 Fluorinated derivatives of aminobutyric acid engage in unique interactions with structural waters within the BPTI/trypsin interface and restore inhibitor activity.
Fluorinated derivatives of aminobutyric acid engage in unique interactions with structural waters within the BPTI/trypsin interface and restore inhibitor activity.
Abstract
Introducing fluorine into molecules has a wide range of effects on their physicochemical properties, often desirable but in most cases unpredictable. The fluorine atom imparts the C–F bond with low polarizability and high polarity, and significantly affects the behavior of neighboring functional groups, in a covalent or noncovalent manner. Here, we report that fluorine, present in the form of a single fluoroalkyl amino acid side chain in the P1 position of the well-characterized serine-protease inhibitor BPTI, can fully restore inhibitor activity to a mutant that contains the corresponding hydrocarbon side chain at the same site. High resolution crystal structures were obtained for four BPTI variants in complex with bovine β-trypsin, revealing changes in the stoichiometry and dynamics of water molecules in the S1 subsite. These results demonstrate that the introduction of fluorine into a protein environment can result in “chemical complementation” that has a significantly favorable impact on protein–protein interactions.
Introduction
Fluorinated organic compounds are the least abundant among naturally occurring organohalides. In spite of this scarcity, fluorine has been recognized as a “magic element” in medicinal chemistry, due to its unique properties. Currently, about 20% of pharmaceuticals and up to 30% of agrochemicals contain at least one fluorine atom. 1,2 It has been shown that the incorporation of fluorine into small molecules affects numerous physicochemical properties and results in “mimic”, “block”, and “steric” effects, as well as changes in lipophilicity.3 In addition, fluorine is an excellent biophysical probe, by virtue of the high sensitivity of the 19F NMR reporter to changes in its environment, and a useful biomedical imaging tool when radioactive 18F is used as a PET tracer. 4–6
A wide variety of amino acid analogues containing fluorine atoms have been incorporated into peptides and proteins,7 but by far the most extensively investigated building blocks are based on those with aliphatic side chains such as leucine (Leu), isoleucine (Ile), and valine (Val) residues, as their synthesis and introduction into polypeptides, in vitro and in vivo, are relatively unproblematic. These building blocks have been shown to, for example, influence the membrane association characteristics of antimicrobial peptides, the thermal stability of model helical bundles and globular proteins, and the kinetics of β-sheet formation. While these effects can often be explained based on relative hydrophobicities and secondary structure propensities, they remain exceedingly difficult to predict because they also depend upon the precise microenvironment experienced by the fluorinated amino acid side chain,8 the number of fluorine atoms it contains, and the number of unnatural residues present in the sequence. 9–14
Studies in our group on coiled-coil peptide model systems were based on the experimentally supported assumption that a trifluoromethyl group and an isopropyl group are comparable regarding steric effects.15 Thus, the unnatural amino acid (2S)-2-aminobutanoic acid (Abu) and three of its fluorinated derivatives, (2S)-2-amino-4-monofluorobutanoic acid (MfeGly), (2S)-2-amino-4,4-difluorobutanoic acid (DfeGly), and (2S)-2-amino-4,4,4-trifluorobutanoic acid (TfeGly), and the analogue (2S)-2-amino-4,4-trifluoropentanoic acid (DfpGly) were tested regarding their impact on thermal stability, binding, and folding. 8,16–18 Within the context of a parallel coiled-coil heterodimer, replacement of a central a or d position of the heptad with one of these building blocks was found to be destabilizing, but to different degrees depending on the position of the substitution: the fluorinated side chains are more destabilizing at the d position than at the a position (up to a 14 °C difference in T M in the case of TfeGly) likely because, as shown by molecular dynamics simulations, the polarized β-methylene groups are oriented toward the hydrophobic core in the former case, and oriented toward solvent in the latter case.8 Phage display-based screening of this model system to identify preferred canonical interaction partners revealed that the thermal stability of these complexes, in which one fluorinated side chain packs against exclusively canonical side chains, can vary up to 10 °C (in the case of DfeGly) when the latter are optimized by randomization.18 DfeGly and TfeGly have also been incorporated into specific sites in model peptides to evaluate the effects that fluorine has on the kinetics of amyloid formation and on proteolytic resistance, and the results of these studies are also difficult to express in terms of simple structure–activity relationships. 16,19,20 That is, the unique chemical properties of fluorine lead to a situation in which its presence in otherwise natural-like amino acid side chains can have unexpected consequences within a protein environment. Because it is currently not possible to judiciously rationally design fluorinated proteins, additional systematic studies are needed to demystify fluorine's impact.
In order to investigate how the unique properties of the C–F bond influence the characteristics of an otherwise fully natural polypeptide, we chose the well-characterized bovine pancreatic trypsin inhibitor (BPTI), also known as aprotinin. This small globular protein has been extensively investigated structurally and functionally. 21–24 It has a relatively broad inhibition profile, blocking serine proteases including chymotrypsin, plasmin, and trypsin, as well as metabolic intermediates and ion channels.25 BPTI is also of clinical importance as an antifibrinolytic agent used to reduce bleeding during cardiac surgery, although it was withdrawn from routine administration in 2008 due to increased risk of thrombotic events.26
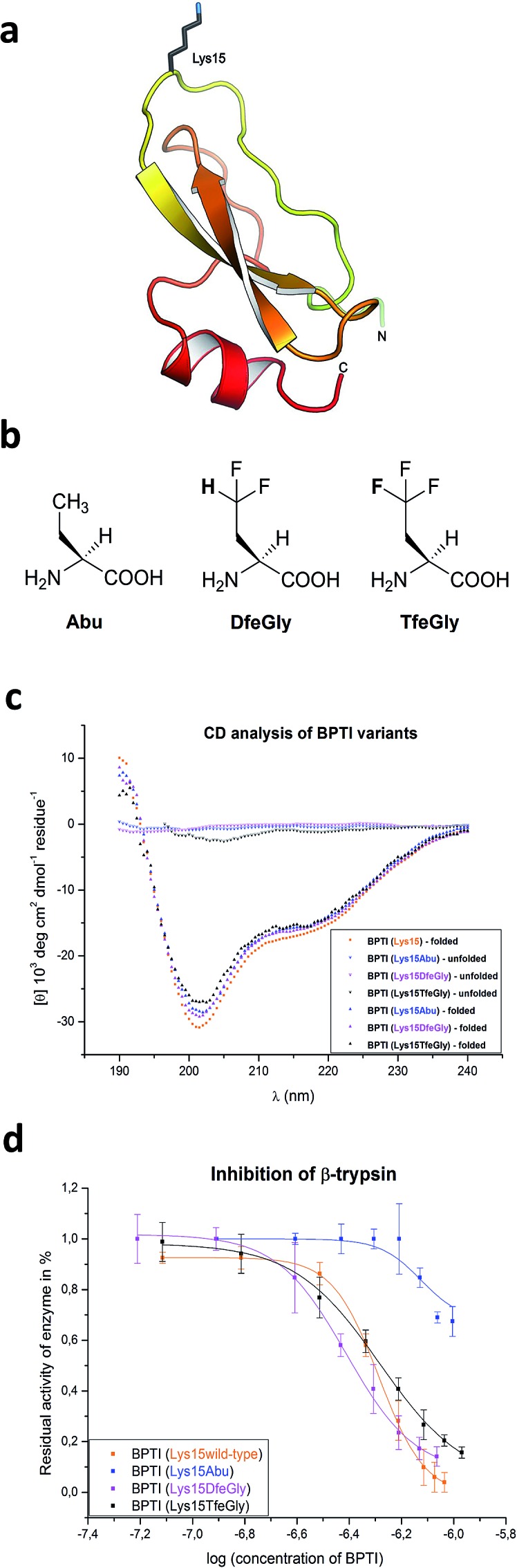
In solution, BPTI folds into a compact pear-shaped structure, comprising an antiparallel β-sheet and two helical regions, that is stabilized by three disulfide bonds (Cys5/55, Cys14/38, Cys30/51). In complex with β-trypsin, residues Pro13 (P3) to Ile19 (P4′) form a binding interface with the S3–S4′ binding sites. 25,27 In particular, the P1–P3 segment forms an anti-parallel β-sheet with residues 214–216 of trypsin, the main chain nitrogen of Ser195 of the catalytic triad (including also His57 and Asp102) is H-bonded to the carbonyl oxygen of P1, but its side chain hydroxyl is not covalently bonded to the P1 carbonyl carbon at the P1–P1′ reaction site. Mutational and structural studies of the side chain of Lys15/P1, fully solvent exposed in BPTI in solution, have demonstrated that it plays an important role in inhibitor binding to the protease (Fig. 1a). 28,29
Fig. 1. (a) Structure of BPTI emphasizing Lys15; (b) chemical structures of the noncanonical amino acids used in this study; (c) CD analysis of synthetic BPTI variants in comparison with commercially available wild-type BPTI; and (d) inhibition of β-trypsin by BPTI variants.
In the current study, we present the total chemical synthesis and characterization of BPTI variants in which Lys15 has been substituted by the noncanonical amino acids Abu, DfeGly, and TfeGly (Fig. 1b). High resolution crystal structures of these mutants of BPTI in complex with trypsin were obtained, revealing how “chemical complementation” involving organic fluorine and structural water molecules can restore inhibitor activity to the Lys15Abu BPTI mutant.
Results and discussion
Synthesis and characterization of mutant BPTIs
Three unnatural variants of BPTI were synthesized by means of solid-phase peptide synthesis and native chemical ligation. 30–32 The yields of the C-terminal peptide fragment and the N-terminal peptide-thioester were comparable to literature values. 30,31 The native chemical ligation proceeded with low efficiency due to the poor solubility of the N-terminal peptide-thioester. It had been previously reported that approximately 50% of BPTI accumulates as dead intermediates due to a kinetic trap within the refolding pathway.33 In our hands, protein renaturation on a smaller scale gave yields of 11–17%. The formation of disulfide bonds was analyzed by ESI-MS (ESI‡).
The overall conformation of the synthetic BPTI variants was analyzed by CD spectroscopy (Fig. 1c). All refolded synthetic mutant BPTIs exhibit CD spectra comparable to the wild-type protein in shape and intensity, indicating that the structural perturbations caused by the Lys15Abu, Lys15DfeGly, and Lys15TfeGly mutations are minimal. In contrast, the unfolded full-length peptides yielded CD spectra that indicate the absence of secondary structure. 34,35
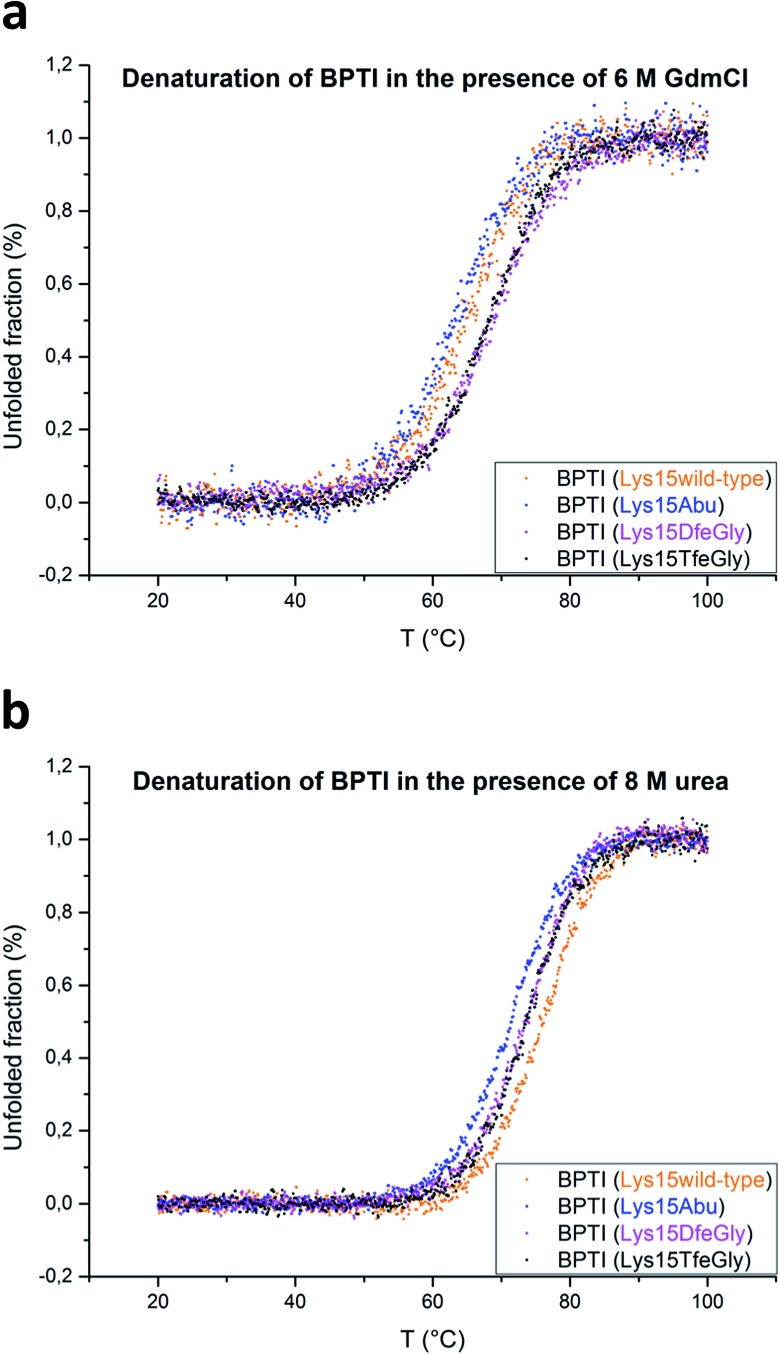
BPTI is a protein with extremely high thermal stability that is also relatively resistant to common protein denaturing agents such as urea and guanidinium chloride (GdmCl). For example, 6 M GdmCl alone, or heating to 100 °C at neutral pH, are conditions that are not sufficient to fully denature the protein. 36–38 Therefore, to analyze the effect of P1 mutation on the thermal stability of BPTI, we conducted denaturation experiments with heat treatment in the presence of denaturants under acidic conditions (Table 1). Because Lys15 has a formal positive charge and the analogues studied here are neutral, two distinct sets of conditions, 6 M GdmCl and 8 M urea, were applied. GdmCl is a salt that has an electrostatic masking effect and is therefore a better reporter for hydrophobic contributions to stability, whereas urea is more suitable for monitoring the contributions of electrostatic effects to stability.39
Table 1. Thermal denaturation parameters for BPTI variants.
| Tm/GdmCl a | ΔG° b | Tm/urea a | ΔG° b | |
| Lys15(wt) | 66.5 | 12.0 | 75.8 | 14.6 |
| Lys15Abu | 63.4 | 11.3 | 71.7 | 13.8 |
| Lys15DfeGly | 68.8 | 12.4 | 74.8 | 14.3 |
| Lys15TfeGly | 68.3 | 12.1 | 73.8 | 14.6 |
aTm (°C) is defined as the temperature at which the fraction of folded species is 0.5. Errors are typically not higher than 0.5 °C.
bΔG° values were calculated for 1 mole at the standard state, errors are typically not higher than 0.2 kcal mol–1.
Under both sets of denaturation conditions, the folded Lys15Abu variant is significantly less stable than Lys15, Lys15DfeGly, and Lys15TfeGly (Fig. 2). The fluorinated side chains have a stabilizing effect that is greater than that of the wild type in the presence of GdmCl, but the opposite is true in the presence of urea, indicating that the positive formal charge of the solvent-exposed Lys contributes to thermal stability to some extent. The Abu data agree well with previous studies of P1 mutants of BPTI, in which differential scanning calorimetry experiments revealed that the presence of hydrophobic aliphatic or aromatic residues at this position is always destabilizing.35 In contrast, the stabilizing effect of DfeGly and TfeGly suggest that these unnatural building blocks do not behave in a manner that can be explained by hydrophobicity arguments alone. Previous experimental and computational studies have revealed that the introduction of fluorine into a methyl group not only leads to an increase in the solvent accessible surface area and hydrophobicity of the group, but also facilitates electrostatic interactions with an aqueous environment due to the highly polarized nature of the C–F bond.40
Fig. 2. Thermal denaturation curves for BPTI variants in the presence of chemical denaturants: (a) 6 M GdmCl, pH 2, or (b) 8 M urea, pH 2.
Inhibitor activity assay
BPTI strongly inhibits serine proteases, including α-chymotrypsin and β-trypsin. In the latter case, it has been shown that the side chain of Lys15/P1 contributes significantly to the binding enthalpy of the inhibitor–enzyme complex, due to both the H-bond network in which it engages by means of its ammonium group (involving Ser190, Asp189, and Val227, among others), and the burial of the alkyl segment –(CH2)4– of Lys15 in the S1 pocket; e.g., the replacement of Lys15 with Gly leads to a reduction in the K a of BPTI for β-trypsin of 109 fold.29
We conducted standard inhibition assays with bovine β-trypsin41 for wild-type BPTI and mutants Lys15Abu, Lys15DfeGly, and Lys15TfeGly (Fig. 1d). Substitution of Lys with Abu at P1 dramatically reduces inhibitor activity: at an inhibitor concentration for which the residual activity of the enzyme is 50% in the case of the Lys15 control, this value is 100% in the case of Lys15Abu. Interestingly, both BPTI variants containing fluorinated analogues at the P1 site restore inhibitor activity: the association constants determined here are 3.88 x 107 M–1 for Lys15DfeGly and 5.20 × 107 M–1 for Lys15TfeGly, similar to the value 5.17 × 107 M–1 obtained for wild-type BPTI (ESI‡). This remarkable observation suggests that the introduction of two or three fluorine atoms into the Abu side chain results in “chemical complementation” that fully restores inhibitor activity to a wild-type level.
High resolution crystal structures of trypsin–BPTI complexes
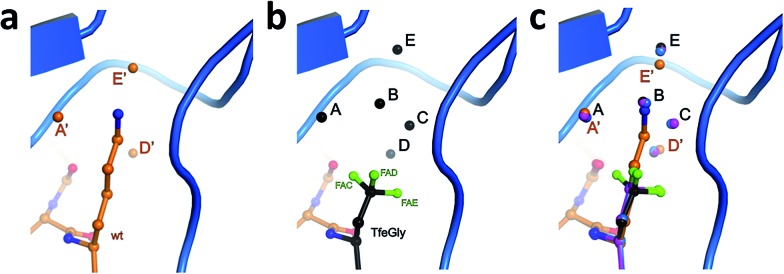
To test the above hypothesis and gain insight into the molecular mechanisms by which the fluorinated residues are able to restore inhibitor activity to the Lys15Abu BPTI mutant, we determined high-resolution crystal structures of wild-type BPTI and all three variants in complex with bovine β-trypsin: wild-type at 1.25 Å, Lys15Abu and Lys15DfeGly at 1.37 Å, and Lys15TfeGly at 1.30 Å. The resolution of the wild-type complex is higher than that of other structures currently available in the Protein Data Bank (PDB). 42,43 All structures were refined to low R/R free values with good stereochemistry (Table S3‡). Root mean square deviations (RMSD, Table 2) indicate negligible differences in both the global conformation of the mutant complexes compared to the wild-type complex, as well as among the mutant complexes, consistent with our CD spectra (Fig. 1c). All β-trypsin–BPTI complexes can be superimposed well, except at the P1 side chain and the water molecules within the S1 pocket (Fig. 3c). These results are consistent with a study published by Smalås et al. in which nine mutants of BPTI, all containing natural amino acids, were crystallized in complex with this enzyme, and all displayed similar global conformations.44
Table 2. RMSD in Å of β-trypsin–BPTI complexes.
| Complexes | β-Trypsin–BPTI |
||
| β-Trypsin | BPTI | BPTI (Pro13 – Arg17) | |
| All | All | All/main chain | |
| Lys15Abu | 0.50 | 0.78 | 0.11/0.06 |
| Lys15DfeGly | 0.38 | 0.90 | 0.09/0.05 |
| Lys15TfeGly | 0.45 | 0.85 | 0.11/0.06 |
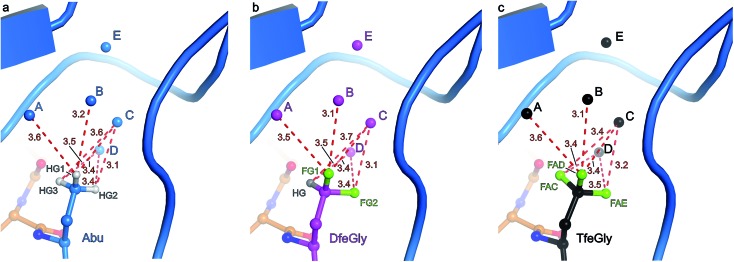
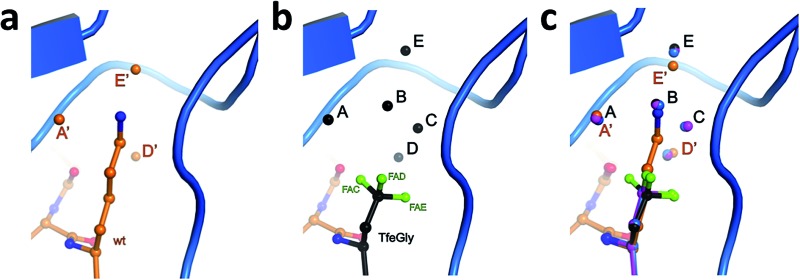
Fig. 3. Structural context of the P1 side chain in the S1 binding pocket. β-Trypsin is shown in cartoon representation and the modified BPTI in stick representation (a) Lys15 and structural water molecules A′, D′, and E′, and (b) Lys15TfeGly and structural water molecules A, B, C, D, and E, and (c) superimposition of P1 side chains and structural water molecules.
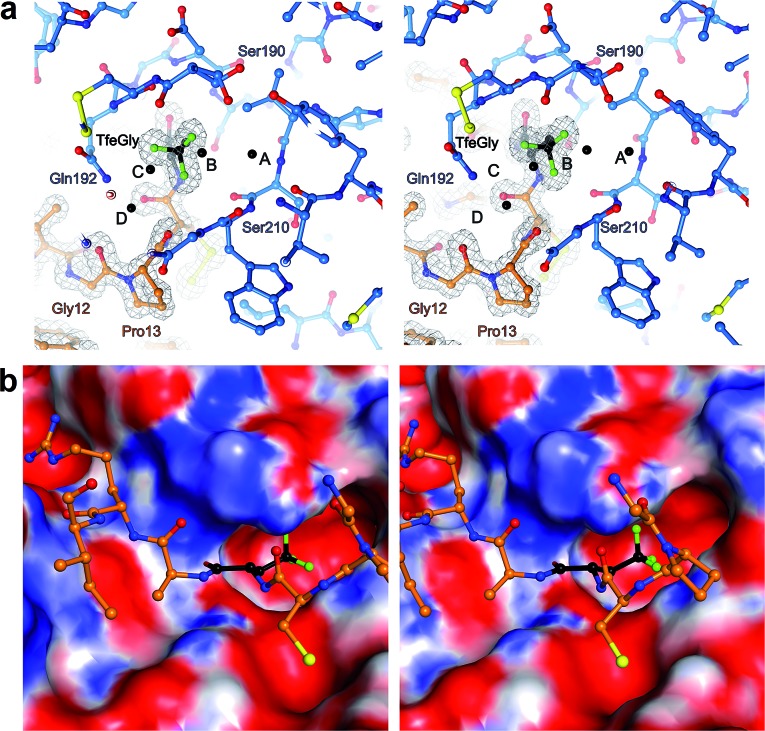
The S1 binding pocket of β-trypsin occupied by Abu and its fluorinated analogues were analyzed in detail. To clarify the nomenclature that is used throughout this section, the pre-existing PDB labels for the chemical groups that are the side chains of Abu, DfeGly, and TfeGly are ABA, OBF, and 3 EG, respectively. Each atom of these functional groups also has its own unique identifier, according to the PDB. In agreement with previous studies, the S1 pocket in all complexes is highly polar (Fig. 4b). 29,44 The electron density around the mutated position 15 of BPTI is well defined, indicating a stable conformation within this enzyme subsite (Fig. 4a). Distances were calculated from each hydrogen or fluorine substituent of the terminal γC of each side chain and broken down into close contacts to atoms of BPTI or β-trypsin (Fig. 5 and Table S5‡) and close contacts to structural waters (Fig. 6 and Table S5‡). Distances between these water molecules and their proximity to atoms belonging to β-trypsin or BPTI (other than the side chain at position 15) are given in Table S6.‡
Fig. 4. (a) Stereoview of Lys15TfeGly occupancy in S1 binding pocket. β-Trypsin is drawn in blue and BPTI in orange. Water molecules are drawn as black spheres. Electron density around BPTI is shown as gray mesh, contoured at 1.8σ (b) stereoview of interacting loop in S1 pocket with S1 pocket represented as an electrostatic potential surface map. Blue coloring indicates positively charged and red coloring indicates negatively charged patches on the surface.
Fig. 5. Distance analysis and top view of the unnatural side chain at position 15 of BPTI depicted in orange within the S1 binding pocket of β-trypsin drawn in blue. Values indicate the distance in Å between the individual hydrogen or fluorine atoms of the terminal γC of the side chain and the ten nearest enzyme atoms (clockwise, Gln192NE, Gln192CD, Gln192CA, Gln192N, Cys191C, Cys191CA, Cys191N, Ser190C, Ser190CA, and Ser190N), as well as the five nearest BPTI atoms (from top to bottom, Xaa15CA, Xaa15N, Cys14C, Cys14CA, and Pro13O): (a) Abu structure with side chain and structural waters A–D in light blue and the three γC hydrogen substituents HG1 (ABAHG1), HG2 (ABAHG2), and HG3 (ABAHG3) in light-gray; (b) DfeGly structure with side chain and structural waters A–D in magenta, the two γC fluorine substituents FG1 (OBFFG1) and FG2 (OBFFG2) in green, and the single γC hydrogen substituent HG (OBFHG) in gray; (c) TfeGly structure with side chain and structural waters A–D in black and the three γC fluorine substituents FAD (3EGFAD), FAE (3EGFAE), and FAC (3EGFAC) in green. Nitrogen atoms are shown in dark blue and oxygen atoms in red.
Fig. 6. Distance analysis and side view of the unnatural side chain at position 15 of BPTI within the S1 binding pocket of β-trypsin. β-Trypsin is shown in cartoon representation and the modified BPTI in stick representation. Values indicate the distance in Å between the individual hydrogen or fluorine atoms of the terminal γC of the side chain and the oxygen atom of the respective structural water molecule A–D: (a) Abu structure with side chain and structural waters A–D in light blue and the three γC hydrogen substituents HG1 (ABAHG1), HG2 (ABAHG2), and HG3 (ABAHG3) in white; (b) DfeGly structure with side chain and structural waters A–D in magenta, the two γC fluorine substituents FG1 (OBFFG1) and FG2 (OBFFG2) in green, and the single γC hydrogen substituent HG (OBFHG) in gray; (c) TfeGly structure with side chain and structural waters A–D in black and the three γC fluorine substituents FAD (3EGFAD), FAE (3EGFAE), and FAC (3EGFAC) in green. Nitrogen atoms are shown in dark blue and oxygen atoms in red.
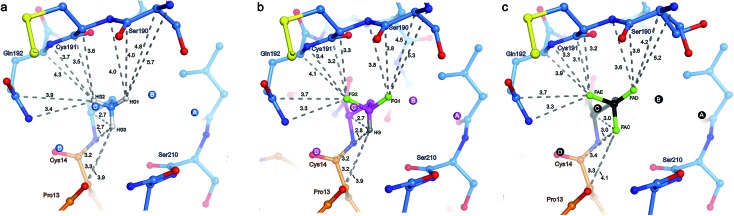
The γCH3, γCHF2, and γCF3 groups of the side chains of Abu, DfeGly, and TfeGly, respectively, have numerous longer range contacts to the S1 subsite of β-trypsin (Fig. 5 and Table S5‡). Atoms ABAHG1, OBFFG1, and 3EGFAD, which are similarly oriented, are in the vicinity of the side chain and carbonyl of Ser190 and the main chain atoms of Cys191, and their contact distances are shorter for DfeGly and TfeGly than for Abu. For example, ABAHG1 is 4.0 Å from Ser190C compared to 3.8 Å and 3.6 Å in the cases of OBFFG1 and 3EGFAD, respectively. In the TfeGly structure, a second conformation (approximately 50% occupancy) is observed for the Ser190 side chain, one in which the H-bond to the hydroxyl of Tyr228 is maintained while the distance between 3EGFAD and Ser190OG increases from 3.3 to 4.6 Å, likely due to electrostatic repulsion between fluorine and oxygen. Atoms ABAHG2, OBFFG2, and 3EGFAE map to one another and are nearest the main chain atoms of Cys191 (3.1–4.1 Å) and the side chain amide and main chain nitrogen of Gln192, 3.3–3.9 Å and 3.3–3.7 Å, respectively.
Considering the studies published by Diederich and coworkers concerning how organic fluorine bonds engage in multipolar interactions with protein backbone fragments, H–Cα–C O, and main chain or side chain amides, we determined the relevant angles to describe how each C–F bond in DfeGly and TfeGly approaches the trypsin main chain atoms of residues Ser190 to Gln192 (Table S5‡).1 Although we observed α4 angles (C–F···CC O) within the range of 70° to 110°, indicating an orthogonal approach, it is difficult to rule out that this may be a consequence of packing; deconvoluting such interactions for an sp3 hybridized group containing multiple fluorine atoms is beyond the scope of this study. In contrast, atoms ABAHG3, OBFHG, and 3EGFAC occupy a region of the S1 pocket in which only two contacts closer than 4.0 Å are observed, to the carbonyl oxygen of Ser210 (3.6–3.7 Å) and to the main chain nitrogen of Trp211 (3.9 Å). These particular atoms of the unnatural side chains at position 15 more closely approach atoms belonging to BPTI itself, namely their own backbone nitrogen, the upstream carbonyl of Cys14, and the backbone oxygen of Pro13. In this case, the only distance that changes significantly across the Abu, DfeGly, and TfeGly series is the one from ABAHG3 and 3EGFAC to Xaa15N (from 2.7 to 3.0 Å), and this difference likely stems from intramolecular electrostatic repulsion in the trifluoromethyl variant. The H-bonds involving Pro13O (with Gly214N), Lys15N (with Ser212O), and Lys15O (with Gly195N and Ser197N) found in the native structure are retained in the mutant complexes.
Three water molecules are found in the S1 binding pocket of the wild-type complex, here referred to as A′, D′, and E′, while five occupy it in the case of all three unnatural complexes, here referred to as A, B, C, D, and E (Fig. 3 and S15‡). Thus, when Lys15 is replaced by Abu, DfeGly, or TfeGly, two additional water molecules occupy the space that is otherwise taken up by the considerably longer side chain of the native P1 residue, a phenomenon that had also been observed in previous work on natural mutants of BPTI, including Lys15Gly and Lys15Thr.15
In agreement with the literature, our wild-type structure shows that A′, D′, and E′ do not interact with one another; instead, A′ and E′ mediate H-bonds between the P1 side chain and the enzyme (Fig. S14 and Table S4‡). A′ corresponds to Sol65244 or DOD1014/E43 from previous reports, and is equidistant, 2.9 Å, from LysNZ and Val227O; E′ had been previously published as Sol65144 or DOD1008/E43, and is 2.8 Å away from LysNZ and Asp189OD2. In contrast, water molecule D′, earlier referred to as Sol65444 is not proximal to the Lys15 side chain, but has a total of four H-bond partners from other sources: three from the enzyme (Gln192NE and Gly214O) and one from position P3 of the inhibitor (Pro13O).
Water molecules A, B, C, D, and E are present in the S1 site of each mutant complex, and are virtually superimposable. B and C represent “new” waters that are not found in the wild-type structure (Fig. 3). A, B, C, and D form what can be thought of as a “hydration shell” or “contour line” around the unnatural side chains in the space that is occupied by the longer side chain of lysine in the wild-type structure (Fig. 6); E is distal from the side chain, located equidistantly “above” B and C (3.3–3.5 Å; Tables S5 and S6). Moving from right to left along this “contour line”, in accordance with the view depicted in Fig. 5, the water molecules are H-bonded to one another as follows: A–B, B–A and –C, C–B and –D, and D–C (Table S6‡).
In all structures, with respect to the enzyme, water molecules A–D have the following environments (Table S6‡): A is in the vicinity of Val227, Ser190, and Trp211; B is close to Asp189 and Ser190; C has contacts to Ser190 and Gly214; D is the only structural water that directly contacts BPTI itself, via Pro13, and is also nearby Gln192 and Gly214; E is proximal to Asp189, Lys220, Gly214, and Ser213. In all structures, with respect to the unnatural side chain, water molecules A and B are closest to atoms ABAHG1, OBFFG1, and 3EGFAD, substituents of the terminal γC group that map to one another (Fig. 6). In contrast, water C is within 3.7 Å of all three substituents of the terminal γC group in all structures, and water D is proximal to two of the three substituents of the terminal γC group in all structures.
Interestingly, water molecule C is closer to the fluorine atom 3EGFAC in the TfeGly structure (3.4 Å) than it is to the hydrogen atom ABAHG3 in the Abu structure (3.6 Å) or the hydrogen atom OBFHG in the DfeGly structure (3.7 Å). Although it cannot be ruled out that this is a consequence of the slight shift in conformation within the side chain that occurs due to the electrostatic repulsion between 3EGFAC and 3EGN, it may also indicate that a favorable interaction, perhaps a weak OH···FC H-bond, exists between the TfeGly side chain and water C. Certainly, there is no evidence of repulsion as was observed for the Ser190 side chain (occupancies here are unity), and a simple argument based on the greater van der Waals radius of fluorine compared to hydrogen would not suffice, as no such phenomenon is seen between water molecule C and atoms OBFFG2 (3.1 Å) or 3EGFAE (3.2 Å), compared to ABAHG2 (3.1 Å); that is, these distances do not decrease when hydrogen is replaced by fluorine.
To summarize the distance analysis of the crystal data (Fig. S16‡), the atoms that are oriented toward the enzyme's main chain (ABAHG1/OBFFG1/3EGFAD and ABAHG2/OBFFG2/3EGFAE) certainly show a trend in which the fluorine atoms (DfeGly and TfeGly) are closer to the enzyme backbone than the hydrogen atoms (Abu). Considering the fact that this is observed for all nearby main chain atoms (Ser190CB, Ser190C, Cys191N, Cys191CA, Cys191C, Gln192N, and Gln192CA), there seems to be evidence for the type of previously described “fluorophilic environment” provided by protein backbone fragments, in this case those of Ser190, Cys191, and Gln192, rather than particular potential hydrogen bonding interactions. For example, the distances between fluorine atoms and the nearby hydrogen atoms of the enzyme were also determined (Table S9 in ESI‡). The closest intermolecular CH···FC contacts are OBFFG2 and 3EGFAE to Cys191HA (2.7 and 2.6 Å, respectively), and 3EGFAC to Trp211HA (2.6 Å). Even if these contacts may be considered to fall within a weak hydrogen-bonding regime, it is unlikely that these specific interactions account for the greater proximity between the side chain constituents of BPTI residue 15 and the enzyme backbone, especially considering that trypsin residue Trp211 is distal to residues Cys191 and Gln192 (i.e. these interactions would not “pull” in the same direction).
As presented in Table 3, the average B-factor of all water molecules in the four complexes is >30 Å2, and the B-factors of the individual structural water molecules in the S1 pocket are much lower, excluding Lys15Abu, 13–23 Å2. The following comparisons are meaningful because of the uniformity in resolution across this data set. The B-factors of A′, D′, and E′ in the wild-type structure (13.5–15.1) are clearly lower than the corresponding values in the Lys15Abu complex (20.7–23.0).
Table 3. Comparison of B-factors given in Å2 of amino acid side chains and water molecules in the S1 binding pocket.
| β-Trypsin–BPTI complexes |
||||
| Wt | Lys15Abu | Lys15DfeGly | Lys15TfeGly | |
| B-factor (Å 2 ) | ||||
| Average B-factor of protein complex | 20.7 | 21.3 | 19.6 | 21.2 |
| All water molecules on average | 31.0 | 33.8 | 31.2 | 32.0 |
| BPTI Pro13 – Arg17 | ||||
| All atoms | 13.0 | 13.2 | 12.4 | 13.4 |
| Main chain | 12.0 | 12.2 | 11.1 | 12.3 |
| Side chain | 14.0 | 14.6 | 13.9 | 14.6 |
| BPTI P1 residues | ||||
| All atoms | 12.1 | 11.9 | 13.2 | 12.9 |
| Main chain | 11.6 | 11.5 | 11.0 | 11.8 |
| Side chain | 12.5 | 12.8 | 15.5 | 13.7 |
| Water molecules in S1 pocket | ||||
| A/A′ | —/13.5 | 21.8/— | 17.5/— | 18.4/— |
| B | — | 27.7 | 20.0 | 20.5 |
| C | — | 31.2 | 22.2 | 20.7 |
| D/D′ | —/15.4 | 23.0/— | 17.8/— | 17.9/— |
| E/E′ | —/14.1 | 20.7/— | 19.1/— | 19.4/— |
Whereas the B-factors for “new” waters B and C are larger (27.7–31.2) in the Lys15Abu structure, these values fall significantly, to 20.0–22.2, when DfeGly and TfeGly are present. Previous crystallographic studies of trypsin-mutant BPTI complexes containing canonical amino acids at the P1 position showed that although the incorporation of bulkier hydrophobic amino acids results in tighter binding than observed for the polar and charged P1 side chain mutants, significantly lower B-factors for structural waters were only observed in the latter category.44
These results suggest that the fluorine atoms in DfeGly and TfeGly interact with these water molecules in such a way that they are more tightly held than they are by the hydrogens in the Abu side chain, an argument that is strengthened by the observation that waters B and C are slightly further away from each other and from water E in the Abu inhibitor variant compared to the fluorinated variants; thus, it seems unlikely that stronger water–water contacts are exclusively responsible for this apparent reduction in dynamics. In light of previous reports that have described the important role of trapped water molecules in protein–ligand interactions,45 and a recent study describing surface fluorination and hydration dynamics in proteins,46 a mechanism in which fluorine interacts with water molecules B and C, which in turn can be thought of as extensions of the enzyme, to restore inhibitor activity, is plausible.
Conclusions
The high resolution crystal structures of the BPTI–trypsin complexes investigated here reveal an intriguing mechanism by which fluorinated amino acids can engage in noncovalent interactions within the context of a protein environment. When the Lys15 residue of wild-type BPTI is substituted with the nonfluorinated hydrophobic amino acid Abu, its inhibitor activity is dramatically reduced and the B-factors of the two new proximal structural waters in the S1 binding site are comparatively high, indicating that the waters are more loosely held. In contrast, when DfeGly or TfeGly are used to replace the P1 residue, the inhibitor activity is restored to the wild-type level and these B-factors are relatively low, indicating that the waters are more tightly held. Our results demonstrate what can be described as “chemical complementation” involving partially fluorinated amino acids and structural water molecules that is only possible due to fluorine's unique impact on an otherwise hydrophobic aliphatic side chain. Such a role for the bioorthogonal element fluorine has not yet been described, and our observations lend support not only to the view that fluorinated amino acids constitute a truly unique family of building blocks for protein engineering, but provide important information that will find applications in peptide and protein engineering, peptide drug design, organocatalysis, and biomaterial development.
Methods
Peptide synthesis
The peptide fragments 1-Xaa15-37 and 38-58 of BPTI were synthesized according to standard Fmoc/tBu chemistry by means of an automated SyroXP-I peptide synthesizer or an Activo P11 synthesizer. The noncanonical amino acids were incorporated into the protein sequence by manual coupling with 1.25 equivalents of the Fmoc-l-amino acid, HOBt, and DIC relative to resin loading. The fully protected peptide fragment of BPTI 1-Xaa15-37 was cleaved from the resin by treatment with trifluoroethanol/HOAc/DCM (v/v/v, 1 : 1 : 8) for two hours, and thioesterification was achieved by incubation with five equivalents of each benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP), p-acetamidothiophenol, and DIEPA in DCM overnight. All peptide fragments were purified by means of RP-HPLC with a C18 column, and purity was verified by analytical RP-HPLC coupled with ESI-MS (ESI‡).
Native chemical ligation and protein refolding
Native chemical ligation between thioester 1-Xaa15-37 and fragment 38-58 was performed by incubation in a solution containing 6 M GdmCl, 0.1 M Na2HPO4, 50 mM 4-mercaptophenylacetic acid (MPAA), and 20 mM tris(2-carbonoxyethyl)phosphine (TECP); peptide fragments were present in the reaction at concentrations of 1–5 mM. The reaction was monitored by means of RP-HPLC/ESI-TOF and the full-length proteins of BPTI variants were purified by means of RP-HPLC with a C18 column. Purified BPTI variants were folded by dissolution in 0.6 M Tris–HCl, pH 8.7, 6 mM EDTA, and 6 M GdmCl followed by rapid six-fold dilution with water. The mixture was exposed to air and stirred for two days. The folding process was monitored by means of RP-HPLC/ESI-TOF (ESI‡).
Global structural analysis and thermal stability determination
CD spectra were recorded on a Jasco J-715 spectropolarimeter at 25 °C. Overall peptide concentrations were 20 μM in 10 mM tris(hydroxymethyl)aminomethane (Tris) buffer at pH 7.4. CD-spectra were obtained in the far-UV range (190–240 nm) using 0.1 cm Quartz Suprasil cuvettes equipped with a stopper. The nitrogen flow rate was set to ∼3.0 L min–1. The spectra were recorded at 0.2 nm intervals with a 2 nm bandwidth and response time of 3 seconds. After baseline correction, the measured ellipticities were converted to molar ellipticities per residue (degree cm2 per dmol per residue) by normalizing for the concentration of peptide, the number of amino acids in the sequence, and the path length.
The thermal stability of BPTI variants was performed by means of CD measurement. Protein samples were dissolved in buffer containing either 10 mM Tris–HCl, pH 2.0, with 6 M GdmCl or 10 mM Tris–HCl, pH 2, with 8 M urea. Melting curves were recorded by monitoring the absorbance at 222 nm while heating at a rate of 3 K min–1, from 20 °C to 100 °C. Melting curves were obtained in triplicate and averaged. Thermodynamic parameters were determined by means of nonlinear least square fitting of the normalized CD melting curves (ESI‡).
Inhibitor activity assay
A standard spectrophotometric inhibitor assay was carried out to determine the inhibitor activity of each BPTI variant (ESI‡). Nα-Benzoyl-l-arginine 4-nitroanilide hydrochloride was used as β-trypsin substrate. The assay was carried out in a buffer containing 0.046 M Tris–HCl and 0.0115 M CaCl2, pH 8.1.
Protein crystallography
β-Trypsin–BPTI complexes were purified by dialysis. Protein crystallization was conducted by means of sitting drop vapor diffusion technique with a commercially available kit (ESI‡). Well-formed crystals were soaked in 30% glycol plus reservoir solution and frozen in liquid nitrogen. Data was collected at Berlin BESSY II, beamline 14-2. X-ray data collection was performed at 100 K. Diffraction data were processed with the XDS47 in space group I222 (Table S3‡). The structure of the wild-type trypsin/BPTI complex was solved by molecular replacement with the coordinates of single polypeptide chains of β-trypsin and BPTI (PDB: ; 2FTL)41 as search models using PHASER.48 The structure of ; 4Y0Z, ; 4Y10, and ; 4Y11 were determined by isomorphous replacement. For calculation of the free R-factor, a randomly generated set of 5% of the reflections from the diffraction data set was used and excluded from the refinement. The structure was initially refined by applying a simulated annealing protocol and in later refinement cycles by maximum-likelihood restrained refinement using PHENIX followed by iterative, manual model building cycles with COOT.49 Model quality was evaluated with MolProbity.50 Figures were prepared using PyMOL.51 The atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under the accession codes ; 4Y0Y, ; 4Y0Z, ; 4Y10, and ; 4Y11.
Acknowledgments
This work was supported with funds from the DFG Graduate School GRK 1582/1 “Fluorine as Key Element”. We accessed beamlines of the BESSY II (Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung II) storage ring (Berlin, Germany) via the Joint Berlin MX-Laboratory sponsored by the Helmholtz Zentrum Berlin für Materialien und Energie, the Freie Universität Berlin, the Humboldt-Universität zu Berlin, the Max-Delbrück Centrum, and the Leibniz-Institut für Molekulare Pharmakologie. The authors are grateful to Dr Mario Salwiczek and Dr Ulla I. M. Gerling for critical reading of the manuscript and fruitful discussions.
Footnotes
†Dedicated to Professor Iwao Ojima on the Occasion of his 70th Birthday.
References
- Müller K., Faeh C., Diederich F. Science. 2007;317:1881. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- Purser S., Moore P. R., Swallow S., Gouverneur V. Chem. Soc. Rev. 2008;37:320. doi: 10.1039/b610213c. [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Taguchi T. and Ojima I., in Fluorine in Medicinal Chemistry and Chemical Biology, John Wiley & Sons, Ltd., 2009, pp. 1–46. [Google Scholar]
- Fielding L. Curr. Top. Med. Chem. 2003;3:39. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- Kitevski-LeBlanc J. L., Prosser R. S. Prog. Nucl. Magn. Reson. Spectrosc. 2012;62:1. doi: 10.1016/j.pnmrs.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Srinivas M., Heerschap A., Ahrens E. T., Figdor C. G., Vries I. J. M. Trends Biotechnol. 2010;28:363. doi: 10.1016/j.tibtech.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwiczek M., Nyakatura E. K., Gerling U. I. M., Ye S., Koksch B. Chem. Soc. Rev. 2012;41:2135. doi: 10.1039/c1cs15241f. [DOI] [PubMed] [Google Scholar]
- Salwiczek M., Samsonov S., Vagt T., Nyakatura E., Fleige E., Numata J., Cölfen H., Pisabarro M. T., Koksch B. Chem.–Eur. J. 2009;15:7628. doi: 10.1002/chem.200802136. [DOI] [PubMed] [Google Scholar]
- Bilgiçer B., Fichera A., Kumar K. J. Am. Chem. Soc. 2001;123:4393. doi: 10.1021/ja002961j. [DOI] [PubMed] [Google Scholar]
- Buer B. C., de la Salud-Bea R., Al Hashimi H. M., G.Marsh E. N. Biochemistry. 2009;48:10810. doi: 10.1021/bi901481k. [DOI] [PubMed] [Google Scholar]
- Holzberger B., Rubini M., Möller H. M., Marx A. Angew. Chem., Int. Ed. 2010;49:1324. doi: 10.1002/anie.200905978. [DOI] [PubMed] [Google Scholar]
- Pal P. P., Bae J. H., Azim M. K., Hess P., Friedrich R., Huber R., Moroder L., Budisa N. Biochemistry. 2005;44:3663. doi: 10.1021/bi0484825. [DOI] [PubMed] [Google Scholar]
- Pendley S. S., Yu Y. B., Cheatham III T. E. Proteins. 2009;74:612. doi: 10.1002/prot.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Ghirlanda G., Petka W. A., Nakajima T., DeGrado W. F., Tirrell D. A. Angew. Chem., Int. Ed. 2001;40:1494. [PubMed] [Google Scholar]
- Koksch B., Michel T., Kaptain P., Quaedflieg P., Broxterman Q. B. Tetrahedron: Asymmetry. 2004;15:1401. [Google Scholar]
- Gerling U. I. M., Salwiczek M., Cadicamo C. D., Erdbrink H., Czekelius C., Grage S. L., Wadhwani P., Ulrich A. S., Behrends M., Haufe G., Koksch B. Chem. Sci. 2014;5:819. [Google Scholar]
- Nyakatura E. K., Reimann O., Vagt T., Salwiczek M., Koksch B. RSC Adv. 2013;3:6319. [Google Scholar]
- Vagt T., Nyakatura E., Salwiczek M., Jackel C., Koksch B. Org. Biomol. Chem. 2010;8:1382. doi: 10.1039/b917205j. [DOI] [PubMed] [Google Scholar]
- Asante V., Mortier J., Wolber G., Koksch B. Amino Acids. 2014;46:2733. doi: 10.1007/s00726-014-1819-7. [DOI] [PubMed] [Google Scholar]
- Smits R., Koksch B. Curr. Top. Med. Chem. 2006;6:1483. doi: 10.2174/156802606777951055. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J., Steigemann W. Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem. 1975;31:238. [Google Scholar]
- Havel T. F., Wüthrich K. J. Mol. Biol. 1985;182:281. doi: 10.1016/0022-2836(85)90346-8. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M. Nature. 1977;267:585. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- Weissman J. S., Kim P. S. Nat. Struct. Mol. Biol. 1995;2:1123. doi: 10.1038/nsb1295-1123. [DOI] [PubMed] [Google Scholar]
- Ascenzi P., Bocedi A., Bolognesi M., Spallarossa A., Colletta M., De Cristofaro R., Menegatticenzi E. Curr. Protein Pept. Sci. 2003;4:231. doi: 10.2174/1389203033487180. [DOI] [PubMed] [Google Scholar]
- Schneeweiss S., Seeger J. D., Landon J., Walker A. M. N. Engl. J. Med. 2008;358:771. doi: 10.1056/NEJMoa0707571. [DOI] [PubMed] [Google Scholar]
- Huber R., Bode W., Kukla D., Kohl U., Ryan C. A. Biophys. Struct. Mech. 1975;1:189. doi: 10.1007/BF00535756. [DOI] [PubMed] [Google Scholar]
- Beckmann J., Mehlich A., Schröder W., Wenzel H. R., Tschesche H. Eur. J. Biochem. 1988;176:675. doi: 10.1111/j.1432-1033.1988.tb14330.x. [DOI] [PubMed] [Google Scholar]
- Krowarsch D., Dadlez M., Buczek O., Krokoszynska I., Smalås A. O., Otlewski J. J. Mol. Biol. 1999;289:175. doi: 10.1006/jmbi.1999.2757. [DOI] [PubMed] [Google Scholar]
- Lu W., Starovasnik M. A., Kent S. B. H. FEBS Lett. 1998;429:31. doi: 10.1016/s0014-5793(98)00547-x. [DOI] [PubMed] [Google Scholar]
- Sewing A., Hilvert D. Angew. Chem., Int. Ed. 2001;40:3395. doi: 10.1002/1521-3773(20010917)40:18<3395::aid-anie3395>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- von Eggelkraut-Gottanka R., Klose A., Beck-Sickinger A. G., Beyermann M. Tetrahedron Lett. 2003;44:3551. [Google Scholar]
- Weissman J. S., Kim P. S. Cell. 1992;71:841. doi: 10.1016/0092-8674(92)90559-u. [DOI] [PubMed] [Google Scholar]
- Ferrer M., Woodward C., Barany G. Int. J. Pept. Protein Res. 1992;40:194. [PubMed] [Google Scholar]
- Krowarsch D., Otlewski J. Protein Sci. 2001;10:715. doi: 10.1110/ps.38101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.-Y., Ballatore A. FEBS Lett. 2000;473:183. doi: 10.1016/s0014-5793(00)01515-5. [DOI] [PubMed] [Google Scholar]
- Makhatadze G. I., Kim K.-S., Woodward C., Privalov P. L. Protein Sci. 1993;2:2028. doi: 10.1002/pro.5560021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses E., Hinz H.-J. J. Mol. Biol. 1983;170:765. doi: 10.1016/s0022-2836(83)80130-2. [DOI] [PubMed] [Google Scholar]
- Monera O. D., Kay C. M., Hodges R. S. Protein Sci. 1994;3:1984. doi: 10.1002/pro.5560031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonov S. A., Salwiczek M., Anders G., Koksch B., Pisabarro M. T. J. Phys. Chem. B. 2009;113:16400. doi: 10.1021/jp906402b. [DOI] [PubMed] [Google Scholar]
- Wenzel H. R. and Tschesche H., in Peptides: Synthesis, Structures, and Applications, ed. B. Gutte, pp. 321–362, Academic Press, New York, 1995. [Google Scholar]
- Hanson W. M., Domek G. J., Horvath M. P., Goldenberg D. P. J. Mol. Biol. 2007;366:230. doi: 10.1016/j.jmb.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K., Yamada T., Kurihara K., Tamada T., Kuroki R., Tanaka I., Takahashi H., Niimura N. Acta Crystallogr., Sect. A: Found. Crystallogr. 2011;67:140. doi: 10.1107/S0907444910053382. [DOI] [PubMed] [Google Scholar]
- Helland R., Otlewski J., Sundheim O., Dadlez M., Smalås A. O. J. Mol. Biol. 1999;287:923. doi: 10.1006/jmbi.1999.2654. [DOI] [PubMed] [Google Scholar]
- Stegmann C. M., Seeliger D., Sheldrick G. M., de Groot B. L., Wahl M. C. Angew. Chem., Int. Ed. 2009;48:5207. doi: 10.1002/anie.200900481. [DOI] [PubMed] [Google Scholar]
- Kwon O.-H., Yoo T. H., Othon C. M., Van Deventer J. A., Tirrell D. A., Zewail A. H. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17101. doi: 10.1073/pnas.1011569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Acta Crystallogr., Sect. A: Found. Crystallogr. 2010;66:125. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C. J. Appl. Crystallogr. 2007;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G., Cowtan K. Acta Crystallogr., Sect. A: Found. Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B., Arendall, III W. B., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. Acta Crystallogr., Sect. A: Found. Crystallogr. 2010;66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L., The PyMOL Molecular Graphics System on World Wide Web, http://www.pymol.org, 2002.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.