Abstract
Mechanosensory transduction for senses such as proprioception, touch, balance, acceleration, hearing and pain relies on mechanotransduction channels, which convert mechanical stimuli into electrical signals in specialized sensory cells1. How force gates mechanotransduction channels is a central question in the field, for which there are two major models. One is the membrane-tension model: force applied to the membrane generates a change in membrane tension that is sufficient to gate the channel, as in the case of bacterial MscL channel and certain eukaryotic potassium channels2-5. The other is the tether model: force is transmitted via a tether to gate the channel. Recent study suggests that NOMPC, a mechanotransduction channel that mediates hearing and touch sensation in Drosophila, is gated by tethering of its ankyrin repeat (AR) domain to microtubules of the cytoskeleton6. Thus, a goal of studying NOMPC is to reveal the underlying mechanism of force induced gating, which could serve as a paradigm of the tether model. NOMPC, a Transient Receptor Potential (TRP) channel and the founding member of the TRPN sub-family7, fulfills all the criteria for a bona fide mechanotransduction channel1,8, and is important for a variety of mechanosensation-related behaviors such as locomotion, touch and sound sensation across different species including C. elegans9, Drosophila8,10-11 and zebrafish12. NOMPC has 29 ARs, the largest number among TRP channels. They are implicated as tether to convey force from cytoskeleton to the channel, thus to mediate mechanosensation6,13-15. A key question is how the long AR domain is organized as a tether that can trigger channel gating. Here we present a de novo atomic structure of NOMPC determined by single particle electron cryo-microscopy (cryo-EM), and discuss how its architecture could provide a means to convey mechanical force to generating an electrical signal within a cell.
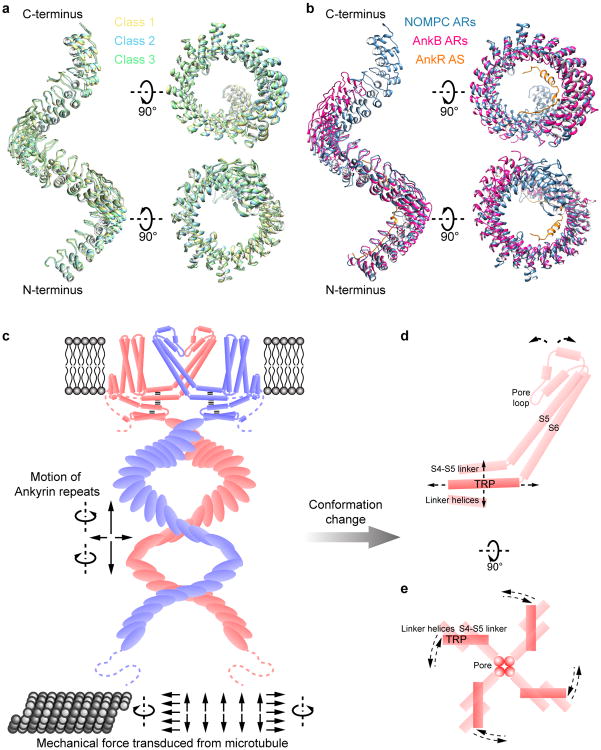
By screening NOMPC homologues, we identified full-length Drosophila NOMPC for structural analysis. Recombinantly expressed NOMPC is functional and purified protein retains microtubule binding activity (Extended Data Fig. 1). Purified and detergent-solubilized protein was either exchanged to amphipols or reconstituted into lipid nanodiscs for structure determination (Extended Data Fig. 2a, b), following procedures previously described16,17. Electron micrographs and two-dimensional (2D) class averages of negatively stained NOMPC showed that the nanodisc sample has better-organized features in the cytoplasmic domain than the amphipol sample (Extended Data Fig. 2c-f). Moreover, estimated resolution and structural features in the map from nanodisc sample are significantly better while the overall architectures are comparable to those from the amphipol sample (Extended Data Figs 3, 4). Based on the 3.6Å resolution map of the channel in nanodiscs (Extended Data Figs 4, 5), we constructed a de novo atomic model of NOMPC. Side chain positions of most residues from Ala359 to Trp1602 (including 22 ARs, namely ARs 8-29) and from Arg1670 to Gly1689 were identified, except for some charged residues exposed to solvent, or located at the contact sites between neighboring AR domains (Fig. 1). In addition, we were able to trace the Cα chain of the first 7 ARs, from Met124 to Ala359. The N-terminal domain prior to the first AR is too flexible to be modeled, and they are predicted to be un-structured.
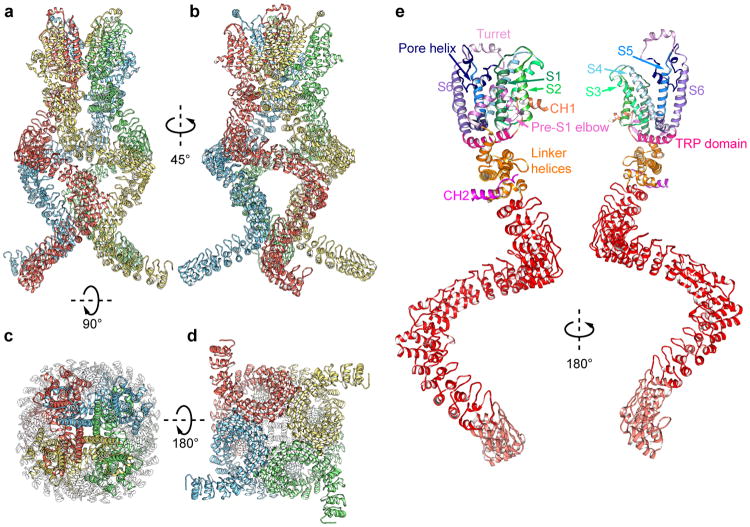
Figure 1. 3D reconstruction of NOMPC.
a–d, NOMPC density map in nanodisc (EMD-8702). Unsharpened (transparent) and sharpened (solid and color coded) maps are shown, from side (a, b), top (c) and bottom (d). e, Diagram illustrating major structural domains of one subunit, color coded to match ribbon diagrams in f. Dashed lines denote regions where the density map is insufficient for model building. ARs 1-7 are colored in pale red because its density is insufficient to position side chains. f, Ribbon diagram showing one NOMPC subunit denoting specific domains (pdb code: 5VKQ).
Strikingly, four AR domains from the tetrameric NOMPC are organized into a quadruple structure with each domain resembling a helical spring (Fig. 1a-d, Extended Data Fig. 6a-d), unlike those in other TRP family proteins such as TRPV116 and TRPA118. The connection between the ARs region and the transmembrane core could be separated into two parts: a stack of linker helices and two helices that span only half of the lipid bilayer from the cytoplasmic side and form an elbow-like structure (Fig. 1f, Extended Data Fig. 6e). We name this new feature as the “pre-S1 elbow”. It packs against the S1 helix and creates a “bay” with S1 and S4 from the same subunit plus S3-S5 from the adjacent subunit to harbor phospholipids (discussed below).
The transmembrane domain of each NOMPC subunit is composed of six transmembrane α-helices (S1–S6) and a re-entrant pore loop located between S5 and S6 (Fig. 1f). Four subunits form a homotetramer through domain swap-interactions (Fig. 1c, Extended Data Fig. 6) as in other TRP channels and voltage-gated potassium (Kv) channels. Following S6, the “signature” TRP domain is sandwiched between the linker helices and the S4-S5 linker (Fig. 1f). A short helix immediately after the TRP domain is packed against S2 and the pre-S1 elbow. Beyond these, the C-terminus is mostly unstructured (Fig. 1f, Extended Data Fig. 6).
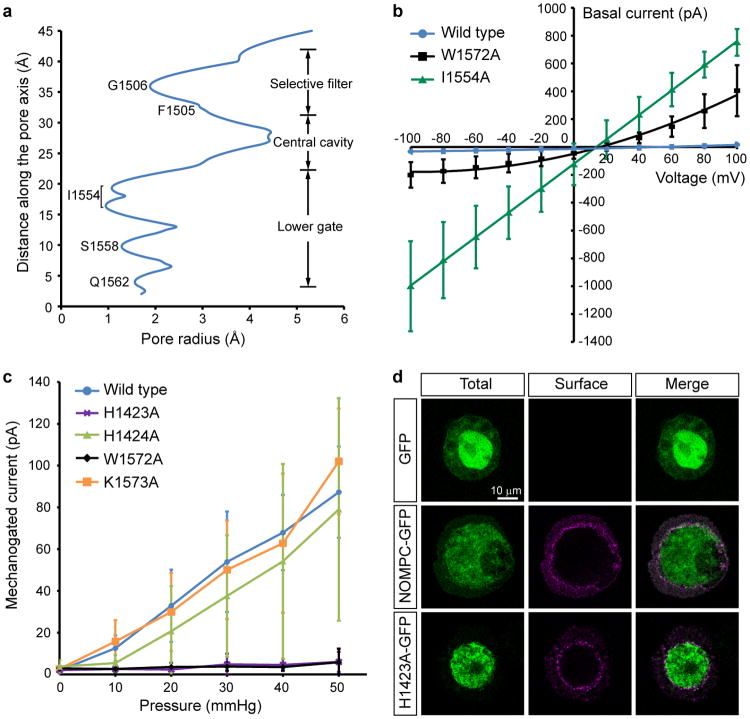
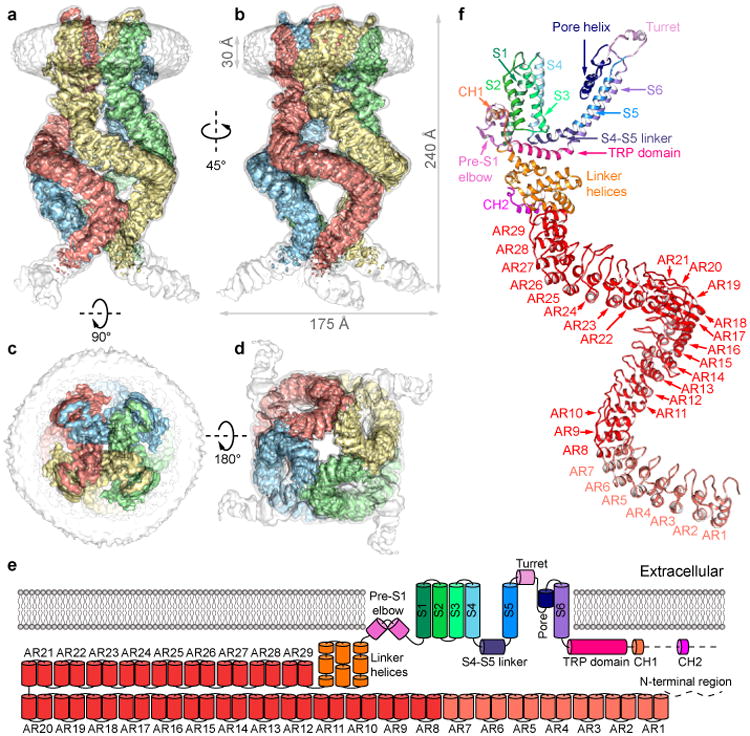
The central pore of NOMPC has two major constrictions, sharing the general architecture of TRP, Nav, Cav and K channels16,19-21. At the upper constriction, the carbonyl oxygens of residues Phe1505 and Gly1506 in the pore loop face the pore (Fig. 2a). The diameter of this constriction (3.8Å defined by opposing van der Waals surfaces (vdW), 7Å defined as the distance between the centers of opposing oxygen atoms) is large enough to accommodate a partially-hydrated cation (Extended Data Fig. 7a), and is comparable to the size of the corresponding region of the open state (∼3.5Å vdW, ∼8Å distance) rather than the closed state (∼1.5Å vdW, ∼4Å distance) of TRPV122. A negatively charged residue (Glu1511) just above the upper constriction presumably attracts cations to the pore (Fig. 2); replacing Glu1511 with a positively charged residue reduces the single channel conductance8.
Figure 2. Architecture of the ion permeation pathway of NOMPC.
a, Solvent-accessible pathway along the ion permeation pore illustrated as blue surface. Residues located at the selectivity filter and lower gate are rendered as sticks. b, Top view of the NOMPC pore region is shown in surface representation, colored by the electrostatic potentials (negative: red; positive: blue).
The lower constriction of the channel adopts a closed architecture (Fig. 2a, Extended Data Fig. 7a) with Ile1554 forming a hydrophobic seal, similar to TRPV1 in the closed state16. Replacing Ile1554 with alanine results in a leaky channel yielding large basal current without mechanical stimulation (Extended Data Fig. 7b), confirming its critical role in preventing ion flow through the pore. The overall arrangement of the central pore of NOMPC is thus similar to the TRPA1 pore, with the upper constriction in a conformation that can accommodate ions while the lower gate is closed18.
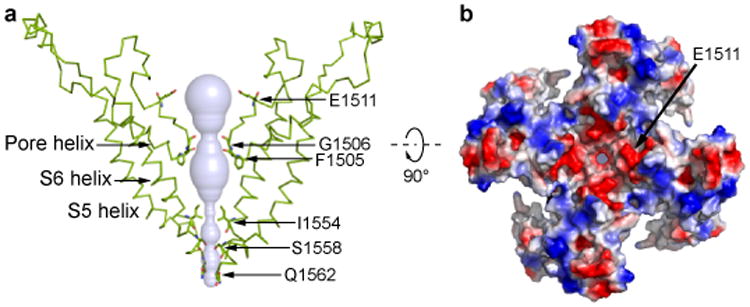
We observed densities that can be attributed to lipids, with the most pronounced densities found in the intracellular leaflet. A tightly bound lipid (Fig. 3a, b) occupies a hydrophobic cleft formed by the pre-S1 elbow, S1, S4 and the S4-S5 linker of one subunit and S5 of its adjacent subunit. The inner surface of this pocket is coated with hydrophobic side chains, creating an ideal environment for the lipid aliphatic chains. Near the solvent-accessible face, His1423 in the S4-S5 linker coordinates the lipid polar headgroup (Fig. 3b). His1423Ala mutant is still expressed on the cell membrane but the channel no longer responds to mechanical stimulation, and mutation of the adjacent His1424 had no effect on channel function (Fig. 3c, Extended Data Fig. 7c, d), suggesting that this specific lipid-protein interaction is functionally important. Density for this lipid also appears in the amphipol map, indicating that this lipid is tightly bound and may represent an endogenous lipid.
Figure 3. Lipid-protein and linker domain interactions.
a, The transmembrane domains of two adjacent subunits are shown in blue and red from the cytosolic face (left) and with the cytosolic domain facing down (right). The lipid bound to the S4-S5 linker of the red subunit is colored green. b, Enlarged view of boxed region of NOMPC shows the density map (green mesh) of the lipid superimposed with an atomic model of the bound phospholipid. The side chain of His1423 from the S4–S5 linker is close (∼2.9Å) to the lipid headgroup, and the adjacent His1424 is further away (∼4.8Å). c, Representative traces of mechanogated current of wild type NOMPC and NOMPC mutants (H1423A, H1424A, K1573A and W1572A) under pressure ranged from 10 mmHg to 50 mmHg with 10 mmHg increment, recorded by outside-out patch clamp. The H1424A and K1573A mutations are negative controls showing that not any random mutation in the S4-S5 linker or TRP domain abolishes mechanogated current of NOMPC. d-g, Enlarged views from the boxed regions showing key interactions between residues from interacting domains. Hydrogen bonds are indicated as dashed lines (d, e). EM density is shown in mesh (f).
The pre-S1 and linker domains that connect the AR domain to the transmembrane core adopt an architecture reminiscent of the stacked helices in TRPA118, but distinct from the known structures of TRPV channels16,23-25. A series of “linker helices” are sandwiched between the transmembrane domain and the AR domain (Fig. 1e, f), creating a network of interactions connecting the S4-S5 linker via TRP domain to the ARs (Fig. 3d-g). As a novel addition to the transmembrane domain, two helices form an inverted “V”, which partially inserts into and then exits the membrane next to (and in sequence immediately before) S1. This “pre-S1 elbow” makes extensive hydrophobic contacts with the S1 helix (Fig. 3d), and at the same time wraps around the cavity occupied by the tightly bound lipid (Fig. 3a, b), holding the lipid in proximity to His1423 of the S4-S5 linker (Fig. 3a, b).
Facing towards the cytoplasm, the S4-S5 linker forms a loop that encircles the side chain of the conserved Trp1572 of the TRP domain (Fig. 3e), creating a pivotal point of contact between the S4-S5 linker and the TRP domain that interacts with the densely-packed linker helices connected to the AR domain (Fig. 3f, g). Substitution of Trp1572 with alanine results in a channel that exhibits a prominent basal current but no longer responds to mechanical stimuli with an increase of current amplitude (Fig. 3c and Extended Data Fig. 7b, c). This tryptophan critical for channel gating is the invariant hallmark of TRP domains conserved in other TRP channel subfamilies26. Its mutation in TRPV3 leads to Olmsted syndrome27, and in TRPV4 causes gain-of-function phenotypes28. Thus, this tryptophan residue is of critical importance in the gating of multiple TRP channels.
On the opposite face of the TRP helix, pointing away from the membrane, a number of residues form complementary hydrophobic and polar contacts with the linker helices below (Fig. 3f). The remaining linker helices sit essentially parallel to the membrane surface, stacking above and below one another through a series of hydrophobic patches on adjacent faces (Fig. 3g). The linker region also contacts a helix from the very C-terminus of NOMPC, with residues from Trp1677 to Gly1689 forming a helix that stacks parallel to the final set of linker helices connected to the ARs (Fig. 3g). The extensive hydrophobic contacts between layers of linker helices provide a continuous connection from the AR domain via TRP domain to the S4-S5 linker and the C-terminus of the S6 helix. This architecture implies that the TRP domain may be the key element in linking the AR domains with the gate of the channel.
The most striking and unique feature of NOMPC is the architecture of its AR domains. Four AR domains that each resembles the shape of a helical-spring bundle together as a quadruplex. Along each subunit, there are four major contact points between adjacent subunits (Fig. 4a), forming two types of inter-subunit interactions that maintain its overall architecture. The first type is between AR24-26 of one subunit and AR16-18 of its neighbor to the left (Fig. 4a, upper panel). Near this position, the quadruplex formed by the four AR domains reaches its largest diameter. The second type is between AR7-10 of one subunit and AR9-12 of its neighbor on its left (Fig. 4a, lower panel), creating a second constriction of the AR quadruplex. Both types of interactions are formed by an array of polar residues from subunits facing each other. These two contact regions also harbor the sites of mutations that affect NOMPC cell surface expression or mechano-gating6. The contact surfaces are relatively broad, spanning several ARs in both subunits. Many residues that appear close enough to interact across subunits are polar, with a number of possible interacting partners on adjacent subunits, potentially allowing different contacts to form and dissolve. Also, side chain densities of about half of the polar residues in these interacting regions are not well resolved, even though most of the non-polar residues that are in these regions but not involved in interaction between AR domains have well resolved side chains. These features suggest that neighboring AR domains could move slightly with respect to one another.
Figure 4. Motion of the AR domain.
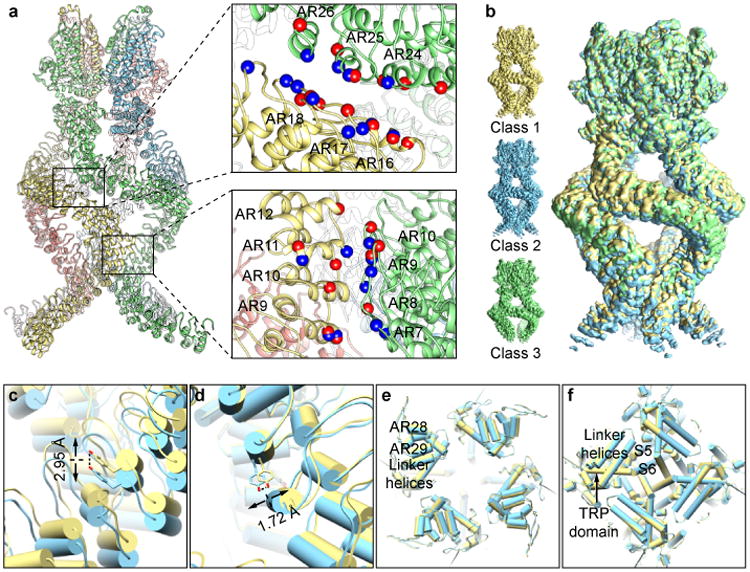
a, Interactions between the adjacent ARs. Residues with charged and polar side chains are indicated as colored balls (negative: red; positive: blue). b, Three sub-classes with different conformation in ARs. c-f, Comparison of class 1 and 2 at different positions along the ARs and linker domain. The ARs shifted vertically along the symmetry axis at the lower junction in the vicinity of AR9 (c) and laterally relative to the symmetry axis above the upper junction, at AR29 (d). While there is no obvious vertical movement at the linker regions, there is a small domain rotation (e and f).
Indeed, a reconstruction combining all particles revealed that the local resolution of the AR domain is lower than that of the transmembrane and linker domains (Extended Data Fig. 4). Further classification of the entire dataset resulted in three major classes (Fig. 4b, Extended Data Fig. 8), with slightly different conformations in the N-terminal ARs (Fig. 4c, d) and linker helices (Fig. 4e) but similar structure of the transmembrane core (Fig. 4f) with the same constriction at Ile1554 that is too narrow to allow the passage of a hydrated ion. Relative to the consensus class (Extended Data Fig. 8), from which the high-resolution structure was determined, the ARs of Class 1 are closer to the transmembrane domain, while the ARs of Class 2 are moved further away (Supplementary Video 1). The linker domain also has a subtle counter-clockwise rotation from Class 1 to 2, when viewed from the cytosolic side (Fig. 4e). From Class 1 to 2, Tyr411 (AR9) moved ∼3Å along the symmetry axis, while Ala1128 (AR29) moved ∼2Å parallel to membrane. It suggests that the downward displacement of the ARs (pulling away from membrane) could cause linker rotation. In Class 3, the linker positions are similar to those in the consensus class, but the distal ARs are displaced outwardly (Supplementary Video 2). Density for the N-terminal (lower) ARs is substantially weaker in this class, and the lower junction between AR domains appears to have widened, indicating that inter subunit interactions are weaker in this region. Superposition of the ARs alone from all classes shows small conformation differences of ARs with overall RMSDs of 0.94Å (class 1 vs 2), 1.27Å (class 1 vs 3) and 1.24Å (class 2 vs 3) (Extended Data Figure 9a). The postulated transitions between three classes (Supplementary Video 1-3) suggest that the entire quadruplex architecture is relatively flexible, raising the possibility that the spring-shaped ARs can transmit the force experienced by the distal N-termini to the linker domain.
It is conceivable that the AR domains could undergo larger deformations than the difference noted between classes 1-3. The 24 ARs of Ankyrin-B co-crystallized with a peptide29 display a spiral substantially different from that seen in NOMPC (Extended Data Fig. 9b), and these 24 ARs are capable of undergoing much larger deformations without unfolding14, consistent with the idea that large, spring-like motions may be possible for the AR domains of NOMPC.
For mechanical gating of NOMPC channel that is conveyed through its AR domain and critically dependent on microtubule integrity6, a movement of S6 that widens the constriction formed by Ile1554 would be required to open the channel. Comparison of the open and closed states of TRPV1 suggests that a rotation of the TRP domain in the plane of the membrane accompanied with movement of the S4-S5 linker correlates with pore opening22,24. A similar movement of the TRP domain in NOMPC would likely be associated with a substantial displacement of the pre-S1 elbow and/or linker helices linked to the AR domain. Notably, superimposition of different classes of NOMPC structures indicates that a rotation of the TRP domain and its associated linker helices may result from movements of the AR domains (Fig. 4e, f). The network of interactions along the linker helices establishes a tight connection from the AR domain to the TRP domain that extends from S6 and packs against the S4-S5 linker, likely coupling a movement of the AR domain to the pore formed by S5, S6 and the intervening re-entrant pore loop.
In cells, the highly conserved AR domain (Extended Data Fig. 10) could in principle be displaced in a variety of ways with respect to the membrane (Extended Data Fig. 9c-e). It appears that a motion of the AR domain greater than what we observed in the three classes would be required to drive channel opening, though we cannot exclude the possibility that some of these closed channel conformations correspond to a desensitized state or a captured intermediate state during gating transitions (Extended Discussion in Methods). While it is unlikely that membrane deformation alone is the driving force in NOMPC gating6, as in the case of other mechanosensitive channels such as Piezo and TRAAK channels3-5, 30, the proximity of a lipid molecule interacting with the functionally important His1423 at the S4-S5 linker suggests that lipid protein interactions may play an important role in channel activity. It will be of interest to explore the conformational changes that underlie mechanogating in future studies. Our structural analysis suggests that the AR domain could act as a spring or a knob, linking mechanical displacement to channel opening.
Methods
Screening of NOMPC orthologues
To identify the best protein for structural analysis, we screened through different NOMPC orthologues from Drosophila, zebrafish and Xenopus for high protein expression and conformational homogeneity (conservativity of these NOMPC orthologues are shown in Exteanded Data Fig. 10). All orthologues were expressed, purified, and analyzed as described in more details below for Drosophila NOMPC. In brief, the orthologues were expressed in HEK293 GnTi- cells, extracted in detergent followed by affinity purification, exchanged into amphipol A8-35, applied to size exclusion chromatography and subsequently examined by negative stain EM. Only Drosophila and zebrafish orthologues were expressed in sufficient amount for protein purification, with Drosophila NOMPC showing narrower peak from size exclusion chromatography as well as more homogeneous protein particles with better-organized features from negative EM. Thus, we selected Drosophila NOMPC for further studies.
Expression and purification of NOMPC
Full-length Drosophila NOMPC was expressed in HEK293 GnTi- cells using the BacMam system as described previously31. In brief, Bacmid for expressing NOMPC tagged with a Strep-GFP tag was generated in DH10Bac E. coli cells by transformation of the modified pFastBac construct with the mammalian CMV promoter. Baculovirus was generated by transfecting Spodoptera frugiperda (Sf9) cells (Thermo Fisher Scientific) cultured at 27 °C in Sf-900 III SFM (Gibco) with the bacmid using the FuGENE transfection reagent (Promega). After that, the baculovirus was amplified twice in Sf9 cells to obtain sufficient virus for large-scale infection. HEK293 GnTi- (ATCC) cells were grown in suspension at 37 °C in ESF SFM Serum-Free Medium (Expression Systems LLC) supplemented with 1% FBS. When the cells reached a density of 1.5-2×106 cells/mL, baculovirus was added to the culture (10-15%, v/v). After 24 hours, the culture was supplemented with 10 mM sodium butyrate to boost the expression, then further incubated at 30 °C for 48 hours before harvest.
Protein purification and sample processing prior to cryo-EM grid preparation was all carried out at 4 °C. Approximately 4 g cell pellet (from ∼0.5 L culture) was resuspended and lysed by stirring for 20 min in 50 mL hypotonic buffer containing 10 mM Bicine pH 8.5, 1 mM EDTA, 1 mM dithiothreitol (DTT) supplemented with 1× complete protease inhibitor cocktail (Roche) and 1 mM phenylmethylsulfonyl (PMSF). The membrane fraction was collected by centrifugation (35,000 ×g, 30 minutes), then homogenized with a Dounce homogenizer in extraction buffer (20 mM Bicine pH 8.5, 500 mM NaCl, 1 mM EDTA, 1× complete protease inhibitor cocktail, 1 mM PMSF). Protein was extracted in 50 mL extraction buffer plus 2% n-Dodecyl-β-D-Maltopyranoside (DDM) and 0.4% Cholesteryl hemisuccinate (CHS) with gentle stirring for 2 hours. Afterward, the lysate was cleared by centrifugation (35,000 ×g, 1 hour). The protein sample in DDM+CHS was then mixed with 2.5 mL CNBr-activated sepharose resin (GE healthcare) coupled with anti-GFP nanobody32 and incubated with inversion overnight. On the second day, the resin was collected on a column by gravity and washed with 10× column volume of wash buffer (WB) containing 20 mM Bicine pH 8.5, 500 mM NaCl, 0.05% DDM, 0.01% CHS, 1 mM EDTA supplemented with 0.1mg/ml 3:1:1 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE): 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG). The washed resin was incubated quiescently with 2.5 mL WB containing 0.2 mg TEV protease overnight to cleave the Strep-GFP tag and release the protein.
Same procedures were followed to purify NOMPC with Strep-GFP tag retained except that Strep-Tactin sepharose (IBA Lifesciences) was used for affinity purification and the protein was eventually eluted by 2.5 mM desthiobiotin (Sigma) in WB.
Amphipol exchange and nanodisc reconstitution
The concentration of retrieved protein was determined by measuring absorbance at 280 nm. For amphipol exchange, the protein sample was mixed with amphipols A8-35 (1:3, w/w) with gentle agitation overnight. Subsequently, 1 mL sample was applied to a Superose-6 column (GE healthcare) equilibrated with column buffer (CB, 20 mM Bicine pH 8.5, 500 mM NaCl, 1 mM EDTA). Further concentration or freeze-thaw of peak collections from size exclusion chromatography causes protein aggregation. The peak fraction corresponding to tetrameric NOMPC at a concentration of ∼0.6 mg/ml was immediately examined by negative stain EM (after 10 fold dilution) and used for cryo-EM grid preparation.
For nanodisc reconstitution, lipid was prepared as described previously17 except that the lipid was rehydrated with water. The protein sample in DDM+CHS was mixed with MSP2N2 and soy PC (Avanti) at a molar ratio of NOMPC monomer:MSP2N2:soy PC=1:4:100. The mixture was incubated with constant rotation for four days. During the first three days, Bio-beads SM2 (Bio-Rad) were supplemented to the mixture once a day (3:100, v/v) to initiate the reconstitution by gradually removing detergents from the system. Afterward, the reconstituted sample was cleared by passing through a mini column, and then applied to a Superose-6 column equilibrated with CB. The peak fractions were collected, concentrated to ∼3 mg/ml using Amicon Ultra filter device (100-kDa MWCO, Millipore), examined by negative stain EM followed by immediate cryo-EM grid preparation.
EM sample preparation and data acquisition
Electron microscopy data was acquired and processed as described previously, with some minor modifications16. First, samples were assessed by negative stain EM to determine suitability for cryoEM33. Briefly, 2.5 μL of sample at 20-50 μg/mL was applied to a glow discharged continuous carbon grid, which was then treated with 0.75% (w/v) uranyl formate. These grids were imaged on a Tecnai T12 microscope (FEI Company) operated at 120 kV and at a nominal magnification of 52,000× using an UltraScan 4000 camera (Gatan), corresponding to a pixel size of 2.02 Å on the specimen.
Grids for cryo-EM were prepared with CP3 (Gatan). C-flat R1.2/1.3 holey carbon grids (Protochips) were glow-discharged for 30s. Then 2.5μL protein sample at a concentration of ∼3mg/mL was applied onto the carbon face of the grids. The grids were blotted with Whatman #1 filter paper on the carbon side and bare plastic on the back side with an offset of -1 for 2.5 seconds at 92% humidity and plunge frozen in liquid ethane cooled by liquid nitrogen. The grids were loaded onto a 300 kV Polara (FEI) with a K2 Summit direct electron detector (Gatan). Data were collected at nominal magnification of 31,000×, corresponding to a physical pixel size of 1.22 Å (0.61 Å super resolution pixel size) on the specimen with a dose rate of 10 electrons per physical pixel per second. Images were recorded with SerialEM34 in super-resolution counting mode and a defocus range of -1.5 to -3.0 μm. A total exposure of 8s was used, with 0.2s subframes (40 total frames) to give a total dose of 54 electrons per Å2 (1.35 electrons per Å2 per subframe).
Cryo-EM grids of NOMPC in amphipols were prepared similarly, and Cryo-EM images were first collected at the Janelia CryoEM facility Titan Krios (FEI Company) microscope operated at 300kV and equipped with a Cs corrector and a post-GIF K2 camera. Cryo-EM images were collected with an energy selection slit of 20 eV and a physical pixel size of 1.35 Å. This yielded an initial reconstruction at ∼4.8 Å (data not shown) from ∼121,000 particles selected from 3,720 images by one round of 2D classification and two rounds of 3D classification in Relion. Subsequent data were collected on the UCSF Polara as above, yielding the amphipol reconstruction shown here.
Image Processing
For negative stain data, Simplified Application Managing Utilities for EM Labs (SAMUEL) scripts were used for image processing, particle picking and 2D classification16. In this package, 2D classification was carried out by ten cycles of correspondence analysis, k-means classification and multi-reference alignment using SPIDER operations ‘CA S’, ‘CL KM’ and ‘AP SH’35. For cryo-EM data, dose-fractionated super-resolution image stacks were drift corrected and binned 2×2 by Fourier cropping using MotionCor2 (ref 36) (after discarding the first two frames). Motion-corrected sums without dose-weighting were used for CTF determination using GCTF37. Motion-corrected sums with dose-weighting were used for all other image processing, including particle picking. Particle picking, 2D classification, 3D classification and refinement procedures were carried out using both RELION1.4 and RELION 2 (ref 38). An ab initio 3D reconstruction was generated from a small initial cryo dataset using the program PRIME implemented in the Simple package39. This reconstruction was then filtered to 60 Å resolution and used as an initial reference model for 3D auto-refinement in RELION. All 3D refinements were carried out following the gold-standard procedure in which the dataset is divided into two half sets and refined independently. 3D reconstruction was first calculated and refined without symmetrization (C1). Since no obvious asymmetric features were seen, C4 symmetry was then applied throughout the refinement. After refinement had converged, the final dataset was subjected to ‘post-processing’ in RELION, in which a soft mask was calculated and applied to the two half-maps before calculating the corrected Fourier shell correlation (FSC) and the resolution was estimated using FSC=0.143 criterion40 on the corrected FSC curves in which the influences of the mask were removed. Temperature-factor estimation and map sharpening were also performed in this step. Local resolution estimates were calculated with unsharpened raw density maps using the ResMap41.
Model building
Ab initio model building was carried out in COOT42. Initial placement of the TM helices was made based on sequence alignment with TRPV1 and TRPA1. A rough model of the N-terminal AR domain was built by positioning individual Ankyrin repeats (pdb code: 4RLV) into EM density for each of the 29 Ankyrin repeats. Sequence register was then determined by assigning aromatic and other residues with bulky sidechains to well-resolved densities in the final sharpened density map. The linker domain, which is largely α -helical, was then built by connecting the two domains, with large, well-resolved side-chains serving as markers. After building by hand, the model was refined in PHENIX43 using 10 cycles of real-space refinement and the electron scattering table. Problematic regions and Ramachandran outliers were then manually corrected in COOT, followed by an additional 10 cycles of real-space refinement in PHENIX. For cross-validation, the final model was refined against one of the half maps generated in RELION. FSC curves were then calculated between the refined model and half map 1 (‘work’, used in test refinement), the refined model and half map 2 (‘free’, not used in test refinement) and the refined model and the summed map. There is no significant gap between ‘work’ and ‘free’ FSC curves, indicating little effect of over-fitting. Additional validation of the model was carried out using MolProbity44 and EMRinger45 (Supplementary Information Table 1). The program CAVER46 was used to calculate the pore profile shown in Figure 3. The program PyMol was used to calculate surface representation of electrostatic potentials.
Microtubule-binding assay by TIRF microscopy
Microtubules were prepared as previously described47. Unlabeled tubulin was mixed with biotinylated tubulin and Alexa-640 labeled tubulin at a ratio of ∼10:2:2 in BRB80 buffer (80 mM PIPES, pH 6.8, 1 mM EGTA, and 1 mM MgCl2). 5 mM GTP was added and the mixture was incubated in a 37°C water bath for 10 minutes. Afterward 20 μM of Taxol was added and the mixture was incubated for an additional 50 minutes at 37°C. Microtubules were spun over a 25% sucrose cushion at 160,000 ×g for 10 minutes in a tabletop centrifuge prior to use.
Flow chambers with attached microtubules were performed as described48. 5mg/ml of biotin-BSA was flowed into a 15uL chamber. After 5 minutes, the chamber was washed with BRB80, and 0.5mg/ml of streptavidin was flowed in. After 5 minutes, the chamber was washed and microtubules were flowed in and incubated for 10 minutes. Afterward the chamber was washed briefly with BRB80 and then assay buffer (30 mM HEPES pH 7.4, 50 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 10% glycerol 0.1 mg/ml Biotin-BSA, 0.5% pluronic acid F-127, and 0.2 mg/ml κ-casein). 70nM of NOMPC-Strep-GFP in amphipol or Strep-GFP was then flowed in and allowed to incubate for 2-3 minutes before washing and imaging. Total internal reflection fluorescence (TIRF) images were acquired on a Nikon Eclipse TE200-E microscope by μManager software49.
NOMPC mutagenesis and cell recording
All point mutations were introduced to nompC gene in the pBluescript vector by site-directed mutagenesis (quick change), and verified by sequencing the entire coding region of nompC.
For patch recording in HEK293 GnTi- cells, nompC in the pFastBac-CMV-GFP vector was transfected with FuGENE transfection reagent (Promega) into HEK293 GnTi- cells plated on glass coverslips. Transfected cells were ready for patch recording after cultured in DMEM medium supplemented with 10% FBS at 30 °C for 36-48 hours. For patch recording in Drosophila S2 cells, wild type and mutant nompC genes were sub-cloned into the pUAST-GFP vector and subsequently co-transfected with pAc-GAL4 into S2 cells using Effectene transfection reagent (QIAGEN). Transfected cells were then cultured at 25 °C in Schneider's Drosophila medium supplemented with 10% FBS. 24-48 hours after transfection, the cells were plated onto glass coverslips coated with 0.2 mg/mL Concanavalin A (Con A, Sigma) 20 minutes prior to recording. GFP positive cells identified under the fluorescence microscope were used for whole-cell and outside-out recordings as described previously6,8. The pipette solution (140 mM potassium Gluconic acid, 10 mM HEPES pH 7.3) and the bath solution (140 mM NaCl, 10 mM HEPES pH 7.3) were adjusted to 295 mOsm and 305 mOsm respectively with sucrose. The electrical resistance of patch electrodes filled with pipette solution is 4-6 MΩ. Mechanical stimuli were given via a High Speed Pressure Clamp (HSPC, ALA-scientific) following established procedures6,8. Mechanogated currents were measured at -60 mV. Electrical signals were sampled at 10 kHz and low-pass filtered at 2 kHz with the Multiclamp 700B amplifier, DIGIDITA 1440A. Data were recorded and processed with Clampex 10.3 (Molecular Devices).
Immunohistochemistry
Non-permeabilized cell staining was performed as previously described7. In brief, an antibody that recognizes an extracellular epitope of NOMPC (rabbit anti-NOMPC-EC7) was diluted (1:100) in serum free medium without detergent and incubated for 30 minutes with transfected S2 cells attached on Con A coated coverslip. Afterward, the cells were washed three times with PBS and fixed in 4% paraformaldehyde for 15 minutes at 4°C. The fixed cells were blocked with 5% normal goat serum, and subsequently incubated with Alexa 555 conjugated anti-rabbit IgG antibody (1:200 dilution, Invitrogen) at room temperature for 30 minutes in PBS containing 0.1% Triton X-100 (PBST). After brief wash with PBST, samples were mounted in Vectashield antifade mounting media (Vector Laboratories). Images were acquired on a Leica SP5 laser scanning confocal microscope.
Extended Discussion
As indicated by our NOMPC structure and electrophysiological data, and comparison with other TRP channels22-24, the movements of transmembrane helices required for the pore to adapt an open conformation would likely be associated with a substantial displacement of the pre-S1 elbow and/or linker helices, which could propagate to the AR domain (or in the case of mechanical gating, this would propagate from the AR domain). Previous studies showed that NOMPC channel gating in response to mechanical stimulation is conveyed through its AR domain and critically dependent on microtubule integrity6. In this study, we show that purified NOMPC also decorates microtubules (Extended Data Figure 1b), likely with its N-terminus interacting with microtubule. Movement of microtubules could then cause AR domain to be displaced in a variety of ways with respect to the membrane: lateral movement, pushing or pulling against the membrane, or even twisting (Extended Data Figure 9c). Conceivably these motions may impact channel gating; it is possible that only one such movement (or a subset of these movements) can cause channel opening.
Our structural analysis suggests that the AR domains possess some degree of flexibility. However, the closed pore conformation of all three classes characterized in this study could indicate that such small motions may not be sufficient to cause channel opening. It appears that a motion of the AR domain greater than what we observed in the three classes would be required to activate the channel. The ability to deform is likely to be an important functional aspect of this unique spring-shaped quadruplex architecture. The linker domain can tolerate small movement without inducing channel activation suggesting that forces below a certain threshold do not lead to channel activation. This may help attenuate response and prevent unwanted channel opening due to small mechanical movements. In addition, this flexibility may allow the channel to deform rather than break outright in response to very strong stimuli. Another possibility is that some of these closed channel conformations correspond to a desensitized state or a captured intermediate state during gating transitions, perhaps analogous to some of the voltage-gated channel structures with the voltage sensor in the activated state and the pore still in the closed (pre-open) state50.
Sensitivity to mechanical stimulation, on the other hand, likely results from the extensive network that connects the cytoplasmic ARs via the linker helices to the TRP domain and the S4-S5 linker, and possibly other structural elements in their vicinity. This bridge between the transmembrane core and AR domains is probably the key to translate ARs movement into displacement of residues constricting the pore, leading to channel opening. The path of force transmission is likely through the TRP domain, and it may also involve the S4-S5 linker, which appears to interact with a lipid molecule.
Data Availability Statement
Cryo-EM density maps of NOMPC in nanodisc is deposited to the Electron Microscopy Data Bank under the accession numbers EMD-8702. Particle image stack after motion correction related to NOMPC in nanodisc has been deposited in the Electron Microscopy Public Image Archive (http://www.ebi.ac.uk/pdbe/emdb/empiar/) under accession number EMPIAR-10093. Atomic coordinate for NOMPC in nanodisc with bound phospholipids is deposited in the Protein Data Bank under the accession number 5VKQ.
Extended Data
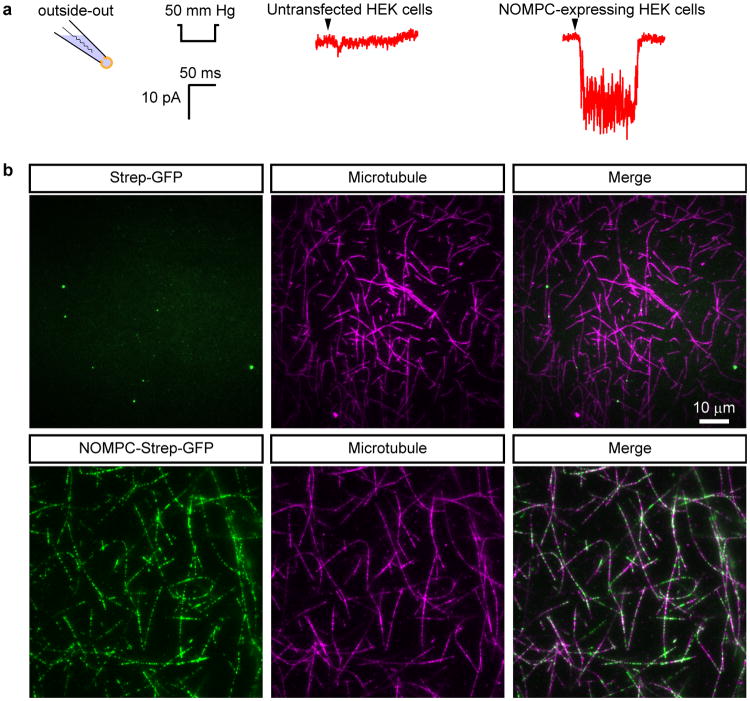
Extended Data Figure 1. Verification of recombinant NOMPC activity.
a, Pressure-induced mechanogated current measured at -60 mV from outside-out patches excised from HEK293 cells untransfected (left) or transfected (right) with the NOMPC construct used for structure determination. b, TIRF microscopy images of surface anchored and fluorescently labeled microtubules (pseudo-colored in magenta) are not decorated by purified Strep-GFP alone (top) but by Strep-GFP-tagged NOMPC (bottom), demonstrating that purified NOMPC is capable of binding microtubules, and this binding is not through interaction between the Strep tag and residual streptavidin on the surface.
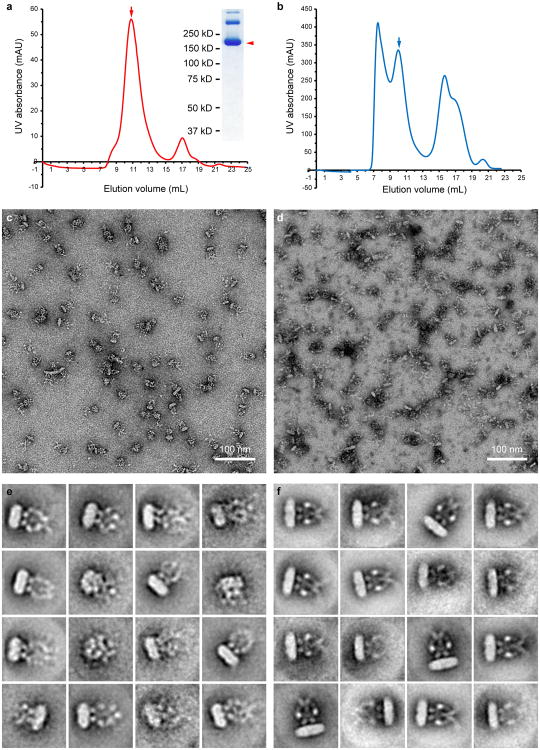
Extended Data Figure 2. Negative stain analysis of purified NOMPC.
a, Size-exclusion chromatography of NOMPC after exchange from DDM into amphipol A8-35. The peak fraction corresponding to NOMPC tetramer (indicated by arrow) was examined by SDS–polyacrylamide gel electrophoresis. The NOMPC monomer (∼190 kD) band is indicated by arrow head. The upper bands may correspond to incompletely disassociated NOMPC oligomers. b, Size-exclusion chromatography of NOMPC reconstituted into lipid nanodisc with MSP2N2. The peak fraction corresponding to NOMPC is indicated by arrow. c, d, Raw micrographs of NOMPC-amphipol (c) and NOMPC–nanodisc (d) samples examined by negative stain EM. Both showed mono dispersed and homogeneous particles. e, f, 2D class averages of NOMPC particles in amphipol (e) or nanodisc (f) by negative stain EM, with the nanodisc sample showing better ordered features than the amphipol sample.
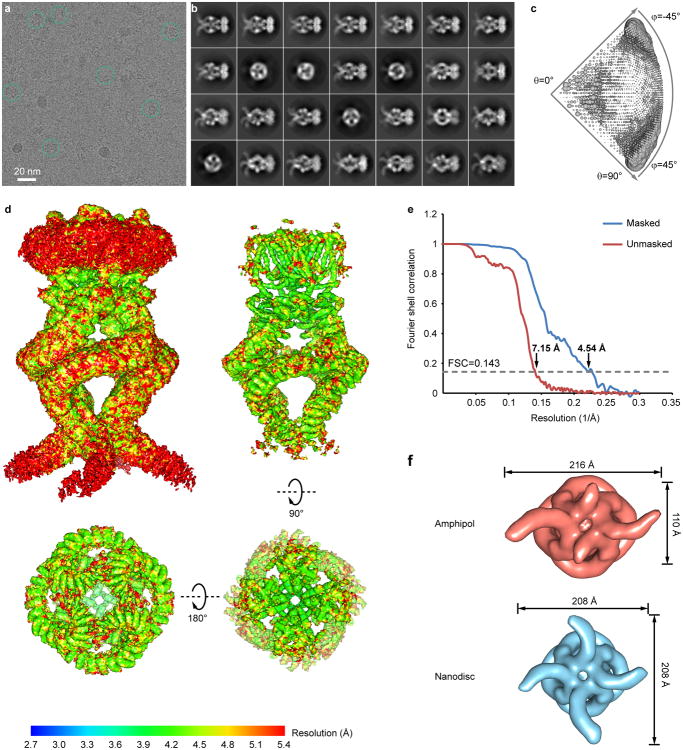
Extended Data Figure 3. Cryo-EM analysis of NOMPC in amphipol.
a, A raw cryo-EM micrograph of NOMPC recorded as described in Methods. b, 2D class averages of the cryo-EM micrographs with a particle box size of 400 pixels (486Å). c, Euler angle distribution of all particles used for the final 3D reconstruction. The size of the sphere is proportional to the amount of particles visualized from that specific orientation. d, Final unsharpened 3D density map of NOMPC-amphipol colored with local resolution at a low isosurface level (top left) to enable visualization of the amphipol layer, and at a high isosurface level in side (top right), top (bottom right) and bottom (bottom left) views. e, FSC curves between two independently refined half maps before (red) and after (blue) post-processing in RELION, indicated with resolutions corresponding to FSC=0.143. f, Density maps of NOMPC in amphipol (top) and in nanodisc (bottom) with a C1 symmetry derived from all particles picked. Comparison of the maps shows NOMPC-nanodisc is closer to a four-fold symmetric structure.
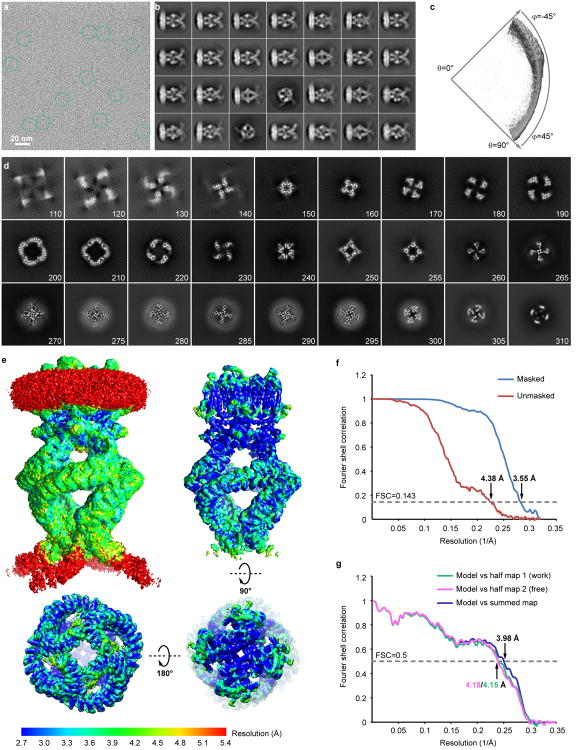
Extended Data Figure 4. Cryo-EM analysis of NOMPC reconstituted in nanodisc.
a, A raw cryo-EM micrograph of NOMPC recorded as described in Methods. b, 2D class averages of the cryo-EM micrographs with a particle box size of 400 pixels (486Å). c, Euler angle distribution of all particles used for the final 3D reconstruction. The size of the sphere is proportional to the amount of particles visualized from that specific orientation. d, Slices through the unsharpened density map at different levels along the channel symmetry axis. The slice numbers starting from the cytoplasmic side are marked. e, Final unsharpened 3D density map of NOMPC-nanodisc colored with local resolution at a low isosurface level (top left) to enable visualization of the lipid bilayer, and at a high isosurface level in side (top right), top (bottom right) and bottom (bottom left) views. f, FSC curves between two independently refined half maps before (red) and after (blue) post-processing in RELION, indicated with resolutions corresponding to FSC=0.143. g, Cross-validation using FSC curves of the density map calculated from the refined model versus half map 1 (work, green), versus half map 2 (free, pink) and versus summed map (blue).
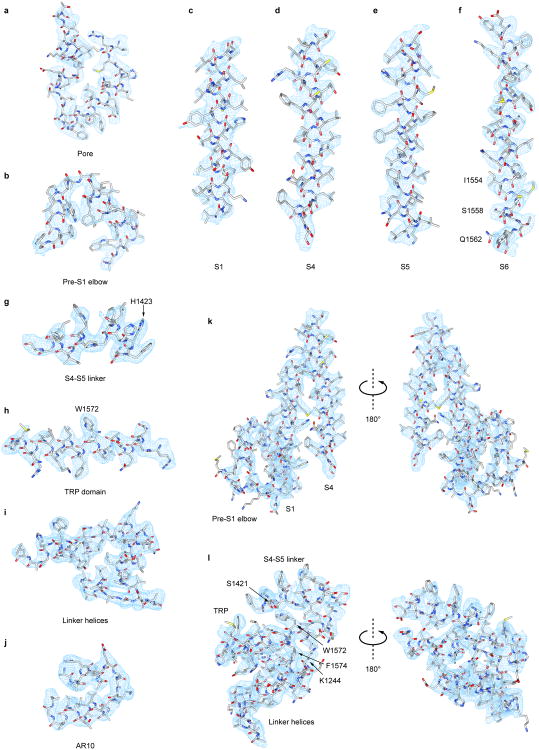
Extended Data Figure 5. Selected segments of cryo-EM density.
a-j, Representative cryo-EM densities of various NOMPC domains as indicated are superimposed on the atomic model. k, Density map of pre-S1 elbow, S1 and S4. l, Density map of stacked S4-S5 linker, TRP domain and linker helices that couple ARs to the pore through domain interactions. All the density maps (a-l) are shown as cyan meshes, and the model is shown as sticks and colored according to atom type (C: light grey; N: blue; O: red; S: yellow).
Extended Data Figure 6. Atomic model of NOMPC.
a-d, Ribbon diagrams of NOMPC atomic model for residues Asn 125–Trp 1602 and Gly1689-Arg1670. The entire AR domain of 29 ARs was resolved. e, Ribbon diagrams showing two views of one NOMPC subunit denoting specific domains
Extended Data Figure 7. Pore profile and channel properties of NOMPC and NOMPC mutants.
a, Pore radius calculated by the CAVER program. b, I-V curves from the steady-state currents of whole cell recording from S2 cells expressing NOMPC or NOMPC mutants suggesting that both W1572A and I1554A mutations resulted in large basal current in the absence of applied pressure (wild type: n=7; W1572A: n=7; I1554A: n=3). Cells expressing I1554A mutant channels displayed very large basal currents and were not amenable to recording from excised patch recording of mechanogated current. c, Dose-dependent curves of pressure-induced mechanogated currents measured at -60 mV from outside-out patches excised from S2 cells expressing NOMPC or NOMPC mutants (n=7). d, Representative images of unpermeabilized staining of S2 cells expressing control proteins or H1423A mutant. The surface staining signal from a number of cells (n>30 for each sample) were visually surveyed under fluorescent microscope. All cells expressing wild type (middle row) and H1423A mutant (bottom row) NOMPC showed similar levels of surface expression, indicating that the H1423A mutant is properly localized to the plasma membrane like the wild type. All error bars (b, c) denote ± standard deviation.
Extended Data Figure 8. 3D classification of NOMPC-nanodisc particles.
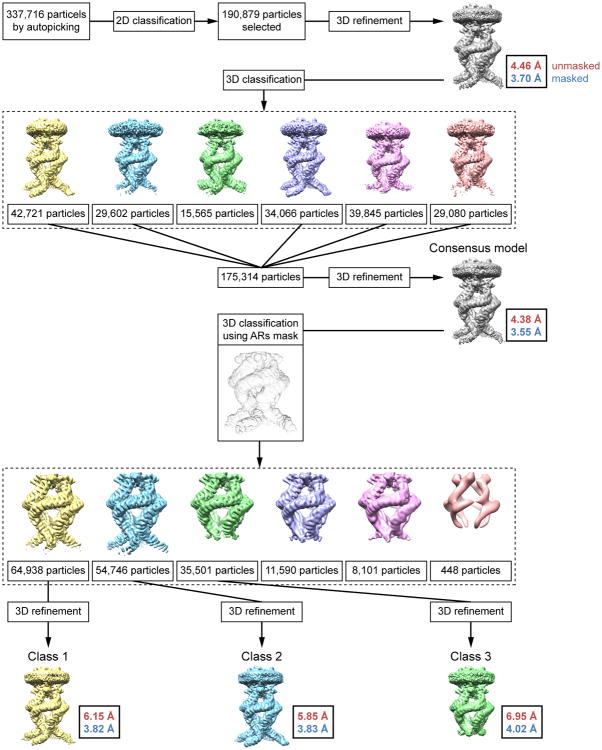
The flowchart of classification procedures with RELION is shown. After 1 round of 2D classification, 190,879 particles were subjected to 3D refinement, yielding a 3.7Å map (C4 imposed). Following 3D classification of these particles, 175,314 particles were selected and refined, yielding the final map at 3.55Å resolution (C4 imposed), which was used to build the ‘consensus model’. Additional classification of these 175,314 particles using a mask to exclude all regions outside the AR domain gave 3 major classes, which were subsequently refined to ∼3.8-4.0Å resolution.
Extended Data Figure 9. Flexibility of AR domains and model of NOMPC mechanogating.
a, Superposition of NOMPC AR domains from three classes are shown in three views as indicated. Overall, the entire AR domains from all three classes overlap well with each other. There are small differences in some ARs, which is caused by slight shift of individual AR as rigid body, suggesting small mobility and plasticity of the AR domain. b, Superposition of NOMPC AR domain (blue) with human Ankyrin-B AR domain (pink) shown in three views as indicated, suggesting that elastic deformation of NOMPC AR domain under pressure could potentially be more dramatic than the shift presumably caused by thermal motion as shown in a. A peptide from the C-terminal region of Ankyrin-R, which was added to stabilize Ankyrin-B ARs for crystallization by forming an auto-inhibitory segment (AS) structure, is also shown here (AnkR AS, in orange). c, Schematic of NOMPC (without precise depiction of domain swap between neighboring subunits) showing the N-terminus tethered to a microtubule. Mechanical force is transduced from the microtubule cytoskeleton to NOMPC, possibly causing lateral movement, extension, compression or torsion of the AR domain. d, e, Movement of the AR domain that immediately precedes the linker helices results in displacement of the TRP domain and S4-S5 linker that are connected to the ends of the pore domain, triggering channel opening.
Extended Data Figure 10. Sequence alignment of NOMPC orthologues.
Sequence homology of NOMPC orthologues were analyzed by clustal omega. The conserved residues are highlighted. The two residues (His1423 and Trp1572), which were shown in this study to be critical for mechanogating, are marked by red triangles. Secondary structure elements are indicated above the sequence.
Supplementary Material
Table 1. Summary of cryo-EM data collection and model refinement.
| Nanodisc | Amphipol | |
|---|---|---|
| Data collection/processing | ||
|
| ||
| Voltage (kV) | 300 | 300 |
|
| ||
| Magnification | 31,000 | 31,000 |
|
| ||
| Defocus range (μm) | -1.5 – -3.0 | -1.5 – -3.0 |
|
| ||
| Pixel size (Å) | 1.2156 | 1.2156 |
|
| ||
| Total electron dose (e-/Å2) | 54 | 54 |
|
| ||
| Exposure time (s) | 8 | 8 |
|
| ||
| Number of images | 1873 | 4135 |
|
| ||
| Number of frames/image | 40 | 40 |
|
| ||
| Initial particle number | 337,716 (autopick) 190,879 (2D select) | 546,733 (autopick) 121,389 (2D select) |
|
| ||
| Final particle number | 175,314 | 52,065 |
|
| ||
| Resolution (unmasked, Å) | 4.38 | 7.15 |
|
| ||
| Resolution (masked, Å) | 3.55 | 4.54 |
|
| ||
| Refinement | ||
|
| ||
| Number of atoms | 40,888 | |
|
| ||
| R.m.s. deviations | ||
|
| ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (0) | 1.03 | |
|
| ||
| Ramachandran | ||
|
| ||
| Favored (%) | 90.60 | |
| Allowed (%) | 9.06 | |
| Outlier (%) | 0.33 | |
|
| ||
| Molprobity score | 1.93 | |
|
| ||
| EMRinger score | 1.88 | |
Acknowledgments
We thank R. Mackinnon, T. Xiao and W. Wang for advice on protein purification, S. Gründer for sharing construct, and members from laboratories of D. Minor, D. Julius, and R. Vale for sharing equipment and reagents. We also thank colleagues in our laboratories for discussions, M Braunfeld and C. Kennedy for technical support, Z. Yu and his colleagues at the HHMI Janelia Cryo-EM Facility for help with data acquisition. This work was supported by grants from the NIH (R01NS069229 to L.Y.J, 5R37NS040929, 1R35NS097227 to Y-N.J, R01GM098672 and S10OD020054 to Y.C.), and by a UCSF Program for Breakthrough Biomedical Research (Y.C.). L.Y.J, Y-N.J and Y.C. are Investigators with the Howard Hughes Medical Institute.
Footnotes
Author Contributions: P.J. and D.B. designed and carried out most biochemical and cryo-EM experiments. Y.G. performed electrophysiology experiments, and W.Z. mutagenesis and initial electrophysiology studies of NOMPC. Z.G. assisted with biochemical experiments. W.H. performed microtubule-binding assay. S.W. helped with cryo-EM experiment. S.M. performed surface staining, T.C. performed mutagenesis. L.Y.J. and Y.N.J. and Y.C supervised experiments and data analysis. P.J., D.B., L.Y.J., Y.N.J. and Y.C. wrote the manuscript. All authors contributed to manuscript preparations.
Author Information: The authors declare no competing financial interests.
References
- 1.Árnadóttir J, Chalfie M. Eukaryotic Mechanosensitive Channels. Annu Rev Biophys. 2010;39:111–137. doi: 10.1146/annurev.biophys.37.032807.125836. [DOI] [PubMed] [Google Scholar]
- 2.Anishkin A, Kung C. Stiffened lipid platforms at molecular force foci. Proc Natl Acad Sci U S A. 2013;110:4886–92. doi: 10.1073/pnas.1302018110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brohawn SG, del Marmol J, MacKinnon R. Crystal Structure of the Human K2P TRAAK, a Lipid- and Mechano-Sensitive K+ Ion Channel. Science. 2012;335:436–441. doi: 10.1126/science.1213808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brohawn SG, Campbell EB, MacKinnon R. Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel. Nature. 2014;516:126–130. doi: 10.1038/nature14013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lolicato M, Riegelhaupt PM, Arrigoni C, Clark KA, Minor DL. Transmembrane helix straightening and buckling underlies activation of mechanosensitive and thermosensitive K2P channels. Neuron. 2014;84:1198–1212. doi: 10.1016/j.neuron.2014.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, et al. Ankyrin Repeats Convey Force to Gate the NOMPC Mechanotransduction Channel. Cell. 2015;162:1391–1403. doi: 10.1016/j.cell.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker RG, Willingham AT, Zuker CS. A Drosophila Mechanosensory Transduction Channel. Science. 2000;287:2229–2234. doi: 10.1126/science.287.5461.2229. [DOI] [PubMed] [Google Scholar]
- 8.Yan Z, et al. Drosophila NOMPC is a mechanotransduction channel subunit for gentle-touch sensation. Nature. 2013;493:221–5. doi: 10.1038/nature11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Feng Z, Sternberg PW, Xu XZ, S A C. elegans stretch receptor neuron revealed by a mechanosensitive TRP channel homologue. Nature. 2006;440:684–687. doi: 10.1038/nature04538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Effertz T, Wiek R, Göpfert MC. NompC TRP channel is essential for Drosophila sound receptor function. Curr Biol. 2011;21:592–597. doi: 10.1016/j.cub.2011.02.048. [DOI] [PubMed] [Google Scholar]
- 11.Cheng LE, Song W, Looger LL, Jan LY, Jan YN. The Role of the TRP Channel NompC in Drosophila Larval and Adult Locomotion. Neuron. 67:373–380. doi: 10.1016/j.neuron.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidi S, Friedrich RW, Nicolson T. NompC TRP channel required for vertebrate sensory hair cell mechanotransduction. Science. 2003;301:96–9. doi: 10.1126/science.1084370. [DOI] [PubMed] [Google Scholar]
- 13.Liang X, et al. A NOMPC-dependent membrane-microtubule connector is a candidate for the gating spring in fly mechanoreceptors. Curr Biol. 2013;23:755–763. doi: 10.1016/j.cub.2013.03.065. [DOI] [PubMed] [Google Scholar]
- 14.Lee G, et al. Nanospring behaviour of ankyrin repeats. Nature. 2006;440:246–249. doi: 10.1038/nature04437. [DOI] [PubMed] [Google Scholar]
- 15.Howard J, Bechstedt S. Hypothesis: A helix of ankyrin repeats of the NOMPC-TRP ion channel is the gating spring of mechanoreceptors [1] Curr Biol. 2004;14:224–226. doi: 10.1016/j.cub.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 16.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–12. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paulsen CE, Armache J, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 20.Ahuja S, et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science. 2015;350:aac5464. doi: 10.1126/science.aac5464. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, et al. Structure of the voltage-gated calcium channel Ca. Nature. 2016;537:191–196. doi: 10.1038/nature19321. [DOI] [PubMed] [Google Scholar]
- 22.Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature. 2013;504:113–8. doi: 10.1038/nature12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saotome K, Singh AK, Yelshanskaya MV, Sobolevsky AI. Crystal structure of the epithelial calcium channel TRPV6. Nature. 2016;534:506–511. doi: 10.1038/nature17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zubcevic L, et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat Struct Mol Biol. 2016;23:1–9. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huynh KW, et al. Structure of the full-length TRPV2 channel by cryo-EM. Nat Commun. 2016;7:11130. doi: 10.1038/ncomms11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venkatachalam K, Montell C. TRP Channels. Annu Rev Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin Z, et al. Exome sequencing reveals mutations in TRPV3 as a cause of Olmsted syndrome. Am J Hum Genet. 2012;90:558–564. doi: 10.1016/j.ajhg.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teng J, Loukin SH, Anishkin A, Kung C. L596–W733 bond between the start of the S4–S5 linker and the TRP box stabilizes the closed state of TRPV4 channel. Proc Natl Acad Sci. 2015;201502366 doi: 10.1073/pnas.1502366112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C, et al. Structural basis of diverse membrane target recognitions by ankyrins. Elife. 2014;3:1–22. doi: 10.7554/eLife.04353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox CD, et al. Removal of the mechanoprotective influence of the cytoskeleton reveals PIEZO1 is gated by bilayer tension. Nat Commun. 2016;7:1–13. doi: 10.1038/ncomms10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–85. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fridy PC, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Booth DS, Avila-Sakar A, Cheng Y. Visualizing Proteins and Macromolecular Complexes by Negative Stain EM from Grid Preparation to Image Acquisition. J Vis Exp. 2011:1–8. doi: 10.3791/3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 36.Zheng SQ, et al. MotionCor2 - anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017:4–9. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elmlund H, Elmlund D, Bengio S. PRIME: Probabilistic initial 3D model generation for single-particle cryo-electron microscopy. Structure. 2013;21:1299–1306. doi: 10.1016/j.str.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barad BA, et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat Methods. 2015;12:943–6. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chovancova E, et al. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLoS Comput Biol. 2012;8:23–30. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science. 2014;345:337–41. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroeder CM, Vale RD. Assembly and activation of dynein-dynactin by the cargo adaptor protein Hook3. J Cell Biol. 2016;214:309–318. doi: 10.1083/jcb.201604002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using manager. Curr Protoc Mol Biol. 2010:1–17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Payandeh J, Scheuer T, Zheng N, Catterall WA. The crystal structure of a voltage-gated sodium channel. Nature. 2011;475:1–7. doi: 10.1038/nature10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps of NOMPC in nanodisc is deposited to the Electron Microscopy Data Bank under the accession numbers EMD-8702. Particle image stack after motion correction related to NOMPC in nanodisc has been deposited in the Electron Microscopy Public Image Archive (http://www.ebi.ac.uk/pdbe/emdb/empiar/) under accession number EMPIAR-10093. Atomic coordinate for NOMPC in nanodisc with bound phospholipids is deposited in the Protein Data Bank under the accession number 5VKQ.