Abstract
The sodium-activated potassium channels Slick (Slo2.1, KCNT2) and Slack (Slo2.2, KCNT1) are paralogous channels of the Slo family of high-conductance potassium channels. Slick and Slack channels are widely distributed in the mammalian CNS and they play a role in slow afterhyperpolarization, generation of depolarizing afterpotentials and in setting and stabilizing the resting potential. In the present study we used a combined approach of (co)-immunoprecipitation studies, Western blot analysis, double immunofluorescence and mass spectrometric sequencing in order to investigate protein–protein interactions of the Slick and Slack channels. The data strongly suggest that Slick and Slack channels co-assemble into identical cellular complexes. Double immunofluorescence experiments revealed that Slick and Slack channels co-localize in distinct mouse brain regions. Moreover, we identified the small cytoplasmic protein beta-synuclein and the transmembrane protein 263 (TMEM 263) as novel interaction partners of both, native Slick and Slack channels. In addition, the inactive dipeptidyl-peptidase (DPP 10) and the synapse associated protein 102 (SAP 102) were identified as constituents of the native Slick and Slack channel complexes in the mouse brain. This study presents new insights into protein–protein interactions of native Slick and Slack channels in the mouse brain.
Keywords: Slo2.1, Slo2.2, SAP 102, DPP 10, TMEM 263, Beta-synuclein
Highlights
-
•
Slick and Slack channels co-localize in the same cellular compartment
-
•
Slick and Slack channels assemble into cellular protein complexes in mouse brain.
-
•
Co-immunoprecipitation followed by mass spectrometric sequencing and Western blotting allowed the identification of novel potential interaction partners of Slick and Slack channels.
1. Introduction
The sodium-activated potassium channels Slick (sequence like an intermediate potassium channel, Slo2.1, KCNT2) and Slack (sequence like a calcium-activated potassium channel, Slo2.2, KCNT1) are structurally highly related and belong to the high-conductance potassium channels of the Slo family. Slick and Slack channels are widely distributed in the rat brain with partial overlap in their expression patterns [1], [2]. In neurons, sodium-activated potassium channels are involved in adapting the firing pattern of neurons, in the generation of the slow afterhyperpolarization (sAHP) and depolarizing afterpotentials (DAP) and in stabilization and setting of the resting membrane potential [3], [4], [5], [6], [7], [8]. The pore-forming alpha subunits of Slick and Slack channels are assembling into tetrameric channels [9], [10]. Alpha subunits of Slick and Slack channels are composed of an intracellular N-terminus, six membrane spanning domains, and a long intracellular C-terminus harboring various functional domains [9], [10].
Native sodium-activated potassium channels are high-conductance outward rectifying potassium channels that are activated upon sodium-influx [11]. Heterologously expressed Slick and Slack channels resemble most of the biophysical properties of native sodium-activated potassium currents. Nonetheless, there are some discrepancies regarding their unitary conductance, sensitivity to internal sodium ions, subconductance states and open probabilities of the channels as well as rundown in excised patches. Such discrepancies were not only observed when comparing heterologously expressed Slick and Slack channels with native currents. In addition, biophysical properties of native sodium-activated potassium currents varied depending on the brain region and/or the cell type examined [11], [12]. Such discrepancies might possibly reflect different isoforms of the underlying channels or channels associating with different endogenous factors and/or with several (regulatory) proteins.
While for the Slick channel no isoforms have been described, five different Slack channel isoforms were identified so far. The Slack channel isoforms only differ in their N-terminal region. The physiological relevance of these Slack channel isoforms has not been investigated yet [13], and most studies exploring various aspects of the Slack channels were analyzing the so-called Slack-B isoform.
Slick and Slack channel diversity may be further increased by the formation of heteromeric Slick and Slack channels. A previous study provided first evidence that Slick alpha-subunits are forming heteromeric channels with Slack-B alpha-subunits in the rat brain. Heterologously expressed Slick/Slack heteromeric channels were shown to have biophysical properties distinct from those of homomeric Slick and Slack channels [14]. The BK channel (big conductance calcium-activated potassium channel) is another potassium channel that is structurally highly related to Slick and Slack channels and thus dedicated to the same family of potassium channels (Slo family). In vitro studies revealed that BK channel alpha subunits do associate with Slack subunits, thereby forming heteromeric potassium channels. However, the existence of such channel complexes has never been proven in vivo [15].
There is growing evidence that Slick and Slack channel activity may be regulated by several cellular signaling pathways, including activation of G-protein coupled receptors linked to activation of protein kinase C (PKC) or protein kinase A (PKA) and by direct phosphorylation by these signaling proteins [14], [16], [17], [18]. Channel activity and gating may also be regulated by binding of endogenous signaling factors to the C-terminal tail of the channel, like NAD+ [19], PIP2 [20] and fragile X mental retardation protein (FMRP) [21], [22] as well as by small changes in cell volume [23].
Moreover, recent studies were suggesting that the Slack channel might interact with the postsynaptic density protein 95 (PSD 95) [24], FMRP [21] and with Glu2/3 subunits of the AMPA receptor [25].
In the present study we aimed to provide new insights into protein–protein interactions of the Slick and Slack channels in mouse brain. In order to address this issue, we performed double immunofluorescence and (co-)immunoprecipitation studies followed by Western blot analysis and mass spectrometric sequencing. Here we report Slick and Slack channels co-assemble into protein complexes in native mouse fore- and midbrain purified synaptic vesicle plasma membranes. Moreover, we provide first evidence for potential novel interaction partners of native Slick and Slack channels.
2. Material and methods
2.1. Animals
C57BL/6J mice were housed and handled in accordance with the guidelines with Austrian law which is in line with the directive of the European Union (2010/63/EU) for the use of laboratory animals. All procedures involving animals were approved by the Austrian Animal Experimentation Ethics Board in compliance with the European Convention for the Protection of Vertebrate Animals Used for Experimental and Other Scientific Purposes. Every effort was taken to minimize the number and suffering of animals used.
2.2. Preparation of purified synaptic plasma membrane vesicles from mouse fore- and midbrain
Preparation of purified synaptic plasma membrane vesicles was performed according to [26]. In brief, 30 male and female C57BL/6J mice were killed by cervical dislocation and fore- and midbrain was excised. Tissue was homogenized in ice-cold homogenization buffer (320 mM sucrose, 10 mM Tris–HCl pH 7.4, 1 mM EDTA, 0.5 mM phenylmethylsulfonylfluoride, PMSF) supplemented with protease inhibitors (complete tablets, Roche). Subsequently, homogenized fore- and midbrain tissue was separated on 7.5%/10% Ficoll gradient. Intact synaptosomes were lysed in 5 mM Tris–HCl supplemented with protease inhibitors. Lysed synaptosomes were spun at 125,000×g for 1 h at 4 °C. Purified synaptic plasma membranes were resuspended in 20 mM Tris–HCl, snap frozen in liquid nitrogen and stored at −80 °C.
2.3. Solubilization of membrane protein
Purified synaptic plasma membrane vesicles from mouse fore- and midbrain were incubated for 30 min at 4 °C in solubilization buffer (150 mM NaCl, 20 mM Tris–HCl pH 7.4, 1 mM EDTA, 1 mM PMSF, 2 μM leupeptin (Sigma-Aldrich), 1.5 μM aprotinin (Sigma-Aldrich), 0.15 μM pepstatin (Sigma-Aldrich), 0.9% n-dodecyl-β-d-maltoside (Calbiochem)) in a protein:detergent ratio of 1:9). Thereafter, soluble protein fraction was separated from unsoluble protein fraction by high-speed centrifugation at 105,000×g for 30 min. Solubilization efficiency was controlled in Western blot analysis. Solubilized protein fraction was used for subsequent immunoprecipitation experiments.
2.4. Immunoprecipitation
Slick and Slack channel specific antibodies were immobilized and cross-linked onto dynabeads protein G (Life Technologies) following manufacture's guidelines and incubated with n-dodecyl-β-d-maltoside (Calbiochem) solubilized synaptic plasma membranes derived from mouse fore- and midbrain overnight at 4 °C. For each experiment, 40 µg of either anti-Slick (clone N11/33, Neuromab) or anti-Slack (clone N3/26, Neuromab) channel antibody was used and 2 mg of synaptic plasma membranes served as starting material for solubilization. Immunoprecipitation experiments using a non-immune antibody of the same IgG subtype raised in chicken (MABC002, Chemicon, Millipore) served as negative control and were run in parallel. Unbound material was removed and collected (flow through). Affinity-purified protein complexes were eluted with Laemmli buffer (reducing agent added after elution). Resulting eluates were analyzed by Western blot and/or by mass spectrometric sequencing.
2.5. Western blot analysis
Western blot analysis was performed as described earlier [26]. In brief, 20 µg of protein samples (starting material, solubilized and unsolubilized protein fractions, and flow through) or 2.5% or 20% of eluates were separated by 4–15% (precasted TGX gel, Biorad) and transferred onto PVDF-membranes. Membranes were blocked in phosphate buffered saline (PBS) containing 0.05% Tween-20 (Roth) and 3% bovine serum albumin (Roth) for 1 h. Subsequently, membranes were incubated in PBS containing 0.05% Tween-20, 3% bovine serum albumin and either mouse monoclonal anti-Slick (1:1000, IgG1, clone N11/33, NeuroMab), mouse monoclonal anti-Slack (1:3000, IgG1, clone N3/26, NeuroMab), mouse polyclonal anti-beta-synuclein (1:100, abcam), rabbit polyclonal anti-DPP 10 (1:500, abcam) or mouse monoclonal anti-SAP 102 (1:2000, IgG1, clone N19/2, Biolegend) antibody for 2.5 h at room temperature. HRP-labeld goat anti-mouse IgG1 (1:100,000, Life Technologies), goat anti-mouse IgG (1:75,000, Dako) or goat anti-rabbit (1:75,000, Dako) were used as secondary antibodies. Western blots were developed using chemiluminescent HRP substrate (Millipore) and subsequently PVDF-membranes were incubated with Amersham hyper film.
2.6. Double immunofluorescence
C57BL/6J mice were sacrificed by cervical dislocation and brains were removed immediately. Brains were snap-frozen in prechilled −50 °C 2-methylbutane (Roth) and cut into coronal 8 µm sections using a cryostat. Sections were thaw mounted onto poly-lysine coated slides (Thermo Scientific). Brain sections were fixed in 1% para-formaldehyde (PFA, Merck) for 5 min at 4 °C. After washing slices in 50 mM Tris–HCl, 150 mM NaCl, 0.2% Triton X-100 (Roth) 3 times for 5 min, slices were blocked in the same solution additionally containing 2% BSA for 1 h at room temperature. For double immunofluorescence, mouse monoclonal anti-Slick (clone N11/33, Neuromab) and anti-Slack channel antibodies (clone N3/26, Neuromab) were directly labeled with Alexa Fluor-488 or Alexa Fluor-594 using the antibody labeling kit from Molecular Probes (A20181 and A20185; Life Technologies). Sections (n=6) were incubated with directly labeled primary antibodies diluted (both 1:200) in 50 mM Tris–HCl, 150 mM NaCl, 0.2% Triton X-100 (Roth) overnight at room temperature. On the next day, brain slices were washed 3 times for 10 min and counterstained with 0.03 µg/ml 4′,6-diamidino-2-phenylindole, dilactate (DAPI, Sigma-Aldrich) for 5 min at room temperature. Thereafter, slices were washed in 50 mM Tris–HCl, 150 mM NaCl, 0.2% Triton X-100 3 times for 10 min and mounted in hard set mounting medium (Vectashield). Confocal imaging was performed using LSM 700 (Zeiss) microscope and ZEN software.
Fluorescence intensity linescan profile was performed using image J software (rel. 1.45) by plotting fluorescence intensity (gray values) versus distance (pixels). Images were split into corresponding color channels (green: Slick channel–Alexa 488, red: Slack channel–Alexa 594). Intensity plots were extracted using identical coordinates and calculated in Excel.
2.7. Mass spectrometric sequencing
For mass spectrometric sequencing, eluates obtained from immunoprecipitation experiments were run on SDS-PAGE minigels and stained with Coomassie blue R250 (Biorad). Protein bands were excised from gel and digested with trypsin from porcine pancreas (Sigma-Aldrich, Vienna, Austria) as previously described [27]. Tryptic digests were analyzed using an UltiMate 3000 nano-HPLC system (Thermo Scientific, Germering, Germany) coupled to a Q Exactive Plus mass spectrometer (Thermo Scientific, Bremen, Germany) equipped with a Nanospray Flex ionization source. The peptides were separated on a homemade fritless fused-silica microcapillary column (75 µm i.d.×280 µm o.d.×10 cm length) packed with 3 µm reversed-phase C18 material (Reprosil). Solvent for HPLC were 0.1% formic acid (solvent A) and 0.1% formic acid in 85% acetonitrile (solvent B). The gradient profile was as follows: 0–2 min, 4% B; 2–55 min, 4–50% B; 55–60 min, 50–100% B, and 60–65 min, 100% B. The flow rate was 250 nL/min.
The Q Exacitve Plus mass spectrometer was operating in the data dependent mode selecting the top 12 most abundant isotope patterns with charge >1 from the survey scan with an isolation window of 1.6 mass-to-charge ratio (m/z). Survey full scan MS spectra were acquired from 300 to 1750m/z at a resolution of 70,000 with a maximum injection time (IT) of 120 ms, and automatic gain control (AGC) target 1e6. The selected isotope patterns were fragmented by higher-energy collisional dissociation (HCD) with normalized collision energy of 25 at a resolution of 17,000 with a maximum IT of 120 ms, and AGC target 5e5.
Data Analysis was performed using Proteome Discoverer 1.4.1.14 (Thermo Scientific) with search engine Sequest. The raw files were searched against the mus musculus database (167,940 entries) extracted from the NCBInr database released on June 2, 2014. Precursor and fragment mass tolerance was set to 10 ppm and 0.02 Da, respectively, and up to two missed cleavages were allowed. Carbamidomethylation of cysteine, and oxidation of methionine were set as variable modifications. Peptide identifications were filtered at 1% false discovery rate.
2.8. Data analysis of proteins identified by mass spectrometric sequencing
Raw data obtained from mass spectrometric analysis were evaluated as follows. As specificity control, immunoprecipitation experiments using a non-immune antibody of the same IgG subtype were run in parallel. In total 3 individual experiments per antibody used served for data evaluation. Proteins were only considered as specific if they were detected in at least two out of three individual experiments, and if they were not detected in any of the control experiments. In addition, only proteins of which at least two unique peptides were detected were considered specific.
2.9. Protein alignments, creation of percent identity matrix and topology prediction of transmembrane protein 263 (TMEM 263)
Protein sequences of different species were obtained from Pubmed. Alignments and creation of percent identity matrix were performed using Clustal Omega (rel. 1.2.1) with default settings. Topology of mouse TMEM 263 was analyzed using the web based protein structure prediction program TMHMM server (rel. 2.0).
Pubmed accession numbers of protein sequences used:
Rattus norvegicus: gi|157823853; Mus musculus: gi|81881684; Homo sapiens gi|74730713; Xenopus laevis gi|82186192; Danio rerio gi|82188197; Bos taurus gi|77736291; Gallus gallus gi|57525414; Pan pansicus gi|675800328
3. Results
3.1. Interaction of Slick and Slick channels in mouse brain
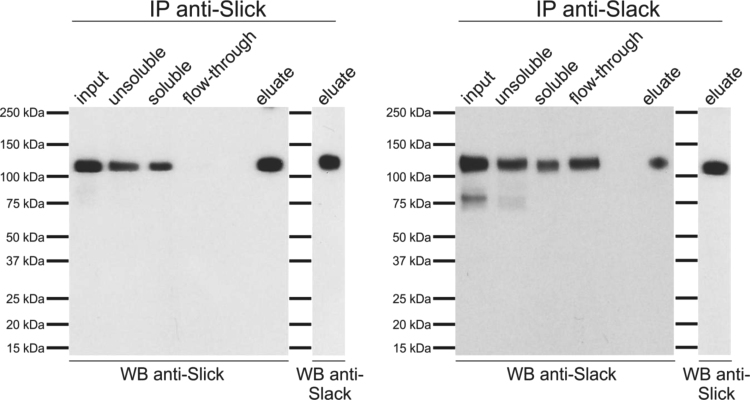
To determine a possible interaction of Slick and Slack channels in mouse fore- and midbrain, we conducted co-immunoprecipitation experiments followed by Western blot analysis. Immunoprecipitation studies were performed using mouse fore- and midbrain purified synaptic plasma membranes solubilized with dodecyl-maltoside (DDM). DDM is a nonionic detergent allowing solubilization of integral membrane proteins and is expected to preserve the native molecular environment of the ion channels. Solubilization efficiency was evaluated by Western blot analysis and was between 40% and 60% of total Slick and Slack protein (Fig. 1). The Slick channel-specific antibody resulted in quantitative precipitation of Slick protein, while the Slack channel-specific antibody precipitated about 40–50% of solubilized Slack protein. After having established a reliable and reproducible immunoprecipitation protocol, we performed co-immunoprecipitation studies using the same antibodies. Co-immunoprecipitation studies revealed that the anti-Slick antibody co-purified Slack protein, and conversely, using the Slack channel-specific antibody we co-immunoprecipitated Slick protein. These results suggest that Slick and Slack channels might interact in a cellular complex in mouse fore- and midbrain.
Fig. 1.
Immunoprecipitation and co-immunoprecipitation studies of Slick and Slack channels. Immunoprecipitation (IP) using DDM-solubilized mouse fore- and midbrain membranes with anti-Slick or anti-Slack antibodies. Different materials were used in Western blots: mouse fore- and midbrain synaptic plasma membranes (input), unsolubilized protein fraction, solubilized protein faction, unbound material (flow-through) and 2.5% of eluted protein fraction (eluate). Slack protein is co-purified by anti-Slick-antibody and vice versa.
3.2. CO-localization of Slick and Slack channels in selected moue brain regions
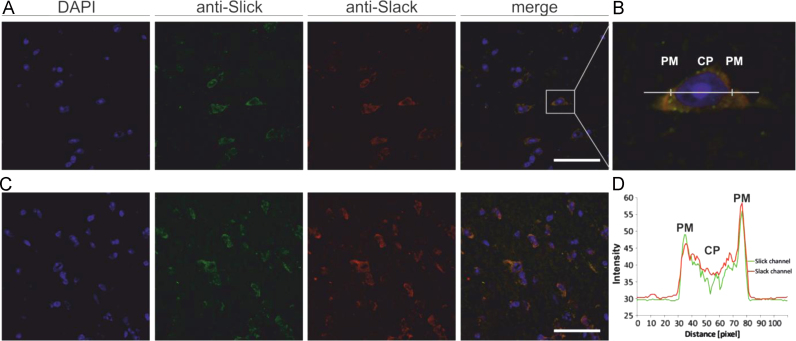
To further investigate a possible interaction of Slick and Slack channels in mouse brain, we performed double-immunofluorescence labeling experiments on mouse brain for both ion channels (n=6). Since both antibodies used were of the same IgG-subtype, we directly labeled them in order to be capable to perform double immunofluorescence stainings with Alexa-flour 488 (anti-Slick) and Alexa-fluor 594 (anti-Slack). Previous studies were suggesting that there is a partial overlap in the distribution pattern of Slick and Slack channels in rat brain, e.g. in the oculomotor and red nucleus [1], [2], [14]. Thus, we have chosen these nuclei as an example for channel co-localization and performed double immunofluorescence stainings in these nuclei of the mouse brain (shown in Fig. 2). Our experiments revealed strong immunostaining for both, Slick and Slack channels in neuronal cell bodies of both nuclei. Fluorescence signal was strongest at the plasma membrane, but was also evident in the cytoplasm. Taken together, these experiments indicated co-localization of Slick and Slack channels at the plasma membranes of neuronal cell bodies of both nuclei.
Fig. 2.
Co-localization of Slick and Slack channels in the red and oculomotor nucleus. Confocal images showing Slick and Slack channel co-localization in A and B magnocellular part of red nucleus and C in oculomotor nucleus. Both nuclei are showing clear somatic staining for both potassium channels. Double immunofluorescence was performed using directly labeled (Slick-Alexa488, shown in green, Slack-Alexa594, shown in red) mouse monoclonal antibodies directed against the C-terminus of respective target. DAPI (blue) was used as counterstain. B Representative staining for Slick and Slack channels in magnocellular part of red nucleus at higher magnification. D Fluoresence intensity linescan profile (as indicated in B) is demonstrating that fluorescence intensity for both channels is highest at the plasma membrane, but some staining is also evident in the cytoplasm. Abbreviations: PM plasma membrane, CP cytoplasm. Scale bars 50 µm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Identification of potential novel Interaction partners of Slick and Slack channels by mass spectrometric sequencing and Western blot analysis
After having established that Slick and Slack channels may co-localize in the same cellular compartment and that they might potentially interact in a cellular complex, we next asked whether additional novel proteins might form complexes with native Slick and Slack channels. In order to address this issue we performed immunoprecipitation experiments as described before and combined them with mass spectrometric sequencing of immuno-purified proteins. Because of current lack of Slick and Slack channel knock-out animals, co-immunoprecipitation experiments with a non-immune antibody of an identical IgG subtype run in parallel served as specificity control. In total, three individual experiments per antibody used served for data evaluation. Proteins were only considered as specific if they were detected in at least two out of three individual experiments and if they were not detected in any of the control experiments. In addition, only those proteins were considered as specific, of which at least two unique peptides were detected. Table 1, Table 2 are providing detailed information on the collected data.
Table 1.
Proteins (co-)purified with the anti-Slick antibody identified by mass spectrometric analysis.
| Protein | Alternative names | Accession # | Individual experiments | # peptides | Protein coverage in % |
|---|---|---|---|---|---|
| Slick | Potassium channel subfamily T member 2; KCNT2, Slo2.1, KCa4.2 | GI: 224028216 | 3/3 | 49 | 51.5 |
| Slack | Potassium channel subfamily T member 1, KCNT1, Slo2.2, KCa4.1 | GI:161168989 | 3/3 | 47 | 45.2 |
| Transmembrane protein 263 | TMEM 263 | GI:81881684 | 2/3 | 2 | 26 |
| Beta-synuclein | SNCB | GI:81879780 | 2/3 | 7 | 52.6 |
| Inactive dipeptidyl-peptidase 10 | DPP 10, Kv4 potassium channel auxiliary subunit | GI:238776842 | 3/3 | 7 | 9.5 |
| Synapse associated protein 102 | SAP 102, disks large homolog 3, DLG 3 | GI:7949129 | 3/3 | 13 | 18 |
Table 2.
Proteins (co-)purified with the anti-Slack antibody identified by mass spectrometric analysis.
| Protein | Alternative names | Accession # | Individual experiments | # peptides | Protein coverage in % |
|---|---|---|---|---|---|
| Slack | Potassium channel subfamily T member 1, KCNT1, Slo2.2, KCa4.1 | GI: 224028216 | 3/3 | 26 | 19.4 |
| Slick | Potassium channel subfamily T member 2; KCNT2, Slo2.1, KCa4.2 | GI:161168989 | 2/3 | 7 | 8.7 |
| Transmembrane protein 263 | TMEM 263 | GI:81881684 | 2/3 | 3 | 40 |
| beta-synuclein | SNCB | GI:81879780 | 2/3 | 7 | 52.6 |
This stringent validation criteria applied to our mass-spectrometry sequencing data allowed us to identify a number of high-confident interaction partners of native Slick and Slack channels. This approach enabled the isolation of Slack protein by an anti-Slick antibody and vice versa, strongly supporting our Western blot data. Interestingly, using the anti-Slick antibody we detected unique peptide sequences specific for the N-terminus of the so-called Slack-B isoform, one out of five previously described Slack channel isoforms, that are all differing in their N-terminal sequence [13].
In addition, we identified the small cytoplasmic protein beta-synuclein and the transmembrane protein 263 (TMEM 263) as novel potential interaction partners of both, Slick and Slack channels (Table 1, Table 2). As of today, little is known about TMEM 263. Thus, we conducted protein sequence alignments of TMEM 263 and additionally, we created a percent identity matrix to predict inter-relatedness of the protein deriving from different species. Sequence alignment using Clustal Omega 2.1 demonstrated that TMEM 263 is highly conserved among species, indicating a substantial role of this protein in different vertebrates (Fig. 3). Human TMEM 263 shares over 96% amino acid sequence identity to mouse and rat TMEM 263, and approximately 74% to zebrafish, 77% to chicken and almost 90% to Xenopus spp. We were also interested in the possible membrane topology of TMEM 263. Web based protein structure prediction programs (TMHMM server 2.0–prediction of transmembrane helices in proteins) suggest that this protein consists of two membrane-spanning domains, an intracellular N- and C-terminus and an extracellular loop (indicated in Fig. 3). The protein has a predicted molecular weight of 9.3 kDa.
Fig. 3.
Alignment of protein sequences of TMEM 263 deriving from different species. Protein alignment was performed using Clustal Omega (1.2.1) multiple sequence alignment program. Transmembrane domain 1 and 2 were identified using the Web based protein structure prediction program TMHMM server 2.0 – prediction of transmembrane helices in proteins.
Interestingly, we detected the integral membrane protein inactive dipeptidyl-peptidase 10 (DPP 10) and the membrane-associated protein synapse associated protein 102 (SAP 102 or disks large homolog 3, DLG 3) as potential novel interaction partners of the Slick channel (Table 1).
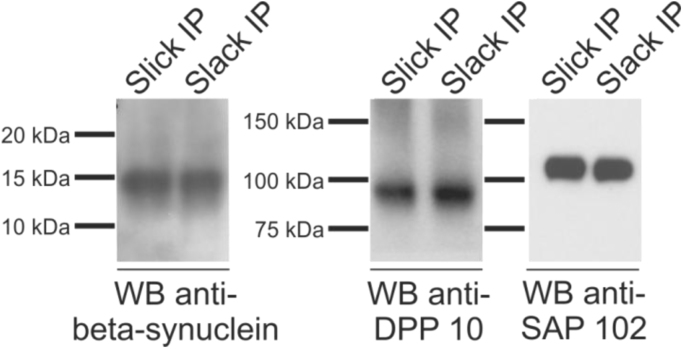
In order to validate our findings obtained from mass spectrometric sequencing, we performed co-immunoprecipitation experiments followed by Western blot analysis using antibodies specific for the potential novel interaction partners. Unfortunately, suitableTMEM 263 specific antibodies were not available. However, both, Slick and Slack channel immunoprecipitation experiments resulted in co-purification of beta-synculein, DPP 10 as well as SAP 102 (shown in Fig. 4). These results are strongly supporting our data obtained from mass spectrometric sequencing. Moreover, in contrast to our data obtained by mass spectrometric sequencing, our Western blot data suggest that the Slack channel is interacting with DPP 10 and SAP 102. Taken together, our results indicate that Slick and Slack channels co-assemble into protein complexes together with beta-synuclein, TMEM 263, DPP 10 and SAP 102 in the mouse fore-and midbrain.
Fig. 4.
Co-immunoprecipitation of novel potential interaction partners of Slick and Slack channels. Immunoprecipitation (IP) using DDM-solubilized mouse fore- and midbrain membranes using anti-Slick or anti-Slack channel antibodies. 20% of eluted protein fraction of both, Slick channel and Slack channel IPs were used for Western blots. Staining with antibodies specific for beta-synuclein (predicted molecular weight 14 kDa), DPP 10 (predicted molecular weight 91 kDa) and SAP 102 (predicted molecular weight 102 kDa) resulted in bands which corresponded well to the predicted molecular weight of the individual proteins.
4. Discussion
In the present study we used a combined approach of double-immunofluorescence, immunoprecipitation studies, Western blot analysis and mass spectrometric sequencing in order to investigate protein-–protein interactions of the native Slick and Slack channels. Our data strongly suggest that Slick and Slack channels are interacting in mouse fore- and midbrain. Moreover, we identified potential new interaction partners of Slick and Slack channels.
In a previous work it was shown that Slick and Slack channels are capable of forming heterotetrameres in vitro with unique biophysical properties. Further, Slick channels have been co-purified from solubilized rat olfactory bulb and brainstem using a Slack-B specific antibody [14]. This Slack channel isoform was found to interact with Slick channels [14]. In this study we have confirmed Slick channel as an interaction partner of Slack-B isoform in mouse brain, indicating that heteromerization of Slick and Slack channels might occur also in this species.
Moreover, in vitro studies suggest that BK channel alpha-subunits are capable to form heteromeric channels with Slack channel alpha-subunits with distinct biophysical and pharmacological properties. However, the existence of such channel complexes has never been proven in vivo [15]. Performing immunoprecipitation studies using a Slack channel specific antibody followed by mass spectrometric sequencing we did not identify the BK channel alpha subunit interacting with Slack channel in mouse fore- and midbrain.
Previous studies focusing on possible protein-protein interactions of the Slack channel in brain have shown that this channel is interacting with the postsynaptic density protein 95 (PSD 95) [24], fragile X mental retardation protein (FMRP) [21] and with Glu2/3 subunits of the AMPA receptor [25]. Following our experimental approach we did not identify these proteins as potential interaction partners of the Slack channel. Different solubilization protocols applied, use of different species (mouse vs. rat) as well as the use of different membrane fractions and antibodies (e.g. PSD 95- and Glu2/3-specific antibodies) in these studies might serve as an explanation.
Using a combination of immunoprecipitation experiments, mass spectrometry and Western blot analysis we identified, for the first time, inactive dipeptidyl-peptidase 10 (DPP 10), synapse associated protein 102 (SAP 102) and beta-synuclein as potential novel interaction partners both, Slick and Slack channels. While Western blotting was capable to detect all three proteins being interacting with Slick and Slack channels, mass spectrometric sequencing failed to directly identify two of these partners (DPP 10 and SAP 102) to be associated with Slack channels. However, they were clearly identified together with Slick channel subunits. This discrepancy might solely reflect different sensitivities of both methods applied. The transmembrane protein 263 (TMEM 263) was identified through mass spectrometric sequencing as novel potential interaction partner of both, Slick and Slack channels. Due to current lack of suitable TMEM 263 specific antibodies, we could not validate this interaction in Western blots.
What functional implications could such an association on the molecular properties of Slick and Slack channels have? We found both potassium channels interacting with TMEM 263. Alignment of protein sequences deriving from different species demonstrated that TMEM 263 is highly conserved, indicating a substantial role of this protein in different vertebrates. Protein structure prediction software suggests that this protein consists of two membrane-spanning domains, an intracellular N- and C-terminus and an extracellular loop. Interestingly, this topological arrangement shares homologies to auxiliary beta-subunits of the aforementioned BK-channel. As to whether TMEM 263 confers properties similar to the BK channel beta-subunit to Slick and Slack channels remains to be determined.
We also identified beta-synuclein interacting with both, Slick and Slack channels. Beta-synuclein has been implicated to play a role in the pathogenesis of neurodegenerative diseases and moreover, in synaptic vesicle endocytosis [28]. This small cytoplasmic protein is detected only in vertebrates and expressed predominately in presynaptic nerve terminals [29].
In addition, we co-purified DPP 10 with both, Slick and Slack channels. DPP 10 was previously shown to associate with alpha subunits of the voltage-gated potassium channels of the Kv4 family (Kv4.1-4.3). This assembly leads to an altered surface expression and changes of biophysical properties of the channel complexes in vitro [30], [31]. Kv4 channels interact with DPP 10 via their S1-S2 segments, a structure also being present in Slick and Slack potassium channels [32].
Moreover, we identified SAP 102 as a high-confident interaction partner of the Slick and Slack channels. SAP 102 is a scaffolding protein typically present at postsynaptic densities. It is involved in trafficking and anchoring of distinct types of glutamate receptors [33] and is associated with inwardly rectifying potassium channels of the Kir2.x family in rat brain [34]. Interactions of SAP 102 and Kir2.x channels are mediated through a PDZ domain of SAP 102 and the C-terminal PDZ class I binding motif of Kir2.x channels [34]. This well preserved PDZ binding motif is also present at the C-terminus of Slick and Slack channels [9], [15], [24].
In the present study we offer new insights in the nano-environment of Slick and Slack channels. Our data presented here could provide the basis for future studies systematically exploring possible functions of newly identified Slick and Slack channel interaction partners. In the future, potential influences of the interaction partners on biophysical or pharmacological properties as well as on channel trafficking or surface expression could be systematically explored by the use of heterologous expression systems. Interaction of some of the newly identified interacting proteins might possibly partly explain the observed discrepancies in the biophysical properties between Slick and Slack channels expressed in heterologous expression systems and native sodium-activated potassium channels as well as previously described heterogeneity of native sodium-activated potassium currents in vivo.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Role of authors
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: S.R., C.S., H.G.K.
Acquisition of data: S.R., L.K., H.H.L.
Analysis and interpretation of data: S.R., L.K., H.H.L.
Drafting of the manuscript: S.R.
Critical revision of the manuscript: S.R., C.S., H.G.K.
Obtained funding: H.G.K.
Administrative, technical, and material support: C.S., H.G.K.
Study supervision: H.G.K.
Acknowledgment
We thank Dr. Romana Gerner and Dr. Alexander Moschen for the use of the confocal microscope. Funding: Austrian Research Fund PhD program SPIN W1206-B05
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.09.024.
Contributor Information
Sandra Rizzi, Email: Sandra.Rizzi@gmx.net.
Christoph Schwarzer, Email: Schwarzer.Christoph@i-med.ac.at.
Leopold Kremser, Email: Leopold.Kremser@i-med.ac.at.
Herbert H. Lindner, Email: Herbert.Lindner@i-med.ac.at.
Hans-Günther Knaus, Email: Hans.G.Knaus@i-med.ac.at.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Bhattacharjee A., Gan L., Kaczmarek L.K. Localization of the Slack potassium channel in the rat central nervous system. J. Comp. Neurol. 2002;454:241–254. doi: 10.1002/cne.10439. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharjee A., von Hehn C.A., Mei X., Kaczmarek L.K. Localization of the Na+-activated K+ channel Slick in the rat central nervous system. J. Comp. Neurol. 2005;484:80–92. doi: 10.1002/cne.20462. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Vives M.V., Nowak L.G., McCormick D.A. Cellular mechanisms of long-lasting adaptation in visual cortical neurons in vitro. J. Neurosci. 2000;20:4286–4299. doi: 10.1523/JNEUROSCI.20-11-04286.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franceschetti S., Lavazza T., Curia G., Aracri P., Panzica F., Sancini G., Avanzini G., Magistretti J. Na+-activated K+ current contributes to postexcitatory hyperpolarization in neocortical intrinsically bursting neurons. J. Neurophysiol. 2003;89:2101–2111. doi: 10.1152/jn.00695.2002. [DOI] [PubMed] [Google Scholar]
- 5.Yang B., Desai R., Kaczmarek L.K. Slack and Slick K(Na) channels regulate the accuracy of timing of auditory neurons. J. Neurosci.: Offic. J. Soc. Neurosci. 2007;27:2617–2627. doi: 10.1523/JNEUROSCI.5308-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Kolaj M., Renaud L.P. Ca2+-dependent and Na+-dependent K+ conductances contribute to a slow AHP in thalamic paraventricular nucleus neurons: a novel target for orexin receptors. J. Neurophysiol. 2010;104:2052–2062. doi: 10.1152/jn.00320.2010. [DOI] [PubMed] [Google Scholar]
- 7.Gao S.B., Wu Y., Lu C.X., Guo Z.H., Li C.H., Ding J.P. Slack and Slick KNa channels are required for the depolarizing afterpotential of acutely isolated, medium diameter rat dorsal root ganglion neurons. Acta Pharmacol. Sin. 2008;29:899–905. doi: 10.1111/j.1745-7254.2008.00842.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu X., Stan Leung L. Sodium-activated potassium conductance participates in the depolarizing afterpotential following a single action potential in rat hippocampal CA1 pyramidal cells. Brain Res. 2004;1023:185–192. doi: 10.1016/j.brainres.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharjee A., Kaczmarek L.K. For K+ channels, Na+ is the new Ca2+ Trends Neurosci. 2005;28:422–428. doi: 10.1016/j.tins.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Salkoff L., Butler A., Ferreira G., Santi C., Wei A. High-conductance potassium channels of the SLO family. Nat. Rev. Neurosci. 2006;7:921–931. doi: 10.1038/nrn1992. [DOI] [PubMed] [Google Scholar]
- 11.Kaczmarek L.K. Slack, Slick and Sodium-Activated Potassium Channels. ISRN neuroscience. 2013;(2013):1–15. doi: 10.1155/2013/354262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igelstrom K.M. Is slack an intrinsic seizure terminator? Neurosci.: Rev. J. Bringing Neurobiol. Neurol. psychiatry. 2013;19:248–254. doi: 10.1177/1073858412446311. [DOI] [PubMed] [Google Scholar]
- 13.Brown M.R., Kronengold J., Gazula V.R., Spilianakis C.G., Flavell R.A., von Hehn C.A., Bhattacharjee A., Kaczmarek L.K. Amino-termini isoforms of the Slack K+ channel, regulated by alternative promoters, differentially modulate rhythmic firing and adaptation. J. Physiol. 2008;586:5161–5179. doi: 10.1113/jphysiol.2008.160861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H., Kronengold J., Yan Y., Gazula V.R., Brown M.R., Ma L., Ferreira G., Yang Y., Bhattacharjee A., Sigworth F.J., Salkoff L., Kaczmarek L.K. The N-terminal domain of Slack determines the formation and trafficking of Slick/Slack heteromeric sodium-activated potassium channels. J. Neurosci.: Offic. J. Soc. Neurosci. 2009;29:5654–5665. doi: 10.1523/JNEUROSCI.5978-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joiner W.J., Tang M.D., Wang L.Y., Dworetzky S.I., Boissard C.G., Gan L., Gribkoff V.K., Kaczmarek L.K. Formation of intermediate conductance calcium activated potassium channels by interaction of Slack and Slo subunits. Nature neuroscience. 1998;1:462–469. doi: 10.1038/2176. [DOI] [PubMed] [Google Scholar]
- 16.Santi C.M., Ferreira G., Yang B., Gazula V.R., Butler A., Wei A., Kaczmarek L.K., Salkoff L. Opposite regulation of Slick and Slack K+ channels by neuromodulators. J. Neurosci.: Offic. J. Soc. Neurosci. 2006;26:5059–5068. doi: 10.1523/JNEUROSCI.3372-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nuwer M.O., Picchione K.E., Bhattacharjee A. cAMP-dependent kinase does not modulate the Slack sodium-activated potassium channel. Neuropharmacology. 2009;57:219–226. doi: 10.1016/j.neuropharm.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Nuwer M.O., Picchione K.E., Bhattacharjee A. PKA-induced internalization of Slack KNa channels produces dorsal root ganglion neuron hyperexcitability. J. Neurosci.: Offic. J. Soc. Neurosci. 2010;30:14165–14172. doi: 10.1523/JNEUROSCI.3150-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamsett T.J., Picchione K.E., Bhattacharjee A. NAD+ activates KNa channels in dorsal root ganglion neurons. J. Neurosci.: Offic. J. Soc. Neurosci. 2009;29:5127–5134. doi: 10.1523/JNEUROSCI.0859-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angeles Tejada M. de los, Jensen L.J., Klaerke D.A. PIP(2) modulation of Slick and Slack K(+) channels. Biochem. Biophys. Res. Commun. 2012;424:208–213. doi: 10.1016/j.bbrc.2012.06.038. [DOI] [PubMed] [Google Scholar]
- 21.Brown M.R., Kronengold J., Gazula V.R., Chen Y., Strumbos J.G., Sigworth F.J., Navaratnam D., Kaczmarek L.K. Fragile X mental retardation protein controls gating of the sodium-activated potassium channel Slack. Nat. Neurosci. 2010;13:819–821. doi: 10.1038/nn.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Brown M.R., Hyland C., Chen Y., Kronengold J., Fleming M.R., Kohn A.B., Moroz L.L., Kaczmarek L.K. Regulation of neuronal excitability by interaction of fragile X mental retardation protein with Slack potassium channels. J. Neurosci.: Offic. J. Soc. Neurosci. 2012;32:15318–15327. doi: 10.1523/JNEUROSCI.2162-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tejada M.A., Stople K., Hammami Bomholtz S., Meinild A.K., Poulsen A.N., Klaerke D.A. Cell volume changes regulate Slick (Slo2.1), but not Slack (Slo2.2) K+ channels. PloS One. 2014;9:e110833. doi: 10.1371/journal.pone.0110833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchino S., Wada H., Honda S., Hirasawa T., Yanai S., Nakamura Y., Ondo Y., Kohsaka S. Slo2 sodium-activated K+ channels bind to the PDZ domain of PSD 95. Biochem. Biophys. Res. Commun. 2003;310:1140–1147. doi: 10.1016/j.bbrc.2003.09.133. [DOI] [PubMed] [Google Scholar]
- 25.Nanou E., Kyriakatos A., Bhattacharjee A., Kaczmarek L.K., Paratcha G., El Manira A. Na+-mediated coupling between AMPA receptors and KNa channels shapes synaptic transmission. Proc. Natl. Acad. Sci. USA. 2008;105:20941–20946. doi: 10.1073/pnas.0806403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sailer C.A., Kaufmann W.A., Marksteiner J., Knaus H.G. Comparative immunohistochemical distribution of three small-conductance Ca2+-activated potassium channel subunits, SK1, SK2, and SK3 in mouse brain. Mol. Cell. Neurosci. 2004;26:458–469. doi: 10.1016/j.mcn.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Sobieszek A., Matusovsky O.S., Permyakova T.V., Sarg B., Lindner H., Shelud’ko N.S. Phosphorylation of myorod (catchin) by kinases tightly associated to molluscan and vertebrate smooth muscle myosins. Arch. Biochem. Biophys. 2006;454:197–205. doi: 10.1016/j.abb.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Vargas K.J., Makani S., Davis T., Westphal C.H., Castillo P.E., Chandra S.S. Synucleins regulate the kinetics of synaptic vesicle endocytosis. J. Neurosci.: Offic. J. Soc. Neurosci. 2014;34:9364–9376. doi: 10.1523/JNEUROSCI.4787-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakes R., Spillantin M.G., Goedert M. Identification of two distinct synucleins from human brain. FEBS Lett. 1994;345:27–32. doi: 10.1016/0014-5793(94)00395-5. [DOI] [PubMed] [Google Scholar]
- 30.Jerng H.H., Qian Y., Pfaffinger P.J. Modulation of Kv4.2 channel expression and gating by dipeptidyl peptidase 10 (DPP10) Biophys. J. 2004;87:2380–2396. doi: 10.1529/biophysj.104.042358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zagha E., Ozaita A., Chang S.Y., Nadal M.S., Lin U., Saganich M.J., McCormack T., Akinsanya K.O., Qi S.Y., Rudy B. DPP 10 modulates Kv4-mediated A-type potassium channels. J. Biol. Chem. 2005;280:18853–18861. doi: 10.1074/jbc.M410613200. [DOI] [PubMed] [Google Scholar]
- 32.Ren X., Hayashi Y., Yoshimura N., Takimoto K. Transmembrane interaction mediates complex formation between peptidase homologues and Kv4 channels. Mol. Cell. Neurosci. 2005;29:320–332. doi: 10.1016/j.mcn.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Elias G.M., Elias L.A., Apostolides P.F., Kriegstein A.R., Nicoll R.A. Differential trafficking of AMPA and NMDA receptors by SAP 102 and PSD 95 underlies synapse development. Proc. Natl. Acad. Sci. USA. 2008;105:20953–20958. doi: 10.1073/pnas.0811025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leonoudakis D., Conti L.R., Anderson S., Radeke C.M., McGuire L.M., Adams M.E., Froehner S.C., Yates J.R., 3rd, Vandenberg C.A. Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)-associated proteins. J. Biol. Chem. 2004;279:22331–22346. doi: 10.1074/jbc.M400285200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material