Abstract
Transcriptional deregulation of oncogenic pathways is a hallmark of cancer and can be due to epigenetic alterations. 5-Hydroxymethylcytosine (5-hmC) is an epigenetic modification that has not been studied in pancreatic cancer. Genome-wide analysis of 5-hmC-enriched loci with hmC-seal was conducted in a cohort of low-passage pancreatic cancer cell lines, primary patient-derived xenografts, and pancreatic controls and revealed strikingly altered patterns in neoplastic tissues. Differentially hydroxymethylated regions preferentially affected known regulatory regions of the genome, specifically overlapping with known H3K4me1 enhancers. Furthermore, base pair resolution analysis of cytosine methylation and hydroxymethylation with oxidative bisulfite sequencing was conducted and correlated with chromatin accessibility by ATAC-seq and gene expression by RNA-seq in pancreatic cancer and control samples. 5-hmC was specifically enriched at open regions of chromatin, and gain of 5-hmC was correlated with up-regulation of the cognate transcripts, including many oncogenic pathways implicated in pancreatic neoplasia, such as MYC, KRAS, VEGFA, and BRD4. Specifically, BRD4 was overexpressed and acquired 5-hmC at enhancer regions in the majority of neoplastic samples. Functionally, acquisition of 5-hmC at BRD4 promoter was associated with increase in transcript expression in reporter assays and primary samples. Furthermore, blockade of BRD4 inhibited pancreatic cancer growth in vivo. In summary, redistribution of 5-hmC and preferential enrichment at oncogenic enhancers is a novel regulatory mechanism in human pancreatic cancer.
Pancreatic ductal adenocarcinoma (aka pancreatic cancer) is a disease of near uniform lethality, and insights into molecular pathogenesis are urgently needed (Vincent et al. 2011). Epigenetic alterations are widespread in cancer and play important roles in cancer growth, and they metastasize through regulation of transcription. Chemical modification of DNA at regulatory regions by DNA methyltransferase enzymes, resulting in generation of a 5-methylcytosine (5-mC) residue, has been extensively studied in cancer and is altered in both global- and site-specific manners. Recently, additional chemical modifications of DNA have been elucidated, including the hydroxymethylcytosine residue, 5-hmC, which is generated from 5-mC through dioxidation by the Ten-eleven translocation (TET) family of proteins. (Tahiliani et al. 2009; Ko et al. 2010; Guo et al. 2011; Pastor et al. 2011). The discovery of loss-of-function mutations in genes encoding TET proteins in various hematological malignancies suggests that defects in the 5-hmC biosynthetic pathway have functional consequences in carcinogenesis (Abdel-Wahab et al. 2009; Figueroa et al. 2010; Bejar et al. 2011; Cimmino et al. 2011; Moran-Crusio et al. 2011). Comparable pathophysiological mechanisms have been implicated in solid tumors like glioblastoma or cholangiocarcinoma, bearing mutations in genes encoding isocitrate dehydrogenase (IDH) 1 or 2, which leads to formation of an oncometabolite that impedes TET function (Lu et al. 2013; Saha et al. 2014). The conclusions emerging from these data suggest that loss of 5-hmC is a cancer-specific “epigenotype” and, in fact, immunohistochemical studies on large panels of human archival cancers for global 5-hmC levels have shown reduced expression (Haffner et al. 2011; Lian et al. 2012). Nonetheless, studies that examine modified DNA moieties at the global scale are likely to miss alterations that might occur at specific genomic loci and have an impact on transcriptional regulation. This is likely to be especially true in cancers such as pancreatic cancer, where genomic alterations of neither TET nor IDH genes have been reported; therefore, cancer cells might retain the ability for site-specific redistribution of chemical moieties to further a growth or survival advantage. Thus, in this study, we conducted a comprehensive genome-wide analysis of 5-hmC in pancreatic cancer and correlated these with transcriptomic alterations in this malignancy.
Results
Widespread 5-hmC alterations are seen in pancreatic cancer
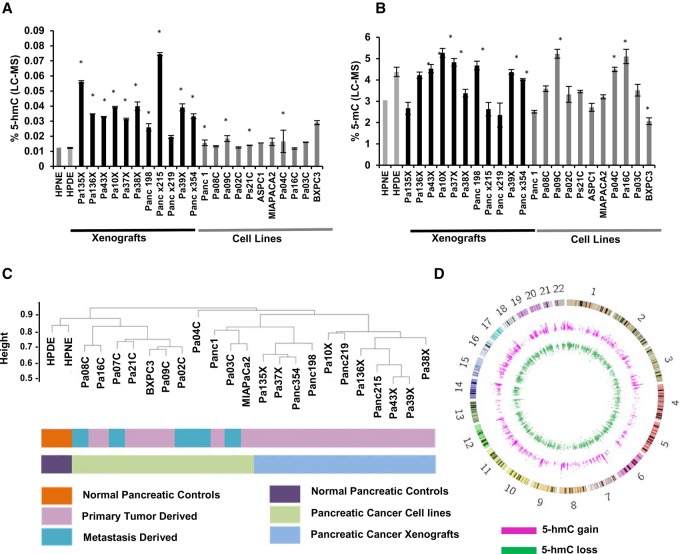
Although prior studies have examined levels of 5-hydroxymethylation in archival tissues by immunohistochemistry (Haffner et al. 2011), there have been no quantitative assessments of global 5-hmC in pancreatic ductal adenocarcinoma (PDAC). Therefore, we first evaluated the status of total 5-hmC in a panel of 11 low-passage PDAC cell lines and 11 patient-derived xenografts (PDXs) (Supplemental Table S1) and compared these levels to immortalized pancreatic control cells by high performance liquid chromatography coupled with tandem mass spectrometry (HPLC-MS/MS). Overall, 5-hmC was detectable in both PDAC and in control non-neoplastic pancreatic epithelial cells, albeit at lower levels than that of 5-mC (Fig. 1A,B). Variability in total 5-hmC was seen across PDAC cell lines, with modestly increased levels when compared to controls (Fig. 1A). 5-hmC content was found to correlate positively with both TET2 and TET3 expression (Supplemental Fig. S1A,B). No correlation between 5-hmC content or tumor (primary or metastatic) stage was observed (Supplemental Fig. S1C). PDAC PDXs had a slightly higher range of 5-hmC amounts, but these also included non-neoplastic murine stromal and inflammatory cells. Next, we analyzed the genome-wide distribution of 5-hmC by using chemical conjugation and affinity purification followed by next-generation sequencing (hMe-Seal) (Song et al. 2011; Mariani et al. 2014). Unsupervised clustering based on global 5-hmC patterns revealed that malignant cell lines and PDXs exhibited similarities in 5-hmC localization, although heterogeneity was seen within these groups that was independent of the stage of the parental tumor. In spite of this heterogeneity, we observed that PDAC cell lines and PDXs clustered distinctly from both non-neoplastic control cells, underscoring key differences in localization of 5-hmC between cancer and normal cells (Fig. 1C). Thus, next, we wanted to study the common 5-hmC alterations that were seen in PDAC cells when compared to controls. Supervised analysis revealed a set of genomic regions that commonly gained and lost 5-hmC residues in cancer versus non-neoplastic pancreatic cells (Fig. 1D). These “differential 5-hmC regions” (DHMRs) were found to occur throughout the genome, illustrating the widespread range of these alterations.
Figure 1.
Altered patterns of hydroxymethylation are seen in pancreatic cancer cell lines and primary xenografts. Total 5-hmC (A) and 5-mC (B) levels were measured by LC-MS in pancreatic cancer cell lines primary xenograft tumors and immortalized healthy pancreatic controls (HPNE, HPDE, gray). Values are expressed as a percentage of total cytosine. (*) P-value <0.05 when compared to HPNE, t-test. (C) Unsupervised clustering based on 5-hmC peaks demonstrates dissimilarity between controls and pancreatic cancer cells. (D) Circos plot (Krzywinski et al. 2009) shows genome-wide distribution of regions that gain and lose 5-hmC in pancreatic cancer cells.
DHMRs in pancreatic cancer occur at regulatory regions of the genome
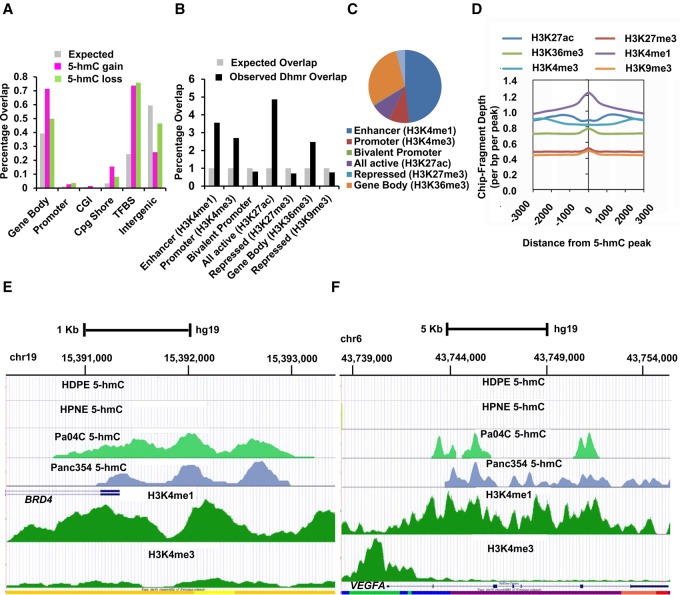
We then mapped the distribution of DHMRs in PDAC, including both gains and losses of 5-hmC signal compared to non-neoplastic pancreatic lines. We found one of the highest frequency of DHMRs, mostly comprised of gains of 5-hmC, occurring at gene bodies, with a significantly lower proportion localized to gene promoters and traditionally defined CpG islands (CGI) (Fig. 2A). A comparable frequency of DHMRs was also found at transcription factor binding sites (TFBS), with nearly equal rates of loss and gain of 5-hmC residues. Other genomic regions harboring significant DHMRs included CpG shores and intergenic sites. The preference of DHMRs at non-CGI loci such as TFBS, in contrast to promoters and traditional CGIs, is consistent with recent findings in TET2-mutant leukemia, wherein differential hypermethylation also occurs preferentially at non-CGIs, including TFBS, at the expense of CGIs (Fig. 2A; Yamazaki et al. 2015). Next, we mapped DHMRs to known histone modifications of the adult pancreatic genome as defined in ENCODE (GSM910576). We observed significant enrichment for DHMRs occurred at enhancers (defined by the H3K4me1 mark), at active promoters (defined by the H3K4me3 mark), at sites of active transcription (defined by the H3K27Ac mark), and at intergenic sites of active genes (defined by the H3K36me3 mark) (Fig. 2B; Pekowska et al. 2011; Rada-Iglesias et al. 2011; Herz et al. 2012b; Pfister et al. 2014). Quantitatively, most DHMR peaks overlapped with pancreatic-specific enhancers (Fig. 2C) and strikingly coincided with H3K4me1 (enhancer) peaks (Fig. 2D). Numerous well-established oncogenes were involved by acquisition of 5-hmC residues at enhancer locations (Fig. 2E,F; Supplemental Fig. S2). For example, the gene encoding the bromodomain-containing transcription factor BRD4 was found to acquire 5-hmC at regulatory regions in PDAC that overlapped with H3K4me1 peaks (Fig. 2E). Notably, no 5-hmC peaks were seen at the BRD4 enhancer in both control cell lines, whereas these were observed in the majority of PDAC cell lines and PDXs (Fig. 2E; Supplemental Fig. S2). Presence of 5-hmC at BRD4 promoter was validated quantitatively by genomic qPCR following 5-hmC pulldown (Supplemental Fig. S2B). Similarly, the gene encoding the angiogenic growth factor VEGFA also acquired 5-hmC residues within the gene body overlapping with known enhancer regions in a majority of neoplastic samples (Fig. 2F; Supplemental Fig. S2).
Figure 2.
Pancreatic cancer cells acquire changes in 5-hmC at regulatory regions of the genome. (A) Common regions of 5-hmC gain and loss in pancreatic cancer occur significantly at gene bodies, promoters, and transcription factor binding sites. Gains/losses compared with expected genomic distribution (P-value <0.05). (B) Overlap of differentially hydroxymethylated regions in cancer (DHMR) with pancreatic histone marks shows enrichment at H3K4me1, H3K4me3, and other regulatory regions. (C) Distribution of differentially hydroxymethylated peaks shows most overlap with H3K4me1 regions. (D) Overlap of 5-hmC peaks with histone modifications shows spatial overlap with H3K4me1 regions. (E) BRD4 gene locus shows acquisition of 5-hmC peaks in pancreatic cancer cell line (Pa04C) and Xenograft (Panc354) around the promoter with strong overlap with H3K4me1 peaks. No 5-hmC peaks were seen in two healthy controls (HDPE, HPNE). (F) VEGFA gene locus shows acquisition of 5-hmC peaks in gene body with strong overlap with H3K4me1 peaks.
Whole-genome analysis of 5-hmC at single-base resolution reveals localization at regions of open chromatin obtained by ATAC-seq
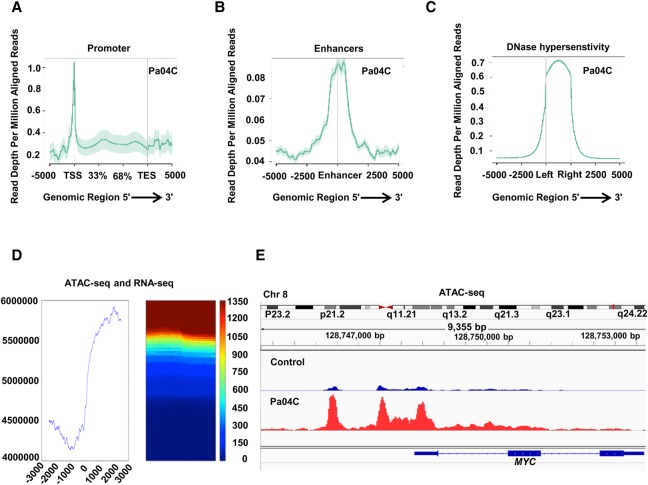
Next, we wanted to analyze 5-hmC and 5-mC at single base pair resolution and determine its association with chromatin architecture. To generate de novo chromatin accessibility maps, we performed ATAC-seq in pancreatic cancer and control cells. ATAC-seq peaks were found to localize at promoters, known enhancers, and DNase hypersensitivity sites, validating the assay (Fig. 3A–C; Supplemental Fig. S3). Correlation of ATAC-seq peaks with gene expression generated by RNA-seq on these cells revealed an association of open chromatin with actively transcribed regions in pancreatic cells (Fig. 3D). Examination of known pancreatic cancer oncogenes also demonstrated prominent ATAC-seq peaks in promoters of MYC (Fig. 3E), KRAS, VEGFA, and BRD4 genes (Supplemental Fig. S4) in pancreatic cancer cells.
Figure 3.
Chromatin accessibility patterns from ATAC-seq in pancreatic cancer define regulatory regions of the genome. (A–C) ATAC-seq peaks in Pa04C pancreatic cancer cells are enriched in promoter regions (A), known enhancers (B), and known DNase hypersentivity regions (C). (D) Correlation of ATAC-seq peaks with RNA-seq shows proximity to highly expressed transcripts. (E) Example of highly expressed MYC oncogene that shows larger ATAC-seq peaks in pancreatic cancer cells when compared to controls.
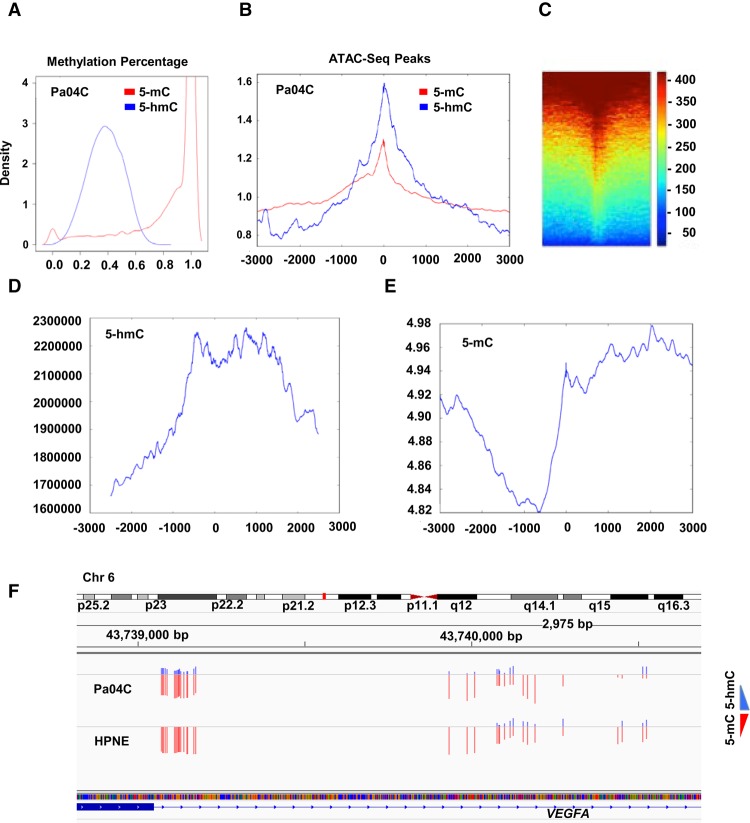
Next, we wanted to compare whole-genome 5-hmC and 5-mC modifications with chromatin accessibility maps that were generated de novo. Oxidative bisulfite sequencing was performed on the same control and pancreatic cancer cells. After validating adequate bisulfite and oxBS conversion (Supplemental Fig. S5; Supplemental Tables S2, S3), a total average of 11 million CpGs were analyzed by deep sequencing for 5-hmC and 5-mC determination. We observed that 5-hmC was seen on 0.006% of CpGs examined in both cancer and control samples and was a significantly less prevalent mark when compared to 5-mC (Supplemental Table S2). On cytosines that contained 5-hmC, the average prevalence of hydroxymethylation on each CpG was 38 ± 12% and was much lower than the methylation percentage (mean of 84 ± 26%) seen on 5mC sites (Fig. 4A). Correlation with ATAC-seq peaks demonstrated that 5-hmC correlated more significantly than 5-mC with open regions of chromatin (Fig. 4B,C) (P < 0.0001, permutation test). Correlation of 5-hmC with expression (RNA-seq) also revealed a positive association of increased 5-hmC at promoters of transcribed genes (Fig. 4D). On the other hand, 5-mC at promoters was found to correlate negatively with gene expression (Fig. 4E). Finally, we examined 5-hmC and 5-mC marks at selected oncogenic regions. The VEGFA gene that is highly expressed in cancer, but not in controls, demonstrated selective gain of 5-hmC in cancer cells (Fig. 4F). Examination of purely 5-mC did not reveal any difference in cytosine methylation between control and cancer cells and does not consequently correlate with expression of the VEGFA. Similar results were seen with other oncogenes examined for 5-hmC and 5-mC, reinforcing that determination of 5-hmC is critical while studying epigenetic control of gene expression (Supplemental Fig. S6).
Figure 4.
5-hmC obtained from OXBS correlates with regions of open chromatin and gene expression. (A) Density plot shows percentage of 5-hmC and 5-mC in CpGs in pancreatic cancer cells. (B) Colocalization of 5-hmC/5-mC with ATAC-seq peaks shows 5-hmC is enriched at areas of open chromatin when compared to 5-mC (P < 0.0001, permutation test on differences in kurtosis). (C) Heatmap shows colocalization of 5-hmC with ATAC-seq peaks. (D,E) Colocalization of 5-hmC loci with RNA-seq shows higher 5-hmC and lower 5-mC at promoters with expressed transcripts. (F) Example of highly expressed VEGFA oncogene, shows similar amounts of 5-mC, but higher amounts of 5-hmC in pancreatic cancer cells when compared to controls.
Differential expression of critical oncogenic pathways in PDAC is associated with alterations of 5-hmC levels
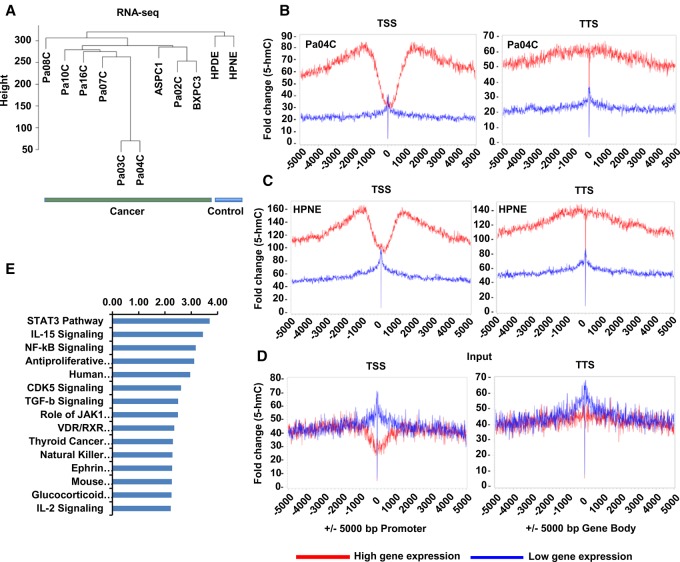
Having shown significant correlation of 5-hmC and gene expression in selected cancer and control cells, we wanted to determine which gene pathways were regulated in concordance with 5-hmC changes in the larger cohort of pancreatic cancer samples. RNA-seq was performed on nine low-passage cancer cell lines and two controls and showed transcriptomic differences between cancer and control samples on unsupervised clustering (Fig. 5A). Correlation with 5-hmC generated by hmc-SEAL on the same cell lines revealed that acquisition of 5-hmC residues at gene promoters and gene bodies correlated significantly with increased expression of the corresponding transcripts (Fig. 5B–D). This was found in both control and cancer samples. To uncover 5-hmC associated transcriptional changes in PDAC, we next determined differentially expressed genes in cancer cells (obtained from RNA-seq of matched samples) that were associated with corresponding changes in 5-hmC. Gene set enrichment analysis showed that 5-hmC-associated transcriptional changes were similar to known cancer-associated gene signatures (Supplemental Table S4; Supplemental Fig. S7). Epigenetically regulated genes in PDAC were also found to be associated with key oncogenic pathways, with STAT3-regulated genes as the top pathway (Fig. 5E; Supplemental Table S5). Furthermore, since we observed a substantial overlap of 5-hmC changes with expression, we analyzed the genomic areas for common motifs and observed that these were also enriched for the STAT3 binding site in addition to other transcription factors (Supplemental Table S6).
Figure 5.
5-hmC correlates with gene expression in pancreatic cancer and regulates oncogenic pathways. (A) Unsupervised clustering based on gene expression profiling by RNA-seq of pancreatic cancer and pancreatic cancer cells are transcriptionally distinct from controls. (B,C) 5-hmC loci were mapped relative to RefSeq transcripts expressed at different levels in pancreatic cells as obtained from RNA-seq analysis. RefSeq transcripts were divided into two bins based on gene expression level and 5-hmC regions centered on transcription start sites or end sites. (B,C) Enrichment of 5-hmC is seen in highly expressed genes in both controls and cancer cells. (D) Input controls are shown. (E) Differentially expressed genes that are associated with corresponding changes in 5-hmC are grouped in functional pathways by the ingenuity pathway tool.
BRD4 and TET3 are overexpressed in pancreatic cancer, and BRD4 expression correlates with acquisition of 5-hmC at enhancer regions
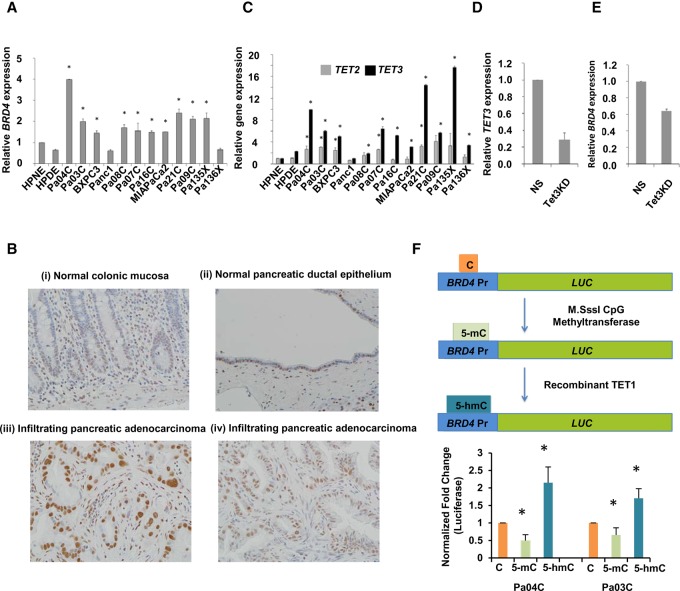
We next determined BRD4 transcript expression by qRT-PCR in pancreatic cancer and control samples and observed overexpression in a majority of cancer cells (Fig. 6A). Overexpression of nuclear BRD4 protein was nearly ubiquitous in tissue microarrays created from archival PDAC; in contrast, minimal nuclear BRD4 expression was observed in non-neoplastic pancreatic ductal epithelium and in other normal tissues, such as colonic epithelium (Fig. 6B). Next, we determined the relative expression of TET demethylases in pancreatic cancer cells. We observed that TET3 was overexpressed in nearly all pancreatic cancer samples (11/12) when compared to TET2 (overexpressed in 5/12) (Fig. 6C). The magnitude of TET3 fold change increase was also greater in pancreatic cancer cells (Fig. 6C). We then used shRNAs to knockdown TET3 expression in Pa04C cells (Fig. 6D) and determined that this led to decreased BRD4 expression in these cells (Fig. 6E). Next, we wanted to determine the effect of in vitro 5-hmC acquisition on BRD4 transcription, as a quantitative measure of BRD4 regulation by 5-hydroxymethylation. The BRD4 promoter region was used to design a reporter that was then either methylated or hydroxymethylated, the latter using in vitro addition of recombinant TET enzyme in two independent PDAC cell lines (Pa03C and Pa04C). We observed that cytosine methylation led to expected decrease in expression (Fig. 6F), whereas 5-hydroxymethylation led to increased expression that was greater than unmodified control, demonstrating that 5-hmC changes directly associated with higher expression of BRD4.
Figure 6.
BRD4 and TET3 are overexpressed in pancreatic cancer, and BRD4 expression correlates with acquisition of 5-hmC at the promoter. (A) qRT-PCR for BRD4 expression shows significant increase in pancreatic cancer cell lines when compared to controls (t-test, P-value <0.05 for all pancreatic cancer samples when compared to HPNE control). (B) IHC for BRD4 shows specific staining in pancreatic cancer cells: normal colonic mucosa with weak staining restricted to basal crypt epithelium (i); normal pancreatic ductal epithelium with patchy nuclear BRD4 labeling (ii); infiltrating pancreatic adenocarcinoma with diffuse strong nuclear BRD4 labeling (iii); and independent example of Infiltrating pancreatic adenocarcinoma with diffuse strong nuclear BRD4 labeling (iv). (C) qRT-PCR for TET2 and TET3 shows higher fold change increase for TET3 in pancreatic cancer cells when compared to controls (t-test, P-value <0.05 for pancreatic cancer samples when compared to HPNE control). (D) qRT-PCR for TET3 shows significant reduction with Pa04C cells with TET3 shRNA when compared to nonsilencing (NS) controls (t-test, P-value <0.05, N = 2). (E) qRT-PCR for BRD4 shows significant reduction with Pa04C cells with TET3 shRNA when compared to nonsilencing (NS) controls (t-test, P-value <0.05, N = 2). (F) Luciferase promoter for BRD4 was methylated and hydroxymethylated in vitro and shows significantly increased expression after 5-hmC acquisition (t-test, P-value <0.05, N = 4).
BRD4 is a therapeutic target in pancreatic cancer
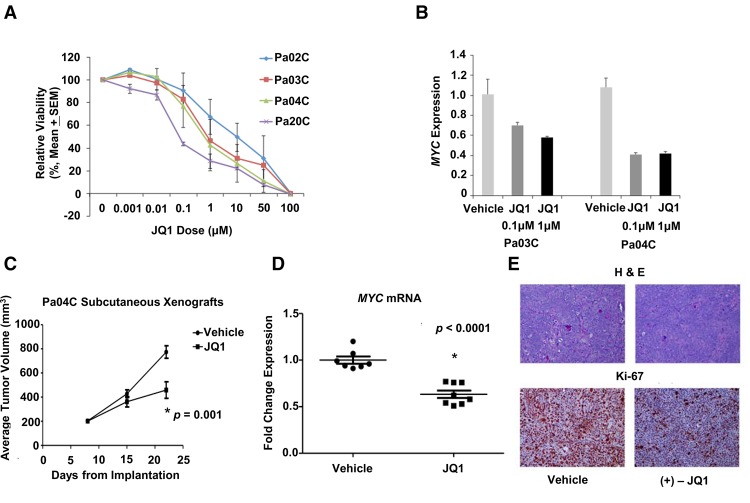
We next tested the functional impact of BRD4 inhibition in PDAC with a small molecule inhibitor JQ1. JQ1 treatment led to dose-dependent decrease in viability of all PDAC cell lines tested (Fig. 7A) and reduced expression of the credentialed BRD4 target, MYC (Fig. 7B). The role of BRD4 inhibition in vivo was tested in subcutaneous Pa04C xenografts (a high BRD4-expressing cell line), which were treated with either vehicle or JQ1 by daily i.p. injection. Treatment with JQ1 significantly inhibited tumor growth (P-value = 0.001) (Fig. 7C) and resulted in significantly decreased MYC transcript expression in the excised tumors (P < 0.0001) (Fig. 7D). Histology on the treated xenografts demonstrated a striking decrease in the proliferating cells upon JQ1 administration, which was confirmed by Ki-67 labeling (Fig. 7E; Supplemental Fig S8).
Figure 7.
BRD4 inhibition leads to abrogation of pancreatic cancer growth in vitro and in vivo. (A) Pancreatic cancer cells were treated with varying doses of BRD4 inhibitor JQ1 and viability was assessed by MTS assay. (B) MYC expression was evaluated after 48 h of treatment with JQ1 in pancreatic cancer cells by qRT-PCR. (C) Athymic nude mice with established bilateral subcutaneous Pa04C xenografts were treated with either vehicle (n = 7 mice) or JQ1 (n = 8 mice) by daily i.p. injection. Treatment with JQ1 significantly inhibited tumor growth (P = 0.001). Mean ± SEM is plotted. (D) Treatment with JQ1 resulted in significantly decreased MYC mRNA expression as measured in excised tumors by qRT-PCR (P < 0.0001). Mean ± SEM is indicated by bars. (E) Ki67 immunohistochemistry and H&E stained sections from representative xenograft per treatment arm. A marked decrease in the proliferation marker Ki67 was observed in xenografts treated with JQ1.
Discussion
Chemical modification of cytosine residues on CpG dinucleotides located within promoters, gene bodies, and intergenic regions has been recognized as an important regulator of transcriptional activity during development and differentiation (Shen and Zhang 2013; Piccolo and Fisher 2014). Although numerous chemical modifications of cytosine have been identified, the 5-hmC residue in particular has garnered considerable attention due to its ability to undergo active or passive demethylation, leading to epigenetic reactivation of transcription. The metazoan TET family of dioxygenases is comprised of three catalytic enzymes (TET1, TET2, and TET3) that mediate the hydroxymethylation step of methylcytosine in an oxoglutarate and Fe(II)-dependent manner (Ito et al. 2010; Ko et al. 2010), and sequencing studies in human neoplasms, as well as in vivo modeling experiments in mice, have established the pathogenic role of loss-of-function TET mutations in various malignancies (Wu and Zhang 2011; Shih et al. 2012; Rampal et al. 2014; Ko et al. 2015). In addition to hematological malignancies, certain solid tumors such as gliomas or cholangiocarcinoma, which do not otherwise display loss-of-function mutations of TET genes, harbor gain-of-function mutations of IDH genes (either IDH1 or IDH2), which lead to production of an oncometabolite (2-hydroxyglurate, 2-HG) that diminishes TET function through competitive blockade of oxoglutarate-dependent catalytic conversion of 5-mC to 5-hmC (Ward et al. 2010; Cairns and Mak 2013). Although a genomic basis for absence of TET function, and hence global reduction in 5-hmC levels, can be attributed to the aforementioned examples, more recent data in archival tissue specimens have suggested that low 5-hmC levels are a generalized “epigenotype” of many solid tumors, even in the absence of TET or IDH coding mutations (Haffner et al. 2011; Lian et al. 2012; Chen et al. 2013). The biochemical basis for this putative loss of TET enzymatic activity in human cancers that do not harbor aberrations at corresponding genomic loci is unclear, but might represent the consequence of alterations in intracellular alpha-ketoglutarate/succinate ratios within the nutrient-deprived milieu of most cancers (Carey et al. 2015). As Thompson and colleagues have shown in embryonic stem (ES) cell models, a high intracellular alpha-ketoglutarate/succinate ratio, in turn impacted by the availability of sufficient extracellular glucose and glutamine, can directly regulate the function of TET enzymes (and thus, maintain genome-wide 5-hmC levels) (Carey et al. 2015). In nutrient-deprived conditions, where ambient glucose and glutamine levels are compromised, and intracellular alpha-ketoglutarate/succinate ratios trend downward, TET function is attenuated, resulting in global reduction of 5-hmC levels. Irrespective of the biochemical mechanism of reduction, our HPL-MS/MS studies do establish that, compared to non-neoplastic pancreatic epithelial cells, there is a modest reduction in 5-hmC levels in PDAC cell lines, and in nearly all cases, these levels are lower than the corresponding 5-mC levels in the same tumor. The global reduction is less obvious in PDAC PDX models; however, these also have accompanying murine stroma and inflammatory cells. Of note, all PDAC cell lines and PDX models used in this study had previously been confirmed to lack coding mutations in either TET or IDH genes (Jones et al. 2008; Biankin et al. 2012). An acknowledged limitation of our study is the absence of primary tumors; however, most primary PDAC are comprised of extensive stromal infiltrates (desmoplasia), which render neoplastic epithelial-specific molecular alterations challenging to discern (Maitra and Hruban 2008). Further, the genome-wide assays utilized in this study are currently not amenable to the scale of limited nucleic acids that would be obtained by microdissection.
In addition to the global assessment of 5-hmC levels, we have performed a mapping of the genome-wide “hydroxymethylome landscape” of PDAC and compared alterations in 5-hmC to that of transcriptomic changes. Irrespective of the extent of global changes in 5-hmC, we found evidence for commonalities in the genome-wide redistribution of 5-hmC residues in PDAC compared to non-neoplastic epithelial cells, resulting in distinct hierarchical clustering of both cell lines and PDXs versus non-neoplastic cells. Of note, our analysis revealed the concentration of these so-called DHMRs occurred mostly at regulatory regions of the genome, and in particular at non-CGIs, such as gene bodies and TFBS. These findings mirror what has been recently reported in the setting of TET2-mutant leukemias for differential 5-mC regions (Yamazaki et al. 2015), underscoring that traditional CGIs are unlikely to be the major regulatory sites for this particular mechanism of transcriptional modulation. More striking was the overlap of DHMRs with specific histone marks in the chromatin, in particular, at enhancers (defined by the H3K4me1 mark), at active promoters (defined by the H3K4me3 mark), at sites of active transcription (defined by the H3K27Ac mark), and at intergenic sites of active genes (defined by the H3K36me3 mark) (Pekowska et al. 2011; Rada-Iglesias et al. 2011; Herz et al. 2012a; Pfister et al. 2014). Overall, these results suggested that redistribution of 5-hmC peaks in PDAC correlates with sites of active transcription; therefore, not surprisingly, many of the regulatory regions containing DHMRs were those linked to known cancer-promoting oncogenes, such as BRD4 or VEGFA. The consequence of redistribution of DHMRs to regulatory regions of actively transcribed genes likely goes beyond the known active or passive demethylation that occurs with chemical conversion of 5-mC to 5-hmC. Recent studies in glioblastoma and lung cancer have also shown that 5-hmC is associated with enhancers (Johnson et al. 2016; Li et al. 2016). Other studies have suggested that 5-hmC may act as an independent activation mark via recruitment of readers such as MECP2 and MBD3 (Yildirim et al. 2011; Mellén et al. 2012). Studies have also shown that 5-hmC is involved in embryonic cell pluripotency and differentiation and can be modulated by metabolic changes (Carey et al. 2015; Etchegaray et al. 2015; McDonald et al. 2017). Interestingly, Godley and colleagues have shown that enrichment of DHMRs occurs at erythroid-specific TFBS during erythroid differentiation and at sites of hypoxia-responsive elements (HREs) in the setting of hypoxia (Madzo et al. 2014; Mariani et al. 2014), presumably directing appropriate erythroid maturation and hypoxia response, respectively. In PDAC cells, we find that DHMRs are similarly enriched at TFBS, including those that have well-established roles in PDAC pathogenesis, such as the “top hit” STAT3, a credentialed oncogenic transcription factor in this neoplasm (Fukuda et al. 2011; Treiber et al. 2011). Overall, our studies underscore that 5hMC redistribution plays a copy number–independent regulatory transcriptional role in cancer.
BRD4 is a bromodomain protein that has been shown to be an important oncogenic transcription factor in multiple cancers, including PDAC (Zuber et al. 2011; Lockwood et al. 2012; De Raedt et al. 2014; Sahai et al. 2014; Mazur et al. 2015; Garcia et al. 2016). Bromodomain inhibitors including JQ1 have been developed that competitively bind to the acetyl-lysine recognition site of BET family bromodomains, displacing BRD4 from nuclear chromatin (Filippakopoulos et al. 2010). Clinical candidates that inhibit bromodomain-family proteins, including BRD4, are being tested in early phase clinical trials (Picaud et al. 2013). In spite of a multitude of studies about BRD4, there is scant knowledge of how expression of this oncogenic transcription factor is regulated in solid tumors outside of the prototype midline NUT carcinoma, where the BRD4 gene undergoes an activating translocation (French 2012). Using PDAC as a prototype solid tumor, we demonstrate that acquisition of 5-hmC at the BRD4 enhancer is a potential regulatory mechanism for oncogenic activation. Inhibition of BRD4 in PDAC xenografts with JQ1 causes in vivo tumor growth inhibition, reiterating the functional impact of acquiring 5-hmC at critical oncogenic regulators on tumor growth.
In summary, our work has, for the first time, defined the hydroxymethylome landscape of PDAC and identified the role that cancer-specific redistribution plays in driving the altered transcriptome of this neoplasm. We identify key chromatin marks that overlap with DHMRs and show the functional consequence of 5-hmC acquisition using BRD4 as a prototype oncogene. Our studies further clarify how cancer cells use subversion of the physiological epigenome to drive carcinogenesis.
Methods
Molecular methods
Cells and reagents
The human PDAC cell lines and PDXs used in this study were established as described (Jones et al. 2008). Cell lines and xenografts used in this study were low-passage cells (fewer than 10 passages) and are listed in Supplemental Table S1. Cells were maintained in Dulbecco's Modified Eagle's Medium (Invitrogen #11965) supplemented with 10% FBS, 1% Pen-Strep, and 5 µg/mL Plasmocin (Invivogen). Pa02C was cultured in RPMI1640 medium (Invitrogen #11875) supplemented with 20% FBS, 1% Pen-Strep, and 5 µg/mL Plasmocin (Invivogen). The human non-neoplastic hTERT-immortalized pancreatic cell line HPNE was obtained from the ATCC and cultured according to published protocols. HPDE is an E6/E7 immortalized human pancreatic ductal epithelial line and was cultured as published previously (Liu et al. 1998). All cell lines were routinely tested and were negative for mycoplasma infection by the MycoAlert Mycoplasma Detection Kit (Lonza). DNA fingerprinting was used to authenticate all cell lines.
BRD4 promoter luciferase
The 1.8-kb upstream promoter region of BRD4 gene was amplified by PCR from human genome and cloned at XhoI and HindIII sites of pGL4.19 reporter plasmid (Promega). Five micrograms of the purified plasmid was treated with M.SssI CpG methylase (New England Biolabs) according to the manufacturer's instructions. The in vitro methylated plasmid was purified and further treated with recombinant Tet1 enzyme (Active Motif). Pa04 and Pa03 cells were cotransfected with 100 ng Control, 5-mC or 5-hmC plasmid with 40 ng Renilla luciferase plasmid with Lipofectamine LTX and PLUS reagent (Invitrogen). Luciferase assay was performed at 24 h post-transfection by Dual-Glo luciferase assay system (Promega). PCR and sequencing primers are in Supplemental Table S1.
qPCR validation of 5-hmC
Real-time qPCR was performed on pulldown and input DNA obtained from CLICK chemistry 5-hmC pulldown (Song et al. 2011). The DNA was amplified by the Whole Genome Amplification Kit (Sigma). A total of 100 pg of DNA was used per reaction using 5× EvaGreen Supermix (Bio-Rad) and primers for selected genes.
BRD4 IHC on TMAs
Immunohistochemisty for BRD4 expression was performed on previously described TMAs created from archival surgically resected PDAC specimens (Swierczynski et al. 2004); the TMAs also contained matched non-neoplastic pancreas tissue on adjacent cores and a variety of control tissues from other organs. Three TMAs, comprising 54 PDAC specimens and matched non-neoplastic pancreatic tissues were used. The primary anti-BRD4 antibody was used at a dilution of 1:100. Only nuclear BRD4 expression was counted as positive expression.
Xenograft studies
All small-animal experiments described conformed to the guidelines of the Animal Care and Use Committee of the Johns Hopkins University. Mice were maintained in accordance with the guidelines of the American Association of Laboratory Animal Care. JQ1 for animal studies was purchased from ChemScene and confirmed in vitro to have the same IC50 as JQ1 obtained from Dr. Bradner prior to use in vivo. Flanks of 5- to 6-wk-old athymic nude mice (Harlan Laboratories) were injected with 5 × 106 Pa04C cells suspended in a total volume of 200 µL of a 1:1 mixture of DPBS and BD Matrigel. Eight days after injection, when visible tumors were established, mice were randomized to receive daily i.p. injections of either vehicle (10% hydroxypropyl β-cyclodextrin in water) or JQ1 (50 mg/kg in 10% hydroxypropyl β-cyclodextrin). Tumor volume (V = ab2/2 where a and b are the long and short dimensions, respectively) and body weight were measured throughout treatment. At the culmination of treatment, visceral organs and tumor tissue were harvested and either fixed in 10% neutral buffered formalin or snap frozen for molecular analysis.
JQ1 cell viability assay
Twenty-four hours prior to drug administration, cells were seeded in 96-well plates in standard media. On the day of treatment, media was aspirated and replaced with standard media containing JQ1 in DMSO (final DMSO concentration 1%). Controls were treated with standard media containing 1% DMSO. Each concentration was performed in triplicate, and each experiment was repeated two independent times. Cells were incubated with JQ1 solution for 48 h, at which point cell viability was measured by CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega) according to the manufacturer's instructions.
qRT-PCR for MYC in JQ1 treated xenografts
Total RNA was isolated from snap-frozen Pa04C xenografts using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. cDNA was generated using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with random primers. qRT-PCR was performed using Fast SYBR Green Master Mix (Applied Biosystems Cat. 4385612) and primers specific to MYC. GUSB was used as an internal loading control.
Ki67 immunohistochemistry
Immunohistochemistry was performed on formalin-fixed paraffin-embedded tissue, using common laboratory techniques. Briefly, the slides were deparaffinized using xylenes and hydrated by a graded series of ethanol washes. Endogenous peroxidase activity was quenched by 20-min incubation in 0.3% H2O2, and antigen retrieval was accomplished by heating the slides in EDTA (pH 8.0) for 35 min at 90°C. Nonspecific binding was blocked by incubation in Serum Free Protein Block (Dako X0909) before incubation with the primary antibody (Ki-67, Cell Marque, Clone SP6, Rabbit Monoclonal, dilution 1:200). Chromogenic detection was enabled using the PowerVision+ Poly-HRP IHC Kit (Immunovision Technologies) following the manufacturer's protocol. Slides were counterstained with Harris-hematoxylin solution. Slides from five independent xenografts per treatment group were stained.
Analysis of global DNA methylation and hydroxymethylation by mass spectrometry
Genomic DNA was hydrolyzed by DNA Degradase Plus (Zymo Research) according to the manufacturer's instructions. Digested DNA was injected onto a UPLC Zorbax Eclipse Plus C18 RRHD column (Agilent Technologies). The analytes were separated by gradient elution using 5% methanol/0.1% formic acid (mobile phase A) and 100% methanol (mobile phase B) at a flow rate of 0.25 mL/min. Mobile phase B was increased from 0 to 3% in 5 min, to 80% in 0.5 min, kept at 80% for 2 min, then switched to initial conditions in 2.5 min. The effluent from the column was directed to the Agilent 6490 Triple Quadrupole mass spectrometer (Agilent Technologies). The following transitions were monitored: m/z 228.1 → 112.1 (C); m/z 242.1 → 126.1 (5-mC); and m/z 258.1 → 142.1 (5-hmC).
Calibration solutions with varying amounts of 5-hmC (0%–3%), 5-mC (0%–10%), and fixed amount of C, were also analyzed together with the samples. The solutions were prepared from a 200-bp DNA standard containing 57 cytosines, which are homogeneous for C, 5-hmC, or 5-mC. Calibration plots of %5-hmC or %5-mC versus MRM Response ratio were constructed based on the data obtained, and %5-hmC is obtained from the ratio of [5-hmC/(5-mC+5-hmC+C)]. Response ratio is the response peak area for 5-hmC or 5-mC divided by the combined peak areas of 5-hmC, 5-mC, and C. The %5-hmC or 5-mC in the samples was determined from the calibration plots.
Genomic and statistical methods
Affinity purification of 5-hmC-enriched sequences by hmC-seal
Genomic DNA (gDNA) was sonicated into 200- to 400-bp-long fragments (Covaris). Briefly, 5-hmC labeling reactions were performed in a 20 µL solution containing 50mM HEPES buffer (pH 7.9), 25 mM MgCl2, 100 ng/µL sonicated gDNA (200–400 bp), 250 µM UDP-6-N3-Glu, and 2.25 µM βGT enzyme. The reactions were incubated for 1 h at 37°C, and following incubation, the labeled DNA was purified by the QIAquick PCR Purification Kit (Qiagen) and eluted in water. The click chemistry reaction was performed by the addition of 150 µM dibenzocyclooctyne modified biotin into the eluted DNA, and the reaction mixture was incubated for 2 h at 37°C. The DNA samples were then purified using the MinElute Reaction Cleanup Kit (Qiagen), and the amount of eluted DNA was determined by Nanodrop UV spectroscope (Thermo). hMe-Seal (affinity pulldown of 5-hmC) was performed as described previously (Song et al. 2011). In total, 20 μg of gDNA was labeled in 30 µL with a biotin linker that contained a disulfide bond. Labeled DNA was pulled down with streptavidin-coated magnetic beads (Invitrogen). After washing, captured DNA was released from beads with 50 mM DTT; excess DTT was removed by chromatography spin column (Bio-Rad), and the DNA was purified in a total volume of 12 μL by the MinElute Reaction Cleanup Kit (Qiagen). The final yield of pulled-down DNA was determined using PicoGreen (Invitrogen).
5-hmC-affinity-sequencing data mapping and analysis
We used the BWA aligner (version 0.6.1) with default parameters to align the sequencing reads for each sample to the NCBI reference human genome sequence (Build 37/hg19). We used hg19 for integrating data from different genomic platforms (RNA-seq, hmC-seal, OXBS, ATAC-seq) that were performed at different times. The hmC-seal and RNA-seq data were collected prior to GRCh38 becoming available. In this paper, the hg19 alignment was primarily used for global analyses and summaries involving multi-kilobase ranges. Since major differences between the hg19 and GRCh38 assemblies involve single-base SNP calling, alternate loci scaffolds, centromeres, and mitochondrial DNA (Schneider et al. 2017), using the GRCh38 assembly would not significantly affect our global integrative analysis and conclusions. Peak calling for each sample was performed using MACS (version 1.4.2) with default parameters.
Whole-genome methylation and hydroxymethylation analysis by oxidative bisulfite sequencing
One microgram genomic DNA from HPNE and Pa04C were sonicated to 100–400 bp by Bioruptor, and 0.5% (w/w) sequencing spike-in control DNA was added thereafter and purified by Ampure XP beads. Spike-in controls were added to the adapted library. Half of the library was subjected to oxidation reaction following the manufacturer's protocol (Cambridge Epigenetix). Both oxidized and nonoxidized samples were then treated with bisulphite conversion reagent. The final PCR was performed according to the manufacturer's guide using 10 cycles of amplification, purified, and sequenced at the Einstein Epigenomics Facility (Booth et al. 2012). Bismark was used to map the sample reads and make methylation calls. At every base location, the 5-mC percentage was estimated by the ratio of nonconverted CpG bases to the total number of bases using the pileup of reads from the OxBS sample. For the estimation of 5-hmC, both BS and OxBS samples were analyzed, and an estimate for the percentage of 5-hmC methylation was calculated by the difference between BS and OXBS conversions.
Since 5-hmC is a less frequent modification, for further stringency in measuring the difference in ratios, we used Fisher's exact test of proportions, using the number of converted and nonconverted reads in the BS and OxBS samples, and selected sites that have a P-value <0.05. 5-mC sites were calculated by the ratio of nonconverted bases to total bases in the OxBS sample with a biologically influenced threshold of 50%. To compare methylation between cancer and control samples, we used Fisher's test and adjusted for multiple comparisons through the Benjamini-Hochberg procedure (Benjamini and Hochberg 1995). Unsupervised clustering of samples based on 5-hmC was based on genomic locations of 5-hmCs peaks and also considered the sum of normalized reads falling in that particular location.
ATAC-seq
A suspension of 50,000 cells from both control (HPNE) and Pancreatic Cancer (Pa04C) cells were harvested, resuspended in cold lysis buffer, and incubated in transposition reaction mixture at 37°C and subjected to ATAC-seq as performed previously (Buenrostro et al. 2015a,b). Briefly, after the reaction was immediately purified by the Qiagen MinElute PCR Purification Kit, PCR was performed on the eluted DNA using barcoded primer and thermal cycle according to the method described above. We used Bowtie 2 (version 2.2.3) with parameters allowing for soft clipping to align the sequencing reads for each sample to the NCBI reference human genome sequence (Build 37/hg19). Peak calling for each sample was performed using MACS2 (version 2.1.0) with default parameters. For discovering differential peaks between samples, one sample was treated as the target, the other as the background control.
Colocalization of 5-hmC and 5-mC with RNA-seq and ATAC-seq
The association of cytosine methylation with RNA expression and chromatin accessibility was examined by taking the 5-mC/5-hmC sites from the OxBS data set and overlaying read depth information from RNA-seq and ATAC-seq data sets in the same samples. Colocalization plots were constructed by centering windows of ±3000 base pairs at the 5-mC and 5-hmC site locations and aggregating the reads lying inside throughout the entire alignment file. For stranded libraries (RNA-seq), we accounted for the directionality of the reads by assigning to the windows front to back when the nearest gene is on the forward strand, and back to front when on the reverse. After all sites were processed, summing the window counts produced a read depth profile, and binning the window counts and sorting by average depth was used to generate a heatmap. To compare the ATAC-seq peaks with 5-mC and 5-hmC, the ATAC-seq reads were normalized by total read count and overlayed. In the colocalization plots, although both 5-hmC and 5-mC show a peak at the center of the window, the spread of the 5-mC profile was wider than that of the 5-hmC profile. To show the significance of this difference statistically, we treated the profiles as probability density functions, and sampled 10,000 points from each distribution. A permutation test (K = 10,000) was run on the difference in kurtosis of the two sampled sets (5-hmC – 5-mC). A significant positive result meant that the 5-hmC profile had a higher kurtosis than the 5-mC profile, which can be interpreted as it having a tighter clustering around the center of the window.
RNA-sequencing
Total RNA was isolated using TRIzol Reagent (Invitrogen), and the integrity of the total RNA was validated using the Agilent 2100 Bioanalyzer. Libraries were generated following the Illumina protocol for preparing samples for sequencing of mRNA, and 1–10 µg of total RNA was used to build libraries for single-read sequencing on the Illumina Hiseq 2000. mRNA was isolated by polyA selection. The mRNA was then fragmented and randomly primed for reverse transcription, followed by second-strand synthesis to create double-stranded cDNA fragments. Ends of the cDNA fragments were repaired with a combination of fill-in reactions and exonuclease activity to produce blunt ends. An “A” base was added to the blunt ends followed by ligation to Illumina sequencing adapters. cDNA fragments ranging from 300 to 500 bps were gel purified after the adapter ligation step. PCR amplified cDNA libraries were quantified on the Agilent 2100 Bioanalyzer and diluted to 10 pM for cluster generation and sequencing. Single-end sequencing was performed for 50 cycles by using the Single Read Cluster Generation Kit (TruSeq SR Cluster Kit v3 - cBot –HS, Cat# GD-401-3001) and Sequencing Kit (TruSeq SBS Kit v3– HS, Cat# FC-401-3002) on an Illumina HiSeq 2000 machine. Sequence reads from RNA-sequencing were aligned to genomic sequences.
RNA-sequencing, data mapping, and analysis
The RNA-sequencing reads were mapped to the reference genome hg19 using TopHat (Trapnell et al. 2009). Gene expression analysis was performed using Cufflinks.
Identification of transcription factor binding site motifs
All 5-hmC-peaks that gained in expression were extracted to perform a transcription factor binding site analysis using HOMER with default parameters for motif identification (Heinz et al. 2010).
Data access
RNA-sequencing, 5-hmC-SEAL, and ATAC-seq sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE97018. The individual submissions are as follows: RNA-seq: GSE97003; 5-hmC-Seal: GSE97014; ATAC-seq: GSE97008 GEO. Pancreatic-specific histone modifications were obtained from the ENCODE (GSM910576) database. All bioinformatics scripts for bioinformatic analysis (Supplemental Table S7) have been deposited at https://github.com/VermaLab/hmcPaper_scripts2.
Supplementary Material
Acknowledgments
We thank the Gottesman Stem Cell Institute and Center of Epigenomics at Albert Einstein College of Medicine, and also the MD Anderson Pancreatic Cancer Moonshot, the Khalifa Bin Zayed Foundation, and the Van Loh family. This work was supported by NIH/NCI U01 CA 196403, and NIH/NCI U01 CA 200468, to A.M.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.222794.117.
References
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. 2009. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 114: 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, Kantarjian H, Raza A, Levine RL, Neuberg D, et al. 2011. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 364: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser B 57: 289–300. [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. 2012. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015a. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. 2015b. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523: 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Mak TW. 2013. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer Discov 3: 730–741. [DOI] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. 2015. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518: 413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Shen F, Huang W, Qi JH, Wang Y, Feng YQ, Liu SM, Yuan BF. 2013. Quantification of 5-methylcytosine and 5-hydroxymethylcytosine in genomic DNA from hepatocellular carcinoma tissues by capillary hydrophilic-interaction liquid chromatography/quadrupole TOF mass spectrometry. Clin Chem 59: 824–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. 2011. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 9: 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Raedt T, Beert E, Pasmant E, Luscan A, Brems H, Ortonne N, Helin K, Hornick JL, Mautner V, Kehrer-Sawatzki H, et al. 2014. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 514: 247–251. [DOI] [PubMed] [Google Scholar]
- Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Gladden A, et al. 2015. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 17: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. 2010. Selective inhibition of BET bromodomains. Nature 468: 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA. 2012. Pathogenesis of NUT midline carcinoma. Annu Rev Pathol 7: 247–265. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, et al. 2011. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 19: 441–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, Heslin MJ, Arnoletti JP, Richardson JH, Chen D, et al. 2016. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene 35: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. 2011. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, Toubaji A, Argani P, Iacobuzio-Donahue C, Nelson WG, et al. 2011. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2: 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garrett AS, Miller C, Casto D, Zhang Y, Seidel C, Haug JS, Florens L, Washburn MP, et al. 2012a. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol Cell Biol 32: 1683–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. 2012b. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 26: 2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KC, Houseman EA, King JE, von Herrmann KM, Fadul CE, Christensen BC. 2016. 5-Hydroxymethylcytosine localizes to enhancer elements and is associated with survival in glioblastoma patients. Nat Commun 7: 13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, An J, Pastor WA, Koralov SB, Rajewsky K, Rao A. 2015. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev 263: 6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu Y, Salz T, Hansen KD, Feinberg A. 2016. Whole-genome analysis of the methylome and hydroxymethylome in normal and malignant lung and liver. Genome Res 26: 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, Xu W, Tan L, Hu Y, Zhan Q, et al. 2012. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 150: 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Furukawa T, Kobari M, Tsao MS. 1998. Comparative phenotypic studies of duct epithelial cell lines derived from normal human pancreas and pancreatic carcinoma. Am J Pathol 153: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. 2012. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci 109: 19408–19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Venneti S, Akalin A, Fang F, Ward PS, Dematteo RG, Intlekofer AM, Chen C, Ye J, Hameed M, et al. 2013. Induction of sarcomas by mutant IDH2. Genes Dev 27: 1986–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madzo J, Liu H, Rodriguez A, Vasanthakumar A, Sundaravel S, Caces DB, Looney TJ, Zhang L, Lepore JB, Macrae T, et al. 2014. Hydroxymethylation at gene regulatory regions directs stem/early progenitor cell commitment during erythropoiesis. Cell Rep 6: 231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Hruban RH. 2008. Pancreatic cancer. Annu Rev Pathol 3: 157–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani CJ, Vasanthakumar A, Madzo J, Yesilkanal A, Bhagat T, Yu Y, Bhattacharyya S, Wenger RH, Cohn SL, Nanduri J, et al. 2014. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep 7: 1343–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sanchez-Rivera FJ, Lofgren SM, Kuschma T, Hahn SA, Vangala D, et al. 2015. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med 21: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, et al. 2017. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet 49: 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellén M, Ayata P, Dewell S, Kriaucionis S, Heintz N. 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151: 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, et al. 2011. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 473: 394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. 2011. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J 30: 4198–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ, Helleday T, Legube G, La Thangue NB, Porter AC, et al. 2014. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 7: 2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picaud S, Da Costa D, Thanasopoulou A, Filippakopoulos P, Fish PV, Philpott M, Fedorov O, Brennan P, Bunnage ME, Owen DR, et al. 2013. PFI-1, a highly selective protein interaction inhibitor, targeting BET Bromodomains. Cancer Res 73: 3336–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo FM, Fisher AG. 2014. Getting rid of DNA methylation. Trends Cell Biol 24: 136–143. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. 2014. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 9: 1841–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, Gurumurthy S, Akbay EA, Sia D, Cornella H, et al. 2014. Mutant IDH inhibits HNF-4α to block hepatocyte differentiation and promote biliary cancer. Nature 513: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai V, Kumar K, Knab LM, Chow CR, Raza SS, Bentrem DJ, Ebine K, Munshi HG. 2014. BET bromodomain inhibitors block growth of pancreatic cancer cells in three-dimensional collagen. Mol Cancer Ther 13: 1907–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. 2017. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27: 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Zhang Y. 2013. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr Opin Cell Biol 25: 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 12: 599–612. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. 2011. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol 29: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczynski SL, Maitra A, Abraham SC, Iacobuzio-Donahue CA, Ashfaq R, Cameron JL, Schulick RD, Yeo CJ, Rahman A, Hinkle DA, et al. 2004. Analysis of novel tumor markers in pancreatic and biliary carcinomas using tissue microarrays. Hum Pathol 35: 357–366. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber M, Neuhöfer P, Anetsberger E, Einwächter H, Lesina M, Rickmann M, Liang S, Kehl T, Nakhai H, Schmid RM, et al. 2011. Myeloid, but not pancreatic, RelA/p65 is required for fibrosis in a mouse model of chronic pancreatitis. Gastroenterology 141: 1473–1485, 1485.e1–7. [DOI] [PubMed] [Google Scholar]
- Vincent A, Omura N, Hong SM, Jaffe A, Eshleman J, Goggins M. 2011. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res 17: 4341–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. 2010. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17: 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. 2011. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J, Jelinek J, Lu Y, Cesaroni M, Madzo J, Neumann F, He R, Taby R, Vasanthakumar A, Macrae T, et al. 2015. TET2 mutations affect non-CpG Island DNA methylation at enhancers and transcription factor-binding sites in chronic myelomonocytic leukemia. Cancer Res 75: 2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147: 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478: 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.