tremethick D.J., Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material