Abstract
The A-type and B-type lamins form a filamentous meshwork underneath the inner nuclear membrane called the nuclear lamina, which is an important component of nuclear architecture in metazoan cells. The lamina interacts with large, mostly repressive chromatin domains at the nuclear periphery. In addition, genome–lamina interactions also involve dynamic association of lamin A/C with gene promoters in adipocytes. Mutations in the human lamin A gene cause a spectrum of hereditary diseases called the laminopathies which affect muscle, cardiac and adipose tissues. Since most mutations in lamin A/C affect skeletal muscle, we investigated lamin–chromatin interactions at promoters of muscle specific genes in both muscle and non-muscle cell lines by ChIP-qPCR. We observed that lamin A/C was specifically associated with promoter regions of muscle genes in myoblasts but not in fibroblasts. Lamin A/C dissociated from the promoter regions of the differentiation specific MyoD, myogenin and muscle creatine kinase genes when myoblasts were induced to differentiate. In the promoter regions of the myogenin and MyoD genes, the binding of lamin A/C in myoblasts inversely correlated with the active histone mark, H3K4me3. Lamin A/C binding on muscle genes was reduced and differentiation potential was enhanced on treatment of myoblasts with a histone deacetylase inhibitor. These findings suggest a role for lamina–chromatin interactions in muscle differentiation and have important implications for the pathological mechanisms of striated muscle associated laminopathies.
Keywords: Nuclear lamina, Lamins, Myogenin, Laminopathies, Muscle differentiation, Genome–lamina interactions
Highlights
-
•
Lamina–chromatin interactions are important for nuclear processes.
-
•
We show lamin A/C binding to promoters of muscle genes in myoblasts.
-
•
Lamin A/C binding is reduced upon myoblast differentiation.
-
•
Lamin A/C binding inversely correlates with active histone marks on muscle genes.
-
•
Our findings suggest that lamin A/C binding to promoters is cell-type specific.
1. Introduction
The nuclear lamina is a filamentous network of proteins that underlies the inner nuclear membrane and extends into the nuclear interior in metazoan cells, and is mainly composed of the lamin proteins. Lamins belong to the type V intermediate filament protein family and are of two types. The A-type lamins, lamin A and C (splice isoforms of the lamin A gene, LMNA) are expressed in most lineage-committed and differentiated cells while the B-type lamins, lamin B1 and B2 (encoded by separate genes) are expressed in all somatic cells. Lamins are the major architectural proteins of the nucleus and are involved in chromatin organisation as well as organisation of various nuclear functions, in addition to providing mechanical support to the nuclear envelope [1], [2], [3], [4].
Mutations in lamins have been linked to a wide spectrum of heritable diseases called laminopathies, which include muscular dystrophies, cardiomyopathies, lipodystrophies and accelerated aging disorders. Laminopathic cells exhibit widespread alterations in chromatin organisation, histone epigenetic marks and gene activation, emphasising the importance of lamins in nuclear chromatin dynamics [1], [2], [3], [4]. The lamina associates with chromatin through large domains termed lamina-associated domains or LADs, which contain mostly inactive genes and are located at the nuclear periphery; lamina–chromatin interactions are likely to be regulated by modified histones and gene regulatory proteins and can be disrupted by inhibitors of histone deacetylases (HDACs) as well as depletion of lamins [5], [6]. In addition to anchoring large regions of chromatin at the nuclear periphery, lamin A/C has also been shown to associate with promoter regions of genes in adipocyte stem cells in a recent study. The association of lamin A/C at adipogenic genes is altered upon adipogenic differentiation, suggesting its importance for lineage commitment [6]. However it is not known whether lamin A/C associates with specific gene promoters in other lineages such as muscle.
Since skeletal muscle is one of the major tissues affected by mutations in lamin A/C, an understanding of lamin A/C–chromatin interactions in muscle cells has important implications for disease mechanisms of laminopathies. In the present study, we report our findings on the interaction of lamin A/C with promoter regions of muscle-specific and non-muscle genes in mouse myoblasts and differentiated myotubes, as well as fibroblasts as an example of a non-muscle cell type.
2. Materials and methods
2.1. Cell culture and antibodies
C2C12 mouse myoblasts were maintained in DMEM supplemented with 20% FBS (growth medium, GM) and induced to differentiate by transfer to DMEM containing 2% horse serum (differentiation medium, DM) for 48 h [7]. NIH3T3 mouse fibroblasts were maintained in DMEM with 10% FBS. For sodium butyrate treatment, proliferating myoblasts were cultured in GM containing 5 mM sodium butyrate (Sigma) for 24 h. A goat polyclonal antibody for lamin A/C (N-18, Santa Cruz Biotechnology) was validated by chromatin immunoprecipitation (ChIP) followed by western blotting as described earlier [8]. A ChIP-grade antibody to H3K4me3 (07-473) was from Millipore Corporation.
2.2. Chromatin immunoprecipitation analysis
ChIP analysis was carried out by standard protocols. Briefly 106 cells were cross-linked with 1% formaldehyde for 10 min at 37 °C followed by quenching with 0.125 M glycine solution. Cells were washed, lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.1) containing protease inhibitors and sonicated to shear chromatin into fragments less than 500 bp in size using Bioruptor (Diagenode). Sheared chromatin was diluted with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1 and 167 mM NaCl containing protease inhibitors), pre-cleared and incubated overnight with lamin A/C antibody (3–4 μg) or unspecific IgG at 4 °C. Following this the antigen–antibody complexes were captured with pre-blocked protein A agarose and washed sequentially with low salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 150 mM NaCl), high salt (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.1, 500 mM NaCl) and LiCl wash buffers (0.25 M LiCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris, pH 8.1) followed by TE buffer (10 mM Tris HCl, 1 mM EDTA, pH 8.0). Antigen–antibody complexes were eluted from agarose using 1 M NaHCO3 and 1% SDS. The input samples and the eluates were reverse cross-linked and proteinase treated. DNA was isolated and purified using the phenol–chloroform–isoamyl alcohol method. Following sodium acetate–ethanol precipitation, DNA was dissolved in 30 μl of water and used for qRT-PCR using 7900HT thermal cycler (Applied Biosystems). Primers specific to different regions of gene promoters were either designed or obtained from previous reports. Each promoter region was analysed in triplicate qRT-PCR reactions per experiment and three independent ChIP experiments were performed. The data is presented as average fold enrichment over IgG, and less than 3-fold enrichment is considered as weak or no binding. Representative IgG ChIP levels have been shown for one experiment in Fig. 1D. Statistical significance was calculated using Student's t-test. List of primers used is provided in the Supplementary Table S1.
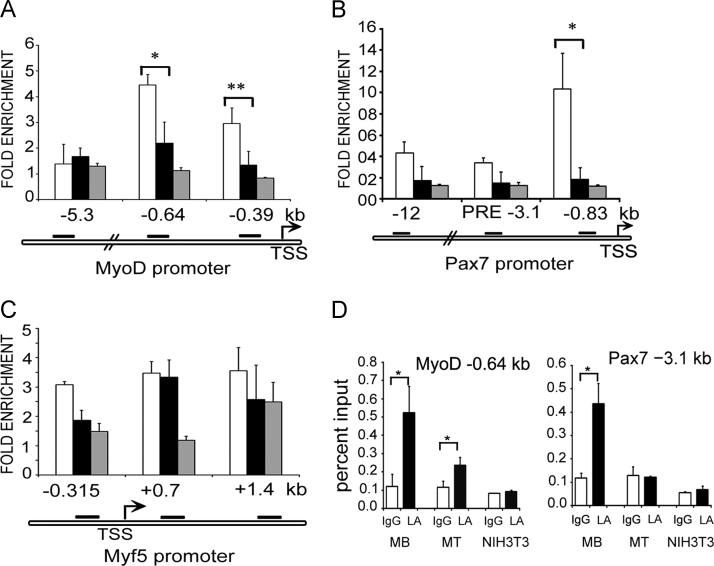
Fig. 1.
Lamin A/C occupancy on promoters of MyoD1, Pax7 and Myf5. ChIP-qPCR analysis with lamin A/C antibody on promoters of myogenic genes in myoblasts (open bars), myotubes differentiated for 48 h (solid bars) and NIH3T3 fibroblasts (grey bars). Fold enrichment over IgG±S.D is represented for (A) MyoD1 promoter, (B) Pax7 promoter and (C) Myf5 promoter. (D) Percentage of input values for unspecific IgG and lamin A/C (LA) obtained at representative promoter regions (MyoD −0.64 kb and Pax7 −3.1 kb) are shown to indicate specificity of lamin A/C ChIP. (p<0.05*, <0.01**).
2.3. Real time transcript analysis
Total RNA was isolated from myoblasts, myotubes and sodium butyrate treated myoblasts using the Trizol method. As per manufacturer's instructions, 1–5 μg of DNase treated RNA was reverse transcribed using Superscript II reverse transcriptase kit (Invitrogen). qRT-PCR was performed as described above. Each gene was analysed in triplicate qRT-PCR reactions per experiment and three independent experiments were performed. RPLP0 (ribosome protein large, P0) was used as internal control as it is reported to be one of the most suitable internal controls for myoblasts [9]. Statistical significance was calculated using the Student's t-test. List of primers used is provided in Table S2.
2.4. Immunofluorescence microscopy
Cells were washed with phosphate buffered saline (PBS), fixed with 3.5% formaldehyde in PBS for 10 min at room temperature, and permeabilised with 0.5% Triton-X 100 (vol/vol) for 6 min at room temperature. After blocking with 0.5% gelatin in PBS for one hour, cells were incubated with primary antibody to myogenin (monoclonal antibody from Developmental Studies Hybridoma Bank, University of Iowa, USA) diluted in PBS for 1–2 h followed by Cy3-conjugated secondary antibody in PBS for an hour. Coverslips were then mounted on glass slides in Vectashield (Vector laboratories, Burlingame, CA, USA) containing 1 μg/ml DAPI to visualise the nucleus. Slides were scanned on a LSM510 META/NLD confocal microscope (Carl Zeiss, Jena, Germany). For quantification of labelled cells, more than n=150 cells were counted per sample in at least three independent experiments.
3. Results
In order to determine whether lamin A/C associates with chromatin in muscle stem cells, we used the C2C12 mouse myoblast cell line which is extensively used as an in vitro model system to study the biology of muscle differentiation [7]. Chromatin was isolated from proliferating myoblasts and after differentiation for 48 h and ChIP assays were performed with lamin A/C antibody and unspecific IgG. Regulatory regions of important muscle regulatory genes as well as non-muscle genes were analysed. Furthermore, to determine the specificity of association of lamin A/C with muscle genes, chromatin from the mouse fibroblast cell line NIH3T3 was also used. We chose to analyse lamin A/C binding at well-documented promoter regions previously reported to be epigenetically regulated, as described below for each gene.
3.1. Association of lamin A/C with promoters of myogenic genes
MyoD is the master regulator of myogenesis and binds to and regulates several loci in myogenic cells [10]. It is expressed in proliferating cells and is upregulated and stabilised in differentiating cells. The MyoD1 gene locus has been shown to migrate from the nuclear periphery in myoblasts to the nuclear interior in differentiating cells [11]. Three regions of the MyoD1 promoter were analysed. The proximal promoter regions (−0.39 and −0.64 kb) [12] showed higher lamin A/C association (3–5-fold) in myoblasts which was significantly reduced in myotubes and was similar to unspecific IgG in NIH3T3 fibroblasts. The MyoD1 enhancer termed the distal regulatory region (DRR) at −5.3 kb, which is known to regulate expression of MyoD during post natal skeletal myogenesis and muscle regeneration [13], did not show significant enrichment of lamin A/C binding in myoblasts, myotubes or fibroblasts (Fig. 1A). Thus the proximal promoter regions of MyoD1 have reduced association with lamin A/C upon onset of differentiation.
The paired-box transcription factor Pax7 is indispensable for muscle satellite cell survival, maintenance and self-renewal [14], [15]. It is expressed in quiescent and proliferating myoblasts and downregulated upon differentiation. Three regions of Pax7 promoter, the proximal promoter region (−0.8 kb), the conserved polycomb response element (PRE) consisting of GAGA rich sequence and YY1 binding motif and the distal enhancer region (−12 kb) [16] were analysed for lamin A/C binding. A 10-fold enrichment of lamin A/C was observed at proximal promoter region (−0.8 kb) of Pax7 in myoblasts but not in myotubes. Other regions did not show significant differences in lamin A/C binding between myoblasts and myotubes and all three regions showed no association with lamin A/C in NIH3T3 fibroblasts (Fig. 1B). At the Myf5 promoter, enrichment of lamin A/C binding was weak (~3-fold), and it did not vary between myoblasts and myotubes at all three reported regulatory regions [17] that were tested (Fig. 1C).
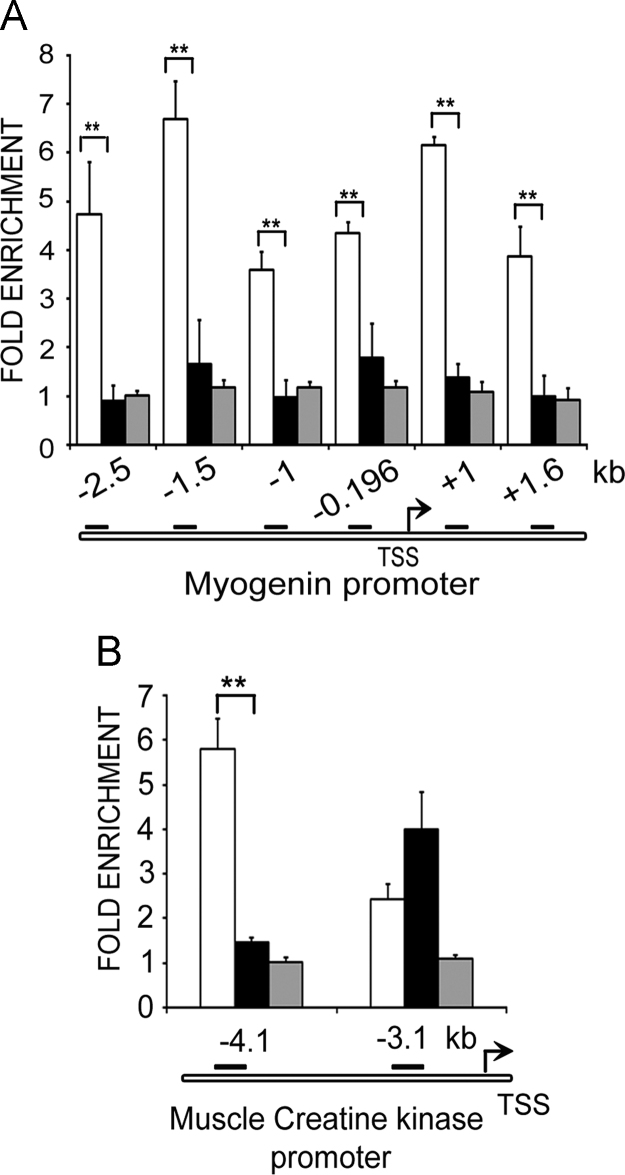
The myogenin and muscle creatine kinase (MCK) genes are muscle-specific and expressed only in differentiated cells [18]. Both these genes are silenced in proliferating myoblasts by histone modifications and DNA methylation [19]. Importantly, mutations in lamin A/C that cause Emery-Dreifus muscular dystrophy impair differentiation and myogenin expression in C2C12 myoblasts [20], [21], and alter epigenetic marks at its promoter region [22]. Six regions of Myog promoter both upstream and downstream of the transcription start site (TSS at −0.196 kb) [19], [23] were analysed for association with lamin A/C. Of these regions −1.5 kb and +1.6 kb are differentiation specific enhancer elements identified in a genome wide ChIP study in C2C12 cell line [19]. All regions showed significantly higher association with lamin A/C in myoblasts (5–7 fold) where myogenin is not expressed. Lamin A/C binding to these regions was not detectable in both myotubes and NIH3T3 fibroblasts (Fig. 2A). Thus a differentiation dependent lamin A/C binding was clearly evident at the Myog promoter. Similarly the Mck enhancer region at −4.1 kb upstream of TSS is a well characterised regulatory region shown to be epigenetically regulated by polycomb complexes during muscle differentiation [23], [24]. At this region more than 5-fold enrichment of lamin A/C was obtained over IgG in myoblasts where MCK is not expressed (Fig. 2B). Lamin A/C binding was not seen in myotubes and also undetectable in NIH3T3 fibroblasts. A similar lamin A/C binding profile was not observed at the non-regulatory region adjacent to the enhancer region at −3.1 kb.
Fig. 2.
Lamin A/C occupancy on promoters of Myog and Mck. ChIP-qPCR analysis with lamin A/C antibody on promoters of differentiation-specific genes in myoblasts (open bars), myotubes differentiated for 48 h (solid bars) and NIH3T3 fibroblasts (grey bars). Fold enrichment over IgG±S.D is represented for promoter regions of (A) Myog and (B) Mck (p<0.01**).
3.2. Association of lamin A/C with non-muscle genes
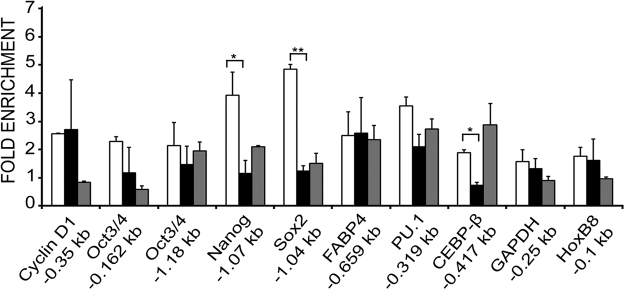
We determined lamin A/C binding at a number of genes that are usually silenced in either differentiated myotubes (cell cycle genes) or in both myoblasts and myotubes (pluripotency and other lineage specific genes). Enrichment of lamin A/C binding was less than 3-fold at the cell cycle gene coding for cyclin D1 and the pluripotency gene Oct3/4. However, regions upstream of TSS of the pluripotency genes Nanog and Sox2 showed 2–3-fold higher binding of lamin A/C in myoblasts when compared to myotubes. Enrichment of lamin A/C binding at promoters of genes expressed in different lineages was less than 3-fold and did not differ between muscle cells and fibroblasts. PU.1/Sfpi is involved in hematopoiesis while FABP4 (fatty acid binding protein 4) and CEBPβ (CCAAT/enhancer binding protein, beta) are involved in adipogenic differentiation and are not expected to be expressed in C2C12 cells. No significant lamin A/C binding was observed at promoters of a house-keeping gene like Gapdh or permanently repressed HoxB8 of the hox cluster of genes (Fig. 3).
Fig. 3.
Lamin A/C occupancy on promoters of non-muscle genes. ChIP-qPCR analysis with lamin A/C antibody on promoters of non-muscle genes in myoblasts (open bars), myotubes differentiated for 48 h (solid bars) and NIH3T3 fibroblasts (grey bars). Fold enrichment over IgG±S.D is represented. (p<0.05*, <0.01**).
3.3. Correlation of lamin A/C binding with epigenetic modifications
In order to determine whether lamin A/C dissociation from promoter regions correlates with deposition of active histone marks, we analysed the profile of active histone mark H3K4me3 across two genes, MyoD1 and Myog during differentiation and compared it with lamin A/C occupancy. At regions proximal to the TSS both in MyoD1 and Myog, levels of H3K4me3 were higher in myotubes when compared to myoblasts (Fig. 4A and B). This correlated with reduced lamin A/C occupancy at these regions in myotubes which was observed in the previous experiment (Figs. 1A and 2A).
Fig. 4.
Binding of histone activation mark H3K4me3 on muscle genes. ChIP-qPCR analysis with antibody to active histone mark H3K4me3 on promoter regions of (A) MyoD1 and (B) Myog in myoblasts (open bars) and myotubes (solid bars). Fold enrichment over IgG±S.D is represented. (p<0.05*, <0.01**).
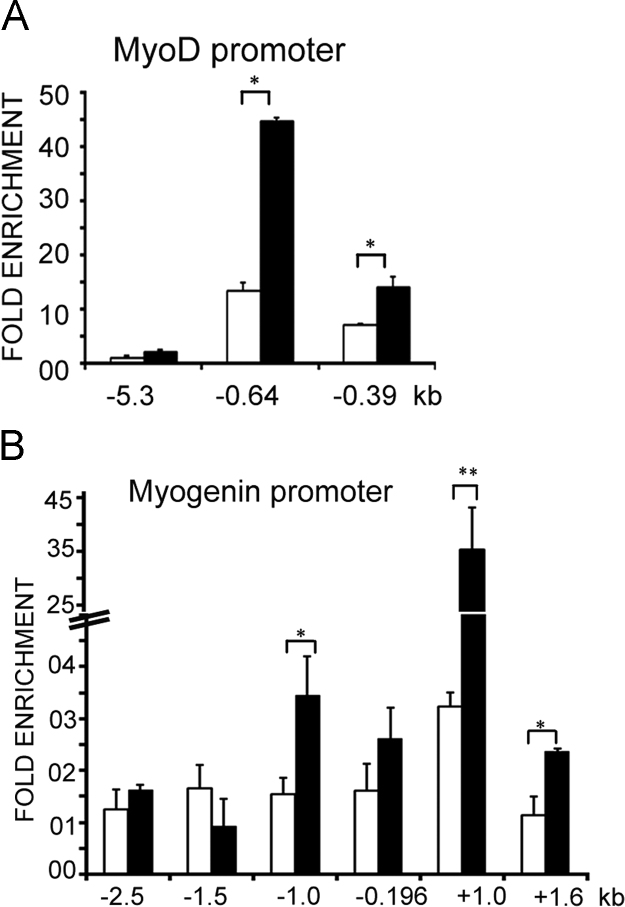
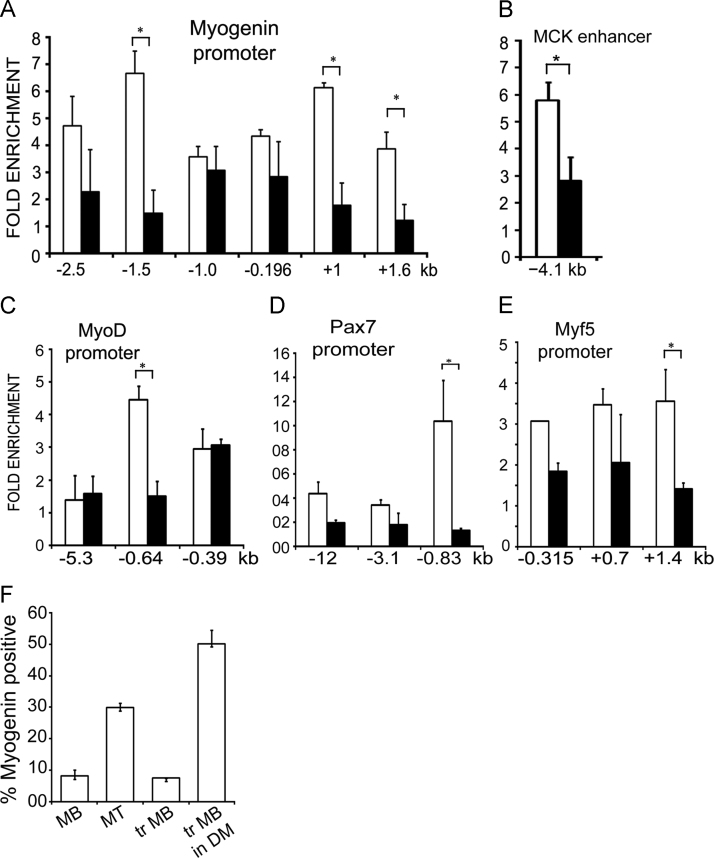
Since our data suggests that lamin A/C may be involved in silencing of differentiation specific genes in myoblasts, we examined whether activation of these genes by inhibition of histone deacetylase activity might cause loss of lamin A/C binding from promoter regions. Treatment of myoblasts with a histone deacetylase inhibitor, sodium butyrate has been previously shown to enhance their differentiation potential [25]. Analysis of Myog promoter regions in sodium butyrate treated myoblasts showed loss of lamin A/C occupancy from −1.5 kb, +1.0 and +1.6 kb promoter regions (Fig. 5A). The Mck enhancer at −4.1 kb, MyoD1 promoter at −0.64 kb and Pax7 promoter at −0.83 kb also showed significant loss of lamin A/C binding (Fig. 5B–D). There was also a decrease in the weak binding of lamin A/C to the +1.4 kb Myf5 promoter (Fig. 5E).
Fig. 5.
Lamin A/C occupancy in myoblasts treated with sodium butyrate. ChIP-qPCR analysis with lamin A/C antibody on promoter regions of muscle genes in untreated myoblasts (open bars) and myoblasts treated with 5 mM sodium butyrate in GM for 24 h (solid bars). Fold enrichment over IgG±S.D is represented for promoters of (A) Myog, (B) Mck, (C) MyoD1, (D) Pax7 and (E) Myf5. (F) Quantification of myogenin expression by immunofluorescence analysis in normal myoblasts (MB), normal myoblasts differentiated to myotubes in DM (MT), myoblasts treated with 5 mM sodium butyrate (tr MB) and then differentiated in DM in the absence of sodium butyrate (tr MB in DM). (p<0.05*).
In order to examine the effects of sodium butyrate on transcription of muscle genes in our experimental system, real time transcript analysis was carried out with myoblasts, sodium butyrate treated myoblasts and myotubes. As seen in Table 1, sodium butyrate treatment increased the transcript levels of the early differentiation marker MyoD1 as well as Pax7, but the levels of myogenin and the late marker MCK were not increased. The transcript levels of Myf5 and the non-muscle specific gene cyclin D1 which do not bind to lamin A/C were also not increased after treatment. Upon differentiation of normal myoblasts, transcript levels of MyoD, myogenin and MCK increased, while cyclin D1 decreased as expected; levels of Pax7 and Myf5 were also not significantly altered. Since expression of myogenin and MCK in myoblasts is normally tightly repressed by different signalling pathways [18], [19], their levels in treated myoblasts were unlikely to increase after treatment. However, sodium butyrate treatment of myoblasts is known to enhance the differentiation capacity of myoblasts after transfer to DM [25]. Consistent with this report, when we analysed myogenin expression in differentiated cells by immunofluorescence assays, we observed a marked increase in myogenin positive cells from ~30% to ~50% after treatment of myoblasts with sodium butyrate (Fig. 5F).
Table 1.
Real time transcript analysis of muscle genes.
| Gene | Myoblasts | Myotubes (48 h) | NIH3T3 | Treated Myoblastsa |
|---|---|---|---|---|
| MyoD | 1 | 3.86±1.01 (*) | NDb | 5.72±1.08 (*) |
| Pax7 | 1 | −1.23±0.18 | ND | 2.91±0.74 (*) |
| Myf5 | 1 | −1.26±0.25 | ND | −1.13±0.28 |
| Myogenin | 1 | 33.80±6.05 (*) | ND | 2.00±0.86 |
| MCK | 1 | 31.27±4.33 (*) | ND | −1.05±0.21 |
| Cyclin D1 | 1 | −3.96±0.40 (*) | −2.72±0.29 | 1.35±0.21 |
Transcript levels ±S.D, normalised to levels in myoblasts are represented.
p<0.05.
Myoblasts treated with 5 mM sodium butyrate.
Not detectable.
4. Discussion
In the present study, we show that lamin A/C associates with distinct promoter regions of muscle-specific genes MyoD1, Myog and Mck in myoblasts but not in fibroblasts, and this interaction is reduced upon differentiation when these genes are upregulated. Lamin A/C binding negatively correlates with deposition of the active histone mark H3K4Me3, and is reduced upon treatment with an HDAC inhibitor.
The lamina interacts with large domains of mostly inactive chromatin in LADs at the nuclear periphery in several cell types [5]. A recent report shows that lamin A/C also specifically associates with short promoter regions of specific genes in a differentiation state dependent manner in adipocytes [6]. These changes in lamin A/C–promoter interactions were observed during adipocyte differentiation and correlated with differential gene expression [6], but were not examined in non-adipocyte cells. Our results indicate that lamin A/C associates with the regulatory regions of muscle genes that are important for induction of muscle differentiation, and that this binding is reduced upon differentiation of myoblasts to myotubes. However, the muscle-specific genes that we tested were not bound by lamin A/C in NIH3T3 fibroblasts suggesting cell-type specificity of these lamin A/C–gene promoter interactions. In addition to promoter proximal regions, we observed that enhancer regions of few genes like Mck and Myog also bind to lamin A/C in a differentiation specific manner. Recent studies have demonstrated alterations in lamin A/C occupancy with changes in histone modification marks at gene regulatory regions, and highlighted the importance of modified histones in genome–lamina interactions [26], [27], [28], [29]. A specific subset of lamin A/C interacting domains or LiDs are enriched in repressive histone marks and have higher gene density compared to lamin B1 interacting domains [30]. A recent study reports that repressive histone marks are replaced by active histone marks on inactive genes associated with lamin A/C in response to insulin signalling, and results in dissociation of lamin A/C followed by gene activation [31]. We found that lamin A/C binding on Myog and MyoD1 promoter regions inversely correlated with the active histone mark H3K4me3, which marked these loci only in myotubes where Myog and MyoD1 are upregulated. Furthermore, inhibition of HDACs by sodium butyrate altered lamin A/C binding at promoter regions of muscle genes, suggesting that an active chromatin structure may not be compatible with lamin A/C binding. We also observed that proximal and enhancer elements of different gene promoters showed differences in levels of lamin A/C occupancy, suggesting that lamin A/C may interact through specific gene sequences or distinct combinations of repressive histone marks; this interaction may occur directly or indirectly through specific binding proteins. Although we cannot rule out the possibility that lamin A/C interactions could be partly stochastic, overall our data suggests that lamin A/C binding is important for repressing the expression potential of differentiation-specific genes in myoblasts.
It has been reported that lamin A/C binding does not strictly correlate with gene silencing and approximately 20% of genes in lamin A/C associated domains are still actively transcribed [6], [32], [33]. We detected increased binding of lamin A/C at the proximal promoter region of Pax7 in myoblasts where it is expressed and lower binding in myotubes where the expression in shut down. We also noted that lamin A/C is bound to the inactive Nanog and Sox2 loci in myoblasts but is dissociated in myotubes and fibroblasts although these loci remain inactive in these cells. The Nanog locus is also bound to lamin A/C in adipose stem cells [6]. Importantly, muscle gene promoters are not bound to lamin A/C in fibroblasts though these genes are not expressed in fibroblasts. These findings raise questions about the precise role of lamin–promoter interactions in different cell types and more extensive analysis on a genome-wide scale with different cell types is required to clarify some of these issues.
Based on our findings, we propose that muscle differentiation cues that regulate the deposition of active histone marks in place of repressive histone marks at promoters of muscle regulatory genes also mediate the dissociation of lamin A/C from these promoters upon differentiation. We further suggest that lamin association with differentiation-specific gene promoters may be important for active repression of these genes in lineage committed stem cells and might help to fine-tune their expression potential. Hence lamin binding at promoters might not be a default repressive mechanism.
Mutations in the human lamin A gene cause striated muscle laminopathies, where diseased cells fail to express muscle-specific genes and are differentiation defective [1], [2], [3], [4]. Our study indicates that lamin A/C–promoter interactions vary in a differentiation dependent manner in muscle cells and these alterations may have roles in gene regulation during differentiation. Understanding these interactions and their consequences are likely to provide insights into disease mechanisms associated with laminopathies.
Acknowledgements
F.A. was supported by a Senior Research Fellowship from the Council of Scientific and Industrial Research, India. V.K. P. is a recipient of the J. C. Bose national fellowship from the Department of Science and Technology, India. Financial support from the Council of Scientific and Industrial Research network project BSC208 for this work is gratefully acknowledged.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.08.021.
Appendix A. Supplementary materials
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
References
- 1.Dechat T., Adam S., Taimen P., Shimi T., Goldman R. Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2010;2:a000547. doi: 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parnaik V.K. Role of nuclear lamins in nuclear organization, cellular signaling, and inherited diseases. Int. Rev. Cell. Mol. Biol. 2008;266:157–206. doi: 10.1016/S1937-6448(07)66004-3. [DOI] [PubMed] [Google Scholar]
- 3.Worman H., Ostlund C., Wang Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010;2:a000760. doi: 10.1101/cshperspect.a000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchinson C.J. B-type lamins in health and disease. Semin. Cell. Dev. Biol. 2014;29:158–163. doi: 10.1016/j.semcdb.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kind J., Van Steensel B. Genome–nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol. 2010;22:320–325. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Lund E., Oldenburg A.R., Delbarre E., Freberg C.T., Duband-Goulet I., Eskeland R., Buendia B., Collas P. Lamin A/C–promoter interactions specify chromatin state-dependent transcription outcomes. Genome Res. 2013;23:1580–1589. doi: 10.1101/gr.159400.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaffe D., Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 8.McCord R., Nazario-Toole A., Zhang H., Chines P., Zhan Y., Erdos M., Collins F., Dekker J., Cao K. Correlated alterations in genome organization, histone methylation, and DNA–lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 2013;23:260–269. doi: 10.1101/gr.138032.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stern-Straeter J., Bonaterra G.A., Hormann K., Kinscherf R., Goessler U.R. Identification of valid reference genes during the differentiation of human myoblasts. BMC Mol. Biol. 2009;10:66. doi: 10.1186/1471-2199-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L., Gentleman R.C., Tapscott S.J. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao J., Fetter R., Hu P., Betzig E., Tjian R. Subnuclear segregation of genes and core promoter factors in myogenesis. Genes Dev. 2011;25:569–580. doi: 10.1101/gad.2021411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tapscott S.J., Lassar A.B., Weintraub H. A novel myoblast enhancer element mediates MyoD transcription. Mol. Cell. Biol. 1992;12:4994–5003. doi: 10.1128/mcb.12.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.L’honore A., Lamb N.J., Vandromme M., Turowski P., Carnac G., Fernandez A. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol. Biol. Cell. 2003;14:2151–2162. doi: 10.1091/mbc.E02-07-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zammit P., Relaix F., Nagata Y., Ruiz A., Collins C., Partridge T., Beauchamp J. Pax7 and myogenic progression in skeletal muscle satellite cells. J. Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 15.Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 16.Palacios D., Mozzetta C., Consalvi S., Caretti G., Saccone V., Proserpio V., Marquez V.E., Valente S., Mai A., Forcales S.V., Sartorelli V., Puri P.L. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKinnell I.W., Ishibashi J., Le Grand F., Punch V.G., Addicks G.C., Greenblatt J.F., Dilworth F.J., Rudnicki M.A. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andres V., Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B.D. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. USA. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favreau C., Higuet D., Courvalin J.-C., Buendia B. Expression of a mutant lamin A that causes Emery-Dreifuss muscular dystrophy inhibits in vitro differentiation of C2C12 myoblasts. Mol. Cell. Biol. 2004;24:1481–1492. doi: 10.1128/MCB.24.4.1481-1492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parnaik V.K., Manju K. Laminopathies: multiple disorders arising from defects in nuclear architecture. J. Biosci. 2006;31:405–421. doi: 10.1007/BF02704113. [DOI] [PubMed] [Google Scholar]
- 22.Hakelien A.-M., Delbarre E., Gaustad K., Buendia B., Collas P. Expression of the myodystrophic R453W mutation of lamin A in C2C12 myoblasts causes promoter-specific and global epigenetic defects. Exp. Cell Res. 2008;314:1869–1880. doi: 10.1016/j.yexcr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 23.Seenundun S., Rampalli S., Liu Q.C., Aziz A., Palii C., Hong S., Blais A., Brand M., Ge K., Dilworth F.J. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lezzi S., Cossu G., Nervi C., Sartorelli V., Puri P.L. Stage-specific modulation of skeletal myogenesis by inhibitors of nuclear deacetylases. Proc. Natl. Acad. Sci. USA. 2002;99:7757–7762. doi: 10.1073/pnas.112218599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lund E., Collas P. Nuclear lamins: making contacts with promoters. Nucleus. 2013;4:424–430. doi: 10.4161/nucl.26865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milon B., Cheng H., Tselebrovsky M., Lavrov S., Nenasheva V., Mikhaleva E., Shevelyov Y., Nurminsky D. Role of histone deacetylases in gene regulation at nuclear lamina. PloS One. 2012;7:e49692. doi: 10.1371/journal.pone.0049692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kind J., Pagie L., Ortabozkoyun H., Boyle S., de Vries S., Janssen H., Amendola M., Nolen L., Bickmore W., van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 29.Melcer S., Hezroni H., Rand E., Nissim-Rafinia M., Skoultchi A., Stewart C.L., Bustin M., Meshorer E. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat. Commun. 2012;3:910. doi: 10.1038/ncomms1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lund E.G., Duband-Goulet I., Oldenburg A., Buendia B., Collas P. Distinct features of lamin A-interacting chromatin domains mapped by ChIP-sequencing from sonicated or micrococcal nuclease-digested chromatin. Nucleus. 2015;6:30–39. doi: 10.4161/19491034.2014.990855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeong K., Kwon H., Lee J., Jang D., Pak Y. Insulin-response epigenetic activation of Egr-1 and JunB genes at the nuclear periphery by A-type lamin-associated pY19-Caveolin-2 in the inner nuclear membrane. Nucleic Acids Res. 2015;43:3114–3127. doi: 10.1093/nar/gkv181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumaran R.I., Spector D.L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S., Solovei I., Brugman W., Gräf S., Flicek P., Kerkhoven R., van Lohuizen M., Reinders M., Wessels L., van Steensel B. Molecular maps of the reorganization of genome–nuclear lamina interactions during differentiation. Mol. Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material