Abstract
Viruses sense environmental cues (such as pH) to engage in membrane interactions for cell entry during infection but how non-enveloped viruses sense pH is largely undefined. Here, we report the structures — at high and low pH conditions — of bluetongue virus (BTV), which enters cells via a two-stage endosomal process. The receptor-binding protein VP2 possesses a zinc-finger and a conserved His866, which may function to maintain VP2 in a metastable state and to sense early-endosomal pH, respectively. The membrane penetration protein VP5 has three domains: dagger, unfurling, and anchoring. Notably, the β-meander motif of the anchoring domain contains a histidine cluster that could sense the late-endosomal pH and four putative membrane-interaction elements. Exposing BTV to low pH detaches VP2 and dramatically refolds the dagger and unfurling domains of VP5. Our biochemical and structure-guided mutagenesis studies support these coordinated pH-sensing mechanisms.
INTRODUCTION
Entry of viruses into host cells to establish infection is a highly coordinated process. The molecular and chemical details of this process are relatively clear for enveloped viruses, such as influenza viruses, human immunodeficiency viruses, herpes viruses, and flaviviruses1,2. By contrast, the mechanisms are not clear for cell entry of non-enveloped viruses, especially those with large complex capsids and multiple conformational stages.
Among large (i.e., >500 Å in diameter) non-enveloped viruses, those in the family Reoviridae are perhaps the most extensively studied. These viruses form the largest and arguably the most diverse family of dsRNA viruses, with capsids consisting of 1, 2, or 3 protein layers and have hosts ranging from plants to insects and humans. The host receptor-binding proteins of these viruses, such as the outer layer proteins of reovirus, rotavirus, bluetongue virus (BTV), and phytoreovirus are largely non-homologous. Some of these viruses enter host cells using two outer-layer proteins3,4. Infection by BTV, an arbovirus of agri-economic significance, is established when the viral core translocates across the endosomal membrane following virus uptake, a process performed by its two outer-layer proteins, VP2 and VP55. This process occurs in two distinctive stages in a coordinated, pH-sensitive manner: first—in the early endosome—VP2 senses pH change (6.5–6.0), detaches itself from VP5 and the virus; and second—in the late endosome—VP5 senses the lowered pH (~5.5) to gain membrane “fusion” activity5,6. However, the molecular details of this dynamic process and coordination of the two outer-layer proteins are unclear in part due to the lack of atomic structures of the outer-layer proteins of BTV.
To reveal how the two outer-layer proteins sense endosomal pH, how they coordinate during cell entry, and how VP5 acquires membrane-penetration activity, we determined the structures of BTV at both physiological and low-pH conditions using cryo electron microscopy (cryoEM), revealing dramatic conformational changes induced by low pH. Atomic modeling, biochemical analyses and structure-guided mutagenesis elucidate the mechanism of the coordinated cell entry process of BTV. This mechanism bears some similarities to that by some enveloped viruses and provides general insights into cell entry by large, non-enveloped viruses.
RESULTS
Overall structure
We recorded cryoEM images (Supplementary Fig. 1a) of intact BTV virions with a direct electron detector operated at super-resolution counting mode and obtained a 3.5 Å resolution structure (Fig. 1, Supplementary Fig. 1b, Supplementary Video 1) by single-particle analysis. The BTV virion contains an outer layer of 60 VP2 trimers and 120 VP5 trimers, a middle layer with 260 VP7 trimers, and an inner layer formed by 120 VP3 monomers (Fig. 1a). Each VP2 trimer binds atop four VP7 trimers (see below). Situated at a 6-coordinated position of the icosahedral lattice, each VP5 trimer bridges across the channel formed by six surrounding VP7 trimers (Fig. 1a and Supplementary Video 1). At 3.5 Å resolution, our cryoEM map resolves most amino acid side chains, permitting atomic modelling for both VP2 and VP5 proteins as detailed below (Fig. 1b,c, Supplementary Videos 2–6, Table 1). The structural features of the middle- and inner-layers, including amino-acid side chains (Supplementary Fig. 1c,d), match the atomic structures of core proteins VP3 and VP7 solved previously by X-ray crystallography7.
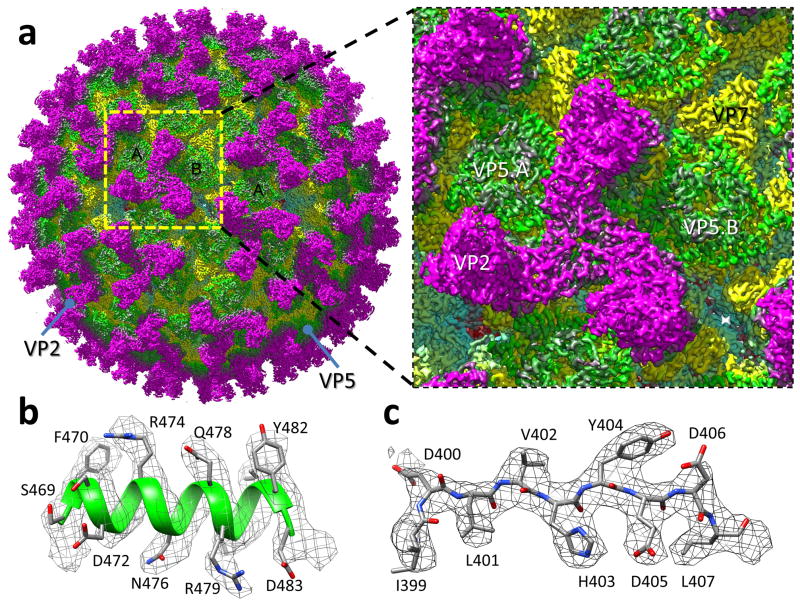
Figure 1. CryoEM reconstruction of the BTV virion.
(a) CryoEM density map of the BTV virion shown as radially-coloured surface representation, and a close-up view of the boxed area containing an asymmetric unit. (b,c) Close-up views of the α17 helix (b) and a β-strand of VP5 (c), showing side-chain density of amino-acid residues in a helix and a loop, respectively. The atomic model is shown as ribbons or sticks superimposed with the density (mesh).
Table 1.
Statistics of atomic model refinement
| VP2 | VP5 | |
|---|---|---|
| Resolution | 3.5Å | 3.5Å |
| Rwork (overall) | 0.18 (40-3.5Å) | 0.16 (40-3.5Å) |
| Rfree (overall) | 0.18 (40-3.5Å) | 0.14 (40-3.5Å) |
| Rwork (highest resolution zone) | 0.28 (4.0-3.5Å) | 0.24 (4.0-3.5Å) |
| Rfree (highest resolution zone) | 0.30 (4.0-3.5Å) | 0.21 (4.0-3.5Å) |
|
| ||
| Ramachandran plot values | ||
|
| ||
| Most favored | 89.54% | 88.66% |
| Generously allowed | 8.42% | 9.49% |
| Disallowed regions | 2.04% | 1.85% |
Structure of the receptor binding protein VP2
VP2 exists as triskelion-like trimers on the outermost layer of the particle. As shown in Figure 2a–c and Supplementary Video 2, each VP2 monomer can be divided into four domains: hub (M1-Y49, G121-C162 and K839-V961), hairpin (D50-V120), body (L163-K190, Y408-T838), and external tip (see below). The hub domain contains a 10-stranded lectin-like β-barrel flanked by three helices outside, as well as both N- and C-termini (denoted respectively by purple and cyan balls in Fig. 2b). Each VP2 trimer is formed through trimerization of the hub domains of three monomers and sits atop the jelly-roll domain of four VP7 trimers, one via its hub domain and three via the body domains of the three monomers (Fig. 1c and Supplementary Video 1). The interaction interface of the VP2 hub domain is highly conserved (central blue region in Fig. 2c), including H866 (91.6% identity in 25 serotypes), suggesting a possible role of the VP2-VP7 interface in VP2 detachment in the early endosome. The hairpin domain consists of a loop, two short α-helices, and a two-stranded β-sheet (green in Fig. 2b). The pyramid-shaped body domain has a β-sheet-rich apex and an α-helix-rich base (Fig. 2b). The apex of the pyramid connects to and interacts with the overlying tip domain.
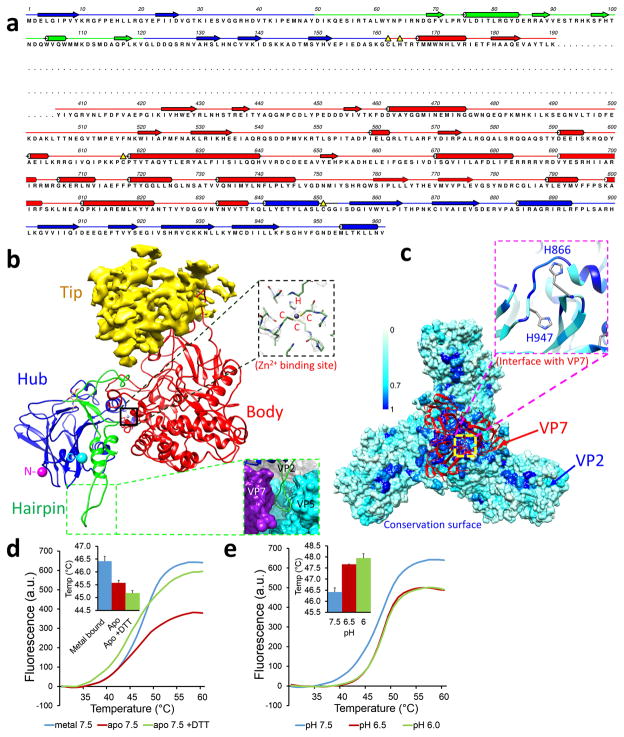
Figure 2. Structure of VP2.
(a) Sequence and secondary structure assignment of VP2. α-Helices are marked by cylinders, β-strands by arrows, loops by thin lines, and the flexible tip domain (P191-K405) by a dashed line. (b) Ribbon model of the VP2 monomer with its three resolved domains: body (red), hairpin (green), and hub (blue). The tip domain is shown as density (yellow). The zinc-finger motif in VP2, a putative Zn2+ binding site, is highlighted by the black box and its density view is shown in the upper inset, while the hairpin loop region interacting with VP7 and VP5 is shown in the lower inset. (c) Conservation surface of the VP2 trimer. The ribbon model (red) shows a VP7 trimer interacting with the hub domains of a VP2 trimer. The inset is a close-up view of the interface on VP2, showing two histidines (H866 and H947). Scale bar: level of conservation from complete (blue) to partial (cyan) to none (white). (d) Effects of divalent metal on the stability of purified VP2. Fluorescence intensities as a function of temperature were measured by differential scanning fluorimetry. Inset: comparison of melting temperatures (midpoints between the upper and lower levels), error bars represent s.d. of n = 3 technical repeats. (e) Similar to (d), but showing the effects of pH on the stability of purified VP2, error bars represent s.d. of n = 3 technical repeats.
The external tip domain contains at least four helices and a β-sheet (yellow density in Fig. 2b) but its density is weaker and at a lower resolution (5–10Å), suggesting flexibility. Consequently, this domain was not modelled and its sequence was deduced to be P191-I407 only by process of elimination. The exposed nature of this domain is consistent with its role as a key determinant of the host antibody response8–11.
We identified a cluster of four residues, CCCH (C162, C617 C851 and H164), that form a well-coordinated tetrahedron – a typical zinc-finger motif – at the interface of the hub and body domains. At the center of this CCCH cluster is a strong density unaccounted for by any amino acid residues (upper inset in Fig. 2b, Supplementary Video 3), which we interpret as a coordinated metal ion expected to be within a zinc-finger motif. Among physiologically available ions, only Zn2+ forms such tetrahedral geometry, and it commonly coordinates with Cysteine and Histidine residues12,13. To confirm that Zn2+ binds to VP2, we expressed and purified recombinant VP2 and subjected it to measurement by inductively-coupled plasma optical emission spectrometry (ICP-OES). This measurement shows an approximately threefold enrichment of Zn2+ in the presence of VP2, suggesting that Zn2+ indeed binds to VP2. Remarkably, the four residues of the CCCH zinc finger of VP2 are completely conserved among all 25 known BTV serotypes, while their VP2 sequences vary greatly, suggesting important functional roles for the zinc-finger.
Since zinc fingers in some proteins are known to control conformations13,14 and BTV VP2 does not bind nucleic acids, we speculate that the zinc-finger motif of VP2 might play a role in controlling conformational changes during cell entry. To measure the effect of Zn2+ binding on the conformation of VP2, we treated VP2 with chelex-100 to remove bound divalent ions. This Apo VP2 was destabilized when compared to untreated VP2, as indicated by the decrease in melting temperatures (Fig. 2d), suggesting that bound Zn2+ plays a stabilizing role. To prevent the formation of a disulfide bond between Cys of the CCCH motif after removal of the bound metal ion, this measurement was repeated in the presence of the reducing agent DTT. Indeed, additional reduction of melting temperature was observed (Fig. 2d), indicating further destabilization of VP2 in a reducing environment. These observations suggest that Zn2+ binding affects VP2 conformation.
Our observations above offer clues as to how VP2 detaches from viral cores in the early endosome5. VP2 detachment could result from disruption of the weak interaction involving the conserved H866 at the VP2-VP7 interface by high salt and low pH15 and perturbation to the zinc finger at low pH16,17. To verify that VP2 alone is pH-sensitive, we subjected purified VP2 to treatments of different pH conditions. The melting temperature of VP2 changed when the pH was shifted from neutral (7.5) to early endosomal pH (6.5 – 6.0), suggesting different physical properties of VP2 at neutral and endosomal pH conditions (Fig. 2e). Taken together, our structural and biochemical data point to a pH-sensing mechanism that involves the CCCH zinc finger, H866, and possibly other elements such as receptor-binding to enable VP2 detachment within the early endosome.
Structure of the penetration protein VP5 at high and low pH
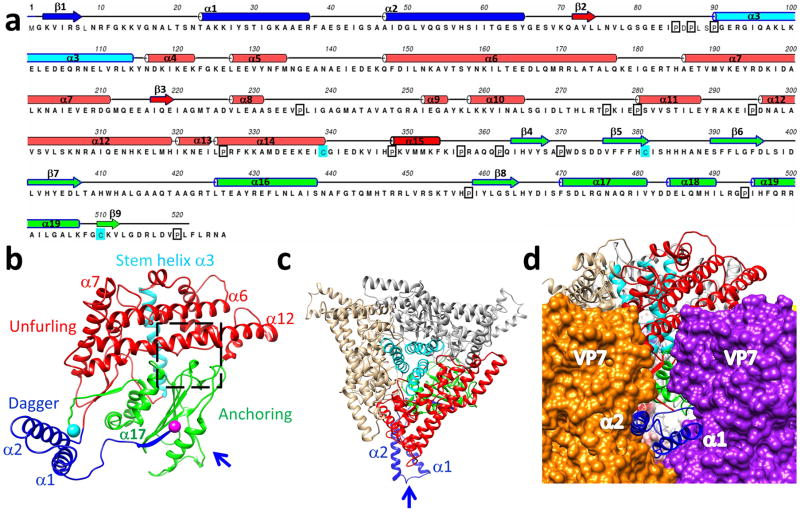
At pH8.8, 521 of the total 526 residues of VP5 were resolved in the 3.5-Å structure, and our atomic model of VP5 contains 19 α-helices and only 2 β-sheets (Fig. 3a-c, Supplementary Video 4). This atomic model confirms the α helix-rich nature of VP5, although the previous provisional sequence mapping18 to the observed helices was incorrect due to insufficient resolution to trace backbone and the differences of the secondary structure predictions in the previous report18. Unlike any known viral fusion and penetration proteins, VP5 has a near-rectangular shape and consists of three domains: dagger (M1-S68), unfurling (K69-F354), and anchoring (I355-A526) (Fig. 3b). The dagger domain is located at the N-terminus and is sequestered in the canyons between two adjacent VP7 trimers, where it is “hidden” from the external surface of the virion (Fig. 3d). The dagger domain interacts with adjacent VP7 molecules through charge complementarity (helix α1) and hydrophobic surfaces (helix α2). Nonetheless, the dagger domain is more flexible than other two domains because the density of this domain was only resolved in four of the six VP5 subunits within each asymmetric unit and we built an atomic model of this domain based on the best resolved subunit. N-terminal residues 1–22, previously shown to permeabilize membranes19, form a β-strand (K3-S7, β1 in Fig.3a,b) and a loop (L8-N22) at physiological pH. The unfurling domain is helix-rich, with two long horizontal helices (α6 & α12 in Fig. 3a,b; termed beam helices) and a stem helix (α3, cyan in Fig. 3b, Supplementary Video 4). Three stem helices in the middle of a VP5 trimer form a coiled coil (cyan helices in Fig. 3c). A small β-sheet was revealed in the unfurling domain with two parallel strands, which augments the third anti-parallel β-strand (C510-V512) (β9 in Fig.3a) from the anchoring domain. The anchoring domain is located in the C-terminal region and contains a highly conserved β-meander motif with 4 antiparallel β-strands (I364-L407) (β4–7 in Fig. 3a). The β-meander motif is not only sandwiched on both sides by four helices (Fig 3b), but also augmented by the fifth, the N-terminal β-strand (β1 in Fig.3a) (K3-R6), thus tethering the dagger domain (Fig 3b). Interestingly, the previously identified WHXL motif (W411-L414), responsible for membrane interactions20, is located on the extension of the β7 strand in the β-meander motif (see below).
Figure 3. Structure of VP5.
(a) Sequence and secondary structure assignment of VP5. α-Helices are shown as cylinders, β-strands as arrows, and loops as lines. (b) Ribbon model of the VP5 monomer colored by anchoring (green), unfurling (red), and dagger (blue) domains. The stem helix (cyan ribbon) of the unfurling domain, with the N- and C-termini indicated as purple and cyan balls respectively. (c) A ribbon model of a VP5 trimer as viewed from outside of the BTV capsid. The same color coding as in (b). The arrow indicates the view direction in (d). (d) The two helices (α1 & α2) of the dagger domain as viewed from along the arrow direction of (c), showing their interactions with two adjacent VP7 trimers.
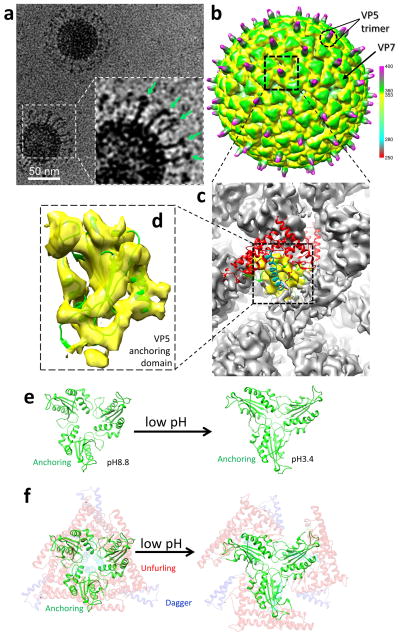
Membrane permeability activity of VP5 was inhibited by VP2 in an experiment using recombinant proteins of VP2 and VP5 engineered with membrane-anchoring tags6. Our atomic structure of the BTV virion reveals the interactions between VP2 and VP5 that might account for such inhibition. First, the loop of the VP2 hairpin domain contains His95 and interacts with the WHXL motif, a loop (352–357) and helix α7 of VP5 (lower inset in Fig. 2b). Second, the hub domain of VP2 interacts with helices α6, α7 and α12 of the VP5 unfurling domain (Fig. 3b). Therefore, we reason that the activation of VP5’s membrane permeability activity requires the initial removal of VP2. Indeed, cryoEM of BTV at low pH conditions (pH3.4 and 5.5) reveals that VP2 dissociates while VP5 remains attached to the VP7 layer but refolds substantially to form a new filamentous barb-like structure which protrudes from the remaining VP5 (Fig. 4a and Supplementary Fig. 1e,f). This barb-like structure is long (~150 Å) and flexible and thus cannot be resolved in the 3D density map at 9Å resolution (Fig. 4b). When the map was filtered to lower resolution (i.e., 15Å resolution, Fig. 4b), the base of the barb-like structure appears at the centre of the remaining VP5 trimer, suggesting that it is likely a filamentous trimer. Rigid body docking of the VP5 atomic model into the remaining VP5 density indicates that the anchoring domain remains attached to the VP7 layer while both the dagger and unfurling domains refold to form the filamentous structure (Fig. 4c,d). Comparing the orientations of the anchoring domains of the two structures indicates that low pH triggers a flower-like opening of the VP5 trimer, which is likely required to relieve restraints and create space to accommodate the simultaneous refolding of the dagger and unfurling domains (Fig. 4e,f).
Figure 4. Structure of VP5 at low pH.
(a) CryoEM image of the BTV virion in pH3.4, showing a dramatic conformational change of VP5. Unfurled domains are indicated by arrows in the inset. (b) cryoEM construction of the BTV virion at acidic pH filtered to 15Å resolution and displayed at threshold of 1.0σ, showing that VP2 is detached and VP5 has undergone large-scale conformation changes, leading to outward protrusion of the dagger and unfurling domains. The radial color scheme is the same as that in the Figure 1a. (c) Close-up view of the region within the dashed box in (b), showing the 9 Å resolution density. The ribbon model of the high-pH VP5 monomer is fitted on the VP5 density (grey and yellow) in the low-pH virion reconstruction. (d) Close-up view of the anchor domain region (yellow in c), showing that the ribbon model of the anchor domain matches the density of BTV in low pH. (e) Orientation change of the anchoring domains in a VP5 trimer when pH is lowered. Each anchoring domain is fitted in the 9Å structure of the BTV in low pH. (f) Same as (e) except that the dagger (faded blue ribbon) and unfurling domains (faded red and cyan ribbon) are also included to illustrate that the VP5 trimer would open like a flower when pH is lowered.
Identification of low pH sensor in VP5
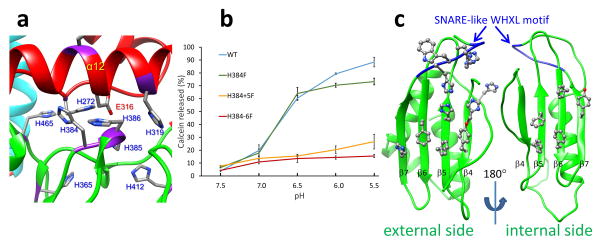
After detachment of the protective VP2, VP5 must sense the low pH environment of the late endosome with a hitherto unknown mechanism to trigger the dramatic refolding described above. We expect the pH sensor to be comprised of histidines since each VP5 monomer contains a total of 19 histidines. Remarkably, 13 of them localize to the anchoring domain, and 5 (H272, 319, 384, 385, and 386) are clustered closely at the interface between the β-meander motif of the anchoring domain and the beam helices of the unfurling domain (Fig. 5a, Supplementary Video 6). Three of these (H272, H319 & H386) are completely conserved among all 25 known BTV serotypes, indicating important roles for these histidines. In addition, a fully conserved Glu316 is also located at the interface, interacting with the histidine cluster (Fig. 5a, Supplementary Video 6). Protonation of these histidine residues at late endosomal pH would change their charge properties, thereby altering the interactions with each other as well as with the Glu316. Therefore, we expect that some or all of these conserved residues could function to sense pH changes in the late endosome during cell entry.
Figure 5. Membrane interaction elements of VP5.
(a) The histidine cluster of VP5. The fully conserved Glu316 is also highlighted. (b) Membrane permeability as measured by calcein release from liposomes as a function of pH for WT, and single- (H384F), double- (H384-5F) and triple- (H384-6F) mutants of the histidine His384–386 cluster of VP5. Error bars represent s.d. of n = 3 technical repeats. (c) External (viewed along the arrow in Figure 3b and internal sides of the core β-meander motif of the anchoring domain, showing a cluster of 5 and 7 aromatic residues on the internal and external side respectively. The SNARE-like WHXL motif (W411-L414) located on the β7 strand is shown in blue.
To confirm the role of the histidines at the interface, we mutated four Histidines to Phenylalanines (as serotype specific Phenylalanine substitutions are seen in this region suggesting this residue is tolerated) in two domains, including (1) the buried H272 in the unfurling domain (single mutation H272F); and (2) single (H384F), double (H384-5F) and triple (H384-6F) histidine(s) in the anchoring domain. Initially a single H272F mutation in the unfurling domain and a single H384F mutation in the anchoring domain were introduced into recombinant protein. The H272F mutation substantially reduced the expression level of VP5, suggesting that this residue plays a critical role in maintaining properly folded VP5. In contrast, H384F mutation was well tolerated. WT protein and the single H384F mutation were tested in a pore formation assay with synthetic liposomes, which mimic the composition of anionic lipids of the endosomal membrane. WT VP5 exhibited membrane disruption activity with maximum pore formation at pH5.5 and the effect in the single (H384F) mutation was only marginal (Fig. 5b). Alignment across serotypes of this histidine cluster displays single substitutions of Histidine to Phenylalanine in the 384 and 385 positions, indicating that a minimum of two Histidines in this region is required for sensing the low pH by VP5. Further, mutation of the conserved H386 in the replicating genome did not hinder the virus recovery by reverse genetics. These results led to the hypothesis that the net charge of this cluster may be a critical factor to VP5 function. Indeed, the pore formation activities for the double mutant (H384-5F, which still preserved the conserved H386) and the triple mutant (H384-6F) were reduced to only ~30% and ~17% of that of the WT VP5 at pH5.5 (Fig. 5b). Taken together, these observations suggest that the H384-386 cluster functions as a pH sensor of VP5 and that the histidine residues in the cluster likely act cooperatively. However, further analysis is required to pinpoint the specific role of each residue in the cluster.
The VP5 structure indicates that the residues H384-386 interact with conserved residues H272, H319 and Glu316 in the unfurling domain (Fig. 5a, Supplementary Video 6). The proximity of their locations implies that disruption of these interactions by protonation would lead to unfurling of VP5 to expose the dagger domain for membrane interaction. Our result of the single mutation H272F also indicates that the change of interactions in this region, either by protonation or by mutagenesis, would greatly impact the stability or conformation of the VP5 protein.
Membrane interaction elements in VP5
Our atomic structure of VP5 resolves the membrane interaction elements previously identified by bioinformatics and biochemical analyses as an N-terminal peptide (M1-S41) and a WHXL lipid interaction motif (W411-L414) usually found in SNARE proteins19,20. M1-S41 is a part of the dagger domain, which is inaccessible to the membrane at physiological pH, but refolds dramatically to project outward at low pH conditions (Fig. 4a,b, Supplementary Fig. 1e,f). The WHXL motif interacts with cholesterol-rich membrane rafts20 and is an extension of the β7 strand of the highly conserved β-meander motif (Fig. 5c, and Supplementary Fig. 2a). Moreover, the β-meander motif possesses three additional membrane-interaction elements that are usually found in bacterial outer-membrane β-barrel proteins21–23: a hydrophobic surface (Supplementary Fig. 2b,c) and two aromatic clusters (Fig. 5c). Indeed, prediction with BOCTOPUS24 identifies six transmembrane β-barrel strands in the anchoring domain, two of which lie in the β-meander motif (Supplementary Fig. 2d). The transmembrane β-strands from multiple copies of VP5 could join together to form a transmembrane β-barrel. Interestingly, rotavirus also contains more than one membrane-interacting element25. Nevertheless, the exact role of the anchoring domain and especially its β-meander motif in membrane interaction remains to be determined. One plausible role for the β-meander motif is to promote oligomerization of the unfurled VP5 in the late endosome to form pores, as observed in the self-assembly of other β-rich structures containing aromatic clusters26.
Interactions between VP2, VP5 and VP7 proteins
VP2 interacts with both VP7 and VP5 on the virion. A VP2 trimer attaches to four VP7 trimers through the bases of its hub, body, and hairpin domains. The bases of three hub domains of a VP2 trimer sitting on the jelly-roll domain of a VP7 trimer and two of the histidines (H866, H947) of VP2 are located at the interacting interface. In addition, the loop of the VP2 hairpin domain also interacts with the jelly-roll domain of the VP7 trimer (inset in Fig. 2b).
VP2 interacts with VP5 at two regions. First, the VP2 hub domain interacts with helices α6, α7, and α12 of the VP5 trimer (Fig. 3b). Second, the loop (A89-H95) of the VP2 hairpin domain interacts with the WHXL motif, a loop (352–357), and the α7 helix of an adjacent VP5 trimer. These relatively weak interactions are consistent with VP2’s facile disengagement during cell entry when VP2 must dissociate to permit the conformational change of VP5.
At physiological pH, VP5 interacts with VP7 extensively through all three of its domains, while at endosomal pH, VP5 interacts with VP7 only through the anchoring domain and its dagger and unfurling domains protrude out.
DISCUSSION
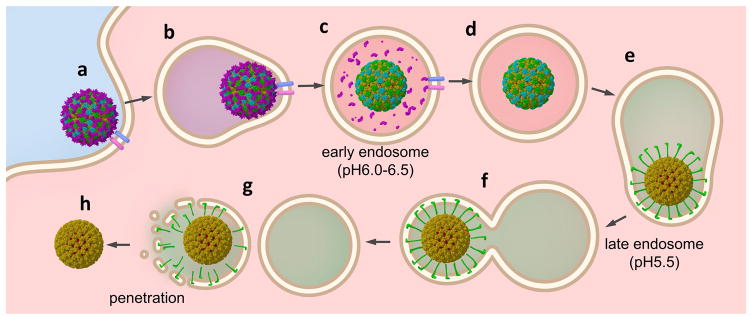
BTV responds to pH changes inside early and late endosomes in a coordinated manner to gain entry into the cytoplasm5. The structures of BTV at high (pH8.8) and low pH conditions (pH3.4 and 5.5) suggest that the highly conserved zinc-finger and His866 in VP2, as well as the His384–386 cluster in VP5, might act as the pH sensors to coordinate detachment of VP2 and subsequent unfurling of VP5. Our functional analysis confirmed the essential role of the His384–386 cluster of VP5 for its pH dependent membrane penetration activity, while His272 of VP5 is required for protein folding and/or structural stability. These data, together with previous biochemical observations, support a model of cell entry as depicted in Figure 6: Upon entrance into the early endosome (Fig. 6a,b), VP2 senses the local pH (6.5–6.0) and detaches from the virion (Fig. 6c); in the late endosome (~pH 5.5) and without VP2’s restraints, VP5 senses the lower pH and extends its unfurling and dagger domains (Fig. 6e) to interact with the membrane while remaining tethered to the viral core by its anchoring domain (Fig. 6e,f), resulting in a membrane-enclosed virus intermediate (Fig. 6f). We hypothesize that the membrane-bound VP5 undergoes further conformational changes to detach from the viral core, disrupt the membrane and consequently release the transcriptionally active core into the cytoplasm (Fig. 6g,h). Though our structural and functional data are consistent with this hypothetical model, the mechanisms of viral entry proposed in this model yet need to be established or refuted. For example, what factor(s) promotes the final detachment of the refolded VP5 from the viral core? What are the functions, if any, of the membrane interaction elements in the anchoring domain of VP5? Does the membrane penetration form a large pore across the membrane or simply disrupt it into fragments (Fig. 6g) in order to translocate the viral core? Answers to these questions should further advance our understanding of the mechanism of membrane penetration employed by non-enveloped viruses.
Figure 6. Schematic diagram of BTV cell entry.
(a) BTV virion binds receptors on the cell surface. (b) Endocytosis. (c) VP2 detaches from the viral core by sensing the local pH (6.0–6.5) in the early endosome. (d) Detached VP2 is likely digested by proteases. (e) VP5 unfurls to protrude the dagger and unfurling domains by sensing the local pH(~5.5) in the late endosome. (f) The refolded VP5 interacts with endosomal membrane. (g) VP5 penetrates the membrane to releases (h) the transcriptionally active viral core into the cytoplasm.
Many other dsRNA viruses, notably mammalian reovirus (MRV) and rotavirus, use one protein for receptor binding and another for membrane penetration; their membrane interaction proteins are protected and must be exposed to cellular factors for membrane penetration4,27,28. Though the structure of the “exposed” MRV penetration protein is unknown, the “exposed” penetration proteins of BTV and rotavirus both are filamentous4,18 and their final detachment from the viral cores likely involves additional triggers29. Despite these similarities, the atomic structures of the receptor-binding and membrane penetration proteins of BTV at high pH reported here bear no similarities to those in other dsRNA viruses4,27, thus representing an extraordinary example of mechanistic diversity among members of this family of viruses, perhaps as a result of arbovirus’ unique route of transmission through arthropod vectors. Other non-enveloped viruses employ a variety of strategies for cell entry with no similarity to the BTV mechanism. For example, acidification within endosomes triggers small RNA viruses, such as picornaviruses, to undergo “molecular plate tectonic” movement to expose myristylated internal proteins30. Entry by non-enveloped DNA viruses may involve partial removal of the penton unit, as in the case of adenoviruses31, or interactions with Golgi and ER components within the cell, as in the case of papillomaviruses32.
Low-pH triggered cell entry processes are well understood for enveloped viruses, as exemplified by the influenza and dengue viruses1,2. Among non-enveloped viruses, the mechanism by which BTV enters cells (Fig. 6) bears the greatest level of similarities to that utilized by enveloped viruses. In a sense, VP5 is similar to some classical viral fusion proteins, such as the influenza HA2 protein, in that these proteins employ Histidine residues as pH sensors and undergo a dramatic conformational change at the late endosomal pH2,33. Along the landscape of cell entry strategies employed by non-enveloped and enveloped viruses, BTV, along with rotavirus, is therefore situated near enveloped viruses; as such, the results presented here should be of general significance to cell entry by both non-enveloped and enveloped viruses.
Online Methods
CryoEM imaging
Virions were purified as previously described18. To prepare cryoEM grids, we applied 2μl of purified BTV sample (pH 8.8) to thin continuous carbon films on lacey grids (Ted Pella Inc.) for 3 minutes, blotted for 9 seconds in an FEI vitrobot with 100% humidity, and then plunged into liquid ethane. CryoEM images were collected at liquid nitrogen temperature in an FEI Titan Krios cryo electron microscope operated at 300kV using parallel illumination. Before data collection, the microscope was carefully aligned, and beam tilt was minimized by coma-free alignment. Images were recorded on a Gatan K2 camera with the super-resolution mode at a magnification of 14,000x. The magnification was calibrated as 24,140x using a catalase crystal sample, giving a pixel size of 1.036Å/pixel on specimen. The dose rate of the electron beam was set to ~9 counts/physical-pix·s on camera and the corresponding dose reading on the electron microscope is ~2.6 e−/Å2·s on specimen due to the miss counted electrons by the K2 camera. The image stacks were recorded at 2 frames/sec for 15 seconds. After drift correction with the UCSF software34, the first 15 frames of each image stack were merged to generate a final image with a total dose of ~20e−/Å2 on specimen.
Data processing for BTV structure at physiological pH
From a total of 3309 recorded micrographs, 1630 were selected and 12,513 images of individual particles were boxed for image processing. The under focus values of these micrographs were determined to range between 0.8μm to 4.5μm using using CTFFIND35. The 2x binned data set was used in the global search while the original data set was used in the local refinement. The orientation and center parameters of each of the 12,513 particle images were first determined by a global search by maximizing the cross-correlation coefficient of the particle image with projections generated from the input density model. After each cycle, three-dimensional (3D) reconstructions were carried out using a GPU-based reconstruction program, G3D36, and the effective resolution was estimated and used as a resolution cutoff in the next alignment cycle until no further improvement could be obtained. Then, we refined these orientation and center parameters iteratively until no further improvement could be obtained. The final map was reconstructed using the top 5,008 (40%) of the 12,513 particles based on their phase residuals using Frealign37. The quality of the maps of the receptor binding protein VP2 and the membrane penetration protein VP5 were further improved by averaging the three VP2 and six VP5 monomers in each asymmetric unit38.
The effective resolution of the whole virion is estimated to be ~3.5 Å based on FSC (≥0.143), and the resolution for the averaged VP2 and VP5 densities is estimated to be better than 3.5 Å based on the R-factor criterion–the R factor given by Phenix 39 reaches 0.5 at 3.5 Å for both VP2 and VP5 densities (Table 1). These estimated resolutions are consistent with the observed structural features of the density maps (Fig. 2 and Supplementary Videos 2–6). The capsid and averaged maps were filtered to the spatial frequency of 1/(3.4 Å) and sharpened with a reverse B-factor of −150Å2. This B-factor was chosen with a trial-and-error method based on optimization of noise level, backbone density continuity, and emergence of side-chain densities. Visualization and segmentation of density maps were done with UCSF Chimera40.
CryoEM and data processing of BTV at low pH
Virions were purified as described18. To prepare cryoEM grids, 2μl purified BTV sample (pH8.8) was applied to thin continuous carbon films. After 3 minutes, the sample was blotted, and 2μl citrate buffer (pH3.4 or 5.5) was immediately applied to the grid for 30 seconds. The grid was blotted for 9 seconds in an FEI Vitrobot with 100% humidity and then plunged into liquid ethane. CryoEM images were collected in an FEI Titan Krios cryo electron microscope operated at 300kV. For the pH3.4 condition, a total of 350 images were recorded on a Gatan 4k x 4k CCD at a calibrated magnification of 79,370x, corresponding to a pixel size of 1.89Å/pix on the specimen. A total of 7474 particles were selected for data processing as described above, and the final 3D map was reconstructed from 4503 particles at a resolution of 9.0Å (FSC=0.143). For the pH5.5 condition, 159 particle images recorded on a Gatan K2 camera were used for a reconstruction at 25Å resolution (FSC=0.143), which appears identical to that of the above pH3.4 structure filtered to the same resolution.
Atomic modeling and model refinement
Based on the averaged density maps of VP2 and VP5, we first built initial Cα and full atom models for VP2 and VP5 with Coot41 without referring to any existing models of other proteins. The initial full atom models were regularized by constraining Ramachandran geometry and secondary structures in Coot41.
These initial full atom models were iteratively refined using structural information of both amplitude and phase (from Fourier transformation of the cryoEM maps) in the following three steps: The first step (automatic) was performed in Phenix39 using Ramachandran restraint. The second step (automatic) was also performed in Phenix to regularize the new model. To regularize the model, hydrogens were added to all atoms of the model from the last refinement, followed by regularization and removal of hydrogens. The latest models were refined iteratively until no further improvement was apparent based on both Ramachandran geometry and R-factors. Then in the third (manual) step, amino acid residues with invalid Ramachandran backbone geometries were identified and manually corrected in Coot. This process of automatic and manual model refinement steps was iterated until no further improvement on both Ramachandran geometry and R-factors was evident.
Protein expression
Both VP2 and VP5 proteins were expressed using a recombinant baculovirus system in Sf9 cells which were purchased from Invitrogen, Cat No. 11496-015 lot No. 1296885 and have been checked for mycoplasma. For both proteins, a 200ml culture of 2 x 106 cells/ml was infected at an MOI of 10. Cells were harvested by centrifugation at 3000 g 48 (VP5) and 65 hours post- infection (VP2). Recombinant VP2 was expressed as an N-terminally tagged HisX6 GST fusion protein with a glycine-serine linker and a TEV cleavage site. Recombinant VP5 was expressed similarly except with an N-terminal HisX6 MBP fusion. Cells were lysed by dounce homogenization (20 strokes) in a lysis buffer (20mM Tris, 150mM NaCl, 20mM imidazole 1% Triton X-100 pH 8.5). Lysate was subsequently clarified by centrifugation at 15000g for 30 minutes. Soluble lysate was purified using an Äkta explorer FPLC unit (GE Healthcare), first utilizing immobilized metal affinity chromatography (IMAC) with a 5ml HisTrap HP column (GE Healthcare) and second, an affinity chromatography using a 1ml GSTrap HP column (GE Healthcare). VP5 and mutants were purified in an analogous manner with a 1ml MBPTrap HP column (GE Healthcare) utilized in the second affinity step. Eluted proteins were concentrated to 200μl and cleaved with 20μg of TEV protease (Sigma Aldrich) overnight at 4°C. Cleaved tags and proteases were removed using IMAC by passing the cleaved protein suspension through a 1ml HisTrap HP column (GE Healthcare) three times. Mutant proteins were produced by site- directed mutagenesis of WT expression constructs.
Differential scanning fluorimetry (DSF)
DSF was performed using an MX3005P Q-PCR system (Aligent technologies) in a manner similar to Neisen et al11. Briefly purified VP2 was reconstituted in DSF buffer (20mM Tris 150mM NaCl pH 7.5) and utilized at a final concentration of 0.2mg/ml with 5x SYPRO orange and additional additives. If stated, temperature was ramped from 25–95 °C at 45 seconds/°C. For metal-binding experiments, VP2 was left untreated (metal bound) or incubated with chelex-100 resin (Sigma Aldrich) for 1 hour (Apo). For pH experiments, protein was acidified to the stated pHs using a pre-determined titration of 0.1 N HCl. Raw data was fitted to a Boltzmann distribution using Prism4 (GraphPad) and the derived melting midpoints (Tm) are plotted.
Calcein loaded liposome production
5mg total lipid with a ratio of 4:2:1 PC:PE:PS (Avanti polar lipids) in chloroform was dried under vacuum to give a solvent-free film. This was hydrated in 1ml of Calcein buffer (50mM calcein, 100mM NaCl, 10mM Na2HPO4 2mM KH2PO4) and freeze-thawed three times. The lipid suspension was then extruded 10 times through a 1μm polycarbonate membrane filter (Whatman) using a Mini Extruder (Avanti polar lipids). Un-encapsulated calcein was removed from the extruded suspension by size exclusion chromatography using sephadex G-50 resin (Sigma Aldrich), while PBS pH 7.5 (liposome buffer) was used as the aqueous phase buffer. Liposomes were stored at 4 °C and used within 1 week of creation.
Pore formation assay
Purified WT and H384-6F VP5 were reconstituted in PBS pH 7.5 containing 1μg/ml of anti gp64 monoclonal antibody B12D5(Ref. 6) (liposme buffer). Calcein-loaded liposomes and VP5 protein were mixed in a flat black 96 well plate (Griener) and incubated at room temperature for 10 minutes at a final concentration of 0.1mg/ml VP5 and 0.1mg/ml total lipid for liposomes. Mixtures were then acidified to the stated pHs using a pre-determined titration of 0.1N HCl and incubated at 37 °C for 20 minutes. Fluorescence was then measured with a SpectraMax® M5 plate reader (Molecular Devices) using top read mode with an Ex/Em 485/535 nm. Percentage calcein release was calculated using the formula R% = (R-R0)/(R100-R0) where R% is percentage release, R is the measured fluorescence, R0 is the measured fluorescence of liposomes acidified with buffer alone and R100 the fluorescence of liposomes with buffer containing 1% triton X-100.
Supplementary Material
Acknowledgments
We thank S. Shivakoti and J. Sun for assistance in cell culture and virus isolation, M. Boyce for advice in sample purification, and I. Jones, L. Nguyen and L. Wang for reading the manuscript. This project is supported in part by grants from the US National Institutes of Health (AI094386 to Z.H.Z.) and the Welcome Trust, UK (to P.R.). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from the US National Institutes of Health (1S10OD018111) and the US National Science Foundation (DBI-1338135).
Footnotes
Accession codes The atomic models and cryoEM density map are deposited to PDB and EMDB with accession number 3J9D (VP2), 3J9E (VP5), EMDB-6444 (whole map, pH8.8), EMDB-6445 (whole map, pH3.4), EMDB-6239 (VP2 averaged map, pH8.8) and EMDB-6240 (VP5 averaged map, pH8.8), respectively.
Author Contributions: ZHZ, PR & XZ designed the experiments. AP expressed proteins and performed the mutagenesis and biochemical experiments. CC and XY grew and isolated viruses. XZ collected and analyzed the cryoEM data and built the atomic models. ZHZ, XZ & PR interpreted the structures and wrote the paper.
The authors declare no competing financial interests.
References
- 1.Ge P, Zhou ZH. Chaperone fusion proteins aid entropy-driven maturation of class II viral fusion proteins. Trends in microbiology. 2014;22:100–106. doi: 10.1016/j.tim.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–8. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nibert ML, Schiff LA, Field BN. Reoviruses and their replication. In: Fields BN, et al., editors. Fields Virology. Vol. 2. Lippincott-Raven; Philadelphia: 2001. pp. 1679–1728. [Google Scholar]
- 4.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO journal. 2011;30:408–16. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Bhattacharya B, Ward TH, Roy P. Trafficking of Bluetongue virus visualized by recovery of tetracysteine-tagged virion particles. Journal of virology. 2014 doi: 10.1128/JVI.01815-14. 01815–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forzan M, Wirblich C, Roy P. A capsid protein of nonenveloped Bluetongue virus exhibits membrane fusion activity. Proc Natl Acad Sci U S A. 2004;101:2100–5. doi: 10.1073/pnas.0306448101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimes JM, et al. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–8. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 8.DeMaula CD, Bonneau KR, MacLachlan NJ. Changes in the outer capsid proteins of bluetongue virus serotype ten that abrogate neutralization by monoclonal antibodies. Virus research. 2000;67:59–66. doi: 10.1016/s0168-1702(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 9.Nunes SF, et al. A Synthetic Biology Approach for a Vaccine Platform against Known and Newly Emerging Serotypes of Bluetongue Virus. Journal of virology. 2014;88:12222–12232. doi: 10.1128/JVI.02183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YY, Johnson TM, Mecham JO, Tam JP, Li JKK. Epitopic mapping of linear and conformation-dependent antigenic determinants on GP5 of five US bluetongue viruses. Virology. 1992;188:530–536. doi: 10.1016/0042-6822(92)90507-l. [DOI] [PubMed] [Google Scholar]
- 11.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 12.Roe RR, Pang YP. Zinc’s Exclusive Tetrahedral Coordination Governed by Its Electronic Structure. Molecular modeling annual. 1999;5:134–140. [Google Scholar]
- 13.Hanas JS, Larabee JL, Hocker JR. Zinc Finger Proteins. Springer; 2005. Zinc finger interactions with metals and other small molecules; pp. 39–46. [Google Scholar]
- 14.Hanas JS, Duke AL, Gaskins CJ. Conformational states of Xenopus transcription factor IIIA. Biochemistry. 1989;28:4083–4088. doi: 10.1021/bi00435a068. [DOI] [PubMed] [Google Scholar]
- 15.Huismans H, van der Walt NT, Cloete M, Erasmus BJ. Isolation of a capsid protein of bluetongue virus that induces a protective immune response in sheep. Virology. 1987;157:172–9. doi: 10.1016/0042-6822(87)90326-6. [DOI] [PubMed] [Google Scholar]
- 16.Esperante SA, et al. Fine Modulation of the Respiratory Syncytial Virus M2–1 Protein Quaternary Structure by Reversible Zinc Removal from Its Cys3-His1 Motif. Biochemistry. 2013;52:6779–6789. doi: 10.1021/bi401029q. [DOI] [PubMed] [Google Scholar]
- 17.Esperante SA, Chemes LB, Sánchez IE, de Prat-Gay G. The Respiratory Syncytial Virus Transcription Antiterminator M2–1 Is a Highly Stable, Zinc Binding Tetramer with Strong pH-Dependent Dissociation and a Monomeric Unfolding Intermediate. Biochemistry. 2011;50:8529–8539. doi: 10.1021/bi200661k. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, et al. Bluetongue virus coat protein VP2 contains sialic acid-binding domains, and VP5 resembles enveloped virus fusion proteins. Proc Natl Acad Sci U S A. 2010;107:6292–7. doi: 10.1073/pnas.0913403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassan SH, Wirblich C, Forzan M, Roy P. Expression and functional characterization of bluetongue virus VP5 protein: role in cellular permeabilization. J Virol. 2001;75:8356–67. doi: 10.1128/JVI.75.18.8356-8367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharya B, Roy P. Bluetongue virus outer capsid protein VP5 interacts with membrane lipid rafts via a SNARE domain. J Virol. 2008;82:10600–12. doi: 10.1128/JVI.01274-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selkrig J, Leyton DL, Webb CT, Lithgow T. Assembly of beta-barrel proteins into bacterial outer membranes. Biochim Biophys Acta. 2014;1843:1542–50. doi: 10.1016/j.bbamcr.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Wimley WC. The versatile beta-barrel membrane protein. Current opinion in structural biology. 2003;13:404–11. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 23.Hong H, Park S, Jimenez RH, Rinehart D, Tamm LK. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. Journal of the American Chemical Society. 2007;129:8320–7. doi: 10.1021/ja068849o. [DOI] [PubMed] [Google Scholar]
- 24.Hayat S, Elofsson A. BOCTOPUS: improved topology prediction of transmembrane beta barrel proteins. Bioinformatics. 2012;28:516–22. doi: 10.1093/bioinformatics/btr710. [DOI] [PubMed] [Google Scholar]
- 25.Elaid S, et al. A peptide derived from the rotavirus outer capsid protein VP7 permeabilizes artificial membranes. Biochim Biophys Acta. 2014;1838:2026–35. doi: 10.1016/j.bbamem.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Biancalana M, Makabe K, Koide A, Koide S. Aromatic cross-strand ladders control the structure and stability of β-rich peptide self-assembly mimics. Journal of molecular biology. 2008;383:205–213. doi: 10.1016/j.jmb.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liemann S, Chandran K, Baker TS, Nibert ML, Harrison SC. Structure of the reovirus membrane-penetration protein, Mu1, in a complex with is protector protein, Sigma3. Cell. 2002;108:283–95. doi: 10.1016/s0092-8674(02)00612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 Å cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–82. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoder JD, et al. VP5* rearranges when rotavirus uncoats. Journal of Virology. 2009;83:11372–7. doi: 10.1128/JVI.01228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergelson J, Coyne C. Picornavirus Entry. In: Pöhlmann S, Simmons G, editors. Viral Entry into Host Cells. Vol. 790. Springer; New York: 2013. pp. 24–41. [Google Scholar]
- 31.Suomalainen M, Greber UF. Uncoating of non-enveloped viruses. Current opinion in virology. 2013;3:27–33. doi: 10.1016/j.coviro.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 32.Day PM, Schelhaas M. Concepts of papillomavirus entry into host cells. Current opinion in virology. 2014;4:24–31. doi: 10.1016/j.coviro.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, et al. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat Struct Mol Biol. 2013;20:105–10. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–90. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Zhang X, Zhou ZH. Low cost, high performance GPU computing solution for atomic resolution cryoEM single-particle reconstruction. Journal of Structural Biology. 2010;172:400–6. doi: 10.1016/j.jsb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.