Abstract
Objective:
The main aim of this scientific report was to investigate a series of phytochemicals in silico and the pharmacology of four plants found at higher altitude in the ginger family, Zingiberaceae (incl. Costaceae) from North-East India, particularly Sikkim. First, the goal was to determine the biological activities of the four herbs (used under Zingiberaceae family) using antioxidant assays to identify the best species. Second, previously reported compounds in litero were subsequently screened for their anticancerous activities using in silico methods.
Materials and Methods:
Using the methanolic extracts of herbs, quantitative detection of phytochemicals such as total phenols and total flavonoids was detected, and the free radical scavenging activity was also studied using 2,2-diphenyl-1-picryl-hydrazyl (DPPH) assay. Docking process was studied, using Discovery Studio version 3.5, to identify suitable molecules at the protein-binding sites through annealing and genetic simulation algorithms. Grids centered on active sites were obtained with spacing of 54 × 55 × 56, and 0.503 grid spacing was calculated. The methods adopted and used in this study were comparisons of Global and Local Search Methods to determine the parameters such as maximum number of 250,000 energy evaluations as well as generations of 27,000, followed by mutation and crossover rates of 0.02 and 0.80. The number of docking runs was set to 10. Molecular dynamics study was done to check the stability of the complex.
Results:
Among all the genus of Zingiberaceae family investigated in this study, Curcuma angustifolia and Hedychium sp. exhibited the highest 537 ± 12.45; 292 ± 9.16 mg gallic acid equivalent/g total polyphenols and 38 ± 1.54; 75 ± 6.75 mg quercetin equivalent/g flavonoids, respectively. Depending on the concentration, the Hedychium sp. extract exerted the highest scavenging activity on DPPH radical (IC50 36.4 μg/mL). In silico result demonstrated that the synergetic effects of β-phellandrene with other compounds might be responsible for its anticancerous activity. β-phellandrene and farnesene epoxide showed bonding with Leu298, Ala302, Met336, Leu339, Leu343, Phe356, Ala302, Glu305, Met340, Leu343, Arg346, Phe356, Ile373, Ile376, Leu380, His475, Leu476, and Leu491.
Conclusion:
Based on the current available literature, this is the first study to understand the interaction of compounds found in the rhizomes of Zingiberaceae family.
SUMMARY
The aqueous methanolic extract of Zingiberaceae family Curcuma angustifolia and Hedychium sp. has potent antioxidant activity as assessed by 2,2-diphenyl-1-picryl-hydrazyl assays
Hedychium sp. is understood to possess more active compounds than other varieties
In silico studies indicated synergetic effects of β-phellandrene and other compounds for its anticancerous activity.
Abbreviations used: CADD: Computer-aided drug designing; ROS: Reactive oxygen species; ADMET: Absorption, distribution, metabolism, and excretion-toxicity; FeCl3: Ferric chloride; DPPH: 2,2-diphenyl-1-picryl-hydrazyl; NaNO2: Sodium nitrite; TCA: Trichloroacetic acid; K2HPO4: Di-potassium hydrogen phosphate; H2O2: Hydrogen peroxide; KH2PO4: Potassium di-hydrogen phosphate, K2Fe (CN)6: Potassium ferricyanide; KOH: Potassium hydroxide; NaOH: Sodium hydroxide; Na2CO3: Sodium carbonate; CH3COONa: Sodium acetate; AlCl3: Aluminum chloride.
Keywords: Zingiberaceae, Curcuma, cancer, CADD, molecular dynamics
INTRODUCTION
Lack of scientific knowledge to find a cure for health ailments leads to the diversion of research toward new strategies such as exploration of traditional ethnobotanical knowledge. Plants constitute an important source of various secondary metabolites.[1] High-altitude medicinal plants acquire a unique collection of secondary metabolites that can act as a potential therapeutic targets for diseases.[2] Bioactive natural inventions with remedial potential have relied on external circumstances and some of these compounds are beyond exploration by traditional means. Therefore, the efficiency of computational tools to facilitate drug discovery are is known and has been recognized even in case of natural products.[1,3,4,5] Computer-aided drug designing (CADD) utilizes the existing information from the ethnobotanical data to provide valuable understanding to address the existing therapeutics and for the development of new nature-derived drugs. In addition, more than a few medicinal plants have been continuously utilized in various conventional medicinal systems since olden days throughout the globe. However, their mechanisms have not yet been explored properly. Therefore, the CADD approaches for plant-based drug discovery would yield immense gain in advancing the world's health population.
Antioxidants are the counterparts that can restrain single oxygen or free radicals prior to any noteworthy oxidant force taking place.[2,6] Increasing knowledge and lookout for a healthier product having fewer preservatives and synthetic additives are motivating research endeavors toward the natural antioxidants. Plant-related dietary products are gaining consideration due to their increasing scientific evidence, having indicated their richness in several phenolic compounds; therefore, preventing pathological conditions such as diabetes, cancer, and neurological diseases.
Zingiberaceae family, an Angiosperm (flowering plants) group which contains 52 plant genera including genus such as Curcuma angustifolia, Curcuma longa, Hedychium sp., and Kaempferia rotunda Linn., is widely distributed in Asian and African regions, known to have diverse pharmacological activities (http://www.theplantlist.org/1.1/browse/A/zingiberaceae/).
Curcuma and Ginger are the most imperative known crops in the hill state of Sikkim, India, at North-East Himalayas (27º 00’ 46” N–28º 07’ 48” N latitudes and 88º 00’ 58” E–88º 55’ 25” E longitudes) next to Citrus sp. and Amomum subulatum. The Sikkimese tribes, namely, Lepcha and Bhutia use them for their religious activities besides culinary and medicinal use.[7] Pradhan and Badola[8] stated 118 medicinal plant species at the high altitude of Sikkim practiced by Lepcha tribe of Dzongu valley, distributed across 71 families and 108 genera. Sharma[9] reported that C. angustifolia plant is used for curing worms and stomach ache. Curcumin isolated from C. longa is well known for its pharmacological properties. The antioxidant, antibacterial, antifungal, cytotoxic, and chemopreventive effects are the well-known pharmacological properties of Hedychium sp. (butterfly ginger) in addition to anti-urolithiasis, anti-angiogenic, neuro-pharmacological, fibrinogenolytic, coagulant, anti-allergic, larvicidal, anthelminthic, analgesic, anti-inflammatory, and hepatoprotective activities.[10] The phytochemistry of Hedychium sp. and K. rotunda Linn. (Indian crocus) is still unknown, which makes these species studying worthwhile. Though the pharmacological properties of these rhizomes have been previously reported, the mechanisms of therapeutic principles are still unknown. Hence, the objective of this study was to investigate the pharmacological properties of four indigenous rhizomes in addition to interaction against an estrogen receptor and a series of previously reported compounds (phytochemicals) in silico found in Zingiberaceae (incl. Costaceae) family from Sikkim, India, reported at a higher altitude.
MATERIALS AND METHODS
Chemicals and reagents
Ascorbic acid, 2,2-diphenyl-1-picryl-hydrazyl (DPPH), ferric chloride, gallic acid, naphthyl ethylenediamine dihydrochloride, quercetin, sodium nitrite (NaNO2), sulphanilamide, and trichloroacetic acid were obtained from HiMedia Laboratories Pvt. Ltd., Mumbai, India. Di-potassium hydrogen phosphate, hydrogen peroxide, methanol, potassium di-hydrogen phosphate, potassium ferricyanide, potassium hydroxide, sodium hydroxide (NaOH), sodium carbonate (Na2CO3), sodium acetate, and sodium nitroprusside were procured from Merck, Mumbai, India, and Folin–Ciocalteu (FC) reagent was purchased from Sisco Research Laboratories Pvt. Ltd., Mumbai, India. Aluminum chloride (AlCl3) and ortho-phosphoric acid were obtained from SD Fine Chemicals Limited, Mumbai, India. All chemicals and solvents were of analytical grade.
Plant materials and extraction
The fresh rhizomes of C. angustifolia, C. longa, Hedychium sp., and K. rotunda Linn. were collected in June 2015 from Sajong, Sikkim, India. The specimen vouchers have been identified by a plant taxonomist and deposited at Sikkim State Council of Science and Technology, Sikkim, India, and the given voucher numbers are GT07, GT08, GT09, and GT10. The washed rhizomes were air-dried and macerated using a grinder. The powdered rhizomes were defatted with 1:1 v/v petroleum ether (b.p. 40°C–60°C) in a Soxhlet apparatus. Then, 20 g of each processed sample was extracted using Soxhlet apparatus with 70% aqueous methanol in 1:15 w/v ratio at 70°C for 4 h in three cycles.[11] The lyophilized extracts were stored at 4°C until used. The samples were dissolved in double-distilled water (DDW) in the required aliquots just before use.
Preliminary phytochemical screening
All four freshly prepared extracts were subjected to preliminary phytochemical analysis according to the standard protocols.[12]
Determination of the yield of plant extract
The yields of evaporated rhizome extracts were calculated using the author's previous reports.[3]
Determination of biochemical constituents
Determination of total phenolic content
FC reagent method[13] with slight alteration was used to assess total phenolic content. A volume of 0.5 mL of the extracts was mixed with 0.5 mL of FC reagent (previously diluted 1:1 with DDW) and incubated at room temperature (RT) for 5 min. Followed by the addition of 1 mL of 20% Na2CO3 solution, the mixture was incubated at RT for 10 min. The absorbance was measured at 730 nm. Gallic acid monohydrate was used as a standard and the total phenolic content was defined as Gram gallic acid equivalents per 100 g of the extract.
Determination of total flavonoid content
The AlCl3 method was used to determine the total flavonoid content of samples.[14] The reactive mixture was prepared by adding the extract with 1.25 mL DDW, followed by 75 μl of NaNO2 (5%). After 5 min of incubation at RT, 0.15 mL of AlCl3 (10%) was added and kept at RT for 6 min. Then, the mixture was treated with 0.5 mL of 1 mM NaOH. Finally, the sample was diluted with 275 μl of DDW followed by incubation at RT for 20 min. The absorbance was measured at 510 nm. The absorbance was measured at 510 nm and the total flavonoid content was determined from quercetin standard curve.
2,2-diphenyl-1-picryl-hydrazyl radical scavenging activity
DPPH method was used to determine the free radical scavenging activity of the sample and the standard, ascorbic acid.[15] A volume of 0.5 mL of 100–1000 μg of the extracts was mixed with freshly prepared 0.5 mL DPPH solution and the final volume was made up to 2 mL. A control sample was the same volume without any extract. Methanol was used as a blank. The reaction mixture is allowed to stand for 30 min in dark. The discoloration was measured at 517 nm. The experiment was performed in triplicates. DPPH scavenging activity (%) was determined using the following equation: DPPH scavenging activity (%) = A0 − A1/A0 × 100
Where A0 is the absorbance of the control and A1 is the sample.
Docking studies
An in silico study was performed to understand the protein–ligand interaction similarly as per our previously designed protocol.[16]
Generation of dataset
Eighty-one compounds of Zingiberaceae (incl. Costaceae) were selected from PubMed (http://pubmed.ncbi.nlm.nih.gov/) and downloaded from PubChem database (http://pubchem.ncbi.nlm.nih.gov/) as ligands considering their chemical and physical properties. The 2D structures of all these compounds were drawn using MarvinSketch, and biological activities were calculated using Molinspiration (www.molinspiration.org) server.
Preparation of the ligands
Drawn ligands were processed during the ligand preparation step for various stages having included addition and deletion of hydrogen atoms and heteroatom.[16,17] In addition, charged group neutralization, low-energy ring conformers, geometry optimization, and saving of output files were performed[5,18] using Discovery Studio (DS) version 3.5 (Biovia, USA). Lipinski's filter of 5 with various parameters such as hydrogen donors, acceptors, log P value, molecular weight, and the reactive filter were followed to discard the ligands with poor pharmacological properties and reactive functional groups.[19]
Preparation of the receptors
The three-dimensional (3D) structure of the receptor (Protein Data Bank [PDB] ID-1U3Q) was downloaded from PDB (http://www.rcsb.org). The X-ray crystal structure of the estrogen receptor has 2.4 Å resolution, R-value free: 0.279, R-value work: 0.227, and is biologically active as well as in a stable state. PDB structure was processed as per the standard accessible protocol of DS version 3.5.[5]
Absorption, distribution, metabolism, and excretion/TOxicity Prediction by Komputer-assisted Technology
The energy-minimized natural compounds were subjected to absorption, distribution, metabolism, and excretion (ADME)/Tox (ADMET) calculations using ADME-TOxicity Prediction by Komputer-assisted Technology (TOPKAT) (DS 3.5). The TOPKAT predicts structurally significant candidates and pharmaceutically relevant properties such as Ames mutagenicity, carcinogenicity, rat oral LD50, rat maximum tolerated dose, rat inhalation toxicity LC50, and Log P of the probable candidates. ADMET descriptor includes various parameters such as human intestinal absorption, CYP2D6 binding, plasma protein binding, blood–brain barrier (BBB) penetration, and aqueous solubility.
Simulation studies
The study was followed to explore the protein–ligand interactions. Docking allows virtually screening compounds and predicting the strongest binders based on their scoring functions. The software used here to do docking simulation was iGEMDOCK against all prepared compounds. This system's total energy alias Gibbs energy (kcal/mol) is calculated and recorded by the system.
LigandFit/Discovery Studio 3.5
Ligand fit is a shape-directed docking method of ligands to bind with the specific active sites of the protein.[20] To initiate docking study in DS 3.5, receptor or target protein was prepared by removing heteroatoms, water molecules. The Chemistry at HARvard Macromolecular Mechanics (CHARMM) force field was applied using the simulation tool DS 3.5. The active site was identified using cavity-based method from receptor cavities. A virtually screened compound after ADME and TOPKAT was subjected to docking process. Top 20 poses/scores were generated for each interaction. Each pose was saved, evaluated, and ranked based on DS consensus scoring function that includes JAIN, Ligscore 1, Ligscore 2, PLP1, PLP2, and potential of mean force. Hydrogen bond interactions between the ligands and active site residues were also assessed.
Molecular dynamic simulation
The final screened compound having the best pose of docked complex from the docking studies (1U3Q-apo protein complex) was subjected to 200 picosecond (ps) of MD simulations using leap-frog verlet integration algorithm in DS 3.5. In addition, SHAKE constraint was introduced to fix all bonds involving hydrogen in the simulation. The executed simulation cascades were minimization with CHARMm force field followed by minimization with steepest descent of 500 steps and conjugate gradient of 500 steps. Consequently, the docked complex is heated upto 300K with 4 ps of simulation time. Finally, production in the canonical ensemble was subjected with equal Tmass and Pmass at 300 k for 200 ps. In addition, spherical cutoff method of electrostatics was implemented to study all nonbonded energy without using periodic boundary condition.
Statistical analysis
All experiments reported were done in triplicates. The Tukey range test was used to check their statistical significance at P < 0.05.
RESULTS AND DISCUSSION
In the modern era, Zingiberaceae family has attained an interest as a potential treatment for many health ailments,[21] including anti-inflammatory disorders[22] and cancer.[23] Till March 2016, only 56 scientific articles were found for the keyword “Zingiberaceae” and “Phytochemistry in the pubmed repository. Most of the species reported were Zingiber sp. (23), Curcuma sp. (9), Kaempferia sp. (3), Hedychium sp. (3), Aframomum sp. (1), Alpinia sp. (1), A. subulatum (1), Boesenbergia sp. (1), Renealmia sp. (1), Siphonochilus sp. (1), and Xiphidium sp. (1) with 73% of all the available publications. However, the authors were not able to find any report with regard to A. purpurata and H. coronarium from North-East India, specifically at high latitude (27.20 N) and longitude (88.40 E). Moreover, there were lots of scientific lacunae in case of any works addressing the phytochemistry of Hedychium sp., K. rotunda Linn., and Curcuma sp. To the authors, it was important to notice that well-documented species of this Zingiberaceae family only represent 5%–10% of the species, and the vast majority belonging to this family remain poorly studied or biochemically unknown. Hence, the present study focused on the phytochemistry and in silico findings of selected species of Zingiberaceae family.
Biochemical investigation
Antioxidants are the molecules to quench the free radicals synthesized in the plants as the inevitable byproducts of turning food into energy and formidable threats faced by them in disease condition.[3] Polyphenols, naturally occurring in our food, are scientifically proven for the major antioxidant activities.[24] The preliminary tests indicated the presence of various constituents such as phenols, saponins, carbohydrates, tannins, flavonoids, and alkaloids in different edible rhizome extracts. The phenols and flavonoids are the most investigated phytochemicals with regard to curing health ailments. Several reports suggested that these phytochemicals are active against diabetes,[1,25] cancer,[16] oxidative diseases,[6,15] and inflammatory diseases.[3,26] The present study aimed to analyze the comparative antioxidative properties of the herbs classified under Zingiberaceae family and their possible anticancer potentials against an estrogen receptor. The extracting solvent used was methanol, and the conditions were based on previous reports.[27,28] Total extract yield was ranging between 1.65% and 3.20% (w/w) for all the four rhizomes [Table 1].
Table 1.
Quantitative phytochemical assay in different rhizomes
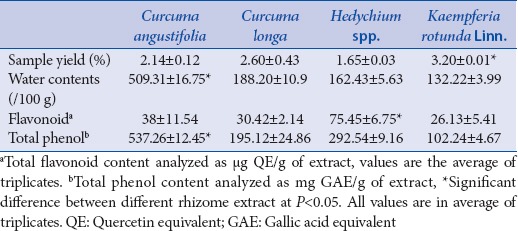
A low-to-high pattern such as K. rotunda Linn. < Hedychium sp. < C. longa < C. angustifolia was seen in case of water contents [Table 1]. Hedychium sp. was found to be the richest in flavonoids, followed by both Curcuma species, though in case of the total phenol, C. angustifolia showed highest phenols followed by Hedychium sp., C. longa, and K. rotunda Linn [Table 1].
Free radical scavenging activity (2,2-diphenyl-1-picryl-hydrazyl method)
All the rhizomes showed free radical scavenging activity as measured by DPPH with the concentrations ranging from 100 to 500 μg/mL [Figure 1]. A dose-dependent approach in DPPH scavenging activity was noticed in the methanolic extract of different rhizomes of Zingiberaceae family. The maximum percentage inhibition was observed at a concentration of 500 μg/mL with 79.5% inhibition by Hedychium sp. (IC50 36.4 μg/mL), whereas that of the standard (ascorbic acid) was found to be 81.76%. The highest radical scavenging activity suggests that all the indigenous rhizomes of Zingiberaceae family might be full of potent-free radical inhibitors, which can act as potential antioxidants.[29] Therefore, as a result, these extracts can scavenge the DPPH*radicals to form constantly reduced DPPH molecules.[27] The methanolic extract of all the four species found to have high DPPH scavenging activities and have already been reported in comparison to the aqueous and acetone extract by other researchers in different plant materials.[15,16,24,27] The DPPH activity in a dose-dependent manner suggested that these rhizomes can serve as a potential targets for several health problems.
Figure 1.
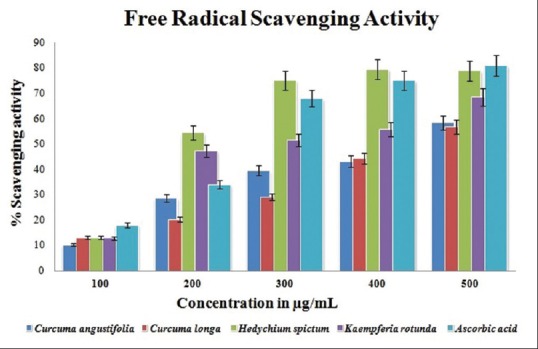
Comparative 2,2-diphenyl-1-picryl-hydrazyl scavenging activity of methanolic extract of different rhizomes of Zingiberaceae family
In silico studies
Molecular docking, a vital tool, has gained a lot of popularity in structural molecular biology and CADD. The aim of the ligand–protein docking is to determine the interaction of a chemical entity (ligand) with a receptor (protein) of already existing 3D structure. First, the purpose of this stochastic interaction study is hypothesized to know a better candidate and second how the drug like molecules (ligands) inhibit the target.
The prediction methods use scoring functions to approximately calculate the binding energies of envisaged ligand–receptor complex. The estimated binding energy (combination of binding constant [Kd] and the Gibbs-free energy [ΔGL]) is carried out by evaluating the most vital physical–chemical phenomena involved in inter-molecular interactions and entropic effects.[29] Based on scoring function, a probable molecule would be ranked.
In this study, we have collected common molecules found in Zingiberaceae family in litero. Lead optimization was done using Lipinski rule of 5 for the molecules reported earlier.[5] Only 71 entities have passed out of 81 compounds.
Lowest energy stabilization is directly proportional to maximum hydrogen bonding interactions with regard to protein–ligand complex and selected as the best pose possible. β-phellandrene (−96.8) has shown the best docking energy score as followed by t-linalool oxide (−87.6) and zingerone (−83.4) [Table 2]. There are previous studies showing β-phellandrene as a potent candidate for cancer studies.[30,31] Song et al.[32] indicated zingerone to ameliorate lipopolysaccharide-induced acute kidney injury by inhibiting toll-like receptor-4 signaling pathway. Chang et al.[33] showed the potency of linalool oxide against cancer cell lines. They reported that linalool can promote gene expression for p53, p21, p27, p16, and p18 genes that can arrest the G0/G1 phase as well as G2/M phase.
Table 2.
Interaction score of the best molecule with bonding pattern using iGemDock
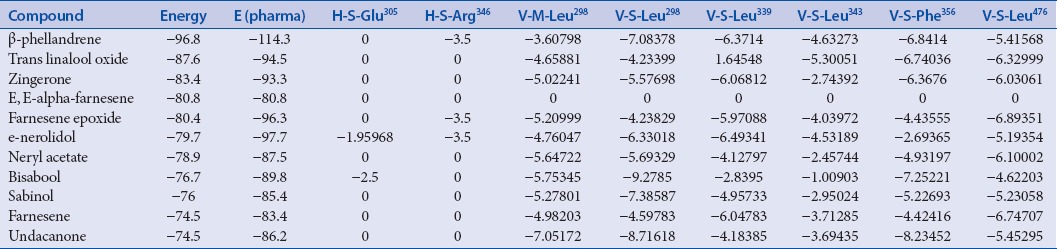
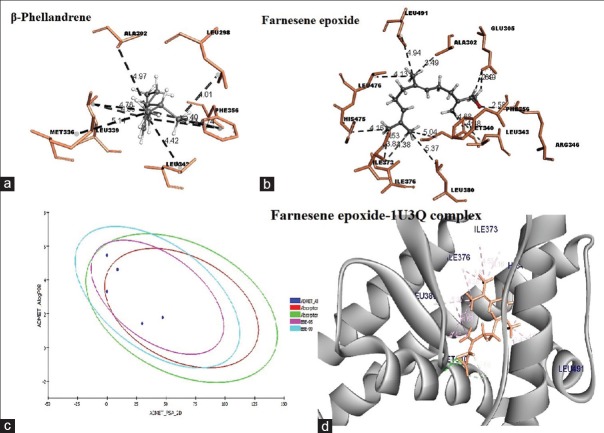
A hydrogen bonding interaction could be pragmatic between the β-phellandrene and the hydroxyl group of Arg346 [Table 2]. Therefore, this could only functional as a weak hydrogen bond acceptor, which is not very physiologically relevant. Few van der Wall interactions are incidental and formed. The major three ligands such as β-phellandrene, trans linalool oxide, and zingerone mutually interacted with amino acids such as Arg346, Leu298, Leu349/343/476, Phe356, and Leu476 [Table 2].
Molecular dynamics (simulations) studies
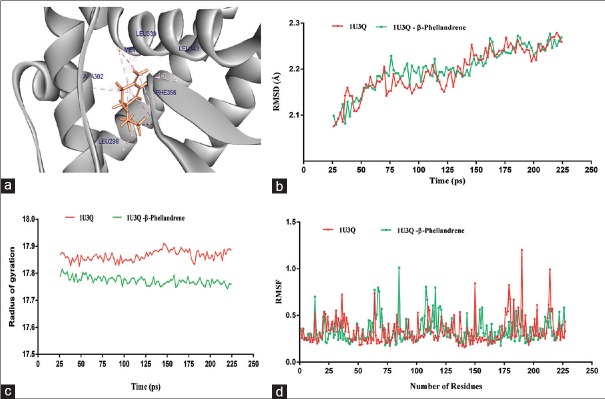
The best pose of 1U3Q – β-phellandrene – complex from virtual screening and LigandFit was selected, and the apo-form of protein (1U3Q) was subjected to 200 ps molecular dynamics (MD) simulations using DS version 3.5. The stability of the protein was analyzed and confirmed by plotting root mean square deviation (RMSD) and radius of gyration (Rg) of protein [Figure 2a]. The RMSD is a measure of the deviation of the conformational stability of the proteins from backbone structure to the early starting structure and fundamental property investigation in MD studies.[34] The RMSD plot of apo-form of protein and 1U3Q – β-phellandrene complex structure has shown deviation between 2.15 and 2.25 Å [Figure 2b]. Root mean square fluctuation (RMSF) of main chain Ca atoms is also calculated with reference to a primary structure for average fluctuation of all residues during the simulation and to locate the flexibility of each amino acid residue along the polypeptide chain. An insight of computed results of RMSF showed that the major alpha-helices are more stable and stabilized [Figure 2c]. Perhaps, slight changes in deviation were observed in C-terminal alpha helices of apo-form of protein and the loop regions are flexible in both structures. The Rg is also calculated to analyze the folding pattern of the protein during simulation[35] and is defined as the root mean square distance of the collection of atoms from their common center of gravity. The results of gyration showed not much of deviation in folding pattern of protein [Figure 2d]. The overall simulation of 1U3Q – β-phellandrene complex showed more stability (−12585.238 kcal/mol) than an apo-form of protein (−12438.166 kcal/mol) variation in energy parameters with respect to time and temperature [Table 3]. Figure 3a–d shows the two best molecules, namely, β-phellandrene and farnesene epoxide interacting with 1U3Q.
Figure 2.
(a) Molecular interaction of β-phellandrene, (b) molecular interaction of farnesene epoxide, (c) absorption, distribution, metabolism, and excretion-Tox plot, (d) 1U3Q-farnesene epoxide complex
Table 3.
Energy parameter variations in different stages of molecular dynamics
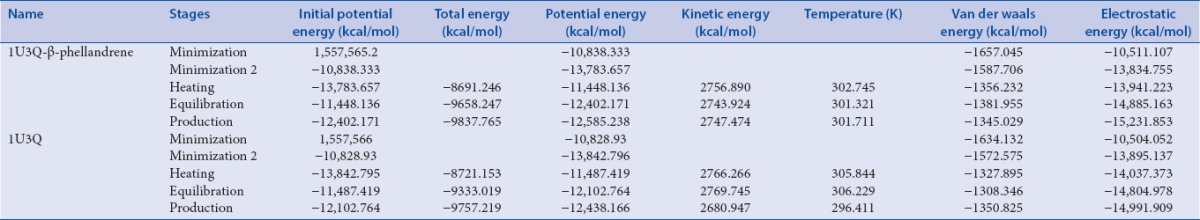
Figure 3.
(a) 1U3Q-β-phellandrene complexes, (b) root mean square deviation plot of 1U3Q-β-phellandrene complexes and 1U3Q-apo protein, during 200 ps simulations. (c) Root mean square fluctuation plot of 1U3Q-β-phellandrene complexes and 1U3Q-apo protein, during 200 ps simulations. (d) Radius of gyration plot of 1U3Q-β-phellandrene complexes and 1U3Q-apo protein
Absorption, distribution, metabolism, and excretion and toxicity study
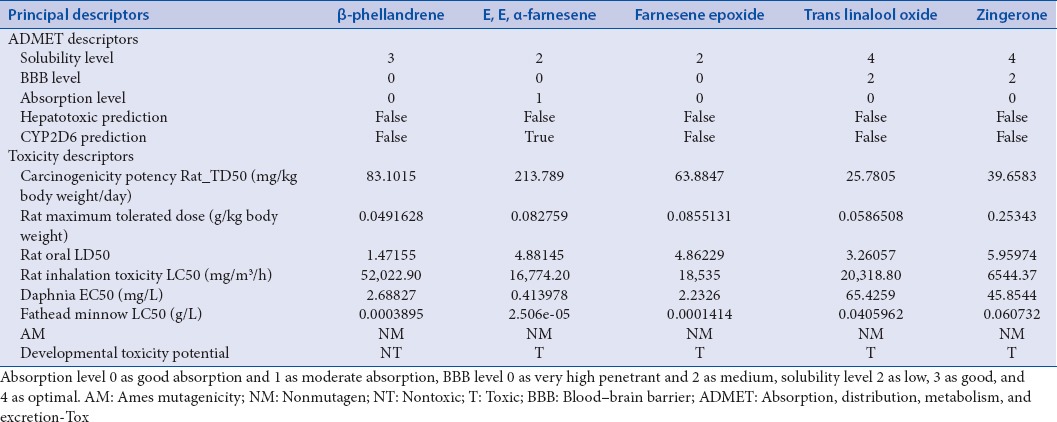
ADME-T plot [Figure 3c] shows 95% and 99% confidence ellipsoids for absorption and BBB, respectively. Farnesene epoxide (55.86) solubility is lower than β-phellandrene (36.61) with good absorption, nonhepatotoxic activity, and high penetration across BBB [Table 1]. Unlike other compounds, β-phellandrene had showed a nontoxic effect in developmental toxicity potential. Hence, authors considered β-phellandrene a better target for estrogen receptor. The two best molecules, i.e., β-phellandrene and farnesene epoxide are forming 11 and 14 favorable nonbond interactions, respectively [Figure 3a and b]. The bond-forming residues in case of compound β-phellandrene are Leu298, Ala302, Met336, Leu339, Leu343, and Phe356 and farnesene epoxide are Ala302, Glu305, Met340, Leu343, Arg346, Phe356, Ile373, Ile376, Leu380, His475, Leu476, and Leu491 [Figure 3a and b]. Figure 3d depicts 1U3Q-farnesene epoxide complex. TOPKAT employs robust and cross-validated Quantitative Structure Toxicity Relationship representation for assessing various measures of toxicity and utilizing the patented optimal predictive space validation method to assist in interpreting the results.[16] Screened compounds showed a nonmutagenic effect when subjected to Ames test [Table 4]. Trans linalool oxide and zingerone showed optimal solubility and were less favorable to cross BBB while E, E alpha-farnesene had a moderate absorption.
Table 4.
Absorption, distribution, metabolism, and excretion and toxicity descriptors for top five drug-like compounds
CONCLUSION AND FUTURE PROSPECTS
To conclude, among all the genus of Zingiberaceae family, Hedychium sp. possess more active compounds than other varieties, followed by K. rotunda Linn., C. angustifolia, and C. longa. This study is an effort to investigate the interaction of Zingiberaceae family compounds and predict the best active compound for anticancerous activity. The bioassay-guided isolation and identification of the bioactive components are still need to be explored. In addition, a detailed research is also required to reveal the structure–activity relationship of these active compounds. The outcome of this research in the aforementioned areas will provide a convincing support for the future clinical uses of these rhizomes in modern medicine.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgements
The authors thank DBT-BIF Facility, Government of India (GOI); BT-Finishing School, GOK, Biotech hub Sikkim, DBT, GOI; and Maharani Lakshmi Ammanni College for Women management, for providing the necessary facility.
REFERENCES
- 1.Middha SK, Goyal AK, Faizan SA, Sanghamitra N, Basistha BC, Usha T, et al. In silico-based combinatorial pharmacophore modelling and docking studies of GSK-3β and GK inhibitors of hippophae. J Biosci. 2013;38:805–14. doi: 10.1007/s12038-013-9367-y. [DOI] [PubMed] [Google Scholar]
- 2.Kasote DM, Katyare SS, Hegde MV, Bae H. Significance of antioxidant potential of plants and its relevance to therapeutic applications. Int J Biol Sci. 2015;11:982–91. doi: 10.7150/ijbs.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Middha SK, Lokesh P, Goyal AK, Prashanth HP, Bhardwaj A, Kamal R, et al. In silico exploration of cyclooxygenase inhibitory activity of natural compounds found in Myrica nagi using LC-MS. Symbiosis. 2016;70:169–78. [Google Scholar]
- 4.Usha T, Goyal AK, Lubna S, Prashanth H, Mohan TM, Pande V, et al. Identification of anti-cancer targets of eco-friendly waste Punica granatum peel by dual reverse virtual screening and binding analysis. Asian Pac J Cancer Prev. 2014;15:10345–50. doi: 10.7314/apjcp.2014.15.23.10345. [DOI] [PubMed] [Google Scholar]
- 5.Usha T, Middha SK, Goyal AK, Karthik M, Manoj D, Faizan S, et al. Molecular docking studies of anti-cancerous candidates in Hippophae rhamnoides and Hippophae salicifolia. J Biomed Res. 2014;28:406–15. doi: 10.7555/JBR.28.20130110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goyal AK, Middha SK, Usha T, Sen A. Ameliorating reactive oxygen species-induced in vitro lipid peroxidation in liver, carbohydrate and DNA damage by Dendrocalamus hamiltonii different leaf extracts. Chiang Mai J Sci. 2016;43:1131–42. [Google Scholar]
- 7.Basistha BC, Handique PJ, Pradhan S. Determination of genetic fidelity of Micropropagate d plants of Zingiber officinale cv- Majhauley of Sikkim Himalaya using RAPD markers. Int J Fund Appl Sci. 2012;1:20–3. [Google Scholar]
- 8.Pradhan BK, Badola HK. Ethnomedicinal plant use by Lepcha tribe of Dzongu valley, bordering Khangchendzonga biosphere reserve, in North Sikkim, India. J Ethnobiol Ethnomed. 2008;4:22. doi: 10.1186/1746-4269-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma A. Traditional processing of Shotti (Curcuma angustifolia Roxb.). - A rhizome based ethnic weaning food. Indian J Tradit Knowl. 2012;11:154–5. [Google Scholar]
- 10.Chan EW, Wong SK. Phytochemistry and pharmacology of ornamental gingers Hedychium coronarium and Alpinia purpurata: A review. J Integr Med. 2015;13:368–79. doi: 10.1016/S2095-4964(15)60208-4. [DOI] [PubMed] [Google Scholar]
- 11.Singh MK, Chandra LR, Bhat D, Arora MS, Nailwal T, Pande V. In vitro antioxidant and free radical scavenging activity of Macrotyloma uniflorum (Gahat dal.). from Kumauni region. Int J Fund Appl Sci. 2012;1:7–10. [Google Scholar]
- 12.Goyal AK, Middha SK, Sen A. Evaluation of DPPH radical scavenging activity, total phenols and antioxidant activities in Indian wild Bambusa vulgaris’Vittata’methanolic leaf extract. J Nat Pharm. 2010;1:40–5. [Google Scholar]
- 13.Ainsworth EA, Gillespie KM. Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin-Ciocalteu reagent. Nat Protoc. 2007;2:875–7. doi: 10.1038/nprot.2007.102. [DOI] [PubMed] [Google Scholar]
- 14.Zhishen J, Mengcheng T, Jianming W. The determination of flavonoid content in mulberry and their scavenging effects on superoxide radicals. Food Chem. 1999;64:555–9. [Google Scholar]
- 15.Goyal AK, Basistha BC, Sen A, Middha SK. Antioxidant profiling of Hippophae salicifolia growing in sacred forests of Sikkim, India. Funct Plant Biol. 2011;38:697–701. doi: 10.1071/FP11016. [DOI] [PubMed] [Google Scholar]
- 16.Usha T, Tripathi P, Pande V, Middha SK. Molecular docking and quantum mechanical studies on pelargonidin-3-glucoside as renoprotective ACE inhibitor. ISRN Comput Biol. 2013;2013:1–4. [Google Scholar]
- 17.Suri C, Naik PK. Elucidating the precise interaction of reduced and oxidized states of neuroglobin with Ubc12 and Cop9 using molecular mechanics studies. Int J Fund Appl Sci. 2012;1:74–7. [Google Scholar]
- 18.Maggiora GM, Mezey PG, Mao B, Chou KC. A new chiral feature in α-helical domains of proteins. Biopolymers. 1990;30:211–4. [Google Scholar]
- 19.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 20.Venkatachalam CM, Jiang X, Oldfield T, Waldman M. Ligand Fit: A novel method for the shape-directed rapid docking of ligands to protein active sites. J Mol Graph Model. 2003;21:289–307. doi: 10.1016/s1093-3263(02)00164-x. [DOI] [PubMed] [Google Scholar]
- 21.Lakhan SE, Ford CT, Tepper D. Zingiberaceae extracts for pain: A systematic review and meta-analysis. Nutri J. 2015;14:50. doi: 10.1186/s12937-015-0038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wohlmuth H, Deseo MA, Brushett DJ, MacFarlane G, Waterman PG, Stevenson LM, et al. Biological activity and novel cytotoxic curcuminoid from Pleuranthodium racemigerum–An Australian Zingiberaceae. Planta Medica. 2007;73:947. [Google Scholar]
- 23.Anasamy T, Abdul AB, Sukari MA, Abdelwahab SI, Mohan S, Kamalidehghan B, et al. A phenylbutenoid dimer, cis-3-(3¢,4¢-dimethoxyphenyl)-4-[(E)-3¢¢¢,4¢¢¢-dimethoxystyryl] cyclohex-1-ene, exhibits apoptogenic properties in T-acute lymphoblastic leukemia cells via induction of p53-independent mitochondrial signalling pathway. Evid Based Complement Altern Med. 2013;2013:1–14. doi: 10.1155/2013/939810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brglez Mojzer E, Knez Hrnčič M, Škerget M, Knez Ž, Bren U. Polyphenols: Extraction methods, antioxidative action, bioavailability and anticarcinogenic effects. Molecules. 2016;21:901. doi: 10.3390/molecules21070901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Middha SK, Usha T, Babu D, Misra AK, Lokesh P, Goyal AK. Evaluation of antioxidative, analgesic and anti-inflammatory activities of methanolic extract of Myrica nagi leaves-an animal model approach. Symbiosis. 2016;70:179–84. [Google Scholar]
- 26.Pande KB, Rizvi SI. Plant polyphenols as dietary antioxidants in human health and disease. Oxid Med Cell Longev. 2009;2:270–8. doi: 10.4161/oxim.2.5.9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surojanametakul V, Satmalee P, Saengprakai J, Siliwan D, Wattanasiritham L. Preparation of curcuminoid powder from turmeric root (Curcuma longa Linn) for food ingredient use. Kasetsart J Nat Sci. 2010;44:123–30. [Google Scholar]
- 28.Devaki M, Nirupama R, Nirupama M, Yajurvedi HN. Protective effect of rhizome extracts of the herb, vacha (Acorus calamus) against oxidative damage: An in vivo and in vitro study. Food Sci Hum Well. 2016;5:76–84. [Google Scholar]
- 29.Jain AN. Scoring functions for protein-ligand docking. Curr Protein Pept Sci. 2006;7:407–20. doi: 10.2174/138920306778559395. [DOI] [PubMed] [Google Scholar]
- 30.Hakkim FL, Al-Buloshi M, Al-Sabahi J. Frankincense derived heavy terpene cocktail boosting breast cancer cell (MDA-MB-231) death in vitro. Asian Pacif J Trop Biomed. 2015;5:824–8. [Google Scholar]
- 31.Piras A, Rosa A, Marongiu B, Atzeri A, Dessì MA, Falconieri D, et al. Extraction and separation of volatile and fixed oils from seeds of Myristica fragrans by supercritical CO2: Chemical composition and cytotoxic activity on CaCo-2 cancer cells. J Food Sci. 2012;77:C448–53. doi: 10.1111/j.1750-3841.2012.02618.x. [DOI] [PubMed] [Google Scholar]
- 32.Song J, Fan HJ, Li H, Ding H, Lv Q, Hou SK, et al. Zingerone ameliorates lipopolysaccharide-induced acute kidney injury by inhibiting toll-like receptor 4 signaling pathway. Eur J Pharmacol. 2016;772:108–14. doi: 10.1016/j.ejphar.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 33.Chang MY, Shieh DE, Chen CC, Yeh CS, Dong HP. Linalool induces cell cycle arrest and apoptosis in leukemia cells and cervical cancer cells through CDKIs. Int J Mol Sci. 2015;16:28169–79. doi: 10.3390/ijms161226089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuzmanic A, Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys J. 2010;98:861–71. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindahl E, Hess B, van der Spoel D. GROMACS 3.0: A package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–17. [Google Scholar]