Abstract
Activities of fast growing human population are altering freshwater ecosystems, endangering their inhabitants and public health. Organic and trace compounds have a high potential for adverse impacts on aquatic organisms in some Great Lakes tributaries. Toxic compounds in tissues of organisms living in contaminated environments change their metabolism and alter cellular components. We measured oxidatively induced DNA damage in the soft tissues of dreissenid mussels to check on the possible contaminant-induced impact on their DNA.
The animals were obtained from archived samples of the National Oceanic and Atmospheric Administration (NOAA) Mussel Watch Program (MWP). Mussels were collected from the harbor of Ashtabula River in Ohio, and a reference area located at the Lake Erie shore. Using gas chromatography-tandem mass spectrometry with isotope dilution, we identified and quantified numerous oxidatively modified DNA bases and 8,5′-cyclopurine-2′-deoxynucleosides. We found significant differences in the concentrations of these potentially mutagenic and/or lethal lesions in the DNA of mussels from the harbor as compared to the animals collected at the reference site. These results align NOAA’s data showing that elevated concentrations of polycyclic aromatic hydrocarbons (PAHs), polychlorinated biphenyls (PCBs), and heavy metals were found in mussels within the harbor as compared to mussels collected in the reference site.
The measured DNA lesions can be used as biomarkers for identifying DNA damage in mussels from polluted and reference sites. Such biomarkers are needed to identify the bioeffects of contaminants in affected organisms, as well as whether remedial actions have proven successful in reducing observed toxic effects.
Keywords: Mussel Watch Program, dreissenid mussels, Great Lakes, oxidatively induced DNA damage, PAHs, PCBs, trace metals, GC-MS/MS
Introduction
The Laurentian Great Lakes region, with its wealth of natural resources and maritime and railroad transportation systems, underwent rapid industrialization and ultimately a legacy of chemical contamination in many of its rivers and harbors.
The National Oceanic and Atmospheric Administration (NOAA), National Centers for Coastal Ocean Science, Mussel Watch Program (MWP), uses dreissenid mussels to monitor chemical contamination in the Great Lakes. In recognizing the need for both chemical and biological monitoring information, the MWP partnered with the National Institute of Standards and Technology (NIST), to conduct a pilot project to implement alternative biomonitoring techniques that complement chemical s concentrations measurements in environmental media (water, sediment, biota). This paper describes an oxidatively induced DNA damage monitoring tool applied to the dreissenid mussel species to distinguish reference site from impacted sites in the Ashtabula River harbor and nearby Lake Erie.
After entering an aquatic system, contaminants can move between media, for example through accumulation in the food chain or through deposition in sediments. Contaminants can cause alterations at different levels of the hierarchy of biological organizations1. Among numerous contaminants, legacy and of emerging concern (CEC), found in the Ashtabula River harbor, PAHs, PCBs and trace elements are of particular concern, with most of them being toxic even at low concentrations1–8. Usually, when high levels of PAHs are present in aquatic systems, abnormal growth of tissue (neoplasms) is observed in fish and other inhabiting animals9. For years, PAHs have been known to be strong inducers of cytochrome P450 (CYP450) activity in fish and other vertebrates10, and are metabolized in living organisms to trigger reactions resulting in oxidatively induced DNA damage11–14. In some circumstances, when parallel presence of non-dioxin-like PCBs and their hydroxyl metabolites occur in the environment15, they can induce DNA damage signaling and enhance the DNA damaging effect of benzo[a]pyrene16. Metabolites of PAHS and PCBs are known to generate oxygen-derived species such as hydroxyl radical (•OH), superoxide radical (O2•−) and hydrogen peroxide (H2O2)14, 15, 17, 18. Among invertebrates the ability to metabolize PAHs differs significantly due to variable levels and activities of the cytochrome P450 dependent oxidases. Mollusks are species with a wide range of metabolic capability19, 20 but clearly showing presence of PAHs metabolites generated by reactions involving oxidative and genotoxic effects21–25. Reactions of •OH cause damage to biological molecules including DNA, proteins and lipids, and thus can lead to increased genetic instability, inflammation, proliferation, reduction of antioxidants, cell death, apoptosis and angiogenesis17, 26. If not repaired, DNA damage may lead to harmful biological effects in living organisms, including mutations, disease and death26.
In aquatic systems, bivalves are recognized as important biomonitors27, 28 because of their ubiquity, abundance, easy sampling and filter -feeding lifestyle. Dreissenid mussels are useful monitoring organisms because they usually dwell in large numbers and are well adapted to naturally stressful environments13, 29–31. Dreissenid mussels in the Great Lakes have been used to monitor chemical contaminant levels for over two decades by the NOAA Mussel Watch Program32, 33 and in recent years for mussel health assessment 32. Here, we report on oxidatively induced DNA damage in dreissenid mussels (Dreissena polymorpha; zebra) collected from the harbor of the Ashtabula River and compare with mussels collected from a reference location in Lake Erie. We identified and quantified numerous DNA lesions including 8,5′-cyclopurine-2′-deoxynucleosides in DNA of dreissenid mussels as potential bioindicators for environmental genotoxicity.
Materials and Methods
Study Area and Sample Collection
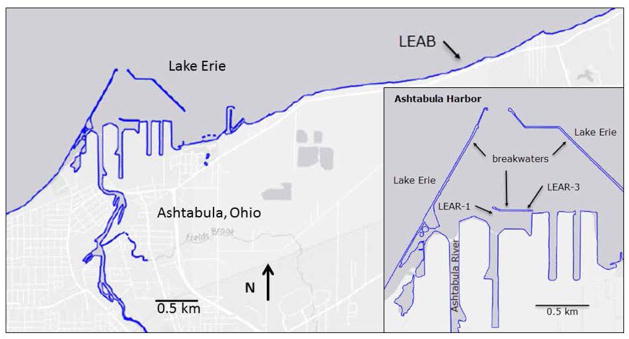
The Ashtabula River flows into Lake Erie and like many rivers of the Great Lakes it received industrial pollution in the years prior to environmental laws and regulations. Chemical contamination from PCBs, PAHs, and heavy metals, led to restrictions on fish and wildlife consumption, degradation of fish and wildlife populations, and loss of fish and wildlife habitat. In 2014 all management actions to clean up the Ashtabula River had been completed. Samples were collected from two sites (LEAR-1 and LEAR-3) outer Ashtabula harbor and from a reference site (LEAB) in Lake Erie approximately 6.5 km east of the Ashtabula River mouth, in 10 m of water and 230 m off-shore (Figure 1) in September 2014. The reference site (LEAB) was established by the NOAA Mussel Watch Program in 1992 and sampled every two years for analysis of over 100 chemical contaminants. Scuba divers removed dreissenid mussels from rock substrate using stainless steel scrapers, placed them in a nylon mesh bag, and upon surfacing transferred them to coolers containing site water.
Fig. 1.
Map of sample collection sites.
Within one hour of collection, the mussels were rinsed free of debris with site water, placed in 5 Ziploc bags in composites of approximately 20 to 30 mussels each. The bagged mussels were then placed in a cooler of dry ice and shipped blind coded to the NIST laboratory, where they were transferred to a freezer at −80 °C until they were thawed for further processing and analyses.
In addition, mussels were sent to a separate laboratory for analysis of chemical contaminants in the soft tissue according to NOAA protocols for sample collection, preparation, and analysis33–35. Methods of PAHs, chlorinated hydrocarbons including PCBs, and trace elements (Cu, Ni, Cr, Fe) analyses are summarized in Supporting Information section, including links and citations for detailed information.
DNA isolation
After removal from the Dewar container, mussels were thawed on ice, then washed with ice-cold deionized water. To minimize the influence of body mass and age of studied animals on the results of measurements of markers of oxidative DNA damage, similar size (≈2 cm) mussels have been selected for each group. Mussel tissues (≈ 100 mg) separated from shells with a scalpel were processed according to the product manual of E.Z.N.A. Mollusc DNA Kit, Omega Bio-tek (Norcross, GA) with modification involving homogenization with Bullet Blender Storm 24 high-throughput bead-mill homogenizer (Next Advance, Averill Park, NY). Tissues were placed in the 1.5 mL Rhino Screw cap tubes (Next Advance, Averill Park, NY) kept on ice, containing 350 μL of ML1 Buffer from the kit and three 2 mm zirconium oxide beads. Tubes were transferred into the Bullet Blenderkept in the refrigerator at 4 °C and processed 2 × 30 s at speed 12 with 30 s break between runs. Subsequently 25 μL of Proteinase K from the kit was added and samples were incubated for 2 h at 60 °C. Then, subsequent steps of the Mollusc DNA Kit protocol were applied. For the final DNA elution, two portions of 100 μL of sterile high-performance liquid chromatography grade water (Sigma-Aldrich, St. Louis, MO) warmed to 70 °C were used. The UV absorbance spectrum of each DNA sample was recorded by absorption spectrophotometry between the wavelengths of 200 nm and 350 nm to ascertain the quality of DNA and to measure the DNA concentration at 260 nm (absorbance of 1 = 50 μg of DNA per milliliter). Aliquots (50 μg) of DNA samples were dried in 1.5 mL deoxyribonuclease-free Eppendorf tubes in a SpeedVac under vacuum and then kept at 80 °C for further analysis.
Gas chromatography-tandem mass spectrometry
Gas chromatography-tandem mass spectrometry (GC-MS/MS) with isotope-dilution was used to identify and quantify modified DNA bases and 8,5′ -cyclopurine-2′-deoxynucleosides. A set of DNA samples was used to identify and quantify oxidatively induced DNA base lesions5 -hydroxy-5-methylhydantoin (5-OH-5-MeHyd), thymine glycol (ThyGly), 5,6-dihydroyuracil (5,6-diOH-Ura),4,6 -diamino-5-formamidopyrimidine (FapyAde), 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyGua) and 8-hydroxyguanine (8-OH-Gua). DNA samples (50 μg each) were supplemented with the aliquots of the stable isotope-labeled analogs of these compounds, i.e., 5-OH-5-MeHyd-13C,15N2, ThyGly-d4, 5,6-diOH-Ura-13C,15N2, FapyAde-13C,15N2, FapyGua-13C,15N2 and 8 -OH-Gua-15N5 as internal standards, which are a part of the NIST Standard Reference Material 2396 Oxidative DNA Damage Mass Spectrometry Standards (for details see http://www.nist.gov/srm/index.cfm and https://www-s.nist.gov/srmors/view_detail.cfm?srm=2396). The samples were dried in a SpeedVac under vacuum, and then dissolved in 50 μL of an incubation buffer consisting of 50 mmol/L phosphate buffer (pH 7.4), 100 mmol/L potassium chloride (KCl), 1 mmol/L ethylenediaminetetraacetic acid (EDTA), and 0.1 mmol/L dithiothreitol. Subsequently, they were incubated with 2 μg of E. coli formamidopyrimidine -DNA glycosylase (Fpg) and 2 μg of E. coli endonuclease III (Nth) at 37 °C for 1 h to release the modified DNA bases from DNA. An aliquot of 100 μL ethanol was added to precipitate DNA and to stop the reaction. After centrifugation, the supernatant fractions were separated, lyophilized and trimethylsilylated as described36. Derivatized samples were analyzed by GC-MS/MS using multiple reaction monitoring (MRM) as described previously36, 37. The mass transitions used for this purpose were: m/z 331→ m/z 316 and m/z 334→ m/z 319 for 5-OH-5-MeHyd and 5-OH-5-MeHyd-13C,15N2, respectively; m/z 432→ m/z 417 and 435→ m/z 420 for 5,6-diOH-Ura and 5,6-diOH-Ura-13C,15N2, respectively; m/z 448→ m/z 259 and m/z 452→ m/z 262 for ThyGly and ThyGly-d4, respectively; m/z 369→ m/z 368 and m/z 372→ m/z 371 for FapyAde and FapyAde-13C,15N2, respectively; m/z 457 → m/z 368 and m/z 460 → m/z 371 for FapyGua and FapyGua-13C,15N2, respectively; m/z 455 → m/z 440 and m/z 460 → m/z 445 for 8-OH-Gua and 8-OH-Gua-15N5, respectively 36. These transitions are based on the known mass spectra of the trimethylsilyl derivatives of modified DNA bases and their fragmentation patterns, which were reported previously (for a review see38).
E. coli Nth was obtained from Dr. Susan Wallace at the University of Vermont. E. coli Fpg was prepared by Dr. Prasad Reddy at NIST39.
Another set of DNA samples (50 μg each) was used for the measurement of 8,5′-cyclopurine-2′-deoxynucleosides, i.e., (5′S)-8,5′-cyclo-2′-deoxyadenosine (S-cdA), (5′R)-8,5′-cyclo-2′-deoxyguanosine (R-cdG) and (5′S)-cyclo-2′-deoxyguanosine (S-cdG). DNA samples were supplemented with the aliquots of the stable isotope-labeled analogs of these compounds The stable isotope-labeled internal standards for modified 2′-deoxynucleosides, i.e., (5′S)-8,5′-cyclo-2′-deoxyguanosine-15N5 (S-cdG-15N5), (5′R)-8,5′-cyclo-2′-deoxyguanosine-15N5 (R-cdG-15N5), (5′S)-8,5′-cyclo-2′-deoxyadenosine-15N5 (S-cdA-15N5) and (5′R)-8,5′-cyclo-2′-deoxyadenosine-15N5 (S-cdA-15N5) were synthesized and isolated as described40, 41. The samples were dried in SpeedVac and then dissolved in 50 μL of 10 mmol/L tris (hydroxymethyl) aminomethane hydrochloride (Tris-HCl) solution (pH 7.5) containing 45 mmol/L zinc chloride (ZnCl2), supplemented with 2.5 μL of 1 mol/L sodium acetate (final pH 6.0). Aliquots of nuclease P1 (2 U), snake venom phosphodiesterase (0.004 U) and alkaline phosphatase (16 U) were added and the samples were incubated at 37 °C for 24 h. After hydrolysis, the samples were filtered using ultrafiltration membranes with a molecular mass cutoff of 3 kDa by centrifugation at 12000 g for 30 min. Filtered samples were lyophilized and then trimethylsilylated as described37. MRM scans were performed with mass transitions m/z 465 → m/z 309 for S-cdA, m/z 470 → m/z 314 for S-dA-15N5, m/z 553 → m/z 397 for R-cdG and S-cdG, and m/z 558 → m/z 402 for R-cdG-15N5 and S-cdG-15N5. These transitions are based on the known mass spectra of the trimethylsilyl derivatives of 8,5′-cyclopurine-2′-deoxynucleosides and their fragmentation patterns, which were reported previously38, 41–44. The optimal (maximum) collision energies of the trimethylsilyl derivatives of S -cdA, R -cdG and S-cdG were determined by varying the collision energy between 5 V and 35 V with 5 V increments. The maximum collision energy for each of these compounds was found to be 15 V, and this was used for the measurements.
Statistical analysis
Six independently prepared DNA samples from 6 different mussels from each collection site were used to identify and quantify the levels of oxidatively induced DNA base lesions and 8,5′-cyclopurine-2′-deoxynucleosides. Statistical analyses of the data were performed using the GraphPad Prism 7.01 software (La Jolla, CA, USA) and nonparametric Kruskal-Wallis and Dunn’s multiple comparisons tests. A p-value < 0.05 was assumed to correspond to statistically significant difference between medians.
Results
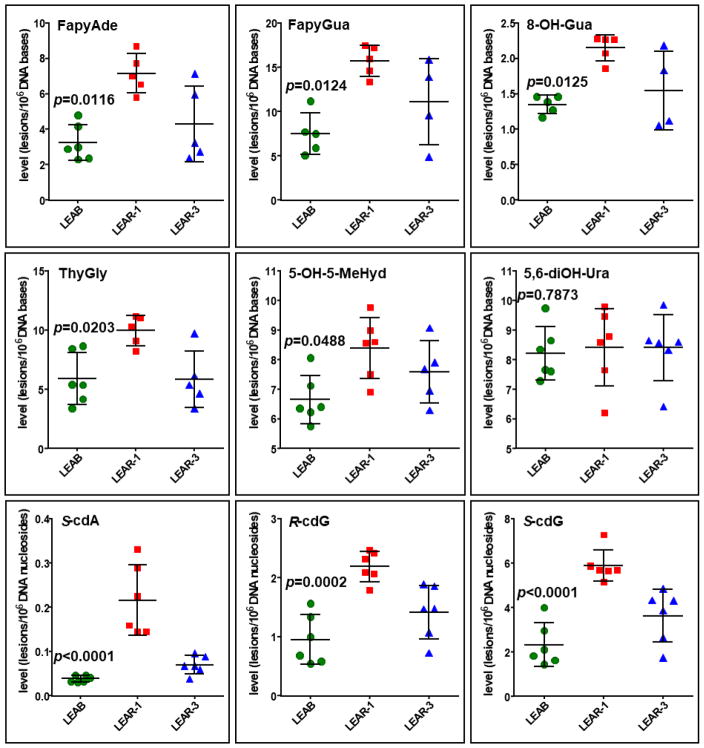
We identified and quantified six DNA base lesions and three 8,5′-cyclopurine-2′-deoxynucleosides in DNA of zebra mussels from the Ashtabula River harbor (LEAR -1 and LEAR-3) and the reference site (LEAB) using GC-MS/MS with isotope-dilution. The identified DNA lesions and their measured concentrations in DNA of the mussels are shown in Figure 2 and Figure 3, respectively. The results show that the mussels from LEAR-1 had significantly greater levels of oxidatively induced DNA bases, except for 5,6-diOH-Ura (p=0.7873), and 8,5′-cyclopurine-2′-deoxynucleosides with the p-values varying from p<0.0001 to 0.0488 than those from LEAB. The greater level of FapyGua in mussels’ DNA than that of 8-OH-Gua suggests possible low level of O2 in their cells/tissues (perhaps reflecting low oxygen level in the river/lake waters). Such phenomenon is known to generate suitable environment for the one-electron reduction of the guanine ring-opened C8-OH-adduct radical, which yields FapyGua at a greater level than 8-OH-Gua45.
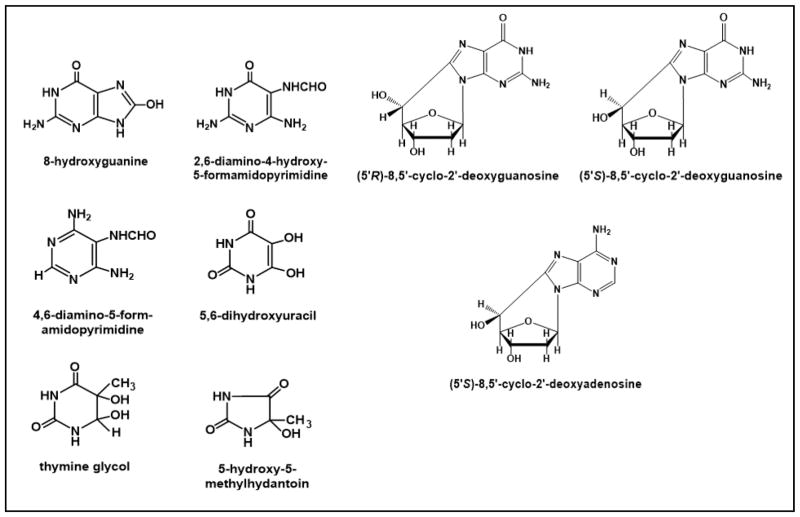
Fig. 2.
Structures of analyzed modified DNA bases and nucleosides.
Fig. 3.
Concentrations of modified DNA bases and nucleosides as measured by GC-MS/MS with isotope dilution; *p values present significant differences of median concentrations of lesions between reference site samples (LARB) and samples from AOC (LEAR-1); no significant difference found for 5,6-diOH-Ura (p=0.7873), Kruskal Wallis Test. Dunns’s multiple comparison test did not demonstrate significant differences between LEAB/LEAR-3 and LEAR-1/LEAR-3 groups. Error bars are standard deviations.
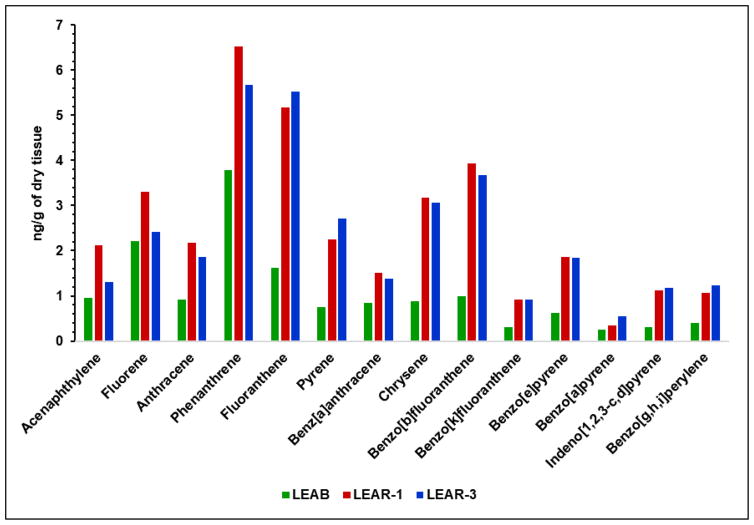
To the best of our knowledge, our work is the first to present application of quantitative mass spectrometric assessment of oxidatively induced DNA base damage in dreissenid mussels from a historically polluted harbor and a corresponding reference area. Previously, NOAA researchers had found elevated concentrations of PAHs and PCBs, including several of the approximately 40000 organic compounds identified as contaminants of emerging concern46, 47, in the outer harbor of Ashtabula River compared to the concentrations observed at relatively clean areas33. Figure 4 shows there presentative concentrations of fourteen PAHs measured in tissues of mussels in three sampling points LEAB, LEAR-1 and LEAR-3. In all cases, the levels of PAHs in LEAR-1 were greater than those in LEAB.
Fig. 4.
Representative concentrations of fourteen PAHs measured in tissues of mussels in sampling points LEAB, LEAR-1 and LEAR-3.
Except for acenaphthylene and fluorene, LEAR-3 also had elevated levels of PAHs when compared to LEAB. LEAR-3 site situated behind breakwater wall and closer to open lake waters, thus not directly exposed as LEAR -1 to sediments transported by the river stream, ship waste and substances eluted by rainwater from tons of coal and minerals stored on harbor banks. Moreover, prevailing winds and lake current move lake water into the harbor inlet and out through the southeast hole in the breakwater. Therefore, significant differences were observed between the total concentration of PAHs found in the sediments collected near sites LEAR-1 and LEAR-3, 4201 μg/kg and 2603 μg/kg, respectively48. Some individual examples of analyzed PAHs show even greater differences, e.g., benzo[a]pyrene, 267.4 μg/kg dry vs. 92 μg/kg dry; pyrene, 646.1 μg/kg dry vs. 235.0 μg/kg dry, and fluorene, 62.3 μg/kg vs. 26.3 μg/kg dry. The same NOAA report also shows striking differences in the results of the toxicity tests for Ashtabula River and harbor stations, conducted near site LEAR-1 and LEAR-3, e.g., Hyalella azteca mean survival (% ±SD), 2.50%±0.46% vs. 43.75%±4.07%, respectively. Average concentrations of PCBs in the sediments where site LEAR-1 were located were also higher than at site LEAR-3: 168 μg/kg dry mass and 58,7 μg/kg dry mass, respectively48. Data showing PAHs concentrations in mussels’ tissues and sediments together with the observed lower level of oxidatively induced DNA damage in animals from LEAR-3 vs. LEAR-1 site suggest possible higher impact on DNA damage of concentration of contaminants absorbed and metabolized through mussels’ digestive and respiratory structures than already accumulated in the tissues. Those findings point also to the sensitivity of the employed technique, which can distinguish between organisms exposed to different concentrations of pollutants; one should notice that the lower concentration of some pollutants has been also found in tissues of mussels at LEAR-3, e.g., acenaphtylene, fluorene, anthracene, phenanthrene and benz[a]anthracene (Fig. 4), which suggests a possible significant impact of those compounds on the concentrations of the markers of oxidatively induced DNA damage. The concentrations of PCBs are shown in Figure 5A as the sums of the concentrations of 39 compounds measured in tissues of mussels in three sampling points.
Fig. 5.
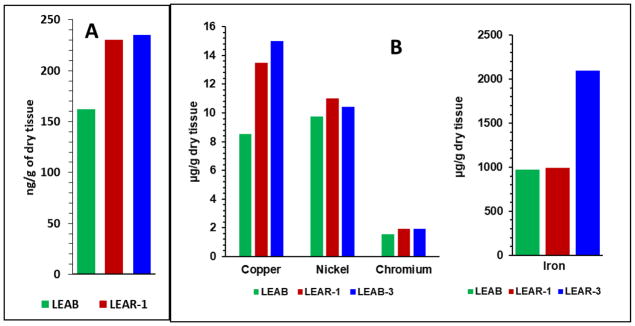
(A) PCBs concentrations shown as the sums of the concentrations of 39 compounds measured in tissues of mussels in sampling points; (B) concentrations of copper, nickel, chromium and iron at the sampling areas.
In this case, too, at least 50 % higher levels of PCBs were observed in LEAR-1 and LEAR-3 than those in LEAB. Figure 5B shows the concentrations of four metals measured at three sampling areas (data presented as from last NOAA report of concentration of those metals in 2011; concentrations of PAHs and PCBs are from 2014, NOAA personal communication: https://coastalscience.noaa.gov/projects/detail?key=179). Only the concentration of Cu was greater in LEAR-1 and LEAR-3 than in LEAB. The concentration of Fe was found to be higher in LEAR-3 than in LEAR-1 and LEAB.
Discussion
Reactions of the highly reactive •OH with DNA constituents generate a plethora of products from all four heterocyclic bases and the sugar moiety of DNA (reviewed in45). Among these products, 8,5′-cyclopurine-2′-deoxynucleosides are unique tandem lesions in that they are formed by abstraction of an H atom by •OH from the C-5′ of 2′-deoxyribose of the nucleoside, followed by C5′ C8-intramolecular cyclization and oxidation. Both (5′-R)- and (5′-S)-diastereomers of 8,5′-cyclopurine-2′-deoxynucleosides are formed (reviewed in45). Many of these DNA lesions have been identified and quantified in DNA in vitro, in cultured mammalian cells, and in human and animal tissues in vivo (reviewed in 45, 49–51). Early studies discovered that PAHs and PCBs cause hepatocellular carcinoma offish from polluted sites and in rodents, and to other detrimental biological effects such as alterations in gene expression and immune response52–62. These findings drew considerable attention to the health of aquatic animals in such environments that may have the potential to affect human health as well. Subsequent studies have suggested that oxidatively induced DNA lesions can be used as highly promising biomarkers for identifying contaminant-induced genomic changes in fish at polluted sites compared to reference sites, and also for determining whether remedial actions were successful in reducing toxic effects3, 63. Thus, a number of DNA lesions have been identified in English sole in a highly industrialized river site at greater concentrations than in a reference site3. In the present work, we hypothesized that other aquatic animals such as dreissenid mussels in the impacted sites around large lakes with expected pollution and near human populations may accumulate oxidatively induced DNA lesions, which may be used as early warning biomarkers for pollution and determining the efficacy of remedial actions.
PAHs and PCBs are known to generate •OH, and other oxygen-derived species such as superoxide radical (O2•−) and hydrogen peroxide (H2O2). The latter two can undergo further transformation into •OH in the presence of transition metal ions such as Fe+3 or Cu +2 ions 17. The DNA lesions identified in the present work are typical products of reactions of •OH with the heterocyclic bases and the sugar moiety of DNA45. If not repaired by cellular repair mechanisms prior to replication, such DNA lesions can lead to mutagenicity and/or lethality and, thus cause genetic instability that may lead to disease processes including carcinogenesis26, 64–68. Specifically, both 8-OH-Gua and FapyGua pair with adenine during replication and lead to G → T transversions69–72. These mutations are the second most common somatic mutations found in human cancers73. FapyAde causes A → T transversions; however, it is weakly mutagenic when compared to 8-OH-Gua and FapyGua 74. ThyGly constitutes a strong block to DNA polymerases and is a lethal lesion and, at best, poorly mutagenic75. 5-OH-5-MeHyd derived from thymine acts in vitro as a strong block for DNA polymerases and may be a lethal lesion in vivo76. Uracil derivatives such as uracil glycol, 5-hydroxyuraciland 5,6-diOH-Ura (isodialuric acid) are formed in DNA by deamination and dehydration of cytosine-derived lesions77. The biological effects of 5,6-diOH-Ura are not known; however, uracil glycol and 5-hydroxyuracilare strongly mutagenic75. In this work, no increase in the level of 5,6-diOH-Ura in the polluted areas was observed when compared to the reference area (Figure 3). Among 8,5′-cyclopurine-2′-deoxynucleosides, S-cdA blocks transcription and several DNA polymerases, and leads to transcriptional mutagenesis and multiple nucleotide deletions78–82. S-cdG blocks replication and leads mainly to G → A transitions, and to G → T transversions to a lesser extent 83. Taken together, the DNA lesions identified in this work with greater levels in the polluted area than in the reference area are highly mutagenic and may thus contribute to the adverse biological effects of PAHs and PCBs.
The increased levels of PAHs and PCBs, and Cu found by NOAA in the two polluted areas when compared to the reference area correspond to the increased levels of a variety of DNA lesions in mussels in the same polluted areas that we report in this work. This fact links, for the first time, the presence of PAHs and PCBs, and metals as possible cause of oxidatively induced DNA damage in terms of numerous DNA lesions in dreissenid mussels. The data suggest that the known mutagenicity or lethality of the identified DNA lesions are highly likely to contribute to carcinogenesis and other disease processes observed in aquatic animals living in polluted areas. We propose the use of oxidatively modified DNA bases and 8,5′-cyclopurine-2′-deoxynucleosides as novel quantitative biomarkers for identifying pollutant-induced changes in DNA of aquatic animals. The discovery that numerous mutagenic or lethal DNA lesions accumulate in mussels in polluted areas adds a novel dimension to previous studies that used a single DNA lesion only. Further evaluation of this monitoring tool is planned with mussels collected from other Great Lakes harbors in agricultural, and industrial watersheds, for comparison with reference sites as part of a larger strategic plan to identify and assess adverse impacts in Great Lakes tributaries.
Acknowledgments
The authors are very grateful to Dr. Susan Wallace, University of Vermont for the gift of E. coli Nth protein and to Dr. Prasad Reddy for isolating and purifying E. coli Fpg protein. Certain commercial equipment or materials are identified in this paper in order to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose. This work was conducted with support from the Great Lakes Restoration Initiative.
References
- 1.Di Leonardo R, Mazzola A, Tramati CD, Vaccaro A, Vizzini S. Highly contaminated areas as sources of pollution for adjoining ecosystems: The case of Augusta Bay (Central Mediterranean) Mar Pollut Bull. 2014;89(1–2):417–426. doi: 10.1016/j.marpolbul.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 2.Knobeloch L, Turyk M, Imm P, Schrank C, Anderson H. Temporal changes in PCB and DDE levels among a cohort of frequent and infrequent consumers of Great Lakes sportfish. Environ Res. 2009;109(1):66–72. doi: 10.1016/j.envres.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Malins DC, Anderson KM, Stegeman JJ, et al. Biomarkers signal contaminant effects on the organs of English sole (Parophrys vetulus) from Puget Sound. Environ Health Perspect. 2006;114(6):823–829. doi: 10.1289/ehp.8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barron MG, Yurk JJ, Crothers DB. Assessment of Potential Cancer Risk from Consumption of PCBs Bioaccumulated in Fish and Shellfish. Environ Health Perspect. 1994;102(6–7):562–567. doi: 10.1289/ehp.94102562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guengerich FP, Shimada T. Oxidation of toxic and carcinogenic chemicals by human cytochrome P-450 enzymes. Chem Res Toxicol. 1991;4(4):391–407. doi: 10.1021/tx00022a001. [DOI] [PubMed] [Google Scholar]
- 6.Dipple A. Polycyclic Aromatic Hydrocarbon Carcinogenesis - an Introduction. Acs Sym Ser. 1985;283:1–17. [Google Scholar]
- 7.Xue W, Warshawsky D. Metabolic activation of polycyclic and heterocyclic aromatic hydrocarbons and DNA damage: a review. Toxicol Appl Pharmacol. 2005;206(1):73–93. doi: 10.1016/j.taap.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin AK, Corsi SR, De Cicco LA, et al. Organic contaminants in Great Lakes tributaries: Prevalence and potential aquatic toxicity. Sci Total Environ. 2016;554–555:42–52. doi: 10.1016/j.scitotenv.2016.02.137. [DOI] [PubMed] [Google Scholar]
- 9.Stegeman JJ, Lech JJ. Cytochrome P-450 monooxygenase systems in aquatic species: carcinogen metabolism and biomarkers for carcinogen and pollutant exposure. Environ Health Perspect. 1991;90:101–109. doi: 10.1289/ehp.90-1519513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagnaire B, Geffard O, Noury P, Garric J. In vivo indirect measurement of cytochrome P450-associated activities in freshwater gastropod molluscs. Environ Toxicol. 2010;25(6):545–553. doi: 10.1002/tox.20515. [DOI] [PubMed] [Google Scholar]
- 11.Michel C, Vincent-Hubert F. DNA oxidation and DNA repair in gills of zebra mussels exposed to cadmium and benzo(a)pyrene. Ecotoxicology. 2015;24(9):2009–2016. doi: 10.1007/s10646-015-1536-3. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira M, Maria VL, Ahmad I, et al. Golden grey mullet and sea bass oxidative DNA damage and clastogenic/aneugenic responses in a contaminated coastal lagoon. Ecotoxicol Environ Saf. 2010;73(8):1907–1913. doi: 10.1016/j.ecoenv.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 13.Vincent-Hubert F, Arini A, Gourlay-France C. Early genotoxic effects in gill cells and haemocytes of Dreissena polymorpha exposed to cadmium, B[a]P and a combination of B[a]P and Cd. Mutat Res. 2011;723(1):26–35. doi: 10.1016/j.mrgentox.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Park JH, Gopishetty S, Szewczuk LM, Troxel AB, Harvey RG, Penning TM. Formation of 8- oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dGuo) by PAH o-quinones: involvement of reactive oxygen species and copper(II)/copper(I) redox cycling. Chem Res Toxicol. 2005;18(6):1026–1037. doi: 10.1021/tx050001a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer WA, Lehmler HJ, Robertson LW, Gupta RC. Oxidative DNA adducts after Cu(2+)- mediated activation of dihydroxy PCBs: role of reactive oxygen species. Free Radic Biol Med. 2009;46(10):1346–1352. doi: 10.1016/j.freeradbiomed.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Anati L, Viluksela M, Strid A, et al. Hydroxyl metabolite of PCB 180 induces DNA damage signaling and enhances the DNA damaging effect of benzo[a]pyrene. Chem Biol Interact. 2015;239:164–173. doi: 10.1016/j.cbi.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. 5. Oxford University Press; 2015. [Google Scholar]
- 18.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95(1):1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McElroy A, Leitch K, Fay A. A survey of in vivo benzo[alpha]pyrene metabolism in small benthic marine invertebrates. Mar Environ Res. 2000;50(1–5):33–38. doi: 10.1016/s0141-1136(00)00054-4. [DOI] [PubMed] [Google Scholar]
- 20.Douben PET. PAHs: An Ecotoxicological Perspective. 1. Vol. 1. Chichester, West SussexPO19 8SQ, England: John Wiley & Sons Ltd; 2003. [Google Scholar]
- 21.Le Goff J, Gallois J, Pelhuet L, et al. DNA adduct measurements in zebra mussels, Dreissena polymorpha, Pallas. Potential use for genotoxicant biomonitoring of fresh water ecosystems. Aquat Toxicol. 2006;79(1):55–64. doi: 10.1016/j.aquatox.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Giannapas M, Karnis L, Dailianis S. Generation of free radicals in haemocytes of mussels after exposure to low molecular weight PAH components: immune activation, oxidative and genotoxic effects. Comp Biochem Physiol C Toxicol Pharmacol. 2012;155(2):182–189. doi: 10.1016/j.cbpc.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 23.Chatel A, Faucet-Marquis V, Pfohl-Leszkowicz A, Gourlay-France C, Vincent-Hubert F. DNA adduct formation and induction of detoxification mechanisms in Dreissena polymorpha exposed to nitro-PAHs. Mutagenesis. 2014;29(6):457–465. doi: 10.1093/mutage/geu040. [DOI] [PubMed] [Google Scholar]
- 24.Song Q, Zheng P, Qiu L, Jiang X, Zhao H. Toxic effects of male Perna viridis gonad exposed to BaP, DDT and their mixture: A metabolomic and proteomic study of the underlying mechanism. Toxicology letters. 2016;240(1):185–195. doi: 10.1016/j.toxlet.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 25.Capo X, Tejada S, Box A, Deudero S, Sureda A. Oxidative status assessment of the endemic bivalve Pinna nobilis affected by the oil spill from the sinking of the Don Pedro. Mar Environ Res. 2015;110:19–24. doi: 10.1016/j.marenvres.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. Washington, D.C: ASM Press; 2006. [Google Scholar]
- 27.Farris JL, Van Hassel JH. Freshwater Bivalve Ecotoxicology. CRC Press; 2006. [Google Scholar]
- 28.Markert BA, Breure AM, Zechmeister HG. Bioindicators and Biomonitors. Elsevier Science; 2003. [Google Scholar]
- 29.Binelli A, Della Torre C, Magni S, Parolini M. Does zebra mussel (Dreissena polymorpha) represent the freshwater counterpart of Mytilus in ecotoxicological studies? A critical review. Environ Pollut. 2015;196:386–403. doi: 10.1016/j.envpol.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Shoults-Wilson WA, Elsayed N, Leckrone K, Unrine J. Zebra mussels (Dreissena polymorpha) as a biomonitor of trace elements along the southern shoreline of Lake Michigan. Environ Toxicol Chem. 2015;34(2):412–419. doi: 10.1002/etc.2825. [DOI] [PubMed] [Google Scholar]
- 31.Schafer S, Hamer B, Treursic B, et al. Comparison of bioaccumulation and biomarker responses in Dreissena polymorpha and D. bugensis after exposure to resuspended sediments. Arch Environ Contam Toxicol. 2012;62(4):614–627. doi: 10.1007/s00244-011-9735-2. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe M, Meyer KA, Jackson TM, Schock TB, Johnson WE, Bearden DW. Application of NMR-based metabolomics for environmental assessment in the Great Lakes using zebra mussel (Dreissena polymorpha) Metabolomics. 2015;11(5):1302–1315. doi: 10.1007/s11306-015-0789-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimbrough K, Johnson WE, Jacob A, et al. Mussel Watch Great Lakes Contaminant Monitoring and Assessment: Phase 1. Silver Spring, MD NOAA Technical Memorandum NOS NCCOS. 2014;180:1–113. [Google Scholar]
- 34.Kimbrough KL, Lauenstein GGE. Major and Trace Element Analytical Methods of the National Status and Trends Program: Update 2000–2006. NOAA Technical Memorandum NOS NCCOS 29, Silverspring, MD. 2007 [Google Scholar]
- 35.Lauenstein GG, Cantillo AY. Analytical methods of the National Status and Trends Program Mussel Watch Project - 1993–1997 Update. NOAA, Technical Memorandum NOS ORCA 130 Silver Spring, MD. 1998 [Google Scholar]
- 36.Reddy PT, Jaruga P, Kirkali G, Tuna G, Nelson BC, Dizdaroglu M. Identification and quantification of human DNA repair protein NEIL1 by liquid chromatography/isotope-dilution tandem mass spectrometry. J Proteome Res. 2013;12(2):1049–1061. doi: 10.1021/pr301037t. [DOI] [PubMed] [Google Scholar]
- 37.Jaruga P, Kirkali G, Dizdaroglu M. Measurement of formamidopyrimidines in DNA. Free Radic Biol Med. 2008;45(12):1601–1609. doi: 10.1016/j.freeradbiomed.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Dizdaroglu M, Coskun E, Jaruga P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic Res. 2015;49:525–548. doi: 10.3109/10715762.2015.1014814. [DOI] [PubMed] [Google Scholar]
- 39.Reddy P, Jaruga P, O’Connor T, Rodriguez H, Dizdaroglu M. Overexpression and rapid purification of Escherichia coli formamidopyrimidine-DNA glycosylase. Protein Expr Purif. 2004;34(1):126–133. doi: 10.1016/j.pep.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 40.Birincioglu M, Jaruga P, Chowdhury G, Rodriguez H, Dizdaroglu M, Gates KS. DNA base damage by the antitumor agent 3-amino-1,2,4-benzotriazine 1,4-dioxide (tirapazamine) J Am Chem Soc. 2003;125(38):11607–11615. doi: 10.1021/ja0352146. [DOI] [PubMed] [Google Scholar]
- 41.Jaruga P, Birincioglu M, Rodriguez H, Dizdaroglu M. Mass spectrometric assays for the tandem lesion 8,5′-cyclo-2′-deoxyguanosine in mammalian DNA. Biochemistry. 2002;41(11):3703–3711. doi: 10.1021/bi016004d. [DOI] [PubMed] [Google Scholar]
- 42.Dizdaroglu M. Free-radical-induced formation of an 8,5′-cyclo-2′-deoxyguanosine moiety in deoxyribonucleic acid. Biochem J. 1986;238:247–254. doi: 10.1042/bj2380247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dirksen ML, Blakely WF, Holwitt E, Dizdaroglu M. Effect of DNA conformation on the hydroxyl radical-Induced formation of 8,5′-cyclopurine-2′-deoxyribonucleoside residues in DNA. Int J Radiat Biol. 1988;54:195–204. doi: 10.1080/09553008814551631. [DOI] [PubMed] [Google Scholar]
- 44.Jaruga P, Dizdaroglu M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst) 2008;7(9):1413–1425. doi: 10.1016/j.dnarep.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Dizdaroglu M, Jaruga P. Mechanisms of free radical-induced damage to DNA. Free Radic Res. 2012;46(4):382–419. doi: 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- 46.Diamond JM, Latimer HA, Munkittrick KR, Thornton KW, Bartell SM, Kidd KA. Prioritizing Contaminants of Emerging Concern for Ecological Screening Assessments. Environ Toxicol Chem. 2011;30(11):2385–2394. doi: 10.1002/etc.667. [DOI] [PubMed] [Google Scholar]
- 47.Klecka G, Persoon C, Currie R. Chemicals of emerging concern in the Great Lakes Basin: an analysis of environmental exposures. Rev Environ Contam Toxicol. 2010;207:1–93. doi: 10.1007/978-1-4419-6406-9_1. [DOI] [PubMed] [Google Scholar]
- 48.Cooksey C, Balthis BW, Fulton MH, Hyland JL, Wirth E. Final Project Report to U.S. EPA under EPA/NOAA Interagency Agreement DW-13–92359501–0. NOAA National Ocean Service; Charleston, SC 29412–9110: 2016. Assessment of Ecological Condition and Stressor Impacts within Great Lakes Rivers and Harbors: Ashtabula River, Ohio; pp. 1–62. [Google Scholar]
- 49.von Sonntag C. Free-Radical-Induced DNA Damage and Its Repair. Hiedelberg: Springer; 2006. [Google Scholar]
- 50.Cadet J, Douki T, Ravanat JL. Oxidatively generated base damage to cellular DNA. Free Radic Biol Med. 2010;49(1):9–21. doi: 10.1016/j.freeradbiomed.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 51.Dizdaroglu M. Oxidatively induced DNA damage: Mechanisms, repair and disease. Cancer Lett. 2012;327:26–47. doi: 10.1016/j.canlet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 52.Dawe CJ, Stanton MF, Schwartz FJ. Hepatic neoplasms in native bottom-feeding fish of deep creek lake, Maryland. Cancer Res. 1964;24:1194–1201. [PubMed] [Google Scholar]
- 53.Mayes BA, McConnell EE, Neal BH, et al. Comparative carcinogenicity in Sprague-Dawley rats of the polychlorinated biphenyl mixtures Aroclors 1016, 1242, 1254, and 1260. Toxicol Sci. 1998;41(1):62–76. doi: 10.1093/toxsci/41.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keyte IJ, Harrison RM, Lammel G. Chemical reactivity and long-range transport potential of polycyclic aromatic hydrocarbons--a review. Chem Soc Rev. 2013;42(24):9333–9391. doi: 10.1039/c3cs60147a. [DOI] [PubMed] [Google Scholar]
- 55.Onozuka D, Yoshimura T, Kaneko S, Furue M. Mortality after exposure to polychlorinated biphenyls and polychlorinated dibenzofurans: a 40-year follow-up study of Yusho patients. Am J Epidemiol. 2009;169(1):86–95. doi: 10.1093/aje/kwn295. [DOI] [PubMed] [Google Scholar]
- 56.Mondon JA, Duda S, Nowak BF. Immune response of greenback flounder Rhombosolea tapirina after exposure to contaminated marine sediment and diet. Mar Environ Res. 2000;50(1–5):443–450. doi: 10.1016/s0141-1136(00)00253-1. [DOI] [PubMed] [Google Scholar]
- 57.Peterson JS, Bain LJ. Differential gene expression in anthracene-exposed mummichogs (Fundulus heteroclitus) Aquat Toxicol. 2004;66(4):345–355. doi: 10.1016/j.aquatox.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 58.MacCubbin AE. DNA adduct analysis in fish: laboratory and field studies. In: Malins DC, Ostrander GK, editors. Aquatic Toxicology: Molecular, Biochemical, and Cellular Perspectives. Baco Raton, FL: Lewis Publishers; 1994. pp. 267–294. [Google Scholar]
- 59.Malins DC, McCain BB, Brown DW, Myers MS, Krahn MM, Chan SL. Toxic chemicals, including aromatic and chlorinated hydrocarbons and their derivatives, and liver lesions in white croaker (Genyonemus lineatus) from the vicinity of Los Angeles. Environ Sci Technol. 1987;21(8):765–770. doi: 10.1021/es00162a006. [DOI] [PubMed] [Google Scholar]
- 60.Roling JA, Bain LJ, Baldwin WS. Differential gene expression in mummichogs (Fundulus heteroclitus) following treatment with pyrene: comparison to a creosote contaminated site. Mar Environ Res. 2004;57(5):377–395. doi: 10.1016/j.marenvres.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 61.Grinwis GC, Vethaak AD, Wester PW, Vos JG. Toxicology of environmental chemicals in the flounder (Platichthys flesus) with emphasis on the immune system: field, semi-field (mesocosm) and laboratory studies. Toxicol Lett. 2000;112–113:289–301. doi: 10.1016/s0378-4274(99)00239-8. [DOI] [PubMed] [Google Scholar]
- 62.Schlezinger JJ, White RD, Stegeman JJ. Oxidative inactivation of cytochrome P-450 1A (CYP1A) stimulated by 3,3′,4,4′-tetrachlorobiphenyl: production of reactive oxygen by vertebrate CYP1As. Mol Pharmacol. 1999;56(3):588–597. doi: 10.1124/mol.56.3.588. [DOI] [PubMed] [Google Scholar]
- 63.Malins DC, Polissar NL, Garner MM, Gunselman SJ. Mutagenic DNA base modifications are correlated with lesions in nonneoplastic hepatic tissue of the English sole carcinogenesis model. Cancer Res. 1996;56(24):5563–5565. [PubMed] [Google Scholar]
- 64.Vogelstein B, Kinzler KW. The Genetic Basis of Human Cancer. New York: McGraw-Hill; 1998. [Google Scholar]
- 65.Friedberg EC. DNA damage and repair. Nature. 2003;421(6921):436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 66.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 67.Beckman RA, Loeb LA. Genetic instability in cancer: theory and experiment. Semin Cancer Biol. 2005;15(6):423–435. doi: 10.1016/j.semcancer.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 68.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nature Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 70.Kuchino Y, Mori F, Kasai H, et al. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 71.Wiederholt CJ, Greenberg MM. Fapy.dG instructs Klenow exo− to misincorporate deoxyadenosine. Journal of the American Chemical Society. 2002;124:7278–7679. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 72.Kalam MA, Haraguchi K, Chandani S, et al. Genetic effects of oxidative DNA damages: comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34(8):2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Delaney MO, Wiederholt CJ, Greenberg MM. Fapy-dA induces nucleotide misincorporation tranlesionally by a DNA polymerase. Angew Chem Int Ed Engl. 2002;41:771–775. doi: 10.1002/1521-3773(20020301)41:5<771::aid-anie771>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 75.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33(1):1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 76.Gasparutto D, Ait-Abbas M, Jaquinod M, Boiteux S, Cadet J. Repair and coding properties of 5-hydroxy-5-methylhydantoin nucleosides inserted into DNA oligomers [In Process Citation] Chem Res Toxicol. 2000;13(7):575–584. doi: 10.1021/tx000005+. [DOI] [PubMed] [Google Scholar]
- 77.Dizdaroglu M, Holwitt E, Hagan MP, Blakely WF. Formation of cytosine glycol and 5,6-dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem J. 1986;235:531–536. doi: 10.1042/bj2350531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kuraoka I, Bender C, Romieu A, Cadet J, Wood RD, Lindahl T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc Natl Acad Sci U S A. 2000;97(8):3832–3837. doi: 10.1073/pnas.070471597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brooks PJ, Wise DS, Berry DA, et al. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J Biol Chem. 2000;275(29):22355–22362. doi: 10.1074/jbc.M002259200. [DOI] [PubMed] [Google Scholar]
- 80.Kuraoka I, Robins P, Masutani C, et al. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase η and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J Biol Chem. 2001;276(52):49283–49288. doi: 10.1074/jbc.M107779200. [DOI] [PubMed] [Google Scholar]
- 81.Marietta C, Brooks PJ. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 2007;8(4):388–393. doi: 10.1038/sj.embor.7400932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marietta C, Gulam H, Brooks PJ. A single 8,5′-cyclo-2′-deoxyadenosine lesion in a TATA box prevents binding of the TATA binding protein and strongly reduces transcription in vivo. DNA Repair (Amst) 2002;1(11):967–975. doi: 10.1016/s1568-7864(02)00148-9. [DOI] [PubMed] [Google Scholar]
- 83.Jasti VP, Das RS, Hilton BA, Weerasooriya S, Zou Y, Basu AK. (5′S)-8,5′-Cyclo-2′-deoxyguanosine Is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry. 2011;50(19):3862–3865. doi: 10.1021/bi2004944. [DOI] [PMC free article] [PubMed] [Google Scholar]