Abstract
Hepatitis C virus (HCV) can infect naïve cells via entry of “cell-free” extracellular virus or direct “cell-to-cell” transmission. Here, we describe an assay for detecting HCV cell-to-cell transmission, using a non-growing cell culture system that avoids confounding effects of cell growth. The assay consists of infecting a small number of cells in a confluent monolayer and then blocking subsequent cell-free extracellular virions with a neutralizing antibody such that only cell-to-cell transmission may occur. Under these conditions, incubation at 37 °C results in the formation of infected cell foci. The extent of cell-to-cell spread can then be determined by counting the number of cells in each focus. The assay may be modified to assess the effects of inhibitors and/or specific cellular genes on cell-to-cell spread of HCV.
Part I. Main protocol: HCV cell-to-cell spread in non-dividing Huh7 cell cultures
Materials and Reagents
Huh7 cells (from Dr. Francis Chisari, The Scripps Research Institute, La Jolla, CA) (Zhong et al., 2005)
-
JFH-1 HCVcc (generated as previously described and quantified by limiting dilution titer assay) (Yu and Uprichard, 2010; Knipe and Howley, 2007)
Note: Other infectious HCVcc clones can be used as well.
Dulbecco’s modified Eagle’s medium (DMEM) (Mediatech, catalog number: MT-10-013-CV)
Fetal bovine serum (FBS) (Hyclone, catalog number: SH30910.03)
Penicillin, streptomycin, L-glutamine (Mediatech, catalog number: MT-30-009-CI)
Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D4540)
1× PBS (Mediatech, catalog number: MT-21-030-CV)
Hydrogen peroxide (H2O2) (Thermo Fisher Scientific, catalog number: H325)
AEC detection substrate (BD Biosciences, catalog number: 551015)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Ezetimibe (Sequoia Research Products, catalog number: SRP04000e) prepared in DMSO at 20 mM
Rabbit anti-human CLDN1 polyclonal antibody (Abcam, catalog number: ab63070)
Rabbit anti-human NPC1L1 polyclonal antibody (Santa Cruz, catalog number: sc-67236)
Human anti-HCV E2 monoclonal antibody MAb AR3A (from Dr. Mansun Law, The Scripps Research Institute, La Jolla, CA) (Law et al., 2008)
Mouse anti-HCV NS5A 9E10 monoclonal antibody (from Dr. Charles Rice, Rockefeller University, New York, NY) (Lindenbach et al., 2005)
HRP-conjugated goat anti-human (Thermo Fisher Scientific, catalog number: 31410), goat anti-mouse (Thermo Fisher Scientific, catalog number: 31430), and goat anti-rabbit (Thermo Fisher Scientific, catalog number: 31460)
Mouse IgG (Santa Cruz, catalog number: sc-2025), rabbit IgG (Santa Cruz, catalog number: sc-2027)
Paraformaldehyde (PFA) (Sigma-Aldrich, catalog number: P6148)
Triton X-100 (Sigma-Aldrich, catalog number: T-8787)
Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A9647)
Lipofectamine RNAiMAX transfection reagent (Life Technologies, catalog number: 13778)
OptiMEM (Life Technologies, Invitrogen™, catalog number: 31985-070)
50 ml centrifuge tubes (Dot Scientific, catalog number: 451-PG)
Complete DMEM (cDMEM) (see Recipes)
Equipment
96-well BioCoat collagen-coated tissue culture plates (Corning, catalog number: 356698)
BioCoat collagen-coated T75 cm2 flask (Corning, catalog number: 356485)
Inverted microscope
Hemocytometer
37 °C, 5% CO2 cell culture incubator
Table top centrifuge (e.g. Sorvall, model: T6000D or Eppendorf, model: 5810 R)
Procedure
- Preparation of non-growing Huh7 cell cultures
- Seed 6,000 Huh7 cells/well in BioCoat collagen-coated 96-well plate in 200 µl/well 10% FBS cDMEM.
- Incubate the plate at 37 °C in 5% CO2 cell culture incubator overnight or until the cells reach 95%–99% confluence.
- Aspirate out or decant plating media and replace with 200 µl/well of 10% FBS cDMEM containing 1% DMSO and continue to incubate cells at 37 °C in 5% CO2 cell culture incubator for 10 days changing the 1% DMSO, 10% FBS cDMEM every other day (media changes should be performed slowly with the pipette tip angled towards the walls of the wells). During the incubation period the cells remain metabolically active, but stop dividing (Sainz and Chisari, 2006). By day 10, each well contains approximately 65,000 cells.
- Initiation of infection
- Dilute HCVcc in 2% FBS cDMEM to 50–100 foci forming units (ffu)/100 µl.
- Add 100 µl of the virus inoculum to each well and incubate at 37 °C in 5% CO2 cell culture incubator overnight (~16 h) to allow initial virus entry.
-
Blocking subsequent “cell-free” extracellular virus entry
This step should be performed no later than 18 h post inoculation to prevent subsequent infection of cells by progeny virus released into the medium from the first round of infected cells.- Remove viral inoculum at 16 h post-infection and rinse gently with 1× PBS.
- Add 10% FBS, 1% DMSO cDMEM containing 10 µg/ml of the HCV E2 antibody MAb AR3A to each well in a total volume of 150 µl /well to neutralize cell-free spread by HCVcc (Barretto et al., 2014; Timpe et al., 2008) (additional factor-specific inhibitors can be added to parallel samples at this point to assess their effect on HCV cell-to-cell spread; see section F below).
- Continue to incubate cultures at 37 °C in 5% CO2 cell culture incubator to allow time for cell-to-cell spread of HCV to occur (e.g. 48– 72 h post-infection).
- Before proceeding to Section D, collect all the culture medium from the sample wells and transfer it to a new 96-well plate for analysis of neutralization as described in section E below (this can be assayed immediately or stored at −80 °C until assay can be performed).
-
Visualizing HCV cell-to-cell spread via immunostaining
Under these conditions, multicellular HCV foci can only be formed by cell-to-cell spread. Hence, the number of HCV-positive cells in individual HCV foci can be used as a readout for cell-to-cell transmission. Here immunostaining is described to visualizing HCV infected cells, but other methods can be used (e.g. Supplemental protocol A).- Fix cells by adding an equal volume of 4% PFA to each well for a final concentration is 2% and incubate at RT for 25 min.
- Decant and rinse the cells three times with 1× PBS.
- Block endogenous peroxidases by incubating with 100 µl/well ice-cold 1× PBS containing 0.3% (v/v) hydrogen peroxide for 5 min at room temperature (RT).
- Decant and rinse 3 times with 1× PBS.
- Permeabilize and block cells for 1 h with 100 µl/well 1× PBS containing 0.5% (v/v) Triton X-100, 3% (w/v) BSA and 10% (v/v) FBS.
- Aspirate off the blocking solution.
- Immediately add 50 µl/well of appropriate HCV-specific primary antibody diluted in 1× PBS containing 0.5% (v/v) Triton X-100 and 3% (w/v) BSA for 1 h at RT. (e.g. The human anti-HCV E2 MAb AR3A can be used at 2.3 µg/ml or the mouse polyclonal serum anti-HCV NS5A 9E10 can be used at a 1:500 dilution.)
- Decant or aspirate off the primary antibody and rinse cells with 1× PBS three times.
- Incubate the cells with an appropriate HRP-conjugated secondary antibody (50 µl/well) for one hour at RT (e.g. we detect bound MAb AR3A with a 1:1,000 dilution of HRP-conjugated anti-human antibody and 9E10 with a 1: 500 dilution of HRP-conjugated anti-mouse).
- Rinse the cells 3 times with 1× PBS.
- Incubate cells with 30 µl/well AEC detection substrate for 30 min at RT.
- Wash cells 3 times with dH2O and add 150 µl/well of a solution of dH2O: glycerol (1:1) for storage. The plates may be stored at 4 °C.
- Quantify and photograph foci using an appropriate microscope. The number of HCV-positive cells per foci is a read-out of HCV cell-to-cell spread (see Figure 1).
-
Neutralization control assay
To ensure that cell-free virus in the medium is neutralized during the assay, the presence of infectious cell-free virus in the medium of experimental cultures should be assayed. These samples are harvested at the end of the spread assay as described in section C4).- Preparation of Huh7 cell cultures.
- Seed 4,000 Huh7 cells/well in 96-well cell culture plate in 200 µl/well 10% FBS cDMEM.
- Incubate the plate at 37 °C in 5% CO2 cell culture incubator overnight.
- Aspirate out plating media and transfer the media samples (~200 µl) onto naïve Huh7 cell monolayers.
- At 24h aspirate off viral inoculum and add 200 µl/well fresh 10% FBS cDMEM.
- At 72 h post-inoculation, fix the cells and perform immunostaining to detect HCV-infected cells (as above).
-
Assess the effect of different factors on HCV cell-to-cell spread
Aside from monitoring the kinetics of HCV cell-to-cell spread, this assay can be used to identify cellular factors involved in or antivirals that inhibit this step of the viral lifecycle.-
Use of Inhibitors.Inhibitors to specific factors or antivirals with unknown mechanisms of action can be added when cell-free virus entry is blocked at step C2. In this case, appropriate controls should also be included as well:
- “Mock” treated negative control samples (e.g. diluent or IgG control) should be included to control for non-specific inhibition.
-
As a positive control for inhibition of cell-to-cell spread, parallel cultures can be treated with an inhibitor that blocks a factor known to be required for HCV cell-to-cell spread (e.g. 20 µg/ml anti-Claudin antibody, 30 µg/ml anti-NPC1L1 antibody or 30 µM of the NPC1L1 inhibitor, Ezetimibe). In the absence of cell division the foci formed in the presence of these HCV spread inhibitors are limited to 1–2 cells.Note: When performing this assay in actively dividing cells (Supplemental protocol B), these controls are particularly helpful in determining the number of cell divisions that occurred during the assay, i.e. the size of the foci in these control samples provide an indication of the extent to which cell division may have contributed to in increased foci size.
-
siRNA knockdown.RNAi is an alternative method for assessing the involvement of cellular genes in HCV cell-to-cell spread. In this case, the assay is carried out as described above after silencing the gene of interest (see Supplemental protocol C). In this case, the following controls should be included:
- To monitor and control for non-specific effects of the transfection.
- Untreated and mock transfected control samples.
- Non-specific siRNA transfected control samples (e.g. siScramble or siGFP).
- As a positive control for inhibition of cell-to-cell spread parallel cultures can be transfected with siRNA targeting a factor known to be required for HCV cell-to-cell spread (e.g. Claudin 1 or NPC1L1).
- During the spread assay, the expression level of the siRNA-targeted gene should be monitored in parallel samples (e.g. RT-qPCR and/or western blot, and/or functional assays, if available).
Note: If silencing begins prior to infection initiation partial inhibition of infection may result in reduced numbers of foci being formed, however the readout for cell-to-cell spread is foci size not foci number thus this technical issue should not prevent analysis of cell-to-cell spread unless the initial infection is blocked completely.
-
-
Example results and Interpretation
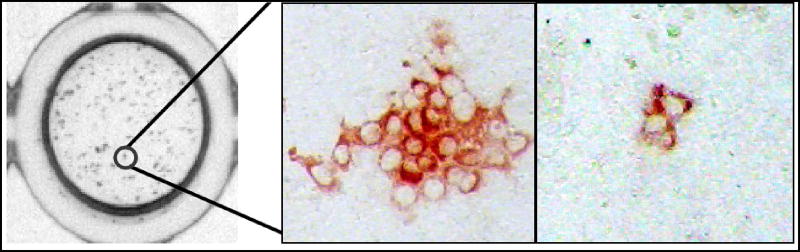
When cell-to-cell spread is blocked, foci of only 1–2 cells in size are observed. Hence, in non-dividing cell cultures foci consisting of 1–2 cells are classified as exhibiting no spread (Figure 1, right panel), while foci with more than 2 cells are interpreted as resulting from of cell-to-cell spread (Figure 1, middle panel). The data can be expressed semi-quantitatively by calculating the percentage of infection events in each well that did not exhibit cell-to-cell spread (i.e. 1–2 cell foci) versus the percentage of infection events in each well that did exhibit cell-to-cell spread (i.e. >2 cell foci). Alternatively, to more quantitatively measure spread (and the effect of different inhibitors/cell factors), the number of cells in each individual focus within a well can be counted.
Figure 1. HCV cell-to-cell spread assay.
Left: Monolayer of Huh7 cells stained for HCV 72 h post infection. Middle: A typical HCV foci formed when cell-to-cell spread is allowed to proceed. Right: HCV foci observed when spread is blocked.
Recipes
-
Complete DMEM (cDMEM)
100 units/ml penicillin
100 mg/ml streptomycin
2 mM L-glutamine
Part II. Supplemental protocols
Supplemental protocol A: Alternative strategy for visualizing HCV positive cells
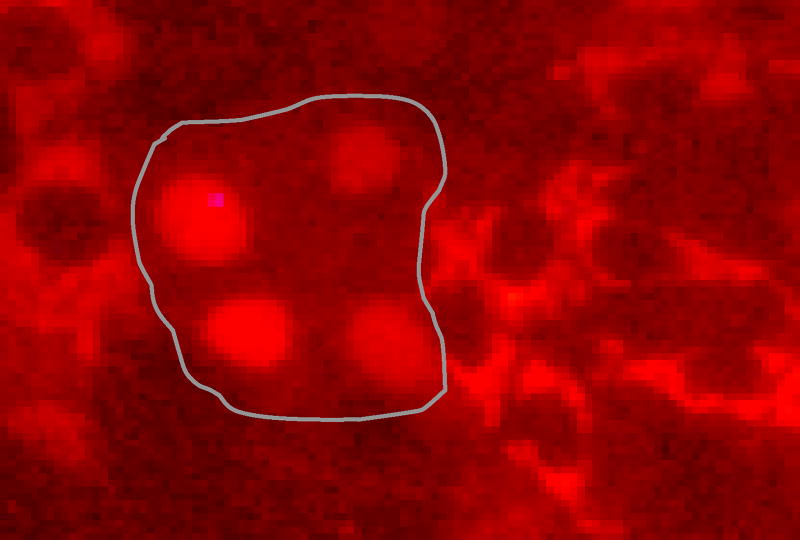
As described above, immunostaining is a convenient method for detecting HCV-positive cells; however other methods may be utilized. For example, if real-time live cell imaging of HCV cell-to-cell spread is desired, the Huh7.5-RFP-NLS-IPS reporter cell line (Jones et al., 2010) can be used. In uninfected cells the RFP-IPS-NLS protein is localized to the mitochondria in the cytoplasm. Upon infection the HCV NS3/4A protease cleaves the RFP-IPS-NLS off the mitochondria allowing the protein to localize to the nucleus. As such, infection of cells can be monitored in real-time based on the relocalization of RFP from the cytoplasm to the nucleus.
Materials and Reagents
Note: The same as in the Main Protocol with the following addition.
Huh7-5.1TRIP-RFP-NLS-IPS (from Dr. Charles Rice, Rockefeller University, New York, NY) (Jones et al., 2010)
Equipment
Note: The same as in the Main Protocol with the following addition.
Inverted fluorescence microscope (e.g. Nikon Corporation, model: TE2000U)
Procedure
Perform experiment as described in the Main protocol except:
Utilize Huh7-5.1TRIP-RFP-NLS-IPS cells instead of the parental Huh7 cells in Main Protocol Section A (Preparation of non-growing Huh7 cell cultures).
Monitor cell-to-cell spread under a microscope based on nuclear localization of RFP signal (instead of immunostaining described in Main protocol Section D) (see Figure 2 example results). Due to increases sensitivity, evidence of cell-to-cell spread in this assay may be detected as early as 12 h post-infection.
Figure 2. Foci consisting of HCV-infected Huh7.5-RFP-NLS-IPS cells (i.e. nuclear RFP) surrounded by uninfected sells (i.e. cytoplasmic RFP).
Supplemental Protocol B: HCV cell-to-cell spread assay in growing cell cultures
For technical reasons, it may sometimes be preferable to perform the assay in growing Huh7 cells (e.g. limiting inhibitor concentrations, toxic effects of sodium azide when higher concentrations of some commercial antibody preparations are used, or more flexibility in timing of siRNA knockdown). In this case, the assay can be performed as described above with the following modifications:
Materials and Reagents
Note: The same as in the Main protocol.
Equipment
Note: The same as in the Main protocol.
Procedure
- Preparation of growing Huh7 cell cultures.
- The day before the assay, seed 12,000 Huh7 cells/well of 96-well tissue culture plate in 10% FBS cDMEM. (Include extra wells for counting cells as indicated below.)
- Incubate the flask at 37 °C in 5% CO2 cell culture incubator overnight and proceed with assay as described in the Main protocol step B.
Accounting for cell division. Because cell division can also contribute to an increase in the number of cells per focus, it is critical to monitor the extent of cell division during the assay. This can be done by counting the number of cells present in triplicate wells at the time of infection and at the end of the assay. The ratio of the number of cells in each well at the end of the assay over the number of cells in each well at the time of infection provides an estimate of the number cell divisions that occurred during the assay. In combination with the positive controls described in the Main protocol step F2, directly monitoring cell amplification can establish the size of foci that could have resulted purely from cell division and thus help control for the effects of cell growth.
Supplemental Protocol C: Reverse siRNA transfection of non-growing Huh7 cells
Materials and Reagents
Note: The same as in the Main protocol.
Equipment
Note: The same as in the Main protocol.
Procedure
- Preparation of non-growing Huh7 cell cultures.
- Seed ~2 × 106 Huh7 cells in a BioCoat T75 cm2 flask in 15 ml of 10% FBS cDMEM.
- Incubate the flask at 37 °C in 5% CO2 cell culture incubator overnight or until the cells reach 95%–99% confluence.
- Replace the medium with 20 ml of 10% FBS cDMEM containing 1% DMSO and continue to incubate cells at 37 °C in 5% CO2 cell culture incubator changing the 1% DMSO, 10% FBS cDMEM every other day.
- RNAiMAX siRNA transfection.
- On day 14–20 post DMSO treatment, trypsinize flask with 5 ml trypsin for 10 min at 37 °C. Neutralize with 7 ml serum containing DMEM without antibiotics.
- Transfer cells to a 50 ml centrifuge tube and spin at 1,000 × g for 5 min in a table top centrifuge.
- Rinse the cells twice in 35 ml OptiMEM, collecting the cell pellet by centrifugation in between. Resuspend cells to 7.8 × 105 cells/ml in OptiMEM.
-
Preparation of transfection mixes (as master mixes or individual transfections in Eppendorf tubes or plate format, as appropriate).For each well of a 96-well plate to be transfected:
- Mix 0.84 µl 10 µM siRNA stock with 30 µl OptiMEM.
- Add 1.2 µl RNAiMax to the siRNA-OptiMEM and gently mix.
- Incubate at RT for 20 min.
- During incubation transfer 30 µl of the transfection mix into appropriate well of a waiting 96 well transfection plate.
- After the 20 min incubation period, add the 90 µl of the cell suspension to each well containing the transfection mix and pipette gently once to mix (the goal is to reestablish the non-growing cell monolayer of ~65,000 cells. Based on the above, each well should receive ~70,000 cells in a final volume of 120 µl with final siRNA concentration of 70 nM allowing for some cell loss).
- Incubate at 37 °C in 5% CO2 cell culture incubator overnight.
- Proceed with infection as described in the Main protocol step B2 (if the silenced gene may be required for cell-free extracellular virus entry, it is recommended to infect the cultures 20–24 h post-transfection before the targeted gene is efficiently silenced).
Acknowledgments
This work was supported by NIH grants R01-AI078881 and R21-AI097809.
Footnotes
In each user’s hands it is important to determine the number of infectious units and time of incubation that will result in the formation of a sufficient number of well-separated foci, which are big enough to clearly observe spread. This can be achieved by performing pilot assays with different volumes of virus inoculum and incubating for various periods of time.
References
- 1.Barretto N, Sainz B, Jr, Hussain S, Uprichard SL. Determining the involvement and therapeutic implications of host cellular factors in hepatitis C virus cell-to-cell spread. J Virol. 2014;88(9):5050–5061. doi: 10.1128/JVI.03241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28(2):167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knipe D, Howley PM. In: Principles of Virology. 5 Williams L, Wilkins, editors. 2007. [Google Scholar]
- 4.Law M, Maruyama T, Lewis J, Giang E, Tarr AW, Stamataki Z, Gastaminza P, Chisari FV, Jones IM, Fox RI, Ball JK, McKeating JA, Kneteman NM, Burton DR. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14(1):25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 6.Sainz B, Jr, Chisari FV. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma-derived cells. J Virol. 2006;80(20):10253–10257. doi: 10.1128/JVI.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, Harris HJ, Schwarz A, Desombere I, Roels GL, Balfe P, McKeating JA. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47(1):17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 8.Yu X, Uprichard SL. Cell-based hepatitis C virus infection fluorescence resonance energy transfer (FRET) assay for antiviral compound screening. Curr Protoc Microbiol. 2010;(Unit 17):15. doi: 10.1002/9780471729259.mc1705s18. Chapter 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102(26):9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]