Abstract
Acetohydroxyacid synthase (AHAS) is the key enzyme in branched chain amino acid biosynthesis pathway. The enzyme activity and properties of a highly thermostable AHAS from the hyperthermophilic bacterium Thermotoga maritima is being reported. The catalytic and regulatory subunits of AHAS from T. maritima were over-expressed in Escherichia coli. The recombinant subunits were purified using a simplified procedure including a heat-treatment step followed by chromatography. A discontinuous colorimetric assay method was optimized and used to determine the kinetic parameters. AHAS activity was determined to be present in several Thermotogales including T. maritima. The catalytic subunit of T. maritima AHAS was purified approximately 30-fold, with an AHAS activity of approximately 160±27 U/mg and native molecular mass of 156±6 kDa. The regulatory subunit was purified to homogeneity and showed no catalytic activity as expected. The optimum pH and temperature for AHAS activity were 7.0 and 85 °C, respectively. The apparent Km and Vmax for pyruvate were 16.4±2 mM and 246±7 U/mg, respectively. Reconstitution of the catalytic and regulatory subunits led to increased AHAS activity. This is the first report on characterization of an isoleucine, leucine, and valine operon (ilv operon) enzyme from a hyperthermophilic microorganism and may contribute to our understanding of the physiological pathways in Thermotogales. The enzyme represents the most active and thermostable AHAS reported so far.
Abbreviations: AHAS, acetohydroxyacid synthase; BCAA, branched chain amino acid; CCE, crude cell extract; CFE, cell-free extract; HTCCE, heat-treated crude cell extract; IB, inclusion body; ilv, isoleucine, leucine, valine; IMAC, immobilized metal affinity chromatography; TmAHAS, Thermotoga maritima acetohydroxyacid synthase; TPP, thiamine pyrophosphate
Keywords: Acetohydroxyacid synthase, Hyperthermophiles, Branched-chain amino acids, Thermotogales
Graphical abstract
Highlights
-
•
First report of AHAS from a hyperthermophilic bacterium.
-
•
Catalytic and regulatory subunits of AHAS of T. maritima was expressed in E. coli.
-
•
Recombinant proteins were purified using a simplified procedure.
-
•
Enzyme represents the most active and thermostable AHAS reported so far.
-
•
Kinetic parameters were determined for the purified recombinant enzyme
1. Introduction
Hyperthermophiles are organisms that exhibit growth temperature optima of 80 °C or above [1], [2]. During the last decade there has been enormous attention towards biotechnological and industrial applications of hyperthermophiles. One promising avenue is to use the organisms (or their metabolites) in bio-processing toward production of value-added commodities (e.g., alcohols). However, advances are hindered by the relatively insufficient understanding of the physiology and metabolic pathways of these organisms [3].
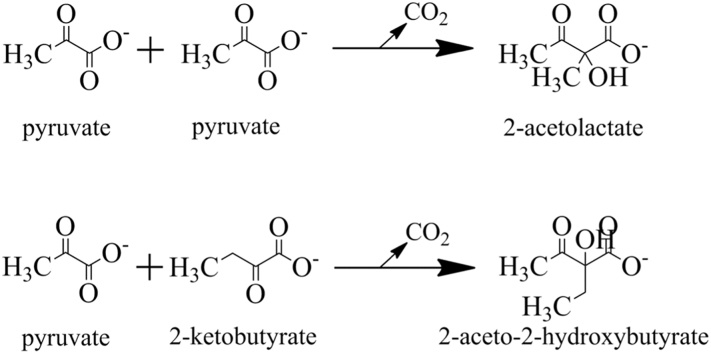
Acetohydroxyacid synthases (AHAS, EC 2.2.1.6) are divided into two classes based on their metabolic/physiological roles, substrate specificity and cofactor (FAD) requirements: anabolic and catabolic AHASs [4], [5]. The anabolic AHAS catalyzes the first common step in the biosynthesis of the branched-chain amino acids (BCAA, valine, leucine, and isoleucine) as well as the precursors derived from the same biosynthetic pathway (e.g. coenzyme A and pantothenate). The enzyme is relatively prevalent in archaea, bacteria, fungi, algae, and plants, but is absent from animals [6], [7], [8]. It catalyzes two parallel reactions during which one molecule of pyruvate is decarboxylated, and the resulting “active aldehyde” is condensed with a second molecule of either pyruvate or 2-ketobutyrate to produce acetolactate (precursor of valine and leucine) and 2-aceto-2-hydroxybutyrate (precursor of isoleucine), respectively (Fig. 1). The catabolic AHAS (also known as acetolactate synthase; ALS) has a single subunit with ~60 kDa in size, and is involved in channeling the excess pyruvate to the less inhibitory product 2-acetolactate (Fig. 1) which can be either decomposed to diacetyl (spontaneously) or be decarboxylated to acetoin by acetolactate decarboxylase [9]. This latter enzyme has only been described from certain bacterial species including Klebsiella, Bacillus species, and some lactic acid bacteria [5], [10].
Fig. 1.
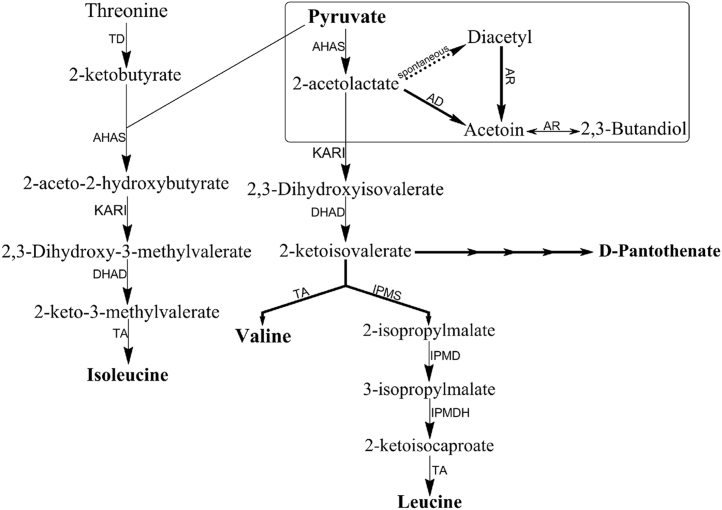
Biosynthesis pathways of branched chain amino acids and the butanediol pathway (Boxed). TD, threonine deaminase (EC 4.3.1.19); KARI, ketol-acid reductoisomerase (EC 1.1.1.86); DHAD, dihydroxyacid dehydratase (EC 4.2.1.9) ; TA, transaminase (EC 2.6.1.42, EC 2.6.1.66, EC 2.6.1.6); IPMS, 2-isopropylmalate synthase (EC 2.3.3.13); IPMD, isopropylmalate dehydratase (EC 4.2.1.33); IPMDH, 3-isopropylmalate dehydrogenase (EC 1.1.1.85); AD, acetolactate decarboxylase (EC 4.1.1.5); AR, acetoin reductase (EC 1.1.1.4).
All of the anabolic AHASs studied so far are composed of two subunits: a larger active subunit known as catalytic subunit (generally 59–66 kDa) and a smaller (catalytically inactive) subunit known as regulatory subunit (generally 9–35 kDa). The regulatory subunit mediates the regulation of the catalytic subunit through the feedback regulation by one or more of the branched chain amino acids [11], [12]. Compared to the catalytic subunit, the holoenzyme (catalytic and regulatory subunits together) generally shows higher activity, which is also evident by the observation that combining separately purified subunits generally results in reconstitution of the holoenzyme with a several-fold increase of activity [13], [14], [15], [16], [17].
Several anabolic AHASs from various organisms have been characterized including different isozymes (isozymes I, II, and III) from Escherichia coli [17], [18], [19], and Salmonella [20], Corynebacterium glutamicum [21], Klebsiella pneumoniae [22], different pathogenic mycobacteria species [23], [24], [25], [26], Shigella sonnei [27] and Haemophilus influenzae [28], Saccharomyces cerevisiae [16], and plants including Arabidopsis (Arabidopsis thaliana) [29], and tobacco (Nicotiana tabacum) [30]. Considering the absence of the branched chain amino acid (BCAA) biosynthesis pathway in animals, the anabolic AHAS has spurred significant interest as a potential candidate for development of new antimicrobial drugs and herbicides (reviewed in Refs. [31] and [32], respectively).
Very few AHASs have been investigated from extremophilic microorganisms. An oxygen-sensitive AHAS from the mesophilic archaeon Methanococcus aeolicus has been characterized [33]. A characteristically similar AHASs has been characterized from the halophilic archaeon Haloferax volcanii [34] and moderately thermophilic bacterium Geobacillus stearothermophilus [35]. However, so far no hyperthermophilic AHAS has been studied.
Thermotoga maritima is the model organism in the order of Thermotogales with a growth temperature range of 55–90 °C and an optimal growth temperature of 80 °C [36]. In the present study, AHAS activity was investigated in Thermotogales. To our knowledge this is the first report on the biochemical characterization of an ilv operon enzyme from a hyperthermophilic bacterium. The genes encoding the hypothetical catalytic and regulatory subunits of AHAS from T. maritima were heterologously expressed in E. coli and the properties of the purified recombinant enzyme were characterized.
2. Materials and methods
2.1. Microorganisms and plasmids
T. maritima , Thermotoga hypogea and Thermotoga neapolitana were obtained from German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and were grown under anaerobic conditions in 20 L glass carboys. T. maritima was grown anaerobically on glucose and yeast extract at 80 °C as described by Huber et al. [36] using a modified procedure as described elsewhere [37]. T. hypogea and T. neapolitana cell biomasses were grown on glucose at 70 °C and 77 °C, respectively using the procedure modified from Fardeau et al. [38] as described by Yang and Ma [39].
The growth was monitored by direct microscopic cell enumeration using a Petroff-Hausser cell counting chamber (1/400 mm2, 0.02 mm deep; Hausser Scientific, Horsham, PA) and a Nikon Eclipse E600 phase-contrast light microscope (Nikon Canada, ON, Canada). The late log-phase cultures were cooled down in ice slurry and centrifuged at 13,000g using a Sharples continuous centrifugation system (Sharples equipment division, PA, USA) at 150–200 ml/min. The resulting biomass was snap-frozen in liquid nitrogen and then stored at −80 °C until use.
E. coli strains DH5α (BRL, CA, USA) was used for recombinant DNA propagation and E. coli BL21 (DE3) Rosetta 2 [F− ompT hsdSB (rB− mB−) gal dcm pRARE27 (CamR)] (Novagen, WI, USA) was used for overexpression of AHAS subunits under the control of T7 polymerase of the plasmid pET30a (+) (Novagen, WI, USA). The recombinant E. coli strains were grown in LB broth (10 g Bacto-Tryptone, 5.0 g yeast extract, 10 g NaCl per liter, pH 7.5) supplemented with kanamycin (30 µg/ml) and chloramphenicol (34 µg/ml) for plasmid maintenance.
2.2. Preparation of cell-free extracts
The frozen biomass was thawed in a pre-degassed flask and then re-suspended in anaerobic lysis buffer (10 mM Tris–HCl, 5% glycerol, 2 mM sodium dithionite (SDT), 2 mM DTT, 0.01 mg/ml DNase, and 0.1 mg/ml lysozyme, pH 7.8) with 1:9 ratio (w/v). The suspension was incubated at 37 °C for 1.5–2 h and then centrifuged at 10,000 x g for 30 min. The supernatant was transferred to a new anaerobic serum bottle and used as cell-free extract (CFE) for further experiments. To study their impact on activity, thiamine pyrophosphate (TPP) and FAD were included into the composition of the lysis buffer with the final concentrations of 0.1 mM and 0.01 mM, respectively.
2.3. Plasmid construction and expression
The genomic DNA from T. maritima was isolated for amplification of the genes encoding the catalytic and regulatory subunits of AHAS. Standard procedures were followed for all DNA isolation and downstream manipulation, competent cell preparation and transformation according to the methods described by Sambrook and Russell [40]. The coding sequence of the putative catalytic subunit (TM0548) was amplified using the following synthetic oligonucleotides: the sense primer 5´-ATACATATGGTTCACGTGAAGATGAAAGG-3´ and antisense primer 5´-TACTCGAGTCTTTCATCACCTCTCCTGCTCT-3´ with cut sites for NdeI and XhoI, respectively (italicized). The gene encoding the regulatory subunit (TM0549) was PCR amplified using the sense primer 5´-ATACATATGACGGACCAGATTCGAGAGC-3' and antisense primer 5'-ATCTCGAGGAATCCCTCCCCTTCTTTTACG-3´ with NdeI and XhoI sites (italicized), respectively. A total of three expression plasmids were constructed for the expression of catalytic (pETTm0548), regulatory (pETTm0549), and both subunits together (simultaneously and in their native order, pETTm0548/9) of TmAHAS-encoding genes, respectively (see Suppl. Fig. S1).
There is a significant difference between codon usage of E. coli and hyperthermophiles. To rectify the codon bias problem, E. coli strain Rosetta 2 BL21 (DE3) was used to supply tRNA for codons AUA, AGG, AGA, CUA, CCC, GGA, and CGG on a chloramphenicol-resistant plasmid. The recombinant AHAS encoding constructs were propagated in E. coli DH5α and subsequently transformed into E. coli BL21 (DE3) Rosetta 2 cells.
The E. coli Rosetta 2 cells harboring the recombinant plasmids were grown in a 4 L flask at 37 °C with shaking at 200 rpm until the OD600 reached 0.6–0.8. The cultures were then induced with 0.4 mM of IPTG. The induced cultures were then incubated at 30 °C overnight. The biomass of the recombinant strain was then harvested by centrifugation at 10,000g for 30 min at 4 °C using a RC6-plus centrifuge in a SLA-3000 rotor (Thermo Scientific, MA, USA). After collection and weighing, the cell-pellets were snap-frozen in liquid nitrogen and stored at −80 °C until use.
2.4. Purification of recombinant catalytic subunit
The purification steps were carried out at room temperature and under anaerobic conditions unless otherwise specified. SDS-PAGE and AHAS activity assay were used to determine the purity of the proteins at each step. Three different preparations were analyzed on the gel depending on the extent of treatment: the crude cell extract (CCE) resulted from lysis of cells in the lysis buffer, the cell-free extract (CFE) was the supernatant from centrifugation of CCE; and heat-treated crude cell extract (HTCCE) was the supernatant resulting from centrifugation of heat-treated CCE at 80 °C for 1 h. The cell lysis was achieved by thawing the cell pellet in lysis buffer. The suspension was incubated at room temperature for 90 min to promote the cell lysis and then 0.01 mg/ml DNase I and 5 mM MgCl2 were added. Incubation continued for another 30 min. The lysate was then heat-treated to denature the native host proteins. The lysate was clarified by centrifugation (10,000 x g for 30 min at 4 °C) and the supernatant was collected.
The catalytic subunit was purified by loading heat treated CFE on a DEAE-Sepharose column, and the AHAS active fractions were eluted at 275–370 mM of sodium chloride, which were pooled and loaded on a hydroxyapatite (HAP) column. The AHAS active fractions were eluted from HAP column at 180–230 mM sodium phosphate concentration. After HAP column the catalytic subunit was pure as judged by SDS-PAGE. Approximately 1 mg purified recombinant protein was obtained from each gram (wet weight) of the E. coli cells.
2.5. Purification of recombinant regulatory subunit
The recombinant regulatory (small) subunit was expressed mostly as insoluble inclusion bodies (IBs) under normal growth conditions. It was then purified by using denaturing IMAC followed by on column renaturation as described previously [41]. To solubilize the IBs, 5 M urea was incorporated into the composition of lysis buffer [10 mM Tris–HCl, 1 mM EDTA, 50 mM sodium chloride, 5% (v/v) glycerol, 0.5 mg/ml lysozyme, 1 mM SDT and 1 mM DTT, 1 mM TPP, and 10 µM FAD, pH 7.8] and buffer A (20 mM Tris–HCl, 0.5 M NaCl, 5 M urea, 10 mM imidazole, 5% glycerol, 1 mM SDT, and 1 mM DTT, pH 8.0). The cells were lysed using the same procedure as for the catalytic subunit.
After the heat precipitation step (80 °C for 1 h), the CFE was loaded on a Ni Sepharose™ High Performance column (GE Healthcare, QC, Canada) equilibrated with buffer A (20 mM Tris–HCl, 0.5 M NaCl, 5 M urea, 10 mM imidazole, 5% glycerol, 1 mM SDT, and 1 mM DTT, pH 8.0). On column refolding was achieved by applying a linear gradient of urea from 5 M (Buffer A) to 0 M (Buffer B: 20 mM Tris–HCl, 0.5 M NaCl, 10 mM imidazole, 5% glycerol, 1 mM SDT, and 1 mM DTT, pH 8.0), and then the recombinant protein was eluted by applying a gradient of imidazole from 10 mM (Buffer B) to 250 mM (Buffer C: 20 mM Tris–HCl, 0.5 M NaCl, 250 mM imidazole, 5% glycerol, 1 mM SDT, and 1 mM DTT, pH 8.0).
2.6. Enzyme activity assay
The AHAS activity assay procedure was a modification of the discontinuous colorimetric method of Singh et al. [42]. The AHAS activity was determined by measuring the production of acetolactate from pyruvate upon its decarboxylation to acetoin or diacetyl under high temperature and acidic condition. The products of decarboxylation would react with the guanidino groups of creatine under alkaline conditions creating a cherry red colored complex, which can be quantified [43]. The enzymatic reactions were carried out in 8 ml vials at 80 °C. The CFEs prepared from E. coli Rosetta 2 strain (the expression host) transformed with an empty blank pET30a vector were used as blank.
The assay mixture (1 ml final volume) containing 100 mM sodium phosphate (pH 7.0), 10 mM MgCl2, 2.5 mM TPP, 50 mM sodium pyruvate, and 10 µM FAD was pre-heated by incubation at 80 °C water bath for 4 min before starting the reaction by adding the enzyme. The final concentration of 2-acetolactate was determined using a calibration curve prepared by linear regression of known concentrations of acetoin processed under same assay conditions. A linear correlation between the activity and the amount of protein in the assay was determined. The assay vials were incubated in the water bath (80 °C) for 30 min after adding the enzyme, and the reaction was quenched by adding sulfuric acid (50% v/v) to a final concentration of 0.85%. The assay solution was then incubated at 60 °C for 15 min to allow transformation of the acetolactate to acetoin (chemical decarboxylation). The amount of the acetoin produced was quantified by adding 0.5% (w/v) creatine (final concentration 0.17%) and 5% (w/v) 1-naphthol in 4 N NaOH (final concentration 1.7%). The mixture was incubated at 60 °C for 15 min and then at room temperature for another 15 min with frequent mixing followed by centrifugation for 2 min at 10,000 x g. The enzyme activity was determined by measuring the absorbance change at 525 nm using a Genesys 10 UV–vis spectrophotometer (Thermo Scientific, MA, USA). One unit of activity was defined as the formation of 1 µmol of acetolactate per min under these conditions.
2.7. Biochemical and biophysical characterization
To investigate the pH dependency, the AHAS activity was assayed at different pH values ranging from 4.0 to 11.0. The pH values expressed throughout this manuscript were adjusted and measured at room temperature. In each case, assays were carried out at 80 °C, using 100 mM buffers as described previously. For pH values between 4 and 5.6, sodium acetate (pKa= 4.8, ΔpKa/°C=0.0002) was used. Sodium phosphate (pKa=7.2, ΔpKa/°C=−0.0028) was used for pH values 6.0, 7.0, and 7.5. EPPS (pKa=8.0, ΔpKa/°C=−0.015) covered pH values 7.5, 8.0, 8.5 and 9.0 and glycine buffer (pKa= 9.6, ΔpKa/°C=−0.0025) was used for the pH values 9.0, 9.5, 10.0, and 10.5. Finally for the pH points of 10.0, 10.5, and 11.0 CAPS buffer (pKa=10.4, ΔpKa/°C=−0.009) was used.
To determine the steady-state kinetic parameters, enzyme assays were performed at optimum pH. The kinetic parameters were determined for pyruvate (5–125 mM), TPP (0.05–4 mM), and FAD (0.05–40 µM) by applying various concentrations of each component while keeping the concentrations of other assay components at saturation. The kinetic parameters were calculated from the best fit of the results to the Michaelis–Menten equation by non-linear regression using SigmaPlot® software (SYSTAT Software Inc., CA, USA).
The oxygen sensitivity of each activity was determined by exposing an enzyme aliquot to ambient atmosphere at 4 ºC by gentle stirring and comparing the enzyme activity at different time courses with the control kept under anaerobic conditions. Both of the enzyme samples were protected from light through the experiments. The temperature dependence of enzyme was determined by measuring the activity at different temperatures ranging from 30 to 95 °C. Thermal stability of the enzyme was determined by incubation of an anaerobic enzyme preparation and determining the residual activity at different time points compared to unheated control. The light sensitivity of the recombinant enzyme was also tested. TmAHAS aliquots were incubated under anaerobic conditions with one exposed to light and the other vial protected from the light by aluminum foil.
2.8. Reconstitution of TmAHAS
The purified catalytic and regulatory subunits of recombinant TmAHAS were combined in various molar ratios, incubated both at low temperature (ice bath) or heat treated (80 ºC) for various periods of time and then were loaded on a size-exclusion chromatography column (2.6×60 cm) of HiLoad Superdex-200 (GE healthcare, QC, Canada) pre-equilibrated with buffer C (50 mM Tris–HCl, 5% glycerol, 100 mM KCl, pH 7.8) at a flow rate of 2 ml/min. The enzyme activity of the reconstituted holoenzyme was then determined.
2.9. Other methods
When necessary the anaerobic procedure were followed as described previously [44]. The protein concentration was determined using the Bradford dye-binding assay [45] with bovine serum albumin (BSA) as the standard. The apparent Mr of the purified catalytic and regulatory subunits were estimated by loading the concentrated individual proteins on a size exclusion chromatography column (2.6×60 cm) of HiLoad Superdex-200 (GE healthcare, QC, Canada) pre-equilibrated with buffer C (50 mM Tris–HCl, 5% glycerol, 100 mM KCl, pH 7.8) at the flow rate of 2 ml/min. The following standards from Pharmacia protein standard kit (Pharmacia, NJ, USA) were applied to the column: blue dextran (2,000,000 Da), thyroglobulin (669,000 Da), ferritin (440,000 Da), catalase (232,000 Da), aldolase (158,000 Da), bovine serum albumin (67,000 Da), ovalbumin (43,000), chymotrypsinogen A (25,000) and ribonuclease A (13,700).
3. Results
3.1. AHAS in Thermotogales
Genes encoding the enzymes involved in the biosynthesis of BCAAs were searched from accessible genome sequences of various Thermotogales. The ilv operon was found to be present in genomes of T. maritima (GenBank accession number NC_000853), Thermotoga thermarum (GenBank accession number NC_015707), T. neapolitana (GenBank accession number NC_011978), Thermotoga naphthophila (GenBank accession number NC_013642), Thermotoga petrophila (GenBank accession number CP000702), and Thermotoga sp. strain RQ2 (GenBank accession number NC_010483) (see Ref. [46, Table 1]). Only the genome of Thermotoga lettingae (GenBank accession number NC_009828) had no recognizable ilv operon, which is in accordance with organism’s auxotrophy for leucine, valine, and isoleucine [47]. The CFEs of T. maritima, T. hypogea, and T. neapolitana contained 100±9, 17.4±2, and 18±1.4 mU/mg of AHAS activity, respectively.
The sequences of the genes encoding AHASs from different microorganisms are quite conserved, and more than 90% amino acid sequence identity is observed among Thermotogales (Suppl. Fig. S2). The only exceptions are the catalytic and regulatory subunits of T. thermarum exhibiting about 47% overall amino acid identity to those of other Thermotoga species (Suppl. Fig. S2). Overall the bacterial and archaeal catalytic subunits can be divided into two different groups (Suppl. Fig. S2A); the regulatory subunits seem not to follow such a trend (Suppl. Fig. S2B).
The catalytic and regulatory subunits of AHAS in bacteria are encoded by ilvB and ilvN, respectively. In T. maritima (and most other Thermotogae), the first gene in the ilv operon is the gene encoding catalytic subunit (ilvB) of AHAS, followed by the gene encoding regulatory subunit (ilvN) and then other genes involved in biosynthesis of the branched chain amino acids in the following order: ketol-acid reductoisomerase (ilvC), dihydroxy-acid dehydratase (ilvD), 2-isopropylmalate synthase (leuA), 3-isopropylmalate dehydratase large and small subunits (leuC and leuD), and 3-isopropylmalate dehydrogenase (leuB) (Fig. 2).
Fig. 2.
Gene organization of the ilv gene cluster in T. maritima. The filled arrows indicate the genes encoding enzymes involved in BCAA biosynthesis. The black arrows are the hypothetical genes encoding the catalytic and regulatory subunits of AHAS. Abbreviations: ilvB: catalytic subunit of acetohydroxyacid synthase (AHASL); ilvN, regulatory subunit of acetohydroxyacid synthase (AHASS); ilvC, ketol-acid reductoisomerase (KARI); ilvD ; dihydroxy-acid dehydratase (DHAD); leuA, 2-isopropylmalate synthase (IPMS); leuC 3-isopropylmalate dehydratase large subunit (IPMDL); and leuD, 3-isopropylmalate dehydratase small subunit (IPMDS).
3.2. Expression of AHAS-encoding genes
The construct pETTm0548 produced soluble recombinant protein. The addition of FAD, TPP, and MgCl2 either to the culture medium or the lysis buffer had no noticeable effect on the expression levels or the solubility of the heterologously expressed catalytic subunit. The regulatory subunit expressed from pETTm0549, on the other hand, showed low solubility and mainly was aggregated in the insoluble fractions. In the case of the construct pETTm0548/9, only the catalytic subunit was expressed as a soluble protein, though the yield of the regulatory subunit was very low, suggesting the possible proteolysis or incomplete translation of the subunit (Ref. [46, Fig. 1]). Altering the expression conditions, namely the temperature and IPTG concentration (0.1–0.5 mM), growing the recombinant strain in the presence of different glycerol concentrations (0–40%), incorporation of TPP, FAD, and MgCl2 (together or individually), as well as expression and cell lysis in the presence of detergents did not improve solubility of the small subunit (data not shown).
3.3. Purification of recombinant TmAHAS subunits
Heat-induced precipitation (heat-treatment) has been used for purification of various recombinant thermostable proteins. During heat-treatment, majority of mesophilic host proteins are denatured and aggregated, rendering the recombinant thermostable protein in soluble fraction [48] (see Ref. [46, Table 2]).
The recombinant catalytic subunit was purified approximately 30-fold to purity (Fig. 3A) with specific activity of 160±27 U/mg (Table 1). The effects of growth temperature (Ref. [46, Figs. 2–4]) and heat-treatment temperatures on yield of the soluble protein and enzyme activity were also optimized (Ref. [46, Figs. 5 and 6]). Incubation of CCEs at either 70 or 80 ºC resulted in enhanced AHAS activity with the highest activity achieved after 1 h of incubation, longer treatments resulted in reduced activity.
Fig. 3.
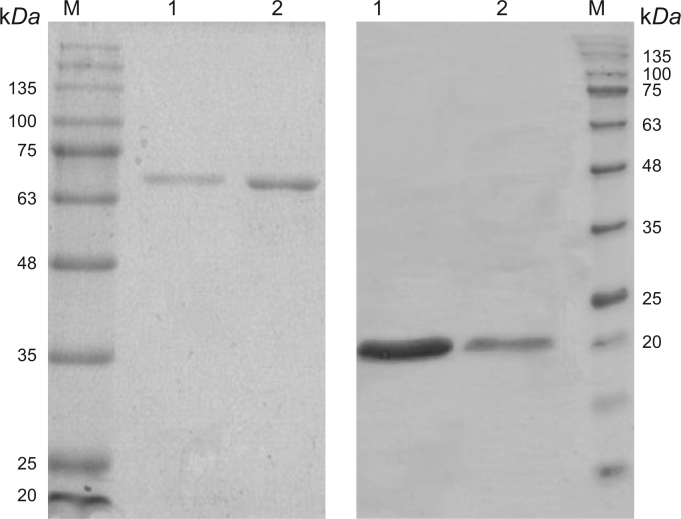
SDS-PAGE analysis of the purified proteins. (A) The catalytic subunit on 12.5% SDS-PAGE and with lane M, pre-stained protein ladder, Lane 1, 2 µg of the purified protein; lane 2, 4 µg of the purified protein. (B) The regulatory subunit of TmAHAS on 15% SDS-PAGE with Lane 1, 8 µg of the purified protein; lane 2, 4 µg of the purified protein; lane M, pre-stained protein ladder.
Table 1.
Purification of recombinant catalytic subunit of TmAHAS.a
| Step | Total protein (mg) | Total activityb (Units) | Specific activity (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Crude extract | 1175 | 6345 | 5.4 | 1 | 100 |
| Heat-treatment | 138 | 4882 | 31.1 | 5.8 | 76.9 |
| DEAE | 53 | 3808 | 70.8 | 13.2 | 60.1 |
| HAP | 21 | 3302 | 163 | 30.2 | 52.1 |
CFE was prepared from 20 g (wet weight) of the recombinant E. coli strain pETTm0548.
Expressed as µmoles of acetolactate produced per min per milligram of the enzyme.
Although expressed as His-tagged recombinant proteins, the purity of the catalytic subunit was improved considerably when it was purified by loading on DEAE and HAP columns instead of IMAC columns (data not shown), resulting in approximately 1 mg purified recombinant protein from each gram (wet weight) of E. coli cells. A typical purification of the catalytic subunit is summarized in Table 1.
Since small subunit has no enzymatic activity, its purity was determined by SDS-PAGE analysis of the chromatography fractions. The small subunit was expressed mostly as insoluble inclusion bodies as judged by SDS-PAGE analysis (data not shown), which was expectable based on a previous report pertaining to expression of the same protein [41]. The re-solubilization of the small subunit was then achieved by purification under denaturing conditions using IMAC and subsequent on-column refolding of the denatured protein (Fig. 3B).
3.4. Molecular weights of the recombinant proteins
The recombinant catalytic subunit (calculated molecular mass was 65,497 Da and estimated molecular mass on SDS-PAGE was 66,398±7,700 Da) was eluted as a single peak and corresponded to molecular mass of 156,830±6,200 Da, indicating a dimeric structure. The regulatory subunit (calculated molecular mass was 20,508 Da and estimated molecular mass on SDS-PAGE gel 22,210±3,540 Da) had a native molecular mass of 37,700±413 Da determined using size-exclusion chromatography, suggesting a dimeric structure for the small subunit. Increasing the salt concentration of the elution buffers from 150 to 500 mM had no effect on protein elution profile.
3.5. Catalytic properties
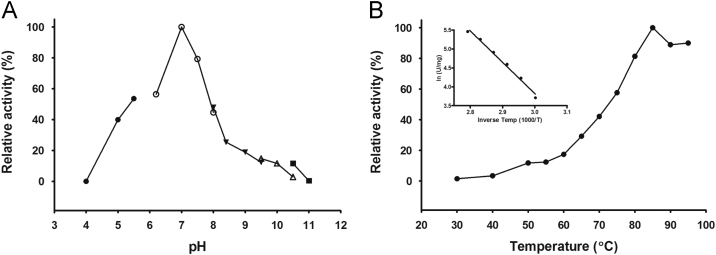
The purified AHAS showed maximum activity at pH 7.0 in sodium phosphate buffer (Fig. 4A). The steady-state kinetic parameters of the catalytic subunit were determined for substrates pyruvate, TPP, and FAD at pH 7.0 and 80 °C (Table 2). It showed the highest specific activity among any native or recombinant AHAS reported so far (see Table 3). The purified enzyme showed the highest activity at 85 °C, which decreased at higher temperatures. Accordingly, the corresponding Arrhenius plot showed a transition point at 85 °C. An Eact value of 69 kJ/mol was calculated for the enzyme over a temperature range of 60–85 °C (Fig. 4B).
Fig. 4.
Effect of pH and temperature on AHAS activity. (A) To determine the optimal pH, the AHAS activity was determined in ● sodium acetate buffer (pH values 4.0, 5.0, and 5.5), ○ sodium phosphate buffer (pH 6.2, 7.0, 7.5, and 8.0), ▼ EPPS (pH 8.0, 8.4, 9.0, and 9.5), ∆ glycine (pH 9.5 and 10), and ■ CAPS (pH 10.5 and 11.0). The relative activities of 100% equals to the highest measured specific activity (114 U/mg) at 80 ºC in sodium phosphate buffer, pH 7.0. (B) To determine the temperature dependence, the reaction vials were incubated at each temperature for 4 min before starting the reaction by adding the enzyme. The inset shows the Arrhenius plot based on the linear part of the plot B (temperatures 60–85 ºC). The relative activity of 100% equals to the highest measured specific activity (235 U/mg) at 85 ºC.
Table 2.
Apparent kinetic parameters for AHAS activity of the catalytic subunit.
| Substrate | Apparent Km (µM) | Apparent Vmax (U/mg) | kcat (s−1) |
|---|---|---|---|
| Pyruvate | 16,400±2000 | 246±7 | 98 |
| TPP | 57±6.0 | 242±4 | 97 |
| FAD | 0.15±0.07 | 134±11 | 54 |
Table 3.
Catalytic and structural properties of AHAS from different organisms.a
| Organisms |
Subunits |
ApparentKmfor |
Specific activity (U/mg)b | Enzyme | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| Catalytic | Regulatory | Pyruvate (mM) | TPP (µM) | Mg2+(µM) | FAD (µM) | ||||
| Mycobacterium avium | 66 | 18 | 2±0.2 | 7.5±0.4 | NR | 0.1 | 4 | R | [25] |
| Haemophilus influenza | 63 | NR | 9.2 | 13.6 | NR | NR | 1.5 | R | [28] |
| Shigella sonnei | 65 | NR | 8.1 | 0.01 | 180 | 0.12 | R | [27] | |
| Mycobacterium tuberculosis | NR | NR | 1.6±0.4 | 1.4±0.3 | NR | NR | 4.6 | R | [24] |
| Escherichia coli Isozyme I | 60.4 | 11.1 | 3.7 | NR | NR | 760 | 0.4 | R | [11], [64] |
| Escherichia coli Isozyme II | 59.3 | 9.6 | 5.0±0.5 2.6 | 0.65±0.03 1.1 | 0.01±0.002 3.8 | 0.17±0.04 NR | 20 52.7 | R | [65,17] |
| Escherichia coli Isozyme III | 63.0 | 17.5 | 11.5±1.4 | 18±3 | 3300±800 | 2.2±0.5 | 2.6 | R | [18], [19] |
| Saccharomyces cerevisiae | ~70 | 40 | 8.6±1.4 | 110 | 280 | 0.3 | 49±1.8 | R | [16] |
| Arabidopsis thalianac | 61 | NR | 11.7±0.6 | 25.3±1.4 | 198±19 | 1.5±0.22 | 7.9 | R | [29], [66] |
| Nicotiana tabacumc | NR | NR | 8.1–12.8 | 80-210 | NR | 1.4–2.6 | 2.8–3.4 | R | [67] |
| Bacillus stearothermophilusc | 62.4 | 18.7 | 8.8±1.2 | 5.5±0.8 | 20±3 | NR | 9.2 | R | [35] |
| Corynebacterium glutamicum | 67 | 15.4 | 8.4 | NR | NR | NR | 0.37 | N | [68], [69] |
| Methanococcus aeolicus | 65 | 19 | 6.8 | 1.6 | 300 | 1.3 | 39.3 | Nd R | [33], [54] |
| Haloferax volcanii | 50 | NR | 25.5±5 | 8.7±0.9 | NR | NR | 0.005 | N | [34] |
| Salmonella typhimurium | 59 | 9.7 | 10.6±0.7 | 1.5±0.2 | 22±4 | 0.8±0.1 | 25.3 | R | [20] |
| Thermotoga maritima | 65.5 | 20 | 16.4±1.9 | 56.5±5.6 | NR | 0.15±0.07 | 134±11 | R | This study |
NR: not reported; R, recombinant; N, native enzyme.
Expressed as µmoles of acetolactate produced per min per milligram of enzyme.
The kinetics parameters are only reported for the catalytic subunit.
The kinetic parameters are reported for the native protein.
The AHAS activity was dependent on the ionic strength of the buffer and diminished considerably (approximately 80%) when assayed in buffers with less than 100 mM concentrations. At concentrations higher than 100 mM the activity levels were quite similar (up to 500 mM was tested), suggesting it might be needed to maintain the functional oligomeric state of the enzyme. The gel-filtration of the purified catalytic subunit at different salt concentrations (150–500 mM) showed no difference in elution profile.
TmAHAS was not oxygen sensitive; however stirring had detrimental effects on activity. When stirred, both aerobic and anaerobic CFE aliquots lost major portion of their activity at almost the same rate resulting in a time required for the loss of its 50% activity (t½) to be about 18 h (data not shown). The purified recombinant AHAS had a t½-value of 30 min and 110 min when moderately stirred in the presence and absence of oxygen, respectively (Ref. [46, Fig. 7A]). When the purified enzyme was kept on ice without stirring and under anaerobic conditions, it was fairly stable and lost only 35–40% activity after 5 days (Ref. [46, Fig. 7]).
There are few reports on light sensitivity of the anabolic AHASs due to “FAD-mediated photo oxidation” [5], [17]. The enzyme assay on exposed and non-exposed preparations showed no difference in the specific activities over 8 h (data not shown). Therefore, it seemed that light exposure had no negative effect on the activity of the TmAHAS.
3.6. Reconstitution of holoenzyme
The purified catalytic and regulatory subunits were mixed under anaerobic conditions and with different molar ratios. The reconstitution of the holoenzyme had evident impact on AHAS activity, which resulted in a 1.5-fold increase in the AHAS activity with a 1:5 ratio of catalytic to regulatory subunit. No further increase was observed by increasing ratios of catalytic to regulatory subunit up to 1:50 (Ref. [46, Fig. 8]).
When the purified large and small subunits were loaded on size-exclusion column separately, each eluted out at their expected elution volumes (Ref. [46, Fig. 8A and B]), corresponding to 160 kDa and 50 kDa, respectively. When catalytic and regulatory subunits were mixed together in a molar ratio of 1:10 and pre-incubated at room temperature for 30 min before being loaded on size-exclusion column, the peak corresponding to catalytic subunit was disappeared and a new peak with higher molecular mass appeared in a position equivalent to an apparent molecular size of the holoenzyme at approximately 230 kDa (Ref. [46, Fig. 8C]), which was suggestive of reconstitution of holoenzyme in higher oligomeric state as expected. It seems then the holoenzyme is composed of two catalytic and two regulatory subunits (α2β2). This is in accordance with findings on some mesophilic AHASs including AHAS III of E. coli [18].
4. Discussion
Thermotogales are able to use both carbohydrates (simple and complex) and peptides as sources of carbon and energy [49], and many of them are capable of de novo synthesis of amino acids required for growth. Anabolic AHASs are required for BCAA biosynthesis. Although they are relatively well studied in plants and a number of mesophilic bacterial pathogens (Table 3), but none has been characterized from any hyperthermophilic microorganism. T. maritima is shown not to require any BCAA in the growth medium, suggesting it is equipped with the enzymes required for the biosynthesis of these amino acids [50]. The genes encoding its AHAS were successfully cloned and overexpressed in E. coli. The purified catalytic subunit of TmAHAS had the highest AHAS specific activity compared to any other AHAS studied so far (Table 3). The regulatory subunit was expressed mostly as inclusion bodies, which is similar to mesophilic ones [41], [51], [52]. However, the inclusion bodies became soluble after the denaturation and renaturation steps.
The activity of the recombinant TmAHAS increased significantly following heat-treatment (Ref. [46, Fig. 6]), indicating the thermal activation of the recombinant protein. Thermal activation has been observed in other heterologously expressed hyperthermophilic proteins. A typical example is the alcohol dehydrogenase (AdhC) from the hyperthermophilic archaeon P. furiosus expressed in E. coli. In this case the homotetrameric recombinant protein does not attain the correct configuration unless a heat-treatment step is applied [53].
Similar to AHASs studied from different organisms which have pH optima between neutral to basic (pH values 7–9), the optimal pH for the activity of the recombinant TmAHAS was found to be 7.0 (Fig. 4A). Although there are reports of oxygen sensitivity of AHAS isolated from the mesophilic archaeon M. aeolicus [33], [54], the enzyme from T. maritima seems to be tolerant towards oxygen. Instability of the native and recombinant AHAS has been reported specially for the fungal anabolic AHASs [5]. The oxygen sensitivity of the AHAS in M. aeolicus, cannot be explained [54], but possibly it is related to the enzyme's high cysteine contents (12 cysteine residues compared to 3 residues in TmAHAS). The optimal temperature of 85 °C found in this study (Fig. 4B) is the highest reported for any AHAS so far. TmAHAS was thermostable when kept under anaerobic conditions without stirring ([46, Fig. 7B]). Further study is required to understand the mechanism of TmAHAS inactivation under stirring.
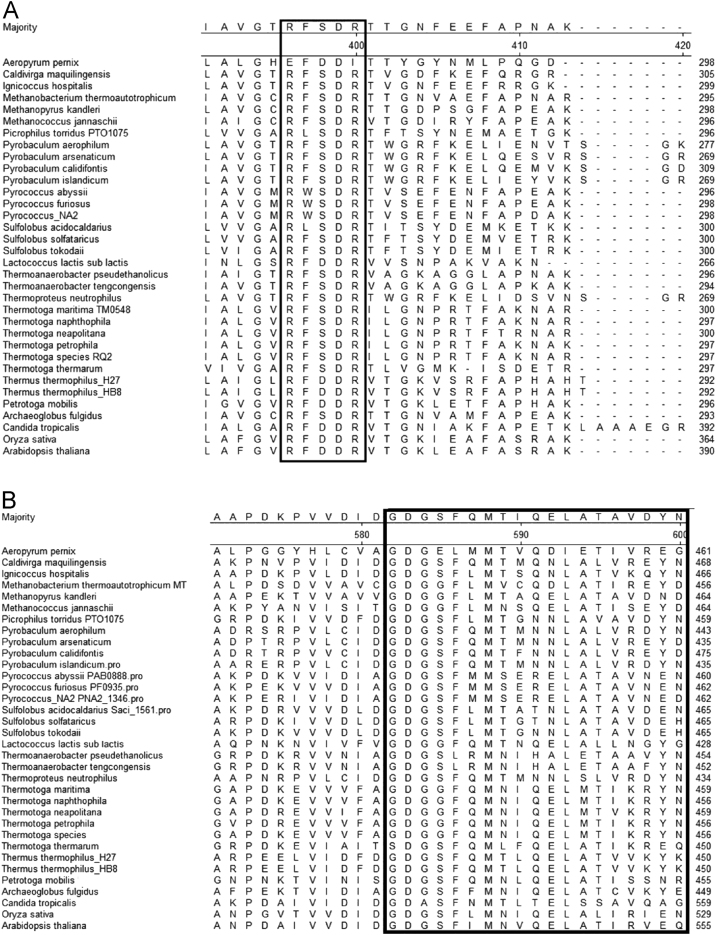
The FAD-dependence is a perplexing feature of the anabolic AHASs, as the enzyme is not catalyzing any redox reaction. The anabolic AHASs contain one molecule of FAD per catalytic site [12]. Based on experiments with FAD-analog molecules [13], it has been suggested that the role of the FAD is merely structural (and not catalytic). This may be further supported by the fact that the catabolic version of the enzyme (ALS) is completely FAD-independent, despite catalyzing the same reaction and having similar crystal structures [4], [55]. A putative FAD-binding motif (RFDDR) has been shown to be associated with FAD-dependence in anabolic AHASs from different mesophilic organisms [56]. The motif is generally conserved as RFSDR in Thermotogales, and RWSDR in Thermococcales but only in members of the genus Pyrococcus that have the gene cluster (Fig. 5A). The apparent Km of FAD for TmAHAS (Table 2) was close to that of most other AHASs characterized (See Table 3). The catalytic subunits of all AHASs contain a typical TPP-binding motif (GDGX24–26N) (Fig. 5B). This highly conserved motif is a common feature of all TPP-dependent enzymes [57].
Fig. 5.
Conserved motifs in AHASs. The highly conserved motifs in AHASs are boxed: (A) The FAD-binding motif (boxed). (B) and TPP-binding motif (boxed). The AHAS amino acid sequences of different thermophilic and mesophilic organisms were aligned using MegAlign software (Lasergene, DNAStar, Madison, USA).
In most anabolic AHASs studied so far, mixing the individually expressed catalytic and regulatory subunits resulted in reconstitution of the holoenzyme with full activity. The reconstitution process is reported to be a rapid and cooperative process and follows the hyperbolic saturation kinetics with a 1:1 stoichiometry [5], [25], [35]. The impact of the regulatory subunit on full activation of the enzyme is usually large but diverse, depending on the source of the enzyme. In case of the AHAS III from E. coli the catalytic subunit alone shows only about 5% of the activity of the holoenzyme [18]. Reconstituted yeast AHAS has an increase of about 7–10-fold in the specific activity [16]. Reconstituted holoenzyme of A. thaliana showed at least 2–3-folds increase over catalytic subunit alone. For all AHASs studied, the cofactor requirement and substrate specificity of the catalytic subunit is similar to the holoenzyme [5], [58].
The enzymatically active unit of the recombinant catalytic subunit of TmAHAS seems to be a dimer. This is in agreement with previous finding that the minimal functional unit of TPP-dependent enzyme has to be a dimer to accommodate the active site in the interface between the two subunits [14], [18], [59], [60]. The recombinant catalytic subunits of AHAS isozyme II from E. coli is such an example [17]. Mixing the purified catalytic and regulatory subunits of TmAHAS led to increased AHAS activity (Ref. [46, Fig. 7]). Such reconstitution of the holoenzyme also resulted in higher oligomeric states, which is similar to those in mesophilic AHASs such as E. coli AHAS III [18]. Because of high propensity of the regulatory subunit for aggregation, it has been suggested that it is involved in the process of oligomerization and enzyme reconstitution [25].
Most of the commercially available amino acids are synthesized through microbial processes, largely through fermentation using the genetically improved strains of E. coli or C. glutamicum [61], [62], [63]. Considering the key role of the AHAS in biosynthesis of the BCAAs, these enzymes are of great importance in the amino acid fermentation industry. The characterization of the TmAHAS may as well provide great potentials towards the metabolic engineering of amino-acid producing organisms for the amino-acid fermentation industry.
Acknowledgment
The authors gratefully acknowledge the efforts of Mr. Alton Wong on growth of the native and recombinant microorganisms.
Author contribution: Experiments were designed by Mohammad S. Eram and Kesen Ma. The experiments were performed by Mohammad S. Eram, Benozir Sarafuddin, and Frank Gong. The paper was prepared by Mohammad S. Eram and Kesen Ma.
Funding: This work was supported by research grant from the Natural Sciences and Engineering Research Council of Canada (NSERC) RGPIN-227233-2009 to K.M.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi:10.1016/j.bbrep.2015.08.014.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Stetter K. History of discovery of the first hyperthermophiles. Extremophiles. 2006;10:357–362. doi: 10.1007/s00792-006-0012-7. [DOI] [PubMed] [Google Scholar]
- 2.Wagner I.D., Wiegel J. Diversity of thermophilic anaerobes. Ann. NY Acad. Sci. 2008;1125:1–43. doi: 10.1196/annals.1419.029. [DOI] [PubMed] [Google Scholar]
- 3.Atomi H., Sato T., Kanai T. Application of hyperthermophiles and their enzymes. Curr. Opin. Biotechnol. 2011;22:618–626. doi: 10.1016/j.copbio.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Duggleby R.G., McCourt J.A., Guddat L.W. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 2008;46:309–324. doi: 10.1016/j.plaphy.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Duggleby R.G., Pang S.S. Acetohydroxyacid synthase. J. Biochem. Mol. Biol. 2000;33:1–36. [Google Scholar]
- 6.Umbarger H.E., Brown B. Isoleucine and valine metabolism in Escherichia coli. J. Biol. Chem. 1958;233:1156–1160. [PubMed] [Google Scholar]
- 7.Radhakrishanan A.N., Snell E.E. Biosynthesis of valine and isoleucine. II. Formation of α-acetolactate and α-aceto-alpha-hydroxybutyrate in Neurospora crassa and Escherichia coli. J. Biol. Chem. 1960;235:2316–2321. [PubMed] [Google Scholar]
- 8.Grimminger H., Umbarger H.E. Acetohydroxy acid synthase I of Escherichia coli: purification and properties. J. Bacteriol. 1979;137:846–853. doi: 10.1128/jb.137.2.846-853.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Störmer F.C. Evidence for induction of the 2,3-butanediol-forming enzymes in Aerobacter aerogenes. FEBS Lett. 1968;2:36–38. doi: 10.1016/0014-5793(68)80094-8. [DOI] [PubMed] [Google Scholar]
- 10.Pang S.S., Duggleby R.G., Schowen R.L., Guddat L.W. The crystal structures of Klebsiella pneumoniae acetolactate synthase with enzyme-bound cofactor and with an unusual intermediate. J. Biol. Chem. 2004;279:2242–2253. doi: 10.1074/jbc.M304038200. [DOI] [PubMed] [Google Scholar]
- 11.Weinstock O., Sella C., Chipman D.M., Barak Z. Properties of subcloned subunits of bacterial acetohydroxy acid synthases. J. Bacteriol. 1992;174:5560–5566. doi: 10.1128/jb.174.17.5560-5566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chipman D.M., Duggleby R.G., Tittmann K. Mechanisms of acetohydroxyacid synthases. Curr. Opin. Chem. Biol. 2005;9:475–481. doi: 10.1016/j.cbpa.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 13.McCourt J.A., Duggleby R.G. Acetohydroxyacid synthase and its role in the biosynthetic pathway for branched-chain amino acids. Amino Acids. 2006;31:173–210. doi: 10.1007/s00726-005-0297-3. [DOI] [PubMed] [Google Scholar]
- 14.Gedi V., Yoon M.-Y. Bacterial acetohydroxyacid synthase and its inhibitors – a summary of their structure, biological activity and current status. FEBS J. 2012;279:946–963. doi: 10.1111/j.1742-4658.2012.08505.x. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y.T., Duggleby R.G. Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochemistry. 2001;40:6836–6844. doi: 10.1021/bi002775q. [DOI] [PubMed] [Google Scholar]
- 16.Pang S.S., Duggleby R.G. Expression, purification, characterization, and reconstitution of the large and small subunits of yeast acetohydroxyacid synthase. Biochemistry. 1999;38:5222–5231. doi: 10.1021/bi983013m. [DOI] [PubMed] [Google Scholar]
- 17.Hill C.M., Pang S.S., Duggleby R.G. Purification of Escherichia coli acetohydroxyacid synthase isoenzyme II and reconstitution of active enzyme from its individual pure subunits. Biochem. J. 1997;327:891–898. doi: 10.1042/bj3270891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vyazmensky M., Sella C., Barak Ze, Chipman D.M. Isolation and characterization of subunits of acetohydroxy acid synthase isozyme III and reconstitution of the holoenzyme. Biochemistry. 1996;35:10339–10346. doi: 10.1021/bi9605604. [DOI] [PubMed] [Google Scholar]
- 19.Sella C., Weinstock O., Barak Z., Chipman D.M. Subunit association in acetohydroxy acid synthase isozyme III. J. Bacteriol. 1993;175:5339–5343. doi: 10.1128/jb.175.17.5339-5343.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schloss J.V., Van Dyk D.E., Vasta J.F., Kutny R.M. Purification and properties of Salmonella typhimurium acetolactate synthase isozyme II from Escherichia coli HB101/pDU9. Biochemistry. 1985;24:4952–4959. doi: 10.1021/bi00339a034. [DOI] [PubMed] [Google Scholar]
- 21.Keilhauer C., Eggeling L., Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H.-L., Wang P.-Y., Wu C.-M., Hwang D.-C., Chang H.-Y. Cloning, sequencing and heterologous expression of a Klebsiella pneumoniae gene encoding an FAD-independent acetolactate synthase. Gene. 1992;117:125–130. doi: 10.1016/0378-1119(92)90500-o. [DOI] [PubMed] [Google Scholar]
- 23.Grandoni J.A., Marta P.T., Schloss J.V. Inhibitors of branched-chain amino acid biosynthesis as potential antituberculosis agents. J. Antimicrob. Chemother. 1998;42:475–482. doi: 10.1093/jac/42.4.475. [DOI] [PubMed] [Google Scholar]
- 24.Zohar Y., Einav M., Chipman D.M., Barak Z. Acetohydroxyacid synthase from Mycobacterium avium and its inhibition by sulfonylureas and imidazolinones. Biochim. Biophys. Acta (BBA): Proteins Proteom. 2003;1649:97–105. doi: 10.1016/s1570-9639(03)00160-2. [DOI] [PubMed] [Google Scholar]
- 25.Choi K.-J., Yu Y.G., Hahn H.G., Choi J.-D., Yoon M.-Y. Characterization of acetohydroxyacid synthase from Mycobacterium tuberculosis and the identification of its new inhibitor from the screening of a chemical library. FEBS Lett. 2005;579:4903–4910. doi: 10.1016/j.febslet.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 26.Singh V., Chandra D., Srivastava B.S., Srivastava R. Biochemical and transcription analysis of acetohydroxyacid synthase isoforms in Mycobacterium tuberculosis identifies these enzymes as potential targets for drug development. Microbiology. 2011;157:29–37. doi: 10.1099/mic.0.041343-0. [DOI] [PubMed] [Google Scholar]
- 27.Lim W.-M., Baig I.J., La I.J., Choi J.-D., Kim D.-E., Kim S.-k, Hyun J.-W., Kim G., Kang C.-H., Kim Y.J., Yoon M.-Y. Cloning, characterization and evaluation of potent inhibitors of Shigella sonnei acetohydroxyacid synthase catalytic subunit. Biochim. Biophys. Acta (BBA): Proteins Proteom. 2011;1814:1825–1831. doi: 10.1016/j.bbapap.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Gedi V., Moon J.-Y., Lim W.-M., Lee M.-Y., Lee S.-C., Koo B.-S., Govindwar S., Yoon M.-Y. Identification and characterization of inhibitors of Haemophilus influenzae acetohydroxyacid synthase. Enzyme Microb. Technol. 2011;49:1–5. doi: 10.1016/j.enzmictec.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Singh B., Schmitt G., Lillis M., Hand J.M., Misra R. Overexpression of acetohydroxyacid synthase from arabidopsis as an inducible fusion protein in Escherichia coli: production of polyclonal antibodies, and immunological characterization of the enzyme. Plant Physiol. Biochem. 1991;97:657–662. doi: 10.1104/pp.97.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon M.-Y., Hwang J.-H., Choi M.-K., Baek D.-K., Kim J., Kim Y.-T., Choi J.-D. The active site and mechanism of action of recombinant acetohydroxy acid synthase from tobacco. FEBS Lett. 2003;555:185–191. doi: 10.1016/s0014-5793(03)01177-3. [DOI] [PubMed] [Google Scholar]
- 31.Pue N., Guddat L.W. Acetohydroxyacid synthase: a target for antimicrobial drug discovery. Curr. Pharm. Des. 2014;20:740–753. doi: 10.2174/13816128113199990009. [DOI] [PubMed] [Google Scholar]
- 32.Zhou Q., Liu W., Zhang Y., Liu K.K. Action mechanisms of acetolactate synthase-inhibiting herbicides. Pestic. Biochem. Physiol. 2007;89:89–96. [Google Scholar]
- 33.Xing R., Whitman W.B. Purification and characterization of the oxygen-sensitive acetohydroxy acid synthase from the archaebacterium Methanococcus aeolicus. J. Bacteriol. 1994;176:1207–1213. doi: 10.1128/jb.176.5.1207-1213.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vyazmensky M., Barak Z., Chipman D.M., Eichler J. Characterization of acetohydroxy acid synthase activity in the archaeon Haloferax volcanii. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 2000;125:205–210. doi: 10.1016/s0305-0491(99)00170-4. [DOI] [PubMed] [Google Scholar]
- 35.Porat I., Vinogradov M., Vyazmensky M., Lu C.-D., Chipman D.M., Abdelal A.T., Barak Ze. Cloning and characterization of acetohydroxyacid synthase from Bacillus stearothermophilus. J. Bacteriol. 2004;186:570–574. doi: 10.1128/JB.186.2.570-574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huber R., Langworthy T.A., König H., Thomm M., Woese C.R., Sleytr U.B., Stetter K.O. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 °C. Arch. Microbiol. 1986;144:324–333. [Google Scholar]
- 37.Yang X., Ma K. Characterization of a thioredoxin-thioredoxin reductase system from the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 2010;192:1370–1376. doi: 10.1128/JB.01035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fardeau M.L., Ollivier B., Patel B.K.C., Magot M., Thomas P., Rimbault A., Rocchiccioli F., Garcia J.L. Thermotoga hypogea sp. nov., a xylanolytic, thermophilic bacterium from an oil-producing well. Int. J. Syst. Bacteriol. 1997;47:1013–1019. doi: 10.1099/00207713-47-4-1013. [DOI] [PubMed] [Google Scholar]
- 39.Yang X., Ma K. Purification and characterization of an NADH oxidase from extremely thermophilic anaerobic bacterium Thermotoga hypogea. Arch. Microbiol. 2005;183:331–337. doi: 10.1007/s00203-005-0777-6. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J., Russell D.W. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring: 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 41.Petkowski J.J., Chruszcz M., Zimmerman M.D., Zheng H., Skarina T., Onopriyenko O., Cymborowski M.T., Koclega K.D., Savchenko A., Edwards A., Minor W. Crystal structures of TM0549 and NE1324—two orthologs of E. coli AHAS isozyme III small regulatory subunit. Protein Sci. 2007;16:1360–1367. doi: 10.1110/ps.072793807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh B.K., Stidham M.A., Shaner D.L. Assay of acetohydroxyacid synthase. Anal. Biochem. 1988;171:173–179. doi: 10.1016/0003-2697(88)90139-x. [DOI] [PubMed] [Google Scholar]
- 43.Westerfeld W.W. A colorimetric determination of blood acetoin. J. Biol. Chem. 1945;161:495–502. [PubMed] [Google Scholar]
- 44.Eram M.S., Oduaran E., Ma K. The bifunctional pyruvate decarboxylase/pyruvate ferredoxin oxidoreductase from Thermococcus guaymasensis. Archaea. 2014;2014:1–13. doi: 10.1155/2014/349379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 46.Eram M.S., Sarafuddin B., Gong F., Ma K. Optimization of expression and properties of the recombinant acetohydroxyacid synthase of Thermotoga maritima. Data in Brief. 2015 doi: 10.1016/j.dib.2015.09.018. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balk M., Weijma J., Stams A.J.M. Thermotoga lettingae sp. nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int. J. Syst. Evol. Microbiol. 2002;52:1361–1368. doi: 10.1099/00207713-52-4-1361. [DOI] [PubMed] [Google Scholar]
- 48.Patchett M., Neal T., Schofield L., Strange R., Daniel R., Morgan H. Heat treatment purification of thermostable cellulase and hemicellulase enzymes expressed in E. coli. Enzyme Microb. Technol. 1989;11:113–115. [Google Scholar]
- 49.Conners S.B., Mongodin E.F., Johnson M.R., Montero C.I., Nelson K.E., Kelly R.M. Microbial biochemistry, physiology, and biotechnology of hyperthermophilic Thermotoga species. FEMS Microbiol. Rev. 2006;30:872–905. doi: 10.1111/j.1574-6976.2006.00039.x. [DOI] [PubMed] [Google Scholar]
- 50.Rinker K.D., Kelly R.M. Effect of carbon and nitrogen sources on growth dynamics and exopolysaccharide production for the hyperthermophilic archaeonThermococcus litoralis and bacterium Thermotoga maritima. Biotechnol. Bioeng. 2000;69:537–547. doi: 10.1002/1097-0290(20000905)69:5<537::aid-bit8>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 51.Vinogradov V., Vyazmensky M., Engel S., Belenky I., Kaplun A., Kryukov O., Barak Ze, Chipman D.M. Acetohydroxyacid synthase isozyme I from Escherichia coli has unique catalytic and regulatory properties. Biochim. Biophys. Acta (BBA): Gen. Subj. 2006;1760:356–363. doi: 10.1016/j.bbagen.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 52.Engel S., Vyazmensky M., Vinogradov M., Berkovich D., Bar-Ilan A., Qimron U., Rosiansky Y., Barak Ze, Chipman D.M. Role of a conserved arginine in the mechanism of acetohydroxyacid synthase. J. Biol. Chem. 2004;279:24803–24812. doi: 10.1074/jbc.M401667200. [DOI] [PubMed] [Google Scholar]
- 53.Kube J., Brokamp C., Machielsen R., van der Oost J., Märkl H. Influence of temperature on the production of an archaeal thermoactive alcohol dehydrogenase from Pyrococcus furiosus with recombinant Escherichia coli. Extremophiles. 2006;10:221–227. doi: 10.1007/s00792-005-0490-z. [DOI] [PubMed] [Google Scholar]
- 54.Bowen T.L., Union J., Tumbula D.L., Whitman W.B. Cloning and phylogenetic analysis of the genes encoding acetohydroxyacid synthase from the archaeon Methanococcus aeolicus. Gene. 1997;188:77–84. doi: 10.1016/s0378-1119(96)00779-2. [DOI] [PubMed] [Google Scholar]
- 55.Tittmann K., Schröder K., Golbik R., McCourt J., Kaplun A., Duggleby R.G., Barak Z., Chipman D.M., Hübner G. Electron transfer in acetohydroxy acid synthase as a side reaction of catalysis. Implications for the reactivity and partitioning of the carbanion/enamine form of (alpha-hydroxyethyl)thiamin diphosphate in a "nonredox" flavoenzyme. Biochemistry. 2004;43:8652–8661. doi: 10.1021/bi049897t. [DOI] [PubMed] [Google Scholar]
- 56.Le D.T., Choi J.D. FAD-independent and herbicide-resistant mutants of tobacco acetohydroxy acid synthase. Bull. Korean Chem. Soc. 2005;26:916–920. [Google Scholar]
- 57.Hawkins C.F., Borges A., Perham R.N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- 58.Chipman D., Barak Ze, Schloss J.V. Biosynthesis of 2-aceto-2-hydroxy acids: acetolactate synthases and acetohydroxyacid synthases. Biochim. Biophys. Acta (BBA): Protein Struct. Mol. Enzymol. 1998;1385:401–419. doi: 10.1016/s0167-4838(98)00083-1. [DOI] [PubMed] [Google Scholar]
- 59.Muller Y.A., Lindqvist Y., Furey W., Schulz G.E., Jordan F., Schneider G. A thiamin diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase. Structure. 1993;1:95–103. doi: 10.1016/0969-2126(93)90025-c. [DOI] [PubMed] [Google Scholar]
- 60.Schellenberger A. Sixty years of thiamin diphosphate biochemistry. Biochim. Biophys. Acta: Proteins Proteom. 1998;1385:177–186. doi: 10.1016/s0167-4838(98)00067-3. [DOI] [PubMed] [Google Scholar]
- 61.M. Ikeda, S. Takeno, Amino acid production by Corynebacterium glutamicum, in: Corynebacterium glutamicum, Springer, 2013, pp. 107–147.
- 62.Ikeda M., Nakagawa S. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 2003;62:99–109. doi: 10.1007/s00253-003-1328-1. [DOI] [PubMed] [Google Scholar]
- 63.Oldiges M., Eikmanns B.J., Blombach B. Application of metabolic engineering for the biotechnological production of L-valine. Appl. Microbiol. Biotechnol. 2014;98:5859–5870. doi: 10.1007/s00253-014-5782-8. [DOI] [PubMed] [Google Scholar]
- 64.Barak Z., Chipman D.M., Gollop N. Physiological implications of the specificity of acetohydroxy acid synthase isozymes of enteric bacteria. J. Bacteriol. 1987;169:3750–3756. doi: 10.1128/jb.169.8.3750-3756.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bar-Ilan A., Balan V., Tittmann K., Golbik R., Vyazmensky M., Hubner G., Barak Ze, Chipman D.M. Binding and activation of thiamin diphosphate in acetohydroxyacid synthase. Biochemistry. 2001;40:11946–11954. doi: 10.1021/bi0104524. [DOI] [PubMed] [Google Scholar]
- 66.Chang A.K., Duggleby R.G. Expression, purification and characterization of Arabidopsis thaliana acetohydroxyacid synthase. Biochem J. 1997;327:161–169. doi: 10.1042/bj3270161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kil M.W., Chang S.I. Expression in Escherichia coli , purification , and characterization of the tobacco sulfonylurea herbicide – resistant recombinant acetolactate synthase and its interaction with the triazolopyrimidine herbicides. Biochem. Mol. Biol. Rep. 1998;31:287–297. [Google Scholar]
- 68.Gollop N., Damri B., Chipman D.M., Barak Z. Physiological implications of the substrate specificities of acetohydroxy acid synthases from varied organisms. J. Bacteriol. 1990;172:3444–3449. doi: 10.1128/jb.172.6.3444-3449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Eggeling I., Cordes C., Eggeling L., Sahm H. Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of α-ketobutyrate to l-isoleucine. Appl. Microbiol. Biotechnol. 1987;25:346–351. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material