Abstract
The seamless ligation cloning extract (SLiCE) method is a novel seamless DNA cloning tool that utilizes homologous recombination activities in Escherichia coli cell lysates to assemble DNA fragments into a vector. Several laboratory E. coli strains can be used as a source for the SLiCE extract; therefore, the SLiCE-method is highly cost-effective.The SLiCE has sufficient cloning ability to support conventional DNA cloning, and can simultaneously incorporate two unpurified DNA fragments into vector. Recently, many seamless DNA cloning kits have become commercially available; these are generally very convenient, but expensive. In this study, we evaluated the cloning efficiencies between a simple and highly cost-effective SLiCE-method and a commercial kit under various molar ratios of insert DNA fragments to vector DNA. This assessment identified that the SLiCE from a laboratory E. coli strain yielded 30−85% of the colony formation rate of a commercially available seamless DNA cloning kit. The cloning efficiencies of both methods were highly effective, exhibiting over 80% success rate under all conditions examined. These results suggest that SLiCE from a laboratory E. coli strain can efficiently function as an effective alternative to commercially available seamless DNA cloning kits.
Abbreviations: CFU, colony-forming units; G6PDH1, glucose-6-phosphate dehydrogenase 1; PCR, polymerase chain reaction; Prx IIE, type II peroxiredoxin E; SLiCE, seamless ligation cloning extract
Keywords: Homologous recombination, Seamless DNA cloning, SLiCE
Graphical abstract
Highlights
-
●
Efficiencies of homemade vs. commercial seamless DNA cloning methods were compared.
-
●
The SLiCE-method functioned at 30–85% efficiency of the In-Fusion cloning method.
-
●
Both seamless DNA cloning methods showed reduced efficiency under excess insert DNA.
1. Introduction
Traditional DNA cloning methods using restriction enzymes and DNA ligases are time consuming because the limited availability of restriction endonuclease sites often interferes with generation of the desired DNA constructs. To avoid such limitations and complicated procedures, various seamless DNA cloning methods have been developed over the last decade [1], [2], [3], [4]. Of these, the seamless ligation cloning extract (SLiCE) method is a novel homemade protocol that utilizes in vitro homologous recombination activities in cell lysates prepared from Escherichia coli [5], [6]. Zhang et al. originally demonstrated that the cell lysates prepared from an E. coli DH10B-derivative modified to express a λ prophage Red/ET recombination system, which was termed the PPY strain, had high DNA cloning activity using short end homology regions between insert and vector DNA fragments [5]. E. coli laboratory strains also contain two endogenous RecA-dependent and -independent pathways of homologous recombination [7], [8], [9], [10], [11]; however, the E. coli RecA− laboratory strain cell lysates did not exhibit high cloning efficiency when utilizing short end homology regions (15–20 bp) [5].
Recently, we reported that the cloning efficiency of cell lysates from E. coli RecA− laboratory strains was improved by harvesting E. coli cells at late log phase and extracting the lysates carefully on ice [6]. This demonstrates that the endogenous RecA-independent recombination activities in E. coli RecA− laboratory strains can function efficiently for the SLiCE-method using short homology lengths (approximately 15−19 bp), without the requirement of exogenously expressing a λ prophage Red/ET recombination system. In addition, SLiCE prepared from an E. coli RecA− laboratory strain could simultaneously incorporate two unpurified insert DNA fragments into vector, indicating highly efficient cloning activity [6]. SLiCE from E. coli laboratory strains is also cost-effective for seamless DNA cloning; however, the use of a commercially available cell lytic reagent, CelLytic B Cell Lysis Reagent (Sigma B7435) has been required for the preparation of SLiCE [5], [6]. Thus, the need for the commercial cell lytic reagent increases the cost of the SLiCE method.
More recently, we found that SLiCE can instead be prepared with buffers containing Triton X-100 [12], which is a commonly available nonionic detergent that is generally used for protein preparations [13], [14], [15], [16]. By using the Triton X-100 buffer, the SLiCE-method becomes an ultra-low cost homemade seamless DNA cloning method [12]. On the other hand, commercially available kits for seamless DNA cloning have been widely used [17], [18], [19], [20], [21], [22], [23], [24]. Although commercial kits are associated with a high cost per reaction, they are generally accepted to be easy to use and efficient. However, the differences in seamless DNA cloning efficiency between a homemade method and commercial kits are not well characterized. Therefore, in this study, we evaluated the efficiency of these methods under various molar ratios of insert DNA fragments to vector DNA.
2. Materials and methods
2.1. Preparation of SLiCE from an E. coli RecA− laboratory strain
SLiCE was prepared from E. coli JM109 using a buffer containing 3% (w/v) Triton X-100 [12]. E. coli JM109 pre-cultured in LB Miller medium (1 mL) at 37 °C were transferred to 2× YT medium (50 mL) in a 100-mL round-bottom, long-neck Sakaguchi shaking flask. The cells were grown at 37 °C in a reciprocal shaker (160 rpm) until the OD600 reached a value of 3.0 (late log phase). The cells were harvested by centrifugation at 5000g for 10 min at 4 °C. The cells were then washed with 50 mL sterilized water (ice-cold), and centrifuged at 5000g for 5 min at 4 °C. The washed cells were recovered with a yield of 0.40 g, and gently resuspended in 1.2 mL 3% (w/v) Triton X-100 in 50 mM Tris–HCl (pH 8.0) and incubated for 10 min at room temperature. The cell lysates were then centrifuged at 20,000g for 2 min at 4 °C. All subsequent procedures were performed on ice. The supernatants were carefully transferred into 1.5-mL microtubes to remove the insoluble materials, and determined the protein concentration as ~0.9 mg/mL by the BCA assay method. An equal volume of ice-cold 80% (v/v) glycerol was added to the supernatant, and mixed gently. The produced SLiCE extracts were snap-frozen in a bath of liquid nitrogen and stored at −80 °C in 40% (v/v, final concentration) glycerol.
2.2. Preparation of insert DNA fragments and linearized vector DNA
The Arabidopsis type II peroxiredoxin E (Prx IIE, 0.6 kilo base pairs (kbp), AT3G52960) [25], [26] and chloroplastic glucose-6-phosphate dehydrogenase 1 (G6PDH1, 1.6 kbp, AT5G35790) [27] genes were used as insert DNA molecules. pET23a (Merck Millipore, Billerica, MA, USA) was used as a vector DNA template. The insert DNA fragments and linearized pET23a DNA (3.7 kbp)were amplified using the primers listed in Table S1 and TksGflex DNA polymerase (Takara-Bio, Otsu, Japan) by polymerase chain reaction (PCR) [6], [12]. Overlap-regions between insert and vector DNA were designed as 15 bp lengths. PCR-amplified insert and vector DNA were treated with DpnI (37 °C, 60 min) to avoid cross-contaminating of methylated-DNA as PCR-template and purified using a FastGene Gel/PCR Extraction Kit (NIPPON Genetics, Tokyo, Japan) following agarose gel electrophoresis.
2.3. SLiCE reaction for seamless DNA cloning
SLiCE buffer (10×; 500 mM Tris–HCl, pH 7.5, 100 mM MgCl2, 10 mM ATP, and 10 mM dithiothreitol) was passed through a 0.2-μm filter, and dispensed in 40 μL aliquots into 0.2 mL 8-strip PCR tubes and stored at −20 °C [6]. The standard SLiCE reaction solution comprised the following components: 10 ng linear vector (PCR amplified), an appropriate amount of insert DNA (1:1–50:1 molar ratio of insert to vector), 1 μL 10× SLiCE buffer, 1 μL SLiCE extract, and sterilized distilled water to a total volume of 10 μL. The SLiCE reaction mixture was incubated at 37 °C for 15 min. The reaction time was adjusted to compare with that used in the In-Fusion cloning method.
2.4. In-Fusion reaction for seamless DNA cloning
The In-Fusion HD Cloning Kit (Clontech, Mountain View, CA, USA) was purchased from Takara-Bio. The standard In-Fusion reaction solution comprised the following components: 10 ng linear vector (PCR amplified), an appropriate amount of insert DNA (1:1–50:1 molar ratio of insert to vector), 2 μL 5× In-Fusion HD enzyme premix, and sterilized distilled water to a total volume of 10 μL. The In-Fusion reaction mixture was incubated at 50 °C for 15 min, according to the instruction manual.
2.5. Transformation of competent cells with SLiCE and In-Fusion solutions
Following incubation, heat-shock transformation was conducted by adding 1 μL of SLiCE or In-Fusion reaction solution into 20 μL ECOS X Competent E. coli DH5α (Nippon Gene, Tokyo, Japan) according to the instruction manual. The transformation efficiency of the competent cells (20 μL) was approximately 2×108 colony-forming units (CFUs)/μg pUC19 DNA. Transformed E. coli cells were plated on LB agar plates containing ampicillin and incubated at 37 °C for 12–16 h.
2.6. Efficiency estimation for SLiCE and In-Fusion cloning methods
The efficiency of seamless DNA cloning was evaluated by two parameters, colony-formation rate and cloning efficiency. Colony formation rate was determined as the number of colonies represented as CFUs per nanogram vector. Cloning efficiencies for the insert DNA were given as the ratio of colonies with an insert of the confirmed correct length as estimated by colony-PCR to the total number of colonies tested [6], [12].
3. Results and discussion
3.1. Estimation of colony formation rate and cloning efficiency for SLiCE and In-Fusion cloning methods
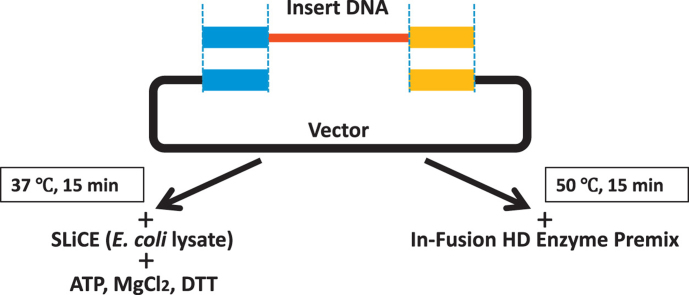
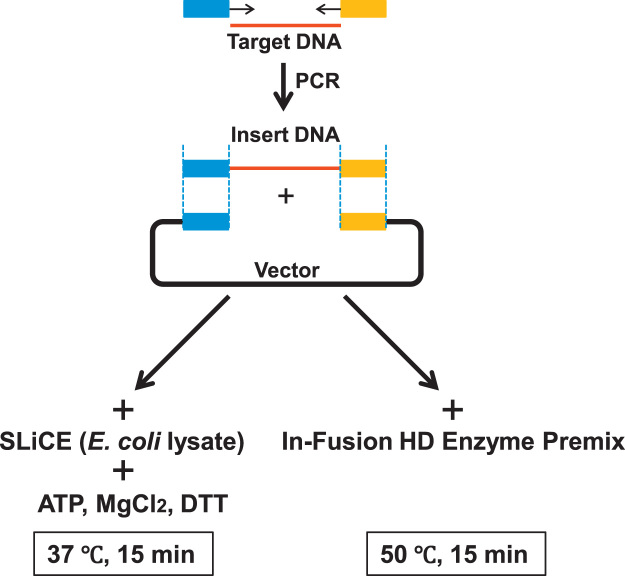
To date, SLiCE has been prepared according to several methods. SLiCE was originally prepared from the E. coli PPY strain, which expresses a λ prophage Red/ET recombination system, using commercially available cell lytic buffer, CelLytic B Cell Lysis Reagent (Sigma B7435) [5]. Recently, we found that SLiCE were able to be prepared from easily available E. coli laboratory strains using a cost-effective Triton X-100 buffer as well as with the commercially available cell lytic buffer [6], [12]. In this study, we used the simplest and most cost-effective SLiCE identified: the extract prepared from the E. coli RecA− laboratory strain JM109 with the 3% Triton X-100 buffer, and compared its cloning efficiency with a commercially available seamless DNA cloning kit. The Clontech In-Fusion HD Cloning Kit, which is widely used for seamless DNA cloning, was selected as being representative of commercial kits [17], [18], [19], [21], [23], [24]. In this comparative experiment, the cloning abilities of both seamless DNA cloning methods were evaluated under various molar ratios of insert DNA fragments to vector DNA by two parameters: colony formation rate and cloning efficiency. The reaction time of SLiCE-cloning was set as 15 min, which is the same as that recommended in the In-Fusion cloning method (Fig.1).
Fig. 1.
A schematic view of two seamless DNA cloning methods. Lower left, SLiCE-cloning from an E. coli RecA− laboratory strain. Lower right, In-Fusion cloning. We utilized 15 bp overlapping sequences in the experiments (short end homologous overlapping sequences were represented as blue and yellow squares)
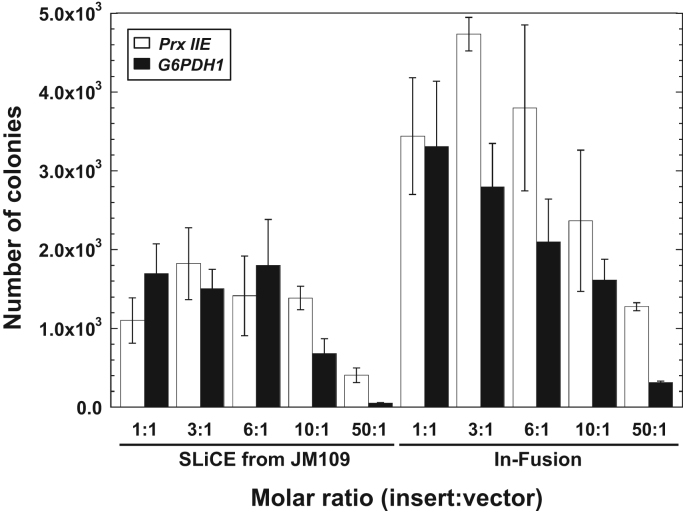
Firstly, we determined the colony formation rate (the number of colonies after transformation), as the efficiency of SLiCE-cloning is largely dependent upon this rate [6]. The In-Fusion cloning method yielded about 2–5×103 colonies/ng vector at 1:1 to 6:1 (insert: vector) ratios (Fig.2, right). For SLiCE-cloning, over 1×103 colonies/ng vector were stably observed over the same range of transformed DNA ratios (Fig. 2, left). These results indicated that the colony formation ability of SLiCE-cloning was 30–85% that of In-Fusion cloning for 1:1 to 6:1 molar ratios of insert to vector. Both rates are sufficient for conventional DNA cloning. An increase of the molar ratio of insert to vector (50:1) markedly reduced the colony formation rate of both cloning methods (Fig. 2). Excess insert DNA might inhibit the precision of the homologous recombination at short end homology regions between insert fragments and vector DNA, as an excess of insert DNA facilitates insert dimer formation.
Fig. 2.
Effects of the molar ratio of insert DNA fragments to vector DNA on transformation efficiency for the two seamless DNA cloning methods. The cloning reactions were performed with the indicated molar ratios of insert: vector. Values for the insert DNA fragments Prx IIE or G6PDH1 are shown. Colony numbers are represented as CFUs per nanogram of vector and reflect the means±standard deviation of three independent experiments [6].
We next confirmed the cloning efficiency of both SLiCE and In-Fusion methods. Previous studies showed that SLiCE-cloning exhibited high fidelity for seamless gene cloning [5], [6]. Therefore, as expected, both methods were able to correctly clone the insert DNA fragments into the vector at high cloning efficiencies; both maintained good efficiencies (>80%) across all input DNA ratio conditions as well (Table 1). A small number of incorrect clones were detected in both methods. In SLiCE method, four of nine incorrect clones were empty vector clones and five incorrect clones had two insert fragments in a vector. For In-Fusion method, one of three incorrect clones was empty vector clone and two incorrect clones had two insert fragments in a vector.
Table1.
Cloning efficiencies of the various molar ratios of insert DNA fragments to vector DNA in the SLiCE and In-Fusion methods.
| Cloning method | Cloning efficiencya |
|
|---|---|---|
| PrxIIE (AT3G52960) | G6PDH1 (AT5G35790) | |
| 0.5 kbp | 1.6 kbp | |
| SLiCE | ||
| 1:1 | 14/16 (87.5%) | 16/16 (100%) |
| 3:1 | 14/16 (87.5%) | 16/16 (100%) |
| 6:1 | 15/16 (93.8%) | 16/16 (100%) |
| 10:1 | 16/16 (100%) | 16/16 (100%) |
| 50:1 | 15/16 (93.8%) | 13/16 (81.3%) |
| In-Fusion | ||
| 1:1 | 16/16 (100%) | 16/16 (100%) |
| 3:1 | 16/16 (100%) | 16/16 (100%) |
| 6:1 | 15/16 (93.8%) | 16/16 (100%) |
| 10:1 | 16/16 (100%) | 16/16 (100%) |
| 50:1 | 16/16 (100%) | 14/16 (87.5%) |
Cloning efficiencies for the insert DNA are represented as the number of clones with the confirmed correct insert length by colony-PCR/number of colonies subjected to colony-PCR.
3.2. Comparison of homemade and commercially available seamless DNA cloning methods
The In-Fusion cloning method is performed using commercially available kit and provided stable cloning activity (Fig. 2). The SLiCE-cloning method also exhibited sufficient high cloning ability for conventional DNA cloning. These results indicate that SLiCE-cloning using extracts from an E. coli laboratory strain is able to serve as an alternative seamless cloning method to the commercial In-Fusion cloning kit. When selecting between homemade reagents or commercially available kits, we generally make the determination considering the time and labor required for preparation of homemade reagents, and the cost of the commercial kits. For seamless DNA cloning, preparation of SLiCE from E. coli laboratory strains reflects a simple and easy protocol, as only two steps are required: growth of an E. coli laboratory strain under normal conditions, and simple cell lysate extraction using lysis buffer [6], [12].
Accordingly, the SLiCE-method presented here would be easy to implement, if care is taken at several points during preparation of SLiCE extract. Firstly, optimum harvest timing (OD600=2.0–3.0) is a critical factor for determining the cloning efficiency. Secondly, SLiCE-preparation must be performed at a low temperature to avoid a decrease in the cloning ability. Thirdly, sonication should not be used as the method for the cell lysate extraction from E. coli, because although sonication is an efficient method for protein extraction from E. coli [28], [29], [30], its use completely abolished SLiCE-cloning ability [12]. Instead, by using a buffer containing Triton X-100 or a commercially available cell lytic reagent, we can maintain SLiCE quality and reproducibility [6], [12]. These results indicate the importance of SLiCE-preparation techniques for maintaining the homologous recombination activity in E. coli cells.
Furthermore, SLiCE-cloning is a convenient and ultra-low cost seamless DNA cloning method for several reasons. The quantity of SLiCE prepared from 50 mL E. coli culture was sufficient to perform over two thousand SLiCE-cloning reactions, and the produced SLiCE extract can be stored in 40% glycerol at −80 °C for long periods if snap-frozen in liquid nitrogen. Additionally, the SLiCE-method presented here does not require a special E. coli strain and a commercial cell lytic buffer, enhancing its accessibility and reducing its cost. As an approximate guide, we calculated the cost of SLiCE prepared from the E. coli RecA− laboratory strain JM109 with a buffer containing Triton X-100 to be ¥0.4 (approximately $0.003) per reaction; over half of this sum is represented by the cost of the plastic tubes and the cost is less than 1/5000 that of the In-Fusion cloning kit. So far, SLiCE-cloning has yielded lower efficiency than In-Fusion cloning, but SLiCE-cloning has the potential for further improvement of the cloning efficiency upon elucidation of the molecular mechanisms of the RecA-independent recombination system in future studies.
Acknowledgments
This work was supported by the MEXT-Supported Program for the Strategic Research Foundation at Private Universities (to K.M.).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.09.005.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Li M.Z., Elledge S.J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 2.Klock H.E., Koesema E.J., Knuth M.W., Lesley S.A. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–994. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 3.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 4.Quan J., Tian J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 2009;4:e6441. doi: 10.1371/journal.pone.0006441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Werling U., Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motohashi K. A simple and efficient seamless DNA cloning method using SLiCE from Escherichia coli laboratory strains and its application to SLiP site-directed mutagenesis. BMC Biotechnol. 2015;15:47. doi: 10.1186/s12896-015-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muyrers J.P., Zhang Y., Buchholz F., Stewart A.F. RecE/RecT and Redα/Redβ initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 9.Lovett S.T., Hurley R.L., Sutera V.A., Jr., Aubuchon R.H., Lebedeva M.A. Crossing over between regions of limited homology in Escherichia coli. RecA-dependent and RecA-independent pathways. Genetics. 2002;160:851–859. doi: 10.1093/genetics/160.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutra B.E., Sutera V.A., Jr., Lovett S.T. RecA-independent recombination is efficient but limited by exonucleases. Proc. Natl. Acad. Sci. USA. 2007;104:216–221. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persky N.S., Lovett S.T. Mechanisms of recombination: lessons from E. coli. Crit. Rev. Biochem. Mol. Biol. 2008;43:347–370. doi: 10.1080/10409230802485358. [DOI] [PubMed] [Google Scholar]
- 12.Okegawa Y., Motohashi K. Evaluation of seamless ligation cloning extract preparation methods from an Escherichia coli laboratory strain. Anal. Biochem. 2015;486:51–53. doi: 10.1016/j.ab.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 13.Kitano K., Motohashi K., Yoshida M., Miki K. A novel approach to crystallizing proteins with temperature-induction method: GrpE protein from Thermus thermophilus. J. Cryst. Growth. 1998;186:456–460. [Google Scholar]
- 14.Motohashi K., Koyama F., Nakanishi Y., Ueoka-Nakanishi H., Hisabori T. Chloroplast cyclophilin is a target protein of thioredoxin. Thiol modulation of the peptidyl-prolyl cis–trans isomerase activity. J. Biol. Chem. 2003;278:31848–31852. doi: 10.1074/jbc.M304258200. [DOI] [PubMed] [Google Scholar]
- 15.Motohashi K., Hisabori T. HCF164 receives reducing equivalents from stromal thioredoxin across the thylakoid membrane and mediates reduction of target proteins in the thylakoid lumen. J. Biol. Chem. 2006;281:35039–35047. doi: 10.1074/jbc.M605938200. [DOI] [PubMed] [Google Scholar]
- 16.Motohashi K., Hisabori T. CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid. Redox Signal. 2010;13:1169–1176. doi: 10.1089/ars.2010.3138. [DOI] [PubMed] [Google Scholar]
- 17.Xu Z., Thomas L., Davies B., Chalmers R., Smith M., Brown W. Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 2013;13:87. doi: 10.1186/1472-6750-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goossens K.V., De Greve H., Willaert R.G. Cloning, expression, and purification of the N-terminal domain of the Flo1 flocculation protein from Saccharomyces cerevisiae in Pichia pastoris. Protein Exp. Purif. 2013;88:114–119. doi: 10.1016/j.pep.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Thomas S., Smits S.H., Schmitt L. A simple in vitro acylation assay based on optimized HlyA and HlyC purification. Anal. Biochem. 2014;464:17–23. doi: 10.1016/j.ab.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Waneskog M., Bjerling P. Multi-fragment site-directed mutagenic overlap extension polymerase chain reaction as a competitive alternative to the enzymatic assembly method. Anal. Biochem. 2014;444:32–37. doi: 10.1016/j.ab.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 21.Fayed B., Younger E., Taylor G., Smith M.C. A novel Streptomyces spp. integration vector derived from the S. venezuelae phage SV1. BMC Biotechnol. 2014;14:51. doi: 10.1186/1472-6750-14-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itkonen J.M., Urtti A., Bird L.E., Sarkhel S. Codon optimization and factorial screening for enhanced soluble expression of human ciliary neurotrophic factor in Escherichia coli. BMC Biotechnol. 2014;14:92. doi: 10.1186/s12896-014-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camilo C.M., Polikarpov I. High-throughput cloning, expression and purification of glycoside hydrolases using Ligation-Independent Cloning (LIC) Protein Exp. Purif. 2014;99:35–42. doi: 10.1016/j.pep.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Davies H.A., Wilkinson M.C., Gibson R.P., Middleton D.A. Expression and purification of the aortic amyloid polypeptide medin. Protein Exp. Purif. 2014;98:32–37. doi: 10.1016/j.pep.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Dietz K.J., Horling F., Konig J., Baier M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J. Exp. Bot. 2002;53:1321–1329. [PubMed] [Google Scholar]
- 26.Brehelin C., Meyer E.H., de Souris J.P., Bonnard G., Meyer Y. Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiol. 2003;132:2045–2057. doi: 10.1104/pp.103.022533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakao S., Benning C. Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J. 2005;41:243–256. doi: 10.1111/j.1365-313X.2004.02293.x. [DOI] [PubMed] [Google Scholar]
- 28.Makyio H., Niwa H., Motohashi K., Taguchi H., Yoshida M. Stabilization of FtsH-unfolded protein complex by binding of ATP and blocking of protease. Biochem. Biophys. Res. Commun. 2002;296:8–12. doi: 10.1016/s0006-291x(02)00830-6. [DOI] [PubMed] [Google Scholar]
- 29.Yamaryo Y., Motohashi K., Takamiya K., Hisabori T., Ohta H. In vitro reconstitution of monogalactosyldiacylglycerol (MGDG) synthase regulation by thioredoxin. FEBS Lett. 2006;580:4086–4090. doi: 10.1016/j.febslet.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 30.Motohashi K., Okegawa Y. Method for enhancement of plant redox-related protein expression and its application for in vitro reduction of chloroplastic thioredoxins. Protein Exp. Purif. 2014;101:152–156. doi: 10.1016/j.pep.2014.07.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material