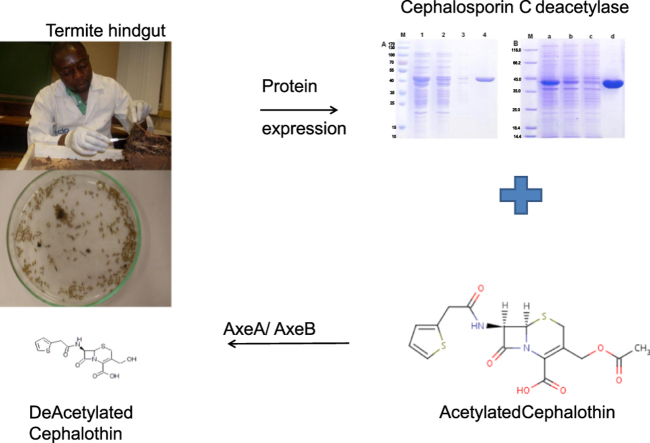
Abstract
In the pharmaceutical industry, de-acetylated cephalosporins are highly valuable starting materials for producing semi-synthetic β-lactam antibiotics. In this study a fosmid metagenome library from termite hindgut symbionts was screened for carboxyl ester hydrolases capable of de-acetylating cephalosporins. Recombinant Escherichia coli clones with esterolytic phenotypes on tributyrin agar plates were selected and further tested for de-acetylating activity against Cephalothin and 7-aminocephalosporanic acid (7-ACA). Two clones displaying de-acetylating activity were sequenced and the corresponding two carboxyl ester hydrolase encoding genes (axeA and axeB) belonging to the carbohydrate esterase family 7 (CE7) were identified. The primary structure of both the axeA and axeB revealed the presence of G-X-S-X-G sequence motif and respective subunit molecular masses of 40 kDa. In addition to de-acetylating cephalosporin based molecules, the two enzymes were also shown to be true esterases based on their preferences for short chain length fatty acid esters.
Keywords: Functional metagenomics, Antibiotic synthesis, Esterases, Deacetylated cephalosporins
Graphical Abstract
Highlights
-
•
Esterases (AxeA and AxeB) contained classical G-X-S-x-G motif and showed deacetylating activities against cephalosporin substrates.
-
•
AxeA and AxeB can be useful in the biocatalytic modification of cephalosporin molecules.
1. Introduction
The de-acetylated cephalosporins are highly valuable starting materials for producing semi-synthetic β-lactam antibiotics in pharmaceutical industry. Chemical processes for the production of de-acetylated cephalosporins involve general hydrolysis of the ester bond under alkaline conditions resulting in low yields and by-products that are toxic to the environment [1]. A white biotechnology process based on the use of ester hydrolysing enzymes has been suggested as an alternative approach to the chemical process for the production of de-acetylated cephalosporins [2]. The de-acetylation of cephalosporins using an enzymatic process offers several advantages including mild reaction conditions of pH, temperature, and high yields [3].
Carboxyl ester hydrolases (3.1.1.1) capable of de-acetylation of cephalosporins have been reported from two different esterase families, namely carbohydrate esterase family 7 (CE7) [4], [5], [6] and family VIII lipolytic enzymes [7]. However, carboxyl ester hydrolases belonging to these two families share no similarities in terms of the amino acid sequence identities and other biochemical properties. For example, the primary structure of family VIII esterases resemble that of class C β-lactamases, peptidases, and penicillin binding proteins [7], [8], while that of CE7 is related to the carbohydrate esterases [6]. Moreover, the substrate specify profile of family VIII esterases reveal a broad specificity range against fatty acid esters (C2-C8) [9], in contrast to carbohydrate esterase family 7 enzymes that show a limited substrate preference to acetate (C2) and propionate (C3) fatty acid esters [10]. Furthermore, family VIII lipolytic esterases generally display monomeric subunits, while CE7 display multi-oligomeric quaternary structures [6].
The P3 section of termites' hindgut is a metabolic engine involved in the degradation of a wide variety of compounds with the aid of obligate symbionts [11]. As a result, hindguts of termite species provide an ideal source for bio-prospecting for novel enzyme genes. In this study, we have screened a previously constructed termite hindgut metagenomic library [12] for esterases/cephalosporin C-deacetylases that can be applied in a chemo-enzymatic synthesis of deacetylated β-lactam molecules. The two identified carboxyl ester hydrolase encoding genes (axeA and axeB) were recombinantly expressed in Escherichia coli and their deacetylating catalytic abilities against acetylated β-lactam substrates were demonstrated.
2. Material and methods
2.1. Metagenomic library screening
The construction and screening of a metagenomic library from termite hindgut symbionts have been described previously [12]. Briefly, following the extraction of a total High molecular weight community DNA from the P3 section of termite hindgut, a large insert DNA library was generated using the copy control fosmid library production kit (Epicentre Biotechnologies, WI, USA). The library was screened for clones displaying esterolytic activity by tributyrin hydrolysis on LB tributyrin agar plates [12]. Esterase genes (AxeA and AxeB) from the positive fosmids were identified by generating pUC19 subclone libraries from fosmids partially digested with Sau3A1. Functional screening of the pUC19 recombinant esterase clones in E. coli was performed on LB agar plates supplemented with tributyrin 1% (v/v) and Gum Arabic 0.1% (w/v), followed by incubation at 37 °C. Esterase positive clones were identified by the presence of zone of clearance around the colony margins.
The nucleotide sequences of the pUC19 derived esterase positive clones were determined using the Sanger-Sequencing method. Open reading frames were identified using the GeneMark gene prediction tool (http://exon.gatech.edu/GeneMark/) and the amino acid sequences determined using the DNA translation tool available on the CLC Combine Workbench software (CLCBIO, Denmark). A similarity search was performed using the Basic Local Alignment Search tool (BLASTP) [13]. Sequence alignments and editing were done using the Bio-Edit software [14].
2.2. Protein expression and purification
The primer pairs, (AxeAF: 5′-ATCCATATGCCGTATATTGATATGC-3′/,AxeAR: 5′-GATCTCGAGTGCCGGATAGCTAACTG-3′) and (AxeBF: 5′-ATCCATATGGCA-CAATTTG-ACCTG-3′/, AxeBR”: 5′-GATCTCGAGGGTGCTTACGCAGCCATG-3′) were designed using available sequence information. The restriction sites (underlined) were incorporated into primers for directional cloning of the two esterase genes (AxeA and AxeB) into the pET20b and pET28a expression vector, respectively. Recombinant proteins were co-expressed with a C-terminus 6x His-tag. Plasmid DNA from pUC19-derived esterase positive clones was used as template for PCR amplification of axeA and axeB genes, using KAPA HiFi DNA Polymerase (KAPA Biosysyems). The resulting PCR products were ligated into pET20b and pET28a vectors restricted with the same enzymes, resulting pET20AxeA and pET28AxeB expression constructs which were used to transform E. coli BL21 (DE3) expression host.
Expression studies were performed using the EnBase technology [15], [16] with EnPresso™ tablet cultivation set (BioSilta, Finland) according to the supplier's recommended instructions. Recombinant protein production was induced with ITPG (1 mM) followed by additional overnight incubation at 20 °C. Cultures were pelleted and cells lysed using B-PER (in phosphate buffer, 50 mM (pH 7.5)) bacterial protein extraction reagent (Pierce, USA) according to the manufacturer's instructions, to release the intracellular proteins. Supernatants were then recovered following centrifugation (22,000g for 30 min).
Purification of recombinant proteins was performed using immobilized metal affinity chromatography with Protino Ni-TED 2000 pre-packed columns (Macherey-Nachel, Germany). Protein supernatants were first buffer exchanged into an equilibration buffer (50 mM NaH2PO4, 300 mM NaCl, pH 8) and AxeA and AxeB proteins were purified following manufacturer's instructions. The protein was eluted with the equilibration buffer containing additional 250 mM imidazole. Elution fractions were transferred into 20 mM Tris–HCl (pH 8) using VIVASPIN 10 kDa cut-off spin columns (Vivascience, U.K.). Protein concentrations were determined by Bradford method [17], using bovine serum albumin (BSA) as a standard. Purified products were analyzed on denaturing sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) [18].
2.3. Determination of native molecular weights
Molecular weights of native AxeA and AxeB proteins were determined with Superdex™ 200 10/30 GL (GE, Healthcare) pre-equilibrated with 50 mM Tris–HCl buffer (pH 7.5) containing 150 mM NaCl. The following molecules were used as reference standards: thyroglobulin (670,000 Da), γ-globulin (160,000 Da), oval bumin (44,000 Da), myoglobin (17,000 Da) and vitamin B12 (135 Da) (Bio-Rad, USA).
2.4. Standard esterase assay
Routine esterase activity assays were performed by a standard assay measuring the release of p-nitrophenol from p-nitrophenyl ester at 410 nm [19] using a Beckman DU850 UV/visible spectrophotometer, equipped with a temperature controller. Unless otherwise described, enzyme activity was measured at 30 °C with 1 mM p-nitrophenyl acetate as the substrate dissolved in 50 mM Tris–HCl, pH 8. The extinction coefficient of p-nitrophenol under these conditions was 13,800 M−1 cm−1.
2.5. Biochemical characterization
2.5.1. Substrate profiling
Substrate specificity of the AxeA and AxeB enzymes was determined using 1 mM p-nitrophenyl esters of various chain lengths: p-nitrophenyl acetate (C2), p-nitrophenyl butyrate (C4), p-nitrophenylcaprylate (C8) and p-nitrophenyllaurate (C12). Described enzyme activity was measured at 30 °C in 20 mM Tris–HCl, pH 7.5 with 1 mM p-nitrophenylesters (dissolved in isopropanol) as the substrate. The extinction coefficient of p-nitrophenol under these conditions was 13,800 M−1 cm−1. Experimental initial velocity data versus substrate concentration, with coefficients of variation of ≤5%, were fitted to the Michaelis–Menten equation.
2.5.2. Temperature optima and stability profiles
The temperature optima of the two enzymes were determined between 30 and 80 °C using esterase standard assay. The thermostability profiles of the AxeA and AxeB were determined by incubating the enzyme at a range of temperatures (30–80 °C) in Tris–HCl (50 mM, pH 7.5) and the residual activity determined at 30-min time intervals using the standard assay.
2.5.3. Activity on cephalosporin based substrates
Deacetylase activity of AxeA and AxeB against β-lactam substrates was determined using high-performance liquid chromatography (HPLC) by measuring de-acetylated forms of Cephalothin and 7-aminocephalosporanic acid. A β-lactamase from Bacillus cereus (Sigma) was used as a positive control. The reaction mixture contained purified enzymes (AxeA and AxeB) or β-lactamase positive control incubated with 1 mM substrate solution in 50 mM Tris–HCl (pH 8) at 30 °C for 1 h. The reaction was stopped by adding 0.2 mL of stop solution (100 mM H2SO4 and 30 mM crotonate). HPLC analyses of antibiotics reaction samples catalysed by either AxeA, AxeB or β-lactamase were carried out using a Hewlett Packard 1100 HPLC (Agilent Technologies Incorporated, Lovedale, CO, USA). The instrument was equipped with a binary pump autosampler, column thermostart, UV diode array detector and ChemStation Chromatography Management software (Reversion B.03.02, Agilent Technologies Incorporated).
Separation of β-lactam hydrolysis products was achieved using a Phenomenex Luna C18 analytical column (150×4.60 mm2, 5 μm inner dimension), coupled with the corresponding guard column (C18, 4×3.0 mm2, Phenomenex, USA). The isocratic mobile phase pumped at a flow rate of 1.00 mL/min consisted of 100 mM ammonium dihydrogen phosphate (HN4H2PO4) buffer, pH 4.50 and Methanol (HPLC grade) mixed at ratio of 95:5 and, 90:10 (% v/v) for 7-aminocephalosporanic acid. Cephalothin separation was carried out using 125 mM NH4H2PO4, pH 4.50 and Methanol (68:32% v/v), flowing at 1.00 mL/min. The mobile phases were prepared fresh, filtered through 0.45 μm membrane filter and degassed in ultrasonic bath for 30 min. Other chromatographic conditions were as follows: column oven temperature, 25 °C; injection volume, 10 μl; detection wavelength, UV 260 nm; and run time of 15 min. The antibiotics in the samples were identified by comparing their peak retention times with that of their standards.
2.5.4. Ultra performance liquid chromatography (UPLC)
The reaction samples of AxeA and AxeB against cephalosporin substrates were chromatographically evaluated on an UPLC high-definition quadrupole time-of-flight MS instrument (UPLC-qTOF SYNAPT G1 HDMS system, Waters, Manchester, UK) fitted with an Acquity HSS T3 C18 column (2.1×150 mm2, 1.7 µm; Waters Corporation). A binary solvent system consisting of eluent A: 0.1% formic acid in water and B: 0.1% formic acid in acetonitrile (Romil Chemistry, UK) was used at a constant column temperature of 60 °C. A 15 min gradient method at a constant flow rate of 0.4 mL/min was used for compound separation, and the conditions were: 5% B over 0.0–1.0 min, 5–95% B over 1.0–11.0 min and 95% B for 1 min. Thereafter, the column was returned to initial conditions at 13 min and allowed to equilibrate for 2 min. Chromatographic separation was monitored using a photodiode array (PDA) detector with a scanning range set between 200 and 500 nm, 1.2 nm bandwidth resolution and a sampling rate of 20 points s−1.
2.5.5. Quadrupole time-of-flight mass spectrometry (Q-TOF-MS)
Post-PDA detection, the reaction products were further detected with the aid of a SYNAPT G1 high definition mass spectrometer operating in positive and negative ionization modes. The MS conditions were as follows: capillary voltage of 2.5 kV, sample cone voltage of 30 V, extraction cone voltage of 4 V, MCP detector voltage of 1600 V, source temperature of 120 °C, desolvation temperature of 450 °C, cone gas flow of 50 L/h, desolvation gas flow of 550 L/h, m/z range of 100–1000, scan time of 0.2 s, interscan delay of 0.02 s, mode set as centroid, lockmass set as leucine enkephalin (556.2771/554.2615 Da), lockmass flow rate of 0.1 mL/min, and mass accuracy window of 0.5 Da. High purity nitrogen gas was used as desolvation, cone and collision gas. The software used to control the hyphenated system was MassLynx Ver. 4.1 (SCN 872).
2.5.6. Accession numbers
The Gene Bank accession numbers of AxeA and AxeB sequences reported in this paper are KF768743 and KF768744, respectively.
3. Results
3.1. Library screening and primary structure analysis
In this study pUC19 subclone libraries were generated from two of the selected active fosmid clones from a previously constructed termite hindgut metagenome library [12]. The fosmid clones were partially digested with Sau3AI and esterase open reading frames (ORFs) were re-screened on tributyrin agar for esterase activity and DNA inserts of active clones were identified with Sanger sequencing. Analysis of the sequences revealed two putative esterase encoding ORFs (ORF1 and ORF2). ORF1 and ORF2 were identified as putative acetyl xylan esterases (Axe) based on BLASTP sequence similarity searches and consequently the two ORFs (ORF1 and ORF2) were named AxeA and AxeB, respectively. The respective GC contents of AxeA and AxeB genes were 43% and 58%. The AxeA ORF was 975 nucleotides in length and encoded a polypeptide of 325 amino acids with a predicted molecular mass of approximately 37 kDa. The AxeB ORF spanned 966 bp encoding 322 residues with a calculated molecular mass of 35 kDa.
Both AxeA and AxeB proteins were identified as acetyl xylanesterases/cepholsporin C deacetalyses from the Genbank database. The AxeA ORF shared 60% sequence identity with the acetyl xylan esterases from Thermoanearobaxterium JW/SL YS485 [Accession no: AAB68821.1] and Paenibacillus sp. JDR-2 [Accession no: YP_003010899.1] as well as cepholsporin C deacetalyse from Thermoanaerobacterium xylanolyticum LX-11[Accession no: YP004471435]. On the other hand AxeB shared 63% sequence identity to acetyl xylan esterases from Xylanimonascellulosilytica [Accession no: YP003327295.1], Cellulomonasflavigena [Accession No: YP003636033.1] and Streptomyces avemitilis [Accession no: NP 822477.1].
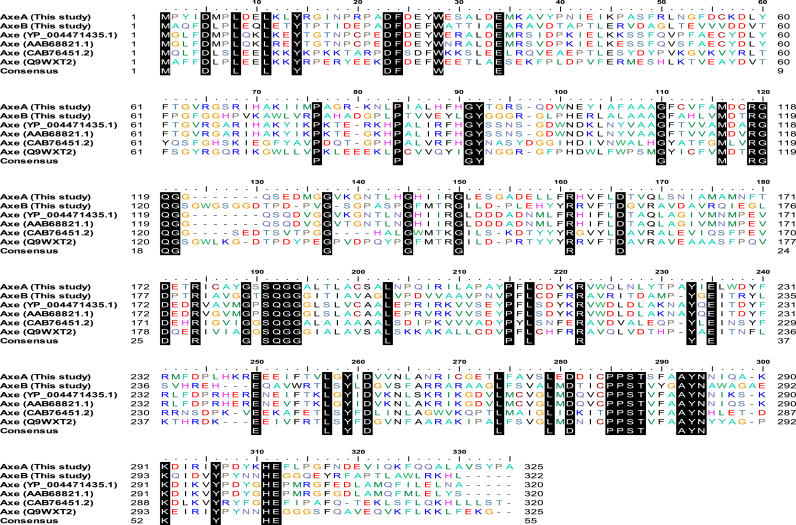
Deduced amino acid sequences for AxeA and AxeB were compared with published primary structures belong to family CE7. The proteins were characterized with a number of conserved sequence motifs common in carbohydrate esterases (Fig. 1). These included the pentapeptide signature (G-x-S-x-G) corresponding motifs (located at 180–185 for AxeA and 185–190 for AxeB (Fig. 1) and the –RGQ– (located at amino acid position 118–120 for AxeA and 119–121 for AxeB) and –HE– (amino acid positions 302–304 for AxeA and 303–305 for AxeB) sequence motifs that are typical for acetyl xylan esterases [3].
Fig. 1.
(A) Multiple alignment of AxeA and AxeB with related family 7 carbohydrate esterases showing the GxSxG and other conserved residues. The acetyl xylan esterases are represented by acetyl xylan esterases from Thermoanearobaxterium JW/SL YS485 (Accession no: AAB68821.1) and Bacillus pumilus (CAB76451). The cepholsporin C deacetalyse from CE-7 are represented by Thermoanaerobacterium xylanolyticum LX-11 (Accession No: YP004471435) and Thermotoga maritima MSB8 (Q9WXT2).
3.2. Protein expression and purification
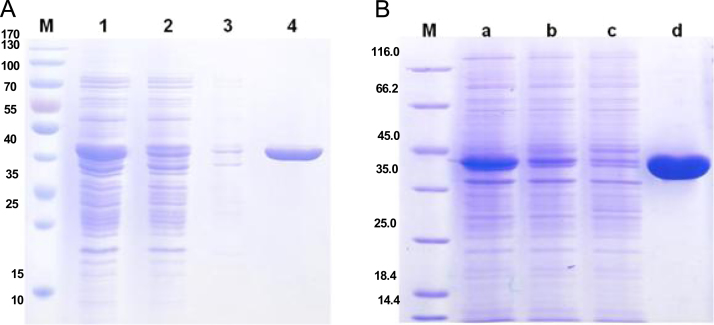
The two C-terminal 6x His-tagged recombinant proteins (AxeA and AxeB) were expressed heterologously using pET20b and pET28a vectors in E. coli BL21 (DE3), respectively and purified using a single step with immobilized metal affinity chromatography (IMAC). The recombinant AxeA and AxeB were purified to near homogeneity with respective 7.7 and 5.3 fold purification increases. The purification yields were 62.5% for AxeA and 38.5% for AxeB. Both the purified AxeA and AxeB fractions showed distinct protein bands of approximately 40 kDa on 12% SDS-PAGE (Fig. 2A and B). The electrophoretically deduced molecular mass of both AxeA and AxeB did not exactly correspond with the calculated molecular masses of 36855.10 and 36378.52 Da from the translated nucleotide sequence.
Fig. 2.
IMAC purification of AxeA and AxeB; (A) AxeA crude sample is represented on lane 1, the flow through and the wash fractions on lanes 2 and 3, respectively and the purified AxeA is on lane 4. (B) Purified AxeB; lane a, b, c and d indicate crude AxeB, flow through, wash and elution fractions, respectively. Lanes M on both gels represents protein ladders.
Native molecular weight determination using analytical size exclusion column chromatography confirmed the multimeric form of these enzymes, with AxeA showing the native molecular mass of 220 kDa while AxeB had a 70 kDa native structure (Fig S1: Supplementary data). The observation suggested that AxeA has a homo hexemaric while AxeB has a dimeric form. The results were consistent with other observations of acetyl xylan enzymes from Carbohydrate esterase (CE) family CE-7 [20], [21].
3.3. Biochemical characterization
3.3.1. Chain length specificity on p-nitrophenyl esters
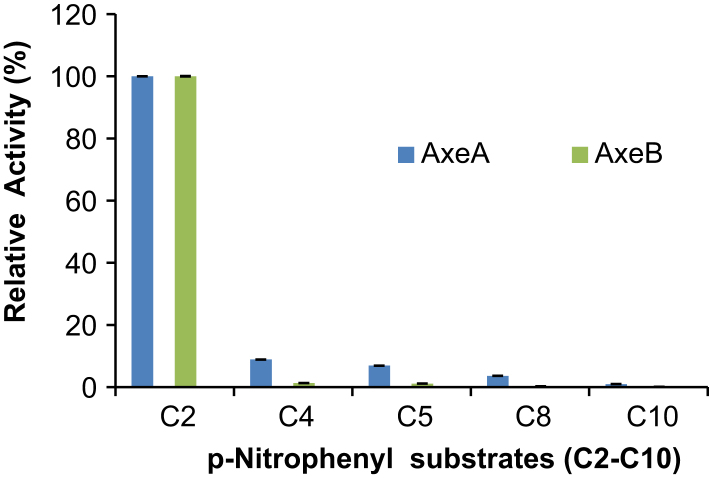
The hydrolytic activities of AxeA and AxeB against different fatty acid esters were investigated using a range of p-nitrophenyl (NP) esters (acetate, C2; butyrate, C4; velarate C5; caproate C8; and caprate, C10). Analysis of AxeA and AxeB hydrolytic patterns against these p-NP esters showed that the activity of these enzymes is limited to p-NP-C2 (Fig. 3). Activity of AxeA and AxeB obtained when using p-NP esters of chain length greater than C2 was 95% lower relative to that of p-NP-C2 (set at 100%).
Fig. 3.
Substrate specificity determination of AxeA and AxeB using p-nitrophenyl esters of various chain lengths: p-nitrophenyl acetate (C2), p-nitrophenyl butyrate (C4), p-nitrophenylcaprylate (C8) and p-nitrophenyllaurate (C12).
The kinetic constants for hydrolysis of the most readily hydrolysed substrate (p-NP-C2) by AxeA and AxeB is summarized in Table 1. AxeA showed higher affinity (KM) towards p-NP-C2 substrate compared to AxeB. Both the catalytic turnover (kcat) and efficiencies of AxeA were consistently higher than those of AxeB.
Table 1.
Purification table and kinetic parameters for hydrolysis of p-nitrophenyl esters by recombinant AxeA and AxeB.
| Enzyme | Fraction | Proteina(Mg) | Activityb(U) | Spec Activity (U mg−1) | Purification fold | Yield (%) | KM(mM) | Vmax(U mg−1) | kcatc(s−1) | kcat/KM(s−1/mM−1) |
|---|---|---|---|---|---|---|---|---|---|---|
| AxeA | Crude | 33 (±0.83) | 341 (±2.7) | 10.4 | 1.0 | 100 | ND | ND | ND | ND |
| Purified | 7.2 (0.1) | 566 (±3.7) | 80.9 | 7.7 | 62.5 | 0.1 | 28.8 | 4.5×10−11 | 4.28E−10 | |
| AxeB | Crude | 105 (±0.4) | 209 (±2.6) | 1.9 | 1.0 | 100 | ND | ND | ND | ND |
| Purified | 55 (±0.5) | 544 (±5.7) | 9.98 | 5.25 | 38.5 | 0.23 | 12.78 | 1.82×10−11 | 7.94E−11 |
Protein concentration was estimated using Bradford Assay.
Activity was assayed using p-nitrophenyl acetate as a substrate.
Kcat was calculated assuming (i) a molecular weight of 40 kDa for both AxeA and AxeB as estimated by SDS-PAGE analysis (The calculation also assumed a single active site per monomeric protein), Final Enzyme amount in all the kinetic assays=3 μg.
3.3.2. Temperature, pH and thermostability profile
Family CE7 carbohydrate esterases include some members that are highly thermostable [22], [23]. The highest sequence identity homologues of AxeA and AxeB identified were a number of biochemically characterized thermostable acetyl xylan esterases [24]. It was therefore of interest to investigate thermostability properties of these enzymes. Both enzymes were thermolabile with a half-life of <1 h at 60 °C (Fig. S2: Supplementary data). The temperature activity profiles showed that both AxeA and AxeB exhibited maximum activity at 40 °C, when using p-nitrophenyl acetate as the substrate (Fig. S3: Supplementary data).
3.3.3. Activity profiles against cephalosporins
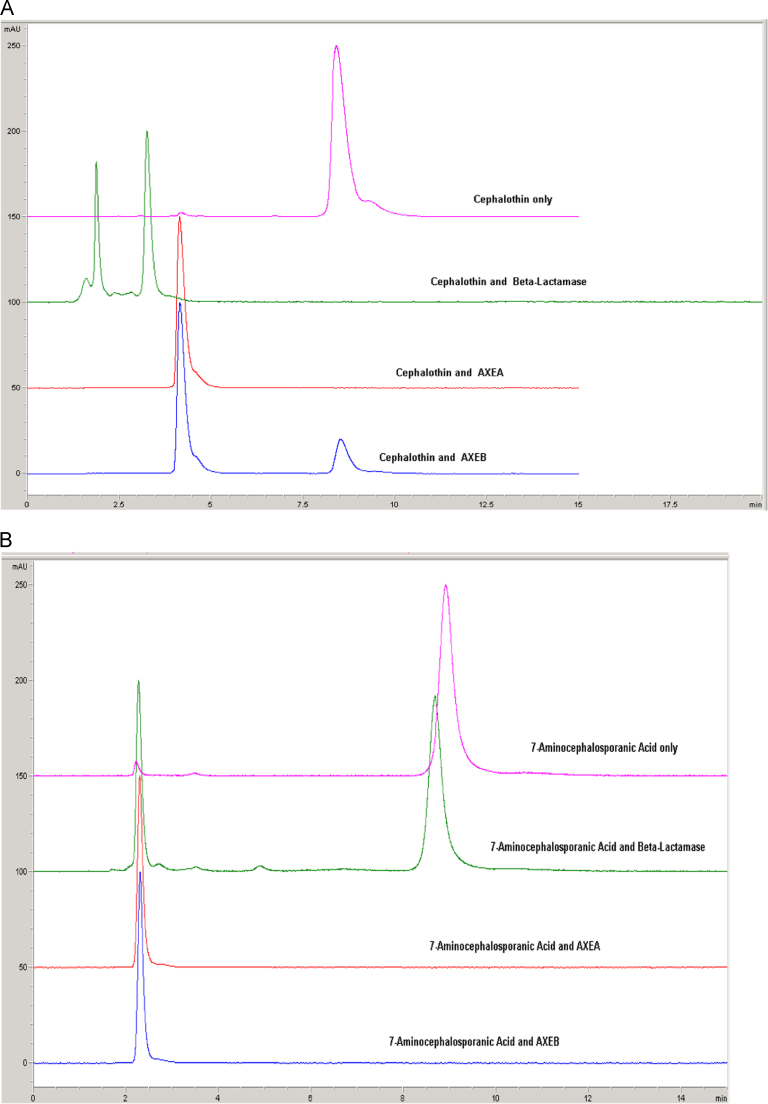
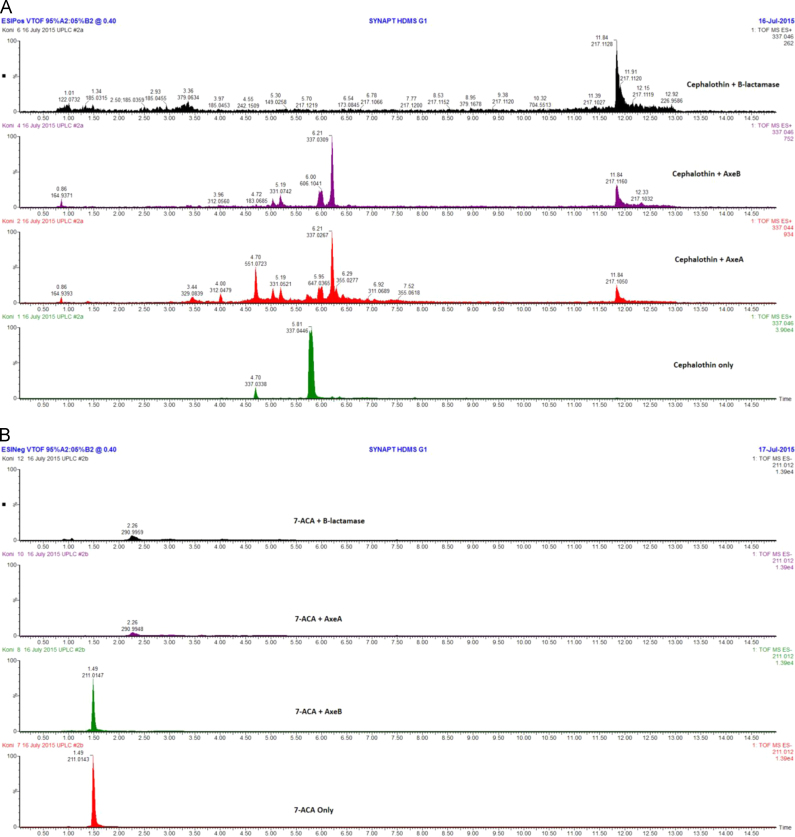
Acetyl xylan esterases/ cephalosporin deacetylases from the carbohydrate esterase family 7 (CE7) exhibit a narrow substrate specificity for acetate esters, but broad substrate specificity for other acetylated substrates including cephalosporins, acelytated carbohydrates and α/β-naphthyl acetate [21], [25]. Therefore AxeA and AxeB were investigated for their ability to remove acetyl groups from β-lactam substrates (Cephalothin and 7-aminocephalosporanic acid). Both enzymes were able to hydrolyse cephalosporin substrates. The average retention times of Cephalothin and 7-aminocephalosporanic acid were 8.415 and 8.888 min, and that of their product reactions were 4.167 and 2.359 min, respectively (Fig. 4A and B). Analysis of the hydrolytic patterns of AxeA and AxeB revealed that the mechanism of hydrolysis by AxeA and AxeB enzymes was different to that of positive control β-lactamase. The reactions of AxeA and AxeB against the Cephalothin and 7-ACA were also analysed by UPLC-TOF MS analysis and data processed using MassLynx software. The base peak intensity chromatograms (BPI) were evaluated in both ionisation polarities and extracted ion chromatograms generated to monitor the presence of Cephalothin and 7-ACA (7-aminocephalosporanic acid). Cephalosporin alcohols were identified from all the reactions that contained the two enzymes, indicating that both Axe A and AxeB selectively hydrolyse the ester bond and not the amide bond (Fig. 5).
Fig. 4.
HPLC analysis of the cephalosporin derivatives reactions performed with AxeA and AxeB using β-lactamase as the positive control, (A) Cephalothin substrate and (B) 7-amino-cephalosporanic acid. Substrates only were also analysed with the reaction products.
Fig. 5.
UPLC-TOF MS analysis of (A) Cephalothin and (B) 7-ACA (7-aminocepha-losporanic acid) hydrolysis by AxeA and AxeB.
4. Discussions
Cephalosporins are β-lactam antibiotics active against a broad spectrum of pathogenic bacteria [26]. The biosynthetic pathway of these molecules is well characterized both genetically and biochemically. Cephalosporin C is the only derivative that occurs naturally and its modification allows production of a series of semi-synthetic cephalosporins. Deacetylated cephalosporins are the starting material for many of the current the semi-synthetic cephalosporins. The production of these compounds can be achieved via an enzymatic process which requires use of cephalosporin C deacetylase enzymes. The enzymes belong to family 7 esterases active on carbohydrate substrates [4], [5] and family VII lipolytic enzymes [7], [8]. In this study, two ORFs encoding esterases AxeA and AxeB were identified from a metagenomic library from termite hindgut prokaryotic symbionts which was previously functionally screened for esterolytic activities [12].
The primary structure of the two ORFs displayed high amino acid sequence similarities to acetyl xylan esterases/cephalosporin C deacetylase. Both acetyl esterases identified in this study belonged to family CE7 of carbohydrate esterase. Esterases from this family are generally characterized by three highly conserved sequence motifs, namely the –RGQ–, –GxSQG– and HE sequence motifs [4]. Multiple sequence alignment revealed corresponding sequence motifs located at position (117–119 aa), (180–185 aa) and (301–302 aa) in the primary structure of AxeA, while in AxeB primary structure those motifs were located at position (118–120 aa), (185–189 aa) and (303–304). Previous studies have revealed that the serine residue within the G-x-S-x-G motif harbours the catalytic serine, which attacks the carbonyl carbon atom of the ester bond; a reaction that yields a tetrahedral intermediate stabilized by a basic His residue and an acidic residue that can either be Asp or Glu [8], [9]. Based on the multiple sequences analysis it was deduced that Ser 182, Asp 272, and His 301, in AxeA and Ser187, Asp273, and His 303 in AxeB form catalytic triad residues. In family CE7, the oxyanion hole that stabilises the tetrahedral intermediates is formed by groups of Tyr residue and Gln within the highly conserved RGQ motif. The putative oxyanion holeresidues were identified as Tyr91 and Gln 117 in AxeA while Tyr 92 and Gln119 in AxeB. Signal peptide prediction analysis of the AxeA and AxeB primary structures indicated the lack of signal peptide sequences, suggesting that both enzymes are expressed as full-length proteins.
Recombinant expression in E. coli and biochemical characterization of several members of family CE7 carbohydrate esterases has been successful [22], [23], [24], [25], [27]. Following expression and optimization studies, both AxeA and AxeB genes were successfully expressed in E. coli BL21 (DE3). The native subunit molecular weight of AxeA was determined to be 220 kDa suggesting a homohexamaric form. In contrast, the native subunit molecular of AxeB was dimeric with molecular mass of 70 kDa. Other proteins from CE7 carbohydrate esterase family have shown to have similar multimeric structures [10], [20], [21], [28].
The overall substrate specificity of carboxylesterases has been attributed to a number of features including differences in the size and overall hydrophobicity or hydrophilicity of the substrate binding pocket [7]. Esterase enzymes only hydrolyse water-soluble short acyl chain esters of less than 10 carbon atoms. Both AxeA and AxeB were only active on the short chain p-nitrophenol esters of acetate and did not hydrolyse esters with acyl chains longer than four carbon atoms. These observations have been reported with other members of family CE-7 esterases [21], [29] and indicate that the substrate binding pockets of both the enzymes could only accommodate a limited carbon chain length. This characteristic of CE-7 enzymes is different from other esterases, which generally have broad substrate specificity and are able to hydrolyse acyl chains longer than four carbons. The enzymes in the CE7 family are good examples of multifunctional proteins, which have typically evolved through the recruitment of different domains that have the required functional diversity [21]. In addition to p-nitrophenyl esters AxeA and AxeB enzymes were able to hydrolyse cephalosporins belonging to the β-lactam class of antibiotics. Analysis of the hydrolytic patterns of AxeA and AxeB revealed that the mechanism of hydrolysis of AxeA and AxeB was different from that of positive control β-lactamase, suggesting that both enzymes are only specific for the ester bond of the β-lactam substrates.
The ability of both AxeA and AxeB to hydrolyse β-lactam based substrate suggests that these enzymes can be useful in the preparation of deacetylated cephalosporins for the production of semi-synthetic β-lactam antibiotics.
Acknowledgements
The authors would like to thank the Council for Scientific and Industrial Research (CSIR) and Technology Innovation Agency of South Africa (TIA) Grant No. (PB99/08) for funding the project and the National Research Foundation of South Africa (NRF) for a Professional Development Program (PDP) fellowship to Dr. Nobalanda Mokoena.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.08.016.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Usher J.J., Lewis M.A., Hughes D.W. Development of cephalosporin C fermentation taking into account the instability of cephalosporin C. Biotechnol. Lett. 1988;10:543–548. [Google Scholar]
- 2.Abbott B., Fukuda D. Physical properties and kinetic behavior of a cephalosporin acetylesterase produced by Bacillus subtilis. Appl. Microbiol. 1975;3:413–419. doi: 10.1128/am.30.3.413-419.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martínez-Martínez I., Montoro-García S., Lozada-Ramírez D.J. A colorimetric assay for the determination of acetyl xylan esterase or cephalosporin C acetyl esterase activities using 7-amino cephalosporanic acid, cephalosporin C, or acetylated xylan as substrate. Anal. Biochem. 2007;369:210–217. doi: 10.1016/j.ab.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Coutinho P.M., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H.J., Davies G., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Royal Society of Chemistry; Cambridge: 1999. pp. 3–12. [Google Scholar]
- 5.Cantarel B.L., Coutinho P.M., Rancurel C. The carbohydrate active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biely P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 2012;30:1575–1588. doi: 10.1016/j.biotechadv.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Arpigny K.L., Jaeger K.E. Bacterial lipolytic enzymes: classification and properties. J. Biochem. 1999;343:177–183. [PMC free article] [PubMed] [Google Scholar]
- 8.Bornscheuer U.T. Microbial carboxyl esterases: classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002;26:73–81. doi: 10.1111/j.1574-6976.2002.tb00599.x. [DOI] [PubMed] [Google Scholar]
- 9.Mokoena N., Mathiba K., Tsekoa T. Functional characterisation of a metageno e derived family VIII esterase with a deacetylation activity on β-lactam antibiotics. Biochem. Biophys. Res. Commun. 2013;437:342–348. doi: 10.1016/j.bbrc.2013.06.076. [DOI] [PubMed] [Google Scholar]
- 10.Lorenz W.W., Wiegel J. Isolation, analysis, and expression of two genes from Thermoanaerobacterium sp. strain JW/SL YS485: a β-xylosidase and a novel acetyl xylan esterase with cephalosporin C deacetylase activity. J. Bacteriol. 1997;179:5436–5441. doi: 10.1128/jb.179.17.5436-5441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arima K., Woodley J. Computational methods for understanding bacterial and archeal genomes. In: Xu Y., Gogatern P., editors. Metagenomics. Imperial College press; London: 2008. pp. 345–350. [Google Scholar]
- 12.Rashamuse K., Mabizela-Mokoena N., Sanyika T.W. Accessing carboxylesterase diversity from termite hindgut symbionts through metagenomics. J. Mol. Microb. Biotechnol. 2012;22:277–286. doi: 10.1159/000342447. [DOI] [PubMed] [Google Scholar]
- 13.Altschul S.F., Madden T.S., Schäffer A.A. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall T.A. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1990;41:95–98. [Google Scholar]
- 15.Krause M., Ukkonen K., Haataja T. A novel fed-batch based cultivation method provides high cell-density and improves yield of soluble recombinant proteins in shaken cultures. Microb. Cell. Fact. 2010;9:1–11. doi: 10.1186/1475-2859-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panula-Perälä J., Siurkus J., Vasala A. Enzyme controlled glucose auto-delivery for high cell density cultivations in microplates and shake flasks. Microb. Cell Fact. 2008;7:7–11. doi: 10.1186/1475-2859-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradford M.M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. J. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Rashamuse K., Burton S., Cowan D. A novel recombinant ethyl ferulate esterase from Burkholderia multivorans. J. Appl. Microbiol. 2007;103:1610–1620. doi: 10.1111/j.1365-2672.2007.03394.x. [DOI] [PubMed] [Google Scholar]
- 20.Tian Q., Song P., Jiang L. A novel cephalosporin deacetylating acetyl xylan esterase from bacillus subtilis with high activity toward cephalosporin C and 7-aminocephalosporanic acid. Appl. Microbiol. Biotechnol. 2013;98:2081–2089. doi: 10.1007/s00253-013-5056-x. [DOI] [PubMed] [Google Scholar]
- 21.Vincent F., Charnock S.J., Verschueren K.H. Multifunctional xylooligosaccharide/cephalosporin C deacetylase revealed by the hexameric structure of the Bacillus subtilis enzyme at 1.9 Å resolution. J. Mol. Biol. 2003;330:593–606. doi: 10.1016/s0022-2836(03)00632-6. [DOI] [PubMed] [Google Scholar]
- 22.Hedge M.K., Gehring A.M., Adkins C.T. The structural basis for the narrow substrate specificity of an acetyl esterase from Thermotoga maritima. Biochim. Biophys. Acta. 1824;2012:1024–1030. doi: 10.1016/j.bbapap.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Levisson M., Han G.W., Deller M.C. Functional and structural characterization of a thermostable acetyl esterase from Thermotoga maritima. Proteins. 2012;80:1545–1559. doi: 10.1002/prot.24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao W., Wiegel J. Purification and characterization of two thermostable acetyl xylan esterases from Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 1995;61:729–733. doi: 10.1128/aem.61.2.729-733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degrassi G., Kojic M., Ljubijankic G. The acetyl xylan esterase of Bacillus pumilus belongs to a family of esterases with broad substrate specificity. Microbiology. 2000;146:1585–1591. doi: 10.1099/00221287-146-7-1585. [DOI] [PubMed] [Google Scholar]
- 26.Medeiros A. Evolution and dissemination of b-lactamases. Clin. Infect. Dis. 1997;24:S19–S45. doi: 10.1093/clinids/24.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 27.Kabel M.A., Yeoman C.J., Han Y. Biochemical characterization and relative expression levels of multiple carbohydrate esterases of the xylanolytic rumen bacterium Prevotella ruminicola 23 grown on an ester-enriched substrate. Appl. Environ. Microbiol. 2011;77:5671. doi: 10.1128/AEM.05321-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takimoto A., Mitsushima K., Yagi S. Purification, characterization and partial amino acid sequences of a novel cephalosporin-C deacetylase from Bacillus subtilis. J. Ferment. Bioeng. 1994;77:17–22. [Google Scholar]
- 29.Alalouf O., Balazs Y., Volkinshtein M. A new family of Carbohydrate esterases is represented by a GDSL/Acetylxylan esterase from Geobacillus stearothermophilus. J. Biol. Chem. 2011;286:41993–42001. doi: 10.1074/jbc.M111.301051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material