Abstract
The stochastic dynamics and regulatory mechanisms that govern differentiation of individual human neural precursor cells (NPC) into mature neurons are currently not fully understood. Here, we used single-cell RNA-sequencing (scRNA-seq) of developing neurons to dissect/identify NPC subtypes and critical developmental stages of alternative lineage specifications. This study comprises an unsupervised, high-resolution strategy for identifying cell developmental bifurcations, tracking the stochastic transcript kinetics of the subpopulations, elucidating regulatory networks, and finding key regulators. Our data revealed the bifurcation and developmental tracks of the two NPC subpopulations, and we captured an early (24 h) transition phase that leads to alternative neuronal specifications. The consequent up-regulation and down-regulation of stage- and subpopulation-specific gene groups during the course of maturation revealed biological insights with regard to key regulatory transcription factors and lincRNAs that control cellular programs in the identified neuronal subpopulations.
Given their defining characteristics to self-renew and give rise to alternative cell fates, human embryonic stem cells (hESC) have become the workhorse to model early human development in vitro (Nicholas et al. 2013; Zhu and Huangfu 2013; Ziller et al. 2015). More recently, exciting advances in self-organizing cell cultures are providing organoids that retain some degree of cellular complexity found in developing tissues (Lancaster and Knoblich 2014; Jo et al. 2016; Qian et al. 2016; Yin et al. 2016). However, the heterogeneity of the cells and the presence of rare cell subtypes, such as those undergoing short-lived cell fate transitions within the mixed population, make it difficult for traditional genomics approaches to identify exquisite spatiotemporal molecular changes that underlie cell fate decisions. Thus, unanswered questions arise regarding whether seemingly identical cells developing within a population exhibit similar intrinsic properties (Jaitin et al. 2015; Stegle et al. 2015; Trapnell 2015; Moris et al. 2016).
Single-cell RNA sequencing (scRNA-seq) analyses have been recently used to identify novel cell types in complex mixtures (Yan et al. 2013; Treutlein et al. 2014; Zeisel et al. 2015; Fuzik et al. 2016; Scialdone et al. 2016), establish developmental kinetics (Kim and Marioni 2013; Deng et al. 2014), and reveal discrete events in transitions between cell states (Buganim et al. 2012; Bendall et al. 2014; Moignard et al. 2015; Trapnell 2015; Olsson et al. 2016). To date, many studies have shown the heterogeneity of neural precursor cells (Johnson et al. 2015; Llorens-Bobadilla et al. 2015) and neurons (Molyneaux et al. 2007; Pollen et al. 2014; Darmanis et al. 2015; Usoskin et al. 2015) in mouse and human brain by scRNA-seq. However, due to complexity of data analysis of cellular dynamics, coupled with the biological variability (birth, death, and differentiation) of individual cells, as well as the presence of technical, environmental, and intracellular noise (Kuznetsov 2001, 2003; Kuznetsov et al. 2002; Kim and Marioni 2013; Kharchenko et al. 2014; Buettner et al. 2015; Daigle et al. 2015; Vu et al. 2016), it remains a challenge to interpret the heterogeneity and dynamics of NPC to neuron transitions (Camp et al. 2015; Bakken et al. 2016; Yao et al. 2017). Given the lack of synchronous development, the molecular patterns that switch on and switch off pathways governing alternative neuronal fate choices (Ming and Song 2011) are not clear. Thus, to dissect the landscape of neural cell development processes, both experimental and computational methodologies are required to identify and track the dynamics of molecular changes within individual cells as they develop (Shalek et al. 2013).
To date, several computational methods have been reported that profile developmental processes, such as Monocle (Trapnell et al. 2014), Wanderlust (Bendall et al. 2014), Wishbone (Setty et al. 2016), SLICER (Welch et al. 2016), Diffusion Pseudotime (Haghverdi et al. 2016), Destiny (Angerer et al. 2016), and SCUBA (Marco et al. 2014). These methods attempt to order cells into smooth continuous spatiotemporal trajectories to model development. However, Destiny lacks unsupervised statistics; Wanderlust typically is perfomed on few genes (<50); and Monocle, Diffusion Pseudotime, Wishbone, and SCUBA are biased (Bacher and Kendziorski 2016; Rizvi et al. 2017) or depend on a few well-known markers to define the bifurcation. Based on topological data analysis (TDA), recently published scTDA (Rizvi et al. 2017) has overcome some of the limitations. However, apart from smooth continuous spatiotemporal trajectories of cell development, there may be other transient developmental processes such as discontinuous cell development and stochastic cell fate changes (Moris et al. 2016). For example, without going through classic intermediate stages, haematopoietic stem cells can give rise to differentiated cells directly (Notta et al. 2016). Thus, compulsively ordering all the cells into smooth trajectories by computational algorithms may miss important biological information. In addition, after defining cell subtypes and mapping developmental trajectories, little has been done to analyze unique devleopmental tracks of different cell subtypes. More and deeper biological meaning could be addressed by (1) seperating the different cell subtypes and cell states of a given mixed population; (2) defining what is the mechanism underlying the specification of different cell subtypes and different cell states; and (3) understanding what are the dynamic differences between distinct subpopualitons that arise during development.
Results
Gene expression analysis by single-cell RNA sequencing of neuronal differentiation
To gain insight into the molecular and cellular mechanisms that govern neurogenesis and develop an approach for studying developmental processes by scRNA-seq, we first sought to discern the gene expression patterns that confer developmental fates on human hindbrain/spinal cord neurons differentiated from neural precursor cells (NPCs). This in vitro model of neurogenesis provided the opportunity to analyze single-cell transcriptome data to define subpopulations of developing human neurons and to tease out the regulatory networks that govern alternative cell fate decisions associated with neurogenesis. Cultures of hESC cultures (H9 cell line) were treated for 8 d with small molecule inhibitors of the GSK3, SMAD, and NOTCH signaling pathways (Li et al. 2011), which gave rise to a seemingly homogenous population of SOX2+ (97.7%) and NESTIN+ (100%) NPCs (Fig. 1A). NPCs (day 0) were shifted to culture media supplemented only with neurotrophic factors, thus permitting multilineage neurogenesis. To reduce technical noise, scRNA-seq was performed on two independent differentiation experiments. After 30 d of nondirected differentiation, the cultures contained primarily TUJ1+ (98.3%) and MAP2+ (85.7%) neurons (Fig. 1A; Supplemental Fig. S1A,B) with predominantly hindbrain/spinal cord specification (Supplemental Fig. S1C), plus a few (<5%) other neuronal subtypes (TH+, 5-HT+, GABA+) (Supplemental Fig. S1B) and very few glial cells. Sequence data of each individual single cell shows that a majority (>90%) of cells expressed hindbrain/spinal cord neural cell markers such as HOX genes (Mazzoni et al. 2013; Philippidou and Dasen 2013; Thompson et al. 2014; Lu et al. 2016), but few cells (<5%) expressed forebrain and midbrain markers (Supplemental Fig. S1C,D). The hindbrain potential of our derived NPC is consistent with a previous report showing that NPC derived in this manner gave rise to hindbrain neurons when transplanted in vivo (Li et al. 2011).
Figure 1.
Transcriptional profiling and statistical properties of gene expression data of the neuronal differentiation process. (A) Immunostaining shows expression of markers for ES cells (NANOG), NPC (SOX2/NESTIN), and neurons (TUJ1). (B) Summary of workflow for capture and quality assessment of single cells at each time point. (C) Box plots show the number of expressed genes for each filtered cell at days 0, 1, 5, 7, 10, and 30 during neuron differentiation. Each dot represents one cell. (D) Summary of 15 pairwise comparisons (upper) that identified 3986 DE genes between any two of six time points at P < 0.05 (Supplemental Table S3). The table shows the numbers of DE genes for each pairwise comparison. (E) After comparing any two time points as shown in D, to enhance pattern detection of DE genes for downstream analysis, curves were drawn based on the gene density and −log10 (P-value) for all DE genes in experiment 1 (orange line) and experiment 2 (green line), respectively (E), from which, 528 genes were obtained (Supplemental Table S4) with fold change >1.5 and P-value <10−7. (F) Hierarchical cluster analysis for all six time points was performed by bootstrapping based on the derived 528 genes from E. Hierarchical cluster analysis was calculated for the cells clustering with the R package “pvclust” using Correlation distance, the Ward clustering method, and the number of bootstrap set to 10,000.
Single cells were isolated at discrete time points (days 0, 1, 5, 7, 10, and 30) with the aim of capturing developmental transition events. The wide time range allowed us to interrogate gene expression dynamics at early stages of neurogenesis and during the subsequent maturation process. We captured 88–96 single cells at each time point followed by library preparation and deep sequencing of transcripts derived from individual cells (Fig. 1B; Supplemental Table S1). To reduce technical noise and to increase the statistical confidence of the single-cell gene-expression profiling, the cells and expressed genes were filtered using quality control criteria (Methods; Supplemental Fig. S2, overview). In total, 8957 genes from 483 cells produced high-quality expression data by scRNA-seq (Supplemental Table S2).
A global comparison of the single-cell expression profiles across all six time points was performed. We found that NPCs (day 0) and differentiated neurons (day 30) expressed fewer genes than the intermediate time points (days 1, 5, 7, and 10) (Fig. 1C), which was confirmed in experiment 2, in which single cells were collected on days 0, 3, 7, and 14 (Supplemental Fig. S3). The gene expression profiles were assessed across all cells at each time point (Supplemental Fig. S4A). Genes expressed in >80% cells at a given time point were considered to be highly penetrant, whereas genes expressed in <20% cells were considered as low penetrance genes. The penetrance plots show that on day 0, only 25% of genes were high penetrance and 15% of genes were low penetrance. Interestingly, 51% of genes were high penetrance and only 3% were low penetrance on day 1, indicating a coordinated gene switch-on process resulting in a less heterogeneous population of cells relative to day 0, and the data from experiment 2 showed similar results (Supplemental Fig. S4B). The dynamics of gene penetrance were concordant with the changes in the total number of expressed genes (Fig. 1C; Supplemental Fig. S3). The dynamics of gene expression associated with cell heterogeneity were confirmed by principal component analysis (PCA) (Supplemental Fig. S5A), and the variance percentage of each principal component was determined (Supplemental Fig. S5B). The PCA shows that cells broadly clustered together based on their time of differentiation. It is also apparent from the PCA analysis that day 1 cells were least heterogeneous and then transcriptional heterogeneity increased, becoming greatest among the neurons at day 30.
Collectively, the single-cell gene expression data show that there was a fair degree of heterogeneity within the starting NPC population and then again at the end stage of differentiation (Supplemental Figs. S4, S5). Interestingly, there appeared to be a rapid constriction in population heterogeneity within one day of the shift to conditions that promoted neural differentiation, which we argue is due to the differentiation program driving cells predominantly toward neuronal development and away from development of astrocytes, oligodendrocytes, and other glial cells.
To explore the gene expression dynamics as NPCs became neurons, 15 pairwise comparisons were performed across all six time points (Fig. 1D). Single-cell differential expression (SCDE) methodology identified 3986 significantly (P < 0.05) differentially expressed genes (DEG) (Supplemental Table S3). Interestingly, for each pairwise comparison, there were peak numbers of DEG at statistical significance of P < 10−7 in experiments 1 and 2 (Fig. 1E; Supplemental Fig. S6A,B). This tight statistical cutoff produced a set of 528 and 279 dynamically expressed genes from experiments 1 and 2, respectively (Supplemental Table S4). Of the derived 279 genes from experiment 2, 71.7% were significantly differentially expressed in experiment 1 (Supplemental Fig. S6C). These data indicated that the tight statistical cutoff provided a highly reliable set of 528 dynamic classifier (DC) genes, which were then used to build an unsupervised hierarchical cluster for all cells across all six time points using a bootstrapping method (Fig. 1F). As expected for a time course of differentiation and as seen in the PCA plots (Supplemental Fig. S5), we found that a majority of cells coming from the same time points grouped together, and groups of cells from adjacent time points were more transcriptionally similar than cells from more widely interrupted time points. The 528 DC genes are potentially useful markers for defining intermediate cell types in hindbrain/spinal cord neuron development (Supplemental Fig. S7).
Identifying and tracking subpopulations of differentiating neurons
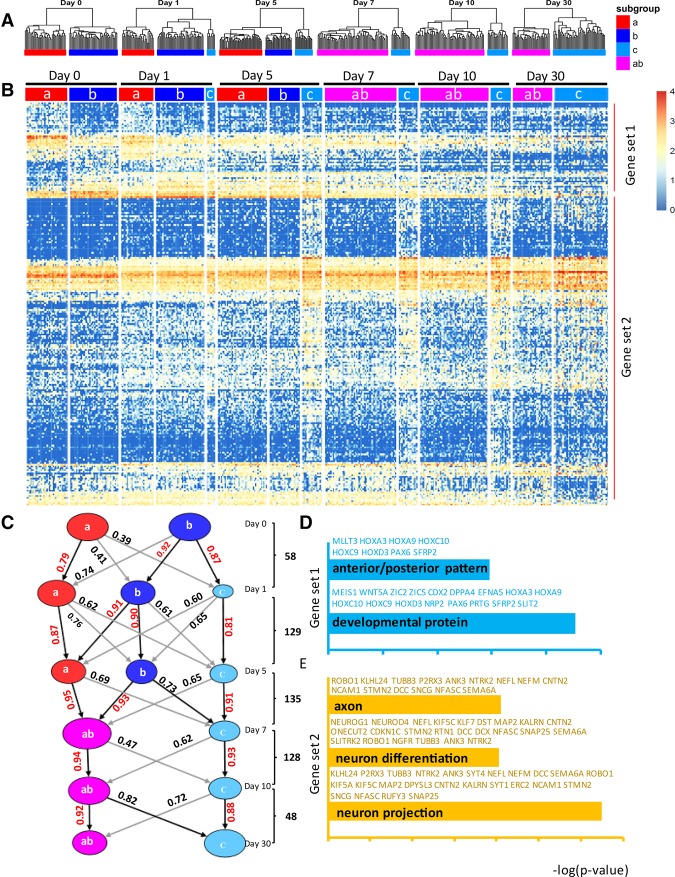
One major advantage of single-cell over bulk-cell analysis is the ability to identify subpopulations that have independent trajectories and thereby determine the gene regulatory networks that uniquely specify the individual subpopulations. We noticed from the hierarchical clustering that mixed populations of cells appeared during the time course (Fig. 1F). We hypothesized that these cells were transient intermediates that would reveal unique developmental lineages that give rise to distinct neurons from progenitors. To identify subpopulations, unsupervised hierarchical clustering was performed for each time point by bootstrapping based on the same 528 DC genes (Fig. 2A). Then all genes differentially expressed between subpopulations within a given time point were identified by SCDE (P < 0.05) (Supplemental Table S5). The analysis revealed two clearly distinguishable subpopulations of similar abundance at day 0. On days 1 and 5, there were three subpopulations, and on days 7, 10, and 30, there were two. To establish the relationship of the subpopulations, we looked for the DEG that were common in subpopulations between any two neighboring time points. For example, there are 58 DEG in common between “a” and “b” subpopulations of day 0 and “a,” “b,” and “c” subpopulations of day 1 (Supplemental Fig. S8; Supplemental Table S6). Similarly, there are 129, 135, 128, and 48 common DEG between subpopulations of days 1 and 5, days 5 and 7, days 7 and 10, and days 10 and 30, respectively (Supplemental Table S6). A heat map was generated based on the expression levels of the common DEG (Fig. 2B). To establish connections of subpopulations between neighboring time points in a statistically rigorous manner, Pearson correlations based on the common DEG between subpopulations of two neighboring time points were determined (Fig. 2C). These analyses established that two subpopulations, called “a” and “b,” at day 0 begat two subpopulations “a” and “b,” respectively, on day 1. The corresponding “a” and “b” lineage cells were also clearly distinguishable on day 5, although the differences between them had diminished. By days 7, 10, and 30, the “a” and “b” subpopulations were no longer clearly distinguishable and collapsed to a single subpopulation, which we termed “ab.” This correlation analysis also indicated that a third subpopulation “c” appeared on day 1, likely derived from the “b” lineage cells. As the “a” and “b” lineages converged into the “ab” lineage, lineage “c” cells became more distinct from the “ab” lineage by day 5. To summarize, our approaches to model developmental trajectories revealed three lineages: the “a” and “b” subpopulations were both present in our starting NPC; the “c” lineage arose from “b” on day 1 and persisted until day 30; the “a” and “b” lineages converged to form a single lineage “ab” by day 7; and two major subpopulations of differentiated neurons emerged from these lineages over the 30-day time course.
Figure 2.
Defining and tracking of subpopulations throughout the time course of differentiation. (A) Hierarchical cluster analysis for each time point was performed by bootstrapping based on the derived 528 DC genes as Figure 1F. (B) A heat map showing common DE genes between cell subpopulations in any two neighboring time points after defining the clusters at each time point. (C) The Pearson correlation coefficients between any two cell subpopulation for two neighboring time points were calculated based on the common DE genes between subpopulations of any two neighboring time points. The common DE genes between neighbor time points are as shown. The correlation values are shown with highest values as indicated (red numbers, black line). Gene ontologies associated with gene set 1 (D) and gene set 2 (E) were generated by David analysis (Bioinformatics 6.7). The top GO terms are shown with representative genes highlighted with blue (gene set 1) and brown (gene set 2). The x-axis indicates the −log (Benjamini P-value).
To gain biological insight, we examined the ontology of genes (gene set 1) that distinguished the “a” from “b” lineages (Fig. 2B; Supplemental Table S7). The Gene Ontology (GO) of gene set 1 identified “anterior/posterior patterning” and “development proteins” as being significantly different in these two lineages (Fig. 2D). The data also showed that another set of DEG (gene set 2) (Fig. 2B; Supplemental Table S7) distinguished “c” lineage cells from “ab” cells, especially at days 5, 7, and 10. The GO of gene set 2 showed enrichment for genes associated with neuronal function (Fig. 2E), indicating that the “c” lineage developed into neurons earlier than the “ab” lineage. GO was also performed for the genes differentially expressed between “ab” and “c” subpopulations of day 30. This showed that “ab” subpopulation expressed genes related to “mitosis signaling” (Supplemental Fig. S9A), indicating that the “ab” subpopulation was comprised of immature neurons. Indeed, flow cytometry showed that some cells (<20%) at day 30 were in G2/M phase and thus were not fully differentiated, post-mitotic neurons (Supplemental Fig. S9B).
Tracking subpopulation trajectories reveals key regulators of neuronal differentiation
We reasoned that tracking the gene expression dynamics of individual subpopulations would allow us to parse developmental processes that would be masked by bulk cell analysis. After building up the connection of the subpopulations from day 0 to day 30 (Fig. 2C), we determined the gene expression patterns unique to the different subpopulations during the progression of the lineages across the time course of differentiation. GO analysis was first performed for genes differentially expressed between all cells of day 0 and day 1 (Fig. 3A, left). We found that the up-regulated genes have “neuron differentiation” and “cell motion” related pathways, whereas “cell cycle” related pathway genes were down-regulated, as expected for neuronal differentiation. Interestingly, the data show specific gene expression dynamics of subpopulations when GO analysis was performed for “a,” “b,” and “c” subpopulations from day 0 to day 1 (Fig. 3A, middle and right); cell cycle pathway genes were not down-regulated in the “a” subpopulation (Fig. 3A, middle), but were down-regulated in “b” and “c” subpopulations of day 1 compared with the “b” subpopulation of day 0 (Fig. 3A, right).
Figure 3.
Subpopulation analysis reveals specific gene expression dynamics. (A) Gene ontologies associated with DEG comparing day 1 and day 0 (left), day 1 “a” and day 0 “a” subpopulation (middle), and day 1 “b” and “c” and day 0 “b” subpopulation (right) were generated by David analysis (Bioinformatics 6.7). The top GO terms are shown. The different GO terms are labeled with orange with representative genes. The x-axis indicates the −log (Benjamini P-value). (B) Dot plots show the representative cell cycle genes (HMGA1, CENPF) and neuron markers (DCX, STMN2) in different subpopulations as indicated for all six time points. The y-axis represents the gene expression level. Each small dot represents a cell, and the large dot indicates the medium expression of a given cell subpopulation. The error bar represents variation of the given gene in a given cell subpopulation. (C) Q-RT-PCR shows the expression level of PAX6 at six time points as shown, and the y-axis represents the fold change of PAX6 mRNA level. (D) The expression level of PAX6 in different cell subpopulations of different time points was shown in dot plots as in B.
We then tracked the dynamic expression of some well-known markers within the identified subpopulations. For example, the expression of cell cycle–related genes, HMGA1 and CENPF, were down-regulated faster and greater in the “c” subpopulation, and the expression of neuronal markers, DCX and STMN2, were up-regulated greater and faster in the “c” subpopulation (Fig. 3B). These unique gene expression dynamics of different subpopulations provide us detailed information regarding neuron development, results that may be obscured by conventional smooth continuous trajectory-based methods. For example, bulk gene expression analysis (Q-RT-PCR) of the neural cell marker PAX6 indicated that it did not change from day 0 to day 10 (Fig. 3C). However, our analysis shows initially low PAX6 expression in the “a” subpopulation that dramatically increased from day 0 to day 1, whereas its expression was consistently high in the “b” subpopulation (Fig. 3D). The accuracy of our scRNA-seq data was confirmed by immunostaining analysis, which showed that 45.2% of cells were PAX6+ on day 0, whereas there were 92.4% and 97.5% PAX6+ cells on days 1 and 5, respectively (Supplemental Fig. S10A,B). Several other subpopulation-specific gene expression patterns were noted, including a cell cycle–related gene (HMGA2), stem cell markers (LIN28A and CDX2), neuronal cell markers (MAP2, KLF7, and HES6), HOX genes (HOXC10, HOXC9, and HOXA3), early B cell factors (EBF1, EBF2, and EBF3), and PAX family members (Supplemental Fig. S11). These data indicate that our subpopulation analyses are sensitive and accurate in identifying unique gene expression dynamics during neuronal differentiation, information that may have been masked by bulk cell analysis or by applying analytical tools that assume continuous smooth trajectories.
Encouraged by our observations of unique gene expression dynamics that allowed us to track subpopulations, we searched for key regulators of the alternative developmental processes. Our hierarchical clustering showed the “c” subpopulation that arose on day 1 was most similar to the “b” subpopulation (Fig. 2A,C). GO analysis revealed that the “b” and “c,” but not “a,” subpopulations displayed significant neuronal features on day 1 (Supplemental Fig. S12). We hypothesized that there would be key genes expressed in “b” and “c” subpopulations that promote neuronal differentiation, whereas other key genes would be expressed in the “a” subpopulation that inhibit neuronal differentiation. As expected, our subpopulation analysis identified previously reported transcription factors and lincRNAs that play important roles during neuronal differentiation. For example, well-known neurogenesis related key genes PAX6 (Osumi et al. 1997, 2008), MEIS1 (Zhang et al. 2002), RMST (Ng et al. 2013), NEUROD1 (Gao et al. 2009; Zhang et al. 2013), NEUROD4 (Ohsawa et al. 2005), and ASCL1 (Ming and Song 2011; Kim and Marioni 2013) were highly expressed in “b” and “c,” but not “a” subpopulations on days 0 and 1 (Fig. 3D; Supplemental Fig. S13). In another example, the neuronal differentiation repressor REST was down-regulated uniquely in “c” subpopulation on days 10 and 30 (Supplemental Fig. S13).
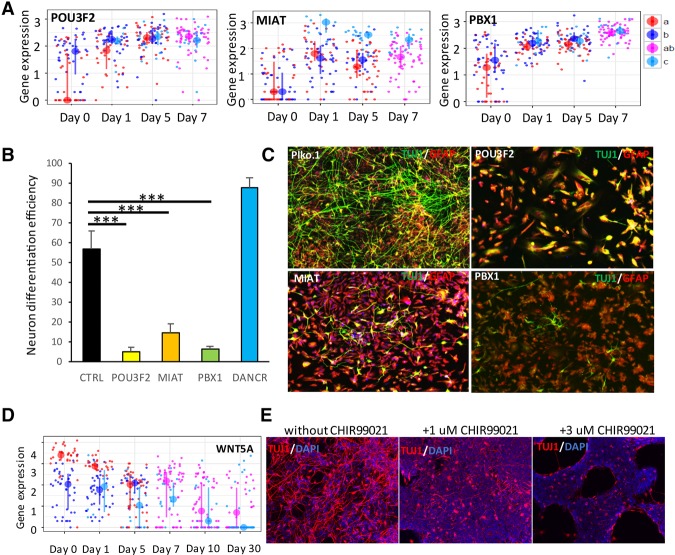
Encouraged by these results, we sought to identify novel transcription factors and lincRNAs that play important roles during neurogenesis. Similar to the gene expression dynamics of PAX6 and ASCL1, our subpopulation analysis showed that POU3F2, PBX1, and MIAT maintained high expression in “b” and “c” subpopulations (Fig. 4A), and their expression significantly increased in the “a” subpopulation after day 0. These dynamic changes of POU3F2 and MIAT were masked in bulk gene expression analysis (Supplemental Fig. S14). To assess the biological importance of these genes, we performed gene disruption studies that showed that knockdown of POU3F2, MIAT, or PBX1 expression significantly blocked neuronal differentiation (Fig. 4B,C), indicating that these genes are critical at early stages of differentiation. Similarly, we found WNT5A, but no other WNTs, was highly expressed in the “a” subpopulation and lowly expressed in the “b” subpopulation on day 0, and its expression in both subpopulations was down-regulated during neuronal differentiation (Fig. 4D; Supplemental Fig. S15). The importance of WNT down-regulation for neurogenesis was confirmed using the WNT signaling activator CHIR99021, which blocked neuronal differentiation (Fig. 4E).
Figure 4.
Subpopulation analyses reveal key regulators. (A) Similar to Figure 3B, the expression levels of genes in different cell subpopulations of different time points is shown in dot plots. (B) Knockdown of gene expression experiments was performed for TFs POU3F2, PBX1, and lincRNA MIAT and DANCR during neuronal differentiation (the empty plko.1 vector served as a negative control). (C) Neuron marker TUJ1 (green) and Astrocyte marker GFAP (red) after 7 d of knock down of these candidates. Immunostained images were quantified by Columbus Analysis System to determine the neuron differentiation efficiency. (D) The expression level of WNT5A in different cell subpopulations of different time points is shown in dot plots. (E) NPCs were cultured in neuron differentiation media with different concentrations of WNT signaling activator CHIR99021. Seven days later, immunostaining was performed for neuron marker TUJ1 (red), and nuclei were stained with DAPI (blue). The concentration of CHIR99021 is as shown.
Elucidating subpopulation-specific gene regulatory networks
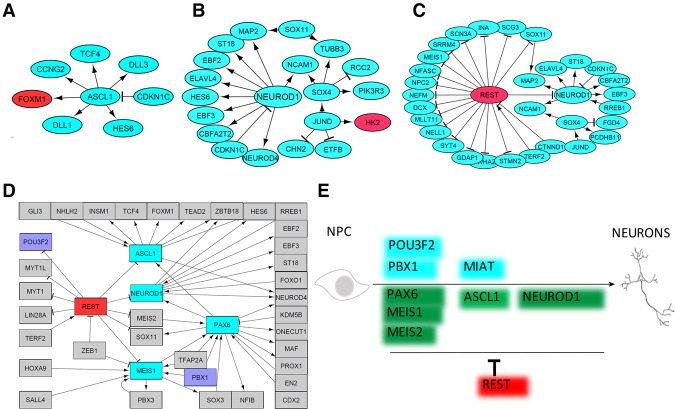
Deciphering gene regulatory networks is useful to understand regulation of complex biological processes, such as cellular lineage specification (Levine and Davidson 2005; Pimanda and Göttgens 2010; Arda et al. 2013; Shubin 2017). We wanted to identify the gene regulatory networks underlying subpopulation development in our in vitro model of neurogenesis. Starting from the scRNA-seq data obtained for each subpopulation, we reconstructed the gene interaction networks using the MetaCore database as a source of experimentally validated interactions. From this information, we derived networks that encompass interactions of known effect (i.e., activation or inhibition) and the directionality of those interactions. This afforded us the opportunity to perform complex modeling of the subpopulation-specific regulatory circuits that control key gene expression programs within each subpopulation. In the regulatory network corresponding to subpopulation “c” on day 1 (Fig. 5A), the interactions among the genes in the most influential regulatory circuit shows that ASCL1 is a hub gene in this network that plays an important role when “c” subpopulation arises on day 1. Similar network analyses were performed for other time points. NEUROD1, identified as a network hub on days 5 and 10, positively regulates other neurogenesis-related genes, such as ELAVL4, ST18, EBF2, NEUROD4, HES6, and MAP2 (Fig. 5B,C). In addition, PAX6 was found to be a hub gene on day 0 (Supplemental Fig. S16A) that regulates MEIS1 and MEIS2. REST, a master negative regulator of neuronal differentiation (Gao et al. 2011; Ng et al. 2013), was found to be at the hub of a network on days 10 and 30, negatively regulating other genes, such as well-known neuron-related genes NEUROD1, DCX, INA, and SOX11 (Fig. 5C; Supplemental Fig. S16B). In total, these gene regulatory network analyses between cell subpopulations of a given time point revealed regulatory pathways comprised of active and inactive hubs that underlie the regulation of neurogenesis.
Figure 5.
Network analyses of subpopulations reveal underlying mechanism. Gene regulatory networks were assembled based on differential gene expression to define key regulatory pathways that control development of the unique “c” subpopulation. Networks were generated for day 1 (A), day 5 (B), and day 10 (C). The genes labeled in red and green are expressed at low and high levels, respectively, in the “c” cell subpopulation relative to the other cell subpopulation at a given time point. (D) A regulatory network analysis was performed for the 195 TFs that were identified as differentially expressed across all the cell subpopulations and all time points. Purple highlighted genes were chosen for functional validation in Figure 4. (E) A simple model for the dynamic regulators of neuronal development. PAX6, MEIS1, MEIS2, ASCL1, and NEUROD1 are well-known key genes during neurogenesis that were identified by our subpopulation analysis; POU3F2, MIAT, and PBX1 were newly identified by our subpopulation analysis and are critical during neurogenesis. REST, which is identified by our subpopulation analysis, is a well-known repressor during neurogenesis.
Our single-cell analysis identified 195 transcription factors that were differentially expressed between subpopulations within the 30-d time course (Supplemental Table S8). A gene regulatory network based on these 195 transcription factors was built (Fig. 5D). The results clearly show that ASCL1, PAX6, NEUROD1, and MEIS1 are key positive regulators, each of which is negatively regulated by REST. Hierarchical clustering (Supplemental Fig. S17) of the expression patterns of these transcription factors in subpopulations together with subpopulation developmental tracking (Figs. 3D, 4A; Supplemental Fig. S13) from days 0 to 30 showed that (1) PAX6, POU3F2, PBX1, MEIS1, and MEIS2 were highly expressed in the “b” subpopulation of days 0 and 1; (2) NEUROD4, ONECUT1, NEUROD1, EBF1, EBF2, EBF3, ASCL1, ST18, and HES6 were higher in “c” subpopulation on all days; and (3) REST expression gradually decreased and showed differential expression in subpopulations on days 10 and 30. Combining the network analyses of cell subpopulations with experimental validation (Figs. 4, 5; Supplemental Figs. S13, S16, S17), a simple model was developed to show the activation and repression of key genes underlying neuronal differentiation (Fig. 5E). Further exploration of the hundreds of factors identified in this study will add new insights to the molecular details that specify alternative human neuronal cell types.
Discussion
With the advent of scRNA-seq, elegant work has been done to identify cell subtypes in mixed populations (Jaitin et al. 2014; Patel et al. 2014; Treutlein et al. 2014; Zeisel et al. 2015; Olsson et al. 2016; Poulin et al. 2016; Dulken et al. 2017). However, it remains a major challenge to understand the mechanisms and dynamics by which distinct cell subtypes arise during development (Trapnell 2015). To understand the transcription processes and identify key steps of transcription regulation that govern developmental processes, suitable experimental and computational methods are needed to exploit fully the application of scRNA-seq. Our scRNA-seq results provided a detailed view of the differentiation processes governing the birthing of neuronal cell subpopulations from NPCs.
Ample evidence shows that that dynamic heterogeneity is functionally relevant to cellular decision making (Moris et al. 2016). For example, dynamically changed NANOG and POU5F1 (also known as OCT4) expression mediate the self-renewal and differentiation of hESC, suggesting some overall control of the heterogeneity (Niwa et al. 2000; Chambers et al. 2007; Kalmar et al. 2009; Singer et al. 2014). Thus, elucidating distinct developmental tracks could help to understand the molecular dynamics that govern alternative cell fate decisions. To do this, one efficient way is to order the cells in a smooth developmental trajectory (Trapnell et al. 2014; Shin et al. 2015), but it remains challenging to distinguish different subtypes and alternate lineages based on standard expression profiling and computational approaches. Instead of compulsively ordering the cells into smooth trajectories, we considered both continuous and discontinuous developmental processes. First of all, since hierarchical cluster analysis has the particular ability to identify rare or transient cell populations (Buettner et al. 2015; Grün et al. 2015; Moris et al. 2016), the individual cells in each time point were first clustered by DEG set identification (Fig. 1E; Supplemental Fig. S6). This allowed us to define two subtypes of NPCs on day 0 and a small cell subpopulation “c” that emerged on day 1 and had a unique transcription profile (Fig. 2A). The hierarchical clustering indicates that there are likely more than two subpopulations, but we chose to be conservative in defining the subpopulation. Analyzing additional single cells at each time point and increasing the number of time points, especially in the first 24 h where we detected many changes, would refine the numbers of subpopulations and improve lineage tracking. A build of connections based on the common DEG of subpopulations between the neighboring time points helped us capture the cell state transitions among “a,” “b,” “c,” and “ab” subpopulations of days 0, 1, and 5 (Fig. 2C).
It has been recently reported that scTDA, which utilizes all expression data in one step regardless of any prior factors, such as time, could uncover asynchronous cell development (Rizvi et al. 2017). In contrast, our analysis first considered time factor. The subpopulations were identified by hierarchical clustering based on pairwise comparisons between time points, which revealed subpopulation developmental tracks over time. Using our data, scTDA identified 204 genes of which only 29 were in our 528 dynamic classifier set (Supplemental Fig. S18A). Although the identified gene lists are different, we do observe a similar, overall developmental trend from day 0 to day 30 (Supplemental Fig. S18B). However, when WNT5A and DCX were used to compare scTDA and our approach (Supplemental Fig. S18C,D), scTDA was unable to show the dynamic expression pattern we revealed for these key genes in different cell subpopulations during neuronal differentiation (Figs. 2A,B, 3B, 4D).
Based on these analyses, important biological information was revealed. First, our data show that the anterior/posterior patterning pathways are differentially expressed among neural precursor cells (day 0) (Supplemental Fig. S19). It is well established that WNT signaling regulates the regional identity along the anterior–posterior axis by specifying HOX genes during neural development (Nordström et al. 2006; Philippidou and Dasen 2013; Moya et al. 2014). However, there are 19 WNT members and 39 HOX members; which WNT member regulated which HOX gene is difficult to discern using bulk population-level studies and sometimes yields contradictory information (Ille and Sommer 2005). Subpopulation analysis of our two independent experiments shows the WNT5A coexpressed with specific HOX genes such as HOXC8, HOXC9, HOXC10, and HOXA3 in neural stem cells (Supplemental Fig. S20A,B), and these HOX genes could pattern neural stem cells into hindbrain and spinal cord neurons (Philippidou and Dasen 2013), which is consistent with fate-mapping studies in vivo (Li et al. 2011). These data suggest that the developmental potential of the NPCs we patterned in vitro are already lineage restricted. Interestingly, it was recently been reported that WNT5A is highly expressed in apical neural progenitor cells and lowly expressed in nonapical neural progenitor cells in human brain (Johnson et al. 2015), indicating that our derived NPC may reflect the heterogeneity of neural progenitor cells in human brain. Therefore, we conclude that the in vitro NPC differentiation process we used mimicked normal developmental processes and thus serves as a useful model.
Second, although key regulators are thought to play an important role to balance self-renewal and differentiation of each individual cell (Pina et al. 2012; Kumar et al. 2014; Nair et al. 2015), current genomic methods are not sensitive enough to reveal these key genes. For example, (1) PAX6, POU3F2, MEIS1, MIAT, and RMST were not observed to significantly change when analyzed by bulk gene expression analysis (Fig. 3C; Supplemental Figs. S14, S21); and (2) some significantly up-regulated TFs and lincRNAs during neuronal differentiation based on bulk cell analysis, such as ELAVL3, RUNX1T1, LINC00461, BCL11A, and MYT1l (Supplemental Fig. S22A), failed to establish their importance during early stages of neurogenesis by our functional validation (Supplemental Fig. S22B,C). Assuming that the gene expression dynamical system could be used to define the state of a cell, heterogeneities can be used to infer the mechanisms of transitions between different states of a developmental system (Moris et al. 2016). It might be an efficient and accurate way to identify key genes by their dynamics of different subpopulation during neuron differentiation. Indeed, our scRNA-seq data identified distinct subpopulations with unique expression dynamics (Figs. 3D, 4A; Supplemental Fig. S13). The functional validation studies show that POU3F2, PBX1, and MIAT are critical for differentiation of hindbrain/spinal cord neurons (Fig. 4B,C). Although it might be expected to see a shift in the bifurcated alternative lineages with gene knockdowns, this was not observed. Additional experiments using tightly regulated, temporally controlled perturbations are required to adequately test this prediction.
Third, 195 transcription factors (Supplemental Table S8) and 138 lincRNAs (Supplemental Table S9) were found differentially expressed in different subpopulations at different time points. The regulatory network analysis based on these TFs provides an interesting view of biological features associated with subtype-specific neuronal differentiation (Fig. 5). For example, ASCL1, NEUROD1, and NEUROD4 are highly expressed in a few cells of “c” subpopulation during an early transition stage (Supplemental Figs. S13, S17). These data demonstrate that subpopulation analysis is sufficiently sensitive to identify key dynamic regulators, even in a few cells or during short transit periods.
Finally, the regulatory network analysis of subpopulations in given time points provides us insight on continuous or discontinuous developmental tracks. For example, ASCL1 is a core gene in the “c” subpopulation of day 1 (Fig. 5A), whereas NEUROD1 is a core gene in the “c” subpopulation of day 5 (Fig. 5B). This would lead one to hypothesize that ASCL1 and NEUROD1 are sequentially active during “c” subpopulation generation (Supplemental Fig. S23A) based on current methods that consider smooth continuous trajectory. However, based on our subpopulation analysis at each time point, one could conclude that either ASCL1 or NEUROD1 is sufficient for cells of the “c” subpopulation generation without sequential activation of ASCL1 and NEUROD1 (Supplemental Fig. S23B,C). There have been indications about this because overexpression of a single transcription factor NEUROD1 (Zhang et al. 2013) or ASCL1 (Pang et al. 2011; Chanda et al. 2014) in hESC is sufficient to generate neurons, whereas forced expression of ASCL1 and NEUROD1 together in hESC is much more efficient (Pang et al. 2011). Instead of building only smooth developmental trajectories, our data provide an unsupervised approach to building specific subpopulation trajectories and enables one to find critical developmental bifurcations and thereby gain meaningful insights into specification of alternative cell fates.
Conclusions
In summary, we developed an experimental design for scRNA-seq time course analysis and computational approaches to dissect the stochastic neuronal differentiation process in vitro. This strategy provides selection, clustering, and tracking of scRNA-seq subtranscriptome profiles at discrete times which can be used for other cell developmental or response-stimulated processes. Our results suggest that (1) NPCs comprising two precursor subtypes drive distinct neuronal cell subpopulation; (2) a 1-d time period after initiation of NPC differentiation is critical for switch-on of the NPCs precursors in both cell subtypes, and we succeeded to capture a relatively small “c” neuronal subpopulation that arises on day 1 and enlarges afterward; (3) dynamic gene expression analysis of subpopulation and functional validation helped us identify key regulators of hindbrain/spinal cord neuronal differentiation; and (4) network analysis based on DEG between subpopulations shed light on continuous and discontinuous neuronal development.
Methods
Cell culture
Human embryonic stem cells (hESCs, H9) were cultured in mTeSR1 complete medium (mTeSR1 [#05851, Stem Cell Technologies]: 5× Supplement [#05852, Stem Cell Technologies] = 4:1). H9 cells were passaged using Dispase (#07923, Stem Cell Technologies) at a dilution of 1:6–1:10.
Neural induction
H9 cells cultured in mTeSR1 complete medium for 1–2 d were then used for neural induction as published (Li et al. 2011). Briefly, 20%–30% confluent H9 cells were treated with CHIR99021, SB431542, and Compound E in neural induction media, changed every 2 d; 7 d later, the cells were split 1:3 by Accutase (# 25-058-CI, Corning) and seeded on matrigel-coated plates. ROCK inhibitor (1254, Tocris) was added (final concentration 10 µM) to the suspension at passaging. Cells were then cultured in neural cell culture medium. These derived cells are neural precursor cells (NPC), which were used for further studies.
Neuronal differentiation
Spontaneous neuronal differentiation was performed as previously described (Li et al. 2011). Briefly, the derived 2 × 105 NPCs were seeded on poly-l-lysine (P4707, Sigma) and laminin (L2020, Sigma)-coated six-well plates in neural cell culture medium. The next day, the cells were cultured in neuron differentiation medium: DMEM/F12(11330-032), Neurobasal (21103-049), 1× N2 (17502-048), 1× B27 (17504-044), 300 ng/mL cAMP (A9501), 0.2 mM vitamin C (A4544-25), 10 ng/mL BDNF (450-02), 10 ng/mL GDNF (450-10) until day 30.
Immunostaining
H9, NPCs, neurons, or cells at different time points during neuronal differentiation were fixed in 4% paraformaldehyde for 15–25 min, then washed twice with PBS and once with PBS plus 0.1% Triton X-100 (PBST) for 5 min. Cells were then incubated in blocking buffer (PBST+ 10% normal serum) for 30–60 min. Before addition of the primary antibody, which was incubated with the cells overnight at 4°C. The next day, cells were washed in PBST and incubated with Alexa Fluor-conjugated secondary antibody (Invitrogen) for 1 h at room temperature. Nuclei were visualized by DAPI staining as in Supplemental Table S10.
Single-cell capture and library preparation
Single cells were captured using standard protocol of C1 single-cell auto prep system (Fluidigm, PN 100-5950 B1). Briefly, neural cells at different time points during neuron differentiation were dissociated by Accutase, then 250,000/mL cell suspension was loaded into the C1 instrument. To prepare single-cell libraries, cDNA products from each single cell were harvested from C1 chip followed by concentration and quality assessment using PicoGreen dsDNA Assay kit (P11496) and Agilent High sensitivity kit (PN 5067-4626). The libraries were generated using Illumina Nextera XT library preparation kit (FC-131-1096, 15032354) after dilution of cDNA to 0.15–0.25 ng/µL.
Lentivirus packaging for RNAi experiments
Candidate genes were knocked down in NPC using plko.1 lentiviral system. Briefly, HEK293T cells were transfected with plko.1, psPAX2, and VSVg at the ratio of 4:3:1. Culture medium was changed 12 h after transfection. Virus was collected after an additional 36 h and used for NPC infection. NPCs were changed to fresh medium. Short hairpin RNAs cloned in PLKO.1 vector are shown in Supplemental Table S11.
Read processing, mapping, gene expression estimating, and quality control
Sequence data were processed and mapped to the human reference genome (hg19) using TopHat (v2.0.11) (Kim et al. 2013) with Bowtie2 (v2.2.1) (Langmead and Salzberg 2012). Gene expression levels were quantified with HTSeq-count (v0.6.1p1) (Anders et al. 2015). The expression counts were performed with the human gene annotation (GENCODE release 19) (Harrow et al. 2012), which yielded 57,820 genes from 553 captured cells. Following the counting of mapped reads, additional quality control criteria were applied to remove low-quality cells and low-expressed genes.
We applied the following criteria for cell quality control:
Total number of mapped reads in each cell >1 million.
Number of genes detected >Q1 − 1.5 × IQR in each time point (Q1 denotes first quartile, IQR denotes interquartile range).
We sequentially applied the following criteria for gene quality control:
Percentage of cells containing at least one read for a given gene in each time point >20% to control number of cells expressing the gene (13,786 genes refer to “gene quality control 2” in Supplemental Fig. S2).
Percentage of cells containing at least five reads for a given gene in each time point >15% to control expression level of the gene (8957 genes, refer to “gene quality control 1” in Supplemental Fig. S2).
After applying these quality criteria, we obtained 8957 expressed genes and 483 cells for downstream analysis. The gene expression levels were normalized using DEseq2 (v 1.13.16) (Love et al. 2014).
Differential expression analysis during neuron differentiation
To identify DEG between neuron development time points, we conducted differential expression analysis using single-cell differential expression (SCDE) (Kharchenko et al. 2014), a Bayesian approach for finding DE genes accounting for frequent dropout events and biological variability within single-cell data. The single-cell data of each time point were compared. Pairwise DE genes were defined as adjusted P-value <0.05. Figure 1D shows the number of DE genes in comparison between days. Our results reveal nonuniform time course variation of the number of DE genes, mostly in the transition period between day 0 and day 1.
After the determination of DE genes, a total of 3986 DE genes were identified in the scRNA-seq during neuron development. To enhance pattern detection of significant DE genes for downstream analysis, 528 DE genes were obtained with fold change >1.5 and P-value <1 × 10−7.
Cell subpopulation analysis
We obtained 528 DE genes as mentioned earlier in differential expression analysis. Hierarchical clustering was used to cluster subpopulations of the cells in each time point. In each time point, two clusters were selected empirically based on the most distinct expression profile of cells except day 1 and day 5. For days 1 and 5, we selected three clusters since transition of cell subpopulations were observed. Finally, we annotated 14 cell subpopulations for downstream analysis.
Next, we performed differential expression analysis between pairs of cell subpopulations in each time point. We first selected genes that were expressed in at least 50% of the cells in any subpopulation. A gene was considered expressed if it had a read count ≥1. For each pair of cell subpopulations in each time point, SCDE was used to identify the number of DE genes with P < 0.05 (gene quality control 2). To identify DE genes that could influence the transition of cell subpopulations between neighbor time points, the common DE genes between two neighbor time points were identified. In total, we found 58, 129, 135, 128, and 48 DE genes which are common in day 0 versus day 1, day 1 versus day 5, day 5 versus day 7, day 7 versus day 10, and day 10 versus day 30, respectively. Using the common DE genes of cell subpopulations between neighboring time points, we identified the transition of expression profiles for cell subpopulations between neighboring time points by Pearson correlation coefficient. The Pearson correlation coefficients were calculated between the cell subpopulation in the earlier time point to the cell subpopulation in neighboring time points. The transition of cell subpopulations was defined by the highest correlation coefficient between time points.
Gene regulatory network analysis
In order to build the gene regulatory networks corresponding to each cell subpopulation, single-cell transcriptomics data was analyzed to identify the DEG between different cell subpopulations of a given time point (P < 0.05). Then the derived DE genes were used to reconstruct the interaction network including high quality gene–gene interaction information. For this purpose, we used the DE gene data and compiled the high-confidence interaction networks querying the MetaCore database from Thomson Reuters, which gave us the possibility of building gene regulatory networks of differentiating cell subpopulations (Crespo et al. 2013a,b; Zickenrott et al. 2016). After building the gene regulatory networks for the cell subpopulations, we identified the most relevant regulatory motifs, including the gene circuits that could have a more influential effect in the regulation of the gene expression patterns characteristic of each cell subpopulation (Zickenrott et al. 2016). The comparative analysis of these regulatory circuits allowed us to identify the regulatory genes that may play a role in the stabilization of the subpopulation phenotype and whose perturbation may have a significant effect and trigger transitions between cell subpopulations during neuronal differentiation.
Data access
The scRNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE102066, and the raw data have been submitted to the Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP097299 and to the NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA360884.
Supplementary Material
Acknowledgments
We thank the sequencing centre of the Genome Institute of Singapore who helped us with sequencing of our samples. We thank Arsen Batagov for help in data analysis and discussions at initial phases of this project. This work was funded by the Genome Institute of Singapore and Bioinformatics Institute and Joint Council Office A*STAR, Singapore.
Author contributions: L.W.S. and V.A.K. proposed and designed this study and obtained funding; J.W. designed and carried out experiments; P.J. and V.A.K. executed computational and bioinformatics works; P.J., J.W., V.E.A., A.B., and V.A.K. provided data analysis and interpretation. L.W.S., V.A.K., and A.D.S. supervised the work. All authors contributed to research and writing the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.223313.117.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C, Buettner F. 2016. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32: 1241–1243. [DOI] [PubMed] [Google Scholar]
- Arda HE, Benitez CM, Kim SK. 2013. Gene regulatory networks governing pancreas development. Dev Cell 25: 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher R, Kendziorski C. 2016. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol 17: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken TE, Miller JA, Ding SL, Sunkin SM, Smith KA, Ng L, Szafer A, Dalley RA, Royall JJ, Lemon T, et al. 2016. A comprehensive transcriptional map of primate brain development. Nature 535: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Davis KL, Amir el AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe'er D. 2014. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 157: 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner F, Natarajan KN, Casale FP, Proserpio V, Scialdone A, Theis FJ, Teichmann SA, Marioni JC, Stegle O. 2015. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 33: 155–160. [DOI] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. 2012. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 150: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. 2015. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci 112: 15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. 2007. Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234. [DOI] [PubMed] [Google Scholar]
- Chanda S, Ang CE, Davila J, Pak C, Mall M, Lee QY, Ahlenius H, Jung SW, Südhof TC, Wernig M. 2014. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep 3: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo I, Krishna A, Le Béchec A, del Sol A. 2013a. Predicting missing expression values in gene regulatory networks using a discrete logic modeling optimization guided by network stable states. Nucleic Acids Res 41: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespo I, Perumal TM, Jurkowski W, del Sol A. 2013b. Detecting cellular reprogramming determinants by differential stability analysis of gene regulatory networks. BMC Syst Biol 7: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle BJ Jr, Soltani M, Petzold LR, Singh A. 2015. Inferring single-cell gene expression mechanisms using stochastic simulation. Bioinformatics 31: 1428–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. 2015. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci 112: 7285–7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q, Ramsköld D, Reinius B, Sandberg R. 2014. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343: 193–196. [DOI] [PubMed] [Google Scholar]
- Dulken BW, Leeman DS, Boutet SC, Hebestreit K, Brunet A. 2017. Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep 18: 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuzik J, Zeisel A, Máté Z, Calvigioni D, Yanagawa Y, Szabó G, Linnarsson S, Harkany T. 2016. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol 34: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, Eisch AJ, Hsieh J. 2009. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci 12: 1090–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Ure K, Ding P, Nashaat M, Yuan L, Ma J, Hammer RE, Hsieh J. 2011. The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J Neurosci 31: 9772–9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A. 2015. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525: 251–255. [DOI] [PubMed] [Google Scholar]
- Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. 2016. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods 13: 845–848. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ille F, Sommer L. 2005. Wnt signaling: multiple functions in neural development. Cell Mol Life Sci 62: 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, et al. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343: 776–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Keren-Shaul H, Elefant N, Amit I. 2015. Each cell counts: hematopoiesis and immunity research in the era of single cell genomics. Semin Immunol 27: 67–71. [DOI] [PubMed] [Google Scholar]
- Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Göke J, Tan ZY, Saw TY, Tan CP, Lokman H, et al. 2016. Midbrain-like organoids from human pluripotent stem cells contain functional dopaminergic and neuromelanin-producing neurons. Cell Stem Cell 19: 248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA. 2015. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci 18: 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Muñoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A. 2009. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol 7: e1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, Scadden DT. 2014. Bayesian approach to single-cell differential expression analysis. Nat Methods 11: 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Marioni JC. 2013. Inferring the kinetics of stochastic gene expression from single-cell RNA-sequencing data. Genome Biol 14: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar RM, Cahan P, Shalek AK, Satija R, DaleyKeyser AJ, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, et al. 2014. Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 516: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov V. 2001. Distribution associated with stochastic processes of gene expression in a single eukaryotic cell. EURASIP J Appl Signal Processing 4: 285–296. [Google Scholar]
- Kuznetsov VA. 2003. Family of skewed distributions associated with the gene expression and proteome evolution. Signal Processing 83: 889–910. [Google Scholar]
- Kuznetsov VA, Knott GD, Bonner RF. 2002. General statistics of stochastic process of gene expression in eukaryotic cells. Genetics 161: 1321–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Knoblich JA. 2014. Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345: 1247125. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Davidson EH. 2005. Gene regulatory networks for development. Proc Natl Acad Sci 102: 4936–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sun W, Zhang Y, Wei W, Ambasudhan R, Xia P, Talantova M, Lin T, Kim J, Wang X, et al. 2011. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci 108: 8299–8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. 2015. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17: 329–340. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Zhong X, Liu H, Hao L, Huang CT, Sherafat MA, Jones J, Ayala M, Li L, Zhang SC. 2016. Generation of serotonin neurons from human pluripotent stem cells. Nat Biotechnol 34: 89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco E, Karp RL, Guo G, Robson P, Hart AH, Trippa L, Yuan GC. 2014. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci 111: E5643–E5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Peljto M, Patel T, Thornton SR, McCuine S, Reeder C, Boyer LA, Young RA, Gifford DK, et al. 2013. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci 16: 1191–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Song H. 2011. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70: 687–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moignard V, Woodhouse S, Haghverdi L, Lilly AJ, Tanaka Y, Wilkinson AC, Buettner F, Macaulay IC, Jawaid W, Diamanti E, et al. 2015. Decoding the regulatory network of early blood development from single-cell gene expression measurements. Nat Biotechnol 33: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. 2007. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci 8: 427–437. [DOI] [PubMed] [Google Scholar]
- Moris N, Pina C, Arias AM. 2016. Transition states and cell fate decisions in epigenetic landscapes. Nat Rev Genet 17: 693–703. [DOI] [PubMed] [Google Scholar]
- Moya N, Cutts J, Gaasterland T, Willert K, Brafman DA. 2014. Endogenous WNT signaling regulates hPSC-derived neural progenitor cell heterogeneity and specifies their regional identity. Stem Cell Rep 3: 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair G, Abranches E, Guedes AM, Henrique D, Raj A. 2015. Heterogeneous lineage marker expression in naive embryonic stem cells is mostly due to spontaneous differentiation. Sci Rep 5: 13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Bogu GK, Soh BS, Stanton LW. 2013. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51: 349–359. [DOI] [PubMed] [Google Scholar]
- Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, et al. 2013. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell 12: 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 24: 372–376. [DOI] [PubMed] [Google Scholar]
- Nordström U, Maier E, Jessell TM, Edlund T. 2006. An early role for WNT signaling in specifying neural patterns of Cdx and Hox gene expression and motor neuron subtype identity. PLoS Biol 4: e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Zandi S, Takayama N, Dobson S, Gan OI, Wilson G, Kaufmann KB, McLeod J, Laurenti E, Dunant CF, et al. 2016. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science 351: aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa R, Ohtsuka T, Kageyama R. 2005. Mash1 and Math3 are required for development of branchiomotor neurons and maintenance of neural progenitors. J Neurosci 25: 5857–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. 2016. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 537: 698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi N, Hirota A, Ohuchi H, Nakafuku M, Iimura T, Kuratani S, Fujiwara M, Noji S, Eto K. 1997. Pax-6 is involved in the specification of hindbrain motor neuron subtype. Development 124: 2961–2972. [DOI] [PubMed] [Google Scholar]
- Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. 2008. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 26: 1663–1672. [DOI] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, et al. 2011. Induction of human neuronal cells by defined transcription factors. Nature 476: 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344: 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. 2013. Hox genes: choreographers in neural development, architects of circuit organization. Neuron 80: 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimanda JE, Göttgens B. 2010. Gene regulatory networks governing haematopoietic stem cell development and identity. Int J Dev Biol 54: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Pina C, Fugazza C, Tipping AJ, Brown J, Soneji S, Teles J, Peterson C, Enver T. 2012. Inferring rules of lineage commitment in haematopoiesis. Nat Cell Biol 14: 287–294. [DOI] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. 2014. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol 32: 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin JF, Tasic B, Hjerling-Leffler J, Trimarchi JM, Awatramani R. 2016. Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci 19: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165: 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi AH, Camara PG, Kandror EK, Roberts TJ, Schieren I, Maniatis T, Rabadan R. 2017. Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development. Nat Biotechnol 35: 551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialdone A, Tanaka Y, Jawaid W, Moignard V, Wilson NK, Macaulay IC, Marioni JC, Göttgens B. 2016. Resolving early mesoderm diversification through single-cell expression profiling. Nature 535: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe'er D. 2016. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 34: 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. 2013. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498: 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. 2015. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17: 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin N. 2017. Gene regulatory networks and network models in development and evolution. Proc Natl Acad Sci 114: 5782–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer ZS, Yong J, Tischler J, Hackett JA, Altinok A, Surani MA, Cai L, Elowitz MB. 2014. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol Cell 55: 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Teichmann SA, Marioni JC. 2015. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 16: 133–145. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Ng L, Menon V, Martinez S, Lee CK, Glattfelder K, Sunkin SM, Henry A, Lau C, Dang C, et al. 2014. A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron 83: 309–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. 2015. Defining cell types and states with single-cell genomics. Genome Res 25: 1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Brownfield DG, Wu AR, Neff NF, Mantalas GL, Espinoza FH, Desai TJ, Krasnow MA, Quake SR. 2014. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509: 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, et al. 2015. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18: 145–153. [DOI] [PubMed] [Google Scholar]
- Vu TN, Wills QF, Kalari KR, Niu N, Wang L, Rantalainen M, Pawitan Y. 2016. ß-Poisson model for single-cell RNA-seq data analyses. Bioinformatics 32: 2128–2135. [DOI] [PubMed] [Google Scholar]
- Welch JD, Hartemink AJ, Prins JF. 2016. SLICER: inferring branched, nonlinear cellular trajectories from single cell RNA-seq data. Genome Biol 17: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, et al. 2013. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20: 1131–1139. [DOI] [PubMed] [Google Scholar]
- Yao Z, Mich JK, Ku S, Menon V, Krostag AR, Martinez RA, Furchtgott L, Mulholland H, Bort S, Fuqua MA, et al. 2017. A single-cell roadmap of lineage bifurcation in human ESC models of embryonic brain development. Cell Stem Cell 20: 120–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O. 2016. Engineering stem cell organoids. Cell Stem Cell 18: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. 2015. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347: 1138–1142. [DOI] [PubMed] [Google Scholar]
- Zhang X, Friedman A, Heaney S, Purcell P, Maas RL. 2002. Meis homeoproteins directly regulate Pax6 during vertebrate lens morphogenesis. Genes Dev 16: 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. 2013. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78: 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Huangfu D. 2013. Human pluripotent stem cells: an emerging model in developmental biology. Development 140: 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickenrott S, Angarica VE, Upadhyaya BB, del Sol A. 2016. Prediction of disease–gene–drug relationships following a differential network analysis. Cell Death Dis 7: e2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Edri R, Yaffe Y, Donaghey J, Pop R, Mallard W, Issner R, Gifford CA, Goren A, Xing J, et al. 2015. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature 518: 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.