Abstract
Guanosine monophosphate synthetase (GMPS), encoded by guaA gene, is a key enzyme for guanine nucleotide biosynthesis in Mycobacterium tuberculosis. The guaA gene from several bacterial pathogens has been shown to be involved in virulence; however, no information about the physiological effect of direct guaA deletion in M. tuberculosis has been described so far. Here, we demonstrated that the guaA gene is essential for M. tuberculosis H37Rv growth. The lethal phenotype of guaA gene disruption was avoided by insertion of a copy of the ortholog gene from Mycobacterium smegmatis, indicating that this GMPS protein is functional in M. tuberculosis. Protein validation of the guaA essentiality observed by PCR was approached by shotgun proteomic analysis. A quantitative method was performed to evaluate protein expression levels, and to check the origin of common and unique peptides from M. tuberculosis and M. smegmatis GMPS proteins. These results validate GMPS as a molecular target for drug design against M. tuberculosis, and GMPS inhibitors might prove to be useful for future development of new drugs to treat human tuberculosis.
Keywords: Guanosine monophosphate synthetase, Nucleotide biosynthesis, GuaA gene, Essential gene, Mycobacterium tuberculosis, Mass spectrometry
Highlights
-
•
The guaA gene is essential for M. tuberculosis H37Rv growth.
-
•
The lethal phenotype of guaA gene disruption was avoided by the ortholog gene from M. smegmatis.
-
•
Multiplexed LC–MS/MS analysis was performed to validate protein expression levels.
-
•
The guaA essentiality was confirmed by gene replacement and quantitative mass spectrometry.
1. Introduction
The causative agent of tuberculosis (TB), Mycobacterium tuberculosis, infects one-third of the world population. The World Health Organization estimates that 9 million new TB cases occurred in 2013, resulting in 1.5 million deaths, of which 360,000 were HIV-positive individuals, and approximately 210,000 were people infected with multidrug-resistant strains [1]. The emergence of drug-resistant strains and the global spread of HIV are aggravating factors related to this infectious disease. Thus, there is a critical need for the development of new drugs and vaccines to decrease TB incidence worldwide.
Understanding the role of enzymes involved in the metabolic pathways of M. tuberculosis is crucial to identify targets for rational drug and vaccine designs. Enzymes involved in the purine biosynthesis pathways have important roles in cellular metabolism, as they provide nucleotides that are essential components of a number of biomolecules [2]. Guanosine monophosphate synthetase (EC 6.3.5.2) (GMPS), encoded by guaA gene (Rv3396c), catalyzes the conversion of xanthosine 5′-monophosphate (XMP) into guanosine 5′-monophosphate (GMP), and is a key enzyme in both purine de novo and salvage pathways of guanine nucleotides [3].
The guaA gene from several bacterial pathogens has been shown to be involved in virulence. Deletion of guaA gene from Francisella tularensis, the infectious agent of tularemia, resulted in a guanine auxotrophic strain that was highly attenuated in a mouse model of infection, and failed to replicate in macrophages [4]. Similarly, the construction of a deletion mutation in the guaBA operon of Shigella flexneri generated a guanine auxotrophic strain effectively attenuated in guinea pig infection model [5]. The guaAB operon was also reported to be essential for survival and infectivity of Borrelia burgdorferi, the causative agent of Lyme borreliosis [6]. Unlike other bacteria, guaA and guaB genes are located in different loci within M. tuberculosis genomic DNA. Both guaB2 (Rv3411c, encoding inosine monophosphate dehydrogenase, IMPDH) and guaA genes were predicted to be essential for M. tuberculosis growth in previous high density mutagenesis studies [7], [8]. The biochemical characterization of M. tuberculosis GMPS (Mtb-GMPS) has been recently published [3]; however, no information on the physiological effect of direct guaA disruption in M. tuberculosis has been described so far.
Therefore, in this study we aimed to evaluate the essentiality of guaA by gene replacement in M. tuberculosis H37Rv strain. The evidence of essentiality was confirmed at protein level via LC–MS/MS through Multidimensional Protein Identification Technology (MudPIT) analysis of M. tuberculosis cellular extracts. Multiplexed quantitative LC–MS/MS analysis with isotopic dimethyl labeling was performed to validate protein expression levels, and to confirm the origin of shared and unique peptides from both gene products.
2. Materials and methods
2.1. Construction of the suicide delivery vector
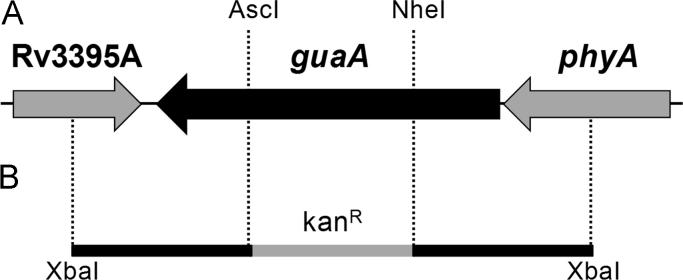
A fragment of 2577 bp containing the guaA gene (1518 bp) with its flanking region was amplified by polymerase chain reaction (PCR) from M. tuberculosis H37Rv genomic DNA, using primers forward (5′-tttttctagaggacccgtcagcacggcgac-3′) and reverse (5′-tttttctagattgtcgacgcgcccacattgca-3′), both containing XbaI restriction sites (underlined). The 2577 bp fragment was subsequently cloned into pUC19 using the XbaI restriction site. The guaA gene was disrupted by the insertion of a kanamycin cassette from pUC4K into unique internal restriction enzymes sites NheI and AscI, releasing a fragment of 719 bp from guaA gene (Fig. 1). Insert was released from pUC19 derivative vector by digestion with XbaI, and subcloned into XbaI linearized pPR27xylE vector [9].
Fig. 1.
Genomic environment of guaA gene in M. tuberculosis (A) and regions cloned into pPR27xylE vector (B). A. Genomic region of guaA gene (1.578 kb) containing unique internal NheI and AscI sites and flanking genes. B. The guaA gene and flanking regions were amplified by PCR from M. tuberculosis H37Rv genomic DNA using primers forward and reverse both containing XbaI restriction sites. The guaA gene was disrupted by the insertion of a kanamycin cassette (kanR) into NheI and AscI sites (guaA::kanR), releasing a fragment of 719 bp. The guaA::kanR fragment was cloned into pPR27xylE vector.
2.2. Construction of the complementing vector
The ortholog guaA gene from Mycobacterium smegmatis mc2155 (MSMEG_1610), flanked by about 200 bp upstream and 100 bp downstream, was amplified by PCR from M. smegmatis genomic DNA using primers forward (5′-tttttctagagccgtccaggtcgaacaggca-3′) and reverse (5′-tttttctagaacccacgagcagcgaacaatt-3′), both containing XbaI restriction sites (underlined), and was cloned into XbaI linearized pNIP40/b, a mycobacteriophage Ms6-derived integrative vector [10].
2.3. Generation of a strain containing disrupted guaA gene
M. tuberculosis H37Rv strain was transformed by electroporation with approximately 2 µg of suicide delivery vector construction. Bacteria were plated on Middlebrook 7H10 10% OADC containing 25 µg/mL kanamycin, and incubated at 32 °C. After 6 weeks, 1% catechol was dropped on colonies to select those containing the plasmid. Six different yellow colonies were picked up from the transformant, and cultivated in Middlebrook 7H9 10% OADC 0.05% tween-80 with 25 µg/mL kanamycin at 32 °C. Individual cultures were plated on Middlebrook 7H10 10% OADC containing 25 µg/mL kanamycin, 2% sucrose, and cultivated at 39 °C. Clones were also plated on solid media containing 20 µg/mL of guanine to evaluate the presence of guanine auxotrophic colonies. After 6 weeks, 1% catechol was dropped on colonies to select those that might be double crossover (DCO) strains. The white colonies were inoculated in liquid media containing 25 µg/mL kanamycin, and cultivated at 37 °C for 3 weeks. Genomic DNA was isolated and PCRs were carried out using gene-specific screening primers forward (5′-gcccgtatggaaatcgactg-3′) and reverse (5′-actatcgcactaaccggcac-3′) to determine whether the wild-type or the disrupted guaA gene was present in the targeted chromosomal region.
2.4. Generation of complemented strain
In order to obtain the complemented strain with the guaA gene from M. smegmatis, M. tuberculosis H37Rv was transformed by electroporation with about 2 µg of the complementing vector construction. Bacteria were plated on Middlebrook 7H10 10% OADC containing 50 µg/mL hygromycin, and incubated at 37 °C. After 3 weeks, a single colony was cultivated in 5 mL Middlebrook 7H9 10% OADC 0.05% tween-80 with 50 µg/mL hygromycin at 37 °C, subcultivated in 50 mL of the same medium, and electrocompetent cells were prepared. The M. tuberculosis strain containing the complemented vector integrated on its genomic DNA was transformed by electroporation with about 2 µg of suicide delivery vector construction. The following steps to obtain the complemented strain were carried out as described above for the generation of a strain containing the guaA gene disrupted, with the addition of 50 µg/mL hygromycin on culture medium.
2.5. Protein extraction
Wild-type H37Rv and complemented strains were grown in 50 mL Middlebrook 7H9 10% OADC 0.05% tween-80 containing the proper antibiotics up to an OD600 of 0.5–0.7. Cellular pellets were washed twice using 10 mM Tris HCl pH 8.0. Cells were resuspended in 1 mL of the same buffer containing protease inhibitor cocktail (Promega), and then transferred to 2 mL lysing matrix B tubes containing 0.1 mm diameter silica beads (MP Biomedicals). Cells were disrupted by using a L-Beader 3 (Loccus Biotecnologia) at a speed setting of 4000 rpm, 10 cycles of 30 s each, cooling between cycles. After lysis, the cell free supernatants were collected by centrifugation at 2300g for 5 min at room temperature. The supernatants were filtered through 0.22 μm Millex Durapore filters (Millipore). Protein concentrations were measured using the bicinchoninic acid assay (Pierce).
2.6. Liquid chromatography and tandem mass spectrometry (LC–MS/MS) analysis
2.6.1. In solution digestion
Chloroform/methanol protein precipitation was performed in cellular extracts (200 µg) from wild-type and complemented strains, according to Wessel and Flügge [11]. Protein pellets were resuspended in 100 mM tris (hydroxymethyl) aminomethane (Tris) HCl pH 8.5 containing 8 M urea, and digested with a protocol adapted from Klammer and MacCoss [12]. Briefly, disulfide bonds were reduced in 5 mM dithiothreitol (DTT) for 20 min at 37 °C and then cysteines were alkylated in 25 mM iodoacetamide (IAM) for 20 min at room temperature in the dark. Urea was diluted to 2 M with 100 mM Tris, and the complex protein extract was digested with trypsin in a ratio of 1:100 enzyme/protein along with 1 mM CaCl2 by overnight incubation at 37 °C. Formic acid was added to finish the reaction (5% v/v, final concentration).
2.6.2. Stable isotope dimethyl labeling
For dimethyl labeling, cellular extracts were submitted to the digestion protocol described above, using triethylammonium bicarbonate (TEAB) buffer instead of Tris. After overnight trypsin digestion, samples were stored at −20 °C without addition of formic acid. Isotopic dimethyl labeling reactions were performed as described by Boersema et al. [13]. Peptides (100 µg) were labeled at free-amines (N-terminus and lysine side chain), by combining isotopic forms of formaldehyde and sodium cyanoborohydride, resulting in mass shifts of +28.0313 Da (Light: CH2O+NaBH3CN), +32.0564 Da (Intermediate: CD2O+NaBH3CN), or +36.0757 Da (Heavy: 13CD2O+NaBD3CN) per incorporated label. Wild-type cellular extract sample (containing Mtb-GMPS) was labeled as “Light”, a mix of equal amounts of wild-type and complemented cellular extracts (containing M. smegmatis GMPS, Msm-GMPS) was labeled as “Intermediate”, and complemented cellular extract sample was labeled as “Heavy”.
2.6.3. LC–MS/MS and MudPIT
Samples were analyzed in technical triplicates via LC–MS/MS. Chromatographic separations were performed on an Eksigent nanoLC Ultra 1D plus autosampler (currently part of AB Sciex) connected to a LTQ-XL Orbitrap Discovery hybrid instrument (Thermo Fisher Scientific) through a nanoeletrospray ion source (Thermo Fisher Scientific). Pre-columns and biphasic MudPIT columns (150 µm ID/360 µm OD fused-silica capillary) were prepared in house by slurry packing 2 cm of C18 reversed phase resin (5 µm ODS-AQ C18 Yamamura Chemical Lab) and 2.5 cm of SCX resin (5 µm Partisphere, Whatman) followed by 2.5 cm of C18 reversed phase material, respectively. Capillary analytical columns (100 µm ID/360 µm OD capillary) were slurry packed with 20 cm of C18 reversed phase packing material behind a 5 µm ID tip.
Five micrograms of either wild-type or complemented cellular extracts were injected through the autosampler at 1 µL/min for 15 min. Mobile phases were water/acetonitrile/formic acid (95:5:0.1) as buffer A, and water/acetonitrile/formic acid (10:90:0.1) as buffer B. Gradient elution was 6 h long at 300 nL/min as follows: 0–5% B in 5 min; 5–25% B in 20 min; 25–50% B in 25 min; 50–80% B in 30 min; 80% B for 5 min; 80–0% B in 1 min; 5% B for 14 min.
MudPIT analysis was carried out according to Wolters et al. [14]. Light, Intermediate and Heavy samples were mixed in equal proportions (25 µg each) and pressure-loaded into a biphasic column. MudPIT consisted of one transfer step followed by 10 buffer C (500 mM ammonium acetate in buffer A) injections (10, 20, 30, 50, 60, 70, 85, 100% C and 90% C/10% B twice) and the reversed phase elution gradients. Transfer step gradient was 1 h long at 300 nl/min as follows: 0–10% B in 3 min; 10–50% B in 39 min; 50–90% B in 5 min; 90% B for 2 min; 90–0% B in 1 min; 0% B for 10 min. Elution gradients were 2 h long at 300 nl/min as follows: 0–5% B in 5 min; 5–25% B in 60 min; 25–50% B in 40 min; 50–80% B in 25 min; 80% B for 5 min; 80–0% B in 1 min; 5% B for 14 min.
Data were collected with one FT full-scan (400–1600 m/z range; 30,000 resolution) followed by data dependent CID MS/MS spectra of the 8 most intense ions in the ion trap, with dynamic exclusion (1 repeat count, 30 s repeat duration, 100 exclusion list size, and 30 s exclusion duration). The mass spectrometer and HPLC were controlled by the Xcalibur data system (Thermo Fisher Scientific).
3. Data analysis
Tandem mass spectra were searched from RAW files with the software Comet [15] in the PatternLab for Proteomics platform [16]. The database contained a non-redundant M. tuberculosis H37Rv reference proteome (Proteome ID UP000001584, www.uniprot.org), the Msm-GMPS sequence (MSMEG_1610, www.uniprot.org), and reverse sequences of all entries. Search space included all fully and semitryptic peptide candidates. Carbamidomethylation of cysteine was considered as static modification. The validity of the peptide spectra matches (PSMs) was assessed using the module Search Engine Processor (SEPro, PatternLab for Proteomics), with a false discovery rate of 1%.
For the dimethyl labeled samples, protein and peptide identification and quantification were carried out in Integrated Proteomics Pipeline-IP2 (Integrated Proteomics Applications, Inc., http://www.integratedproteomics.com). Tandem mass spectra were extracted from RAW files using RawXxtract 1.9.9.2 [17] and searched with ProLuCID v.1.3.1 [18], including fully and semitryptic peptide candidates, using carbamidomethylation of cysteine as static modification, and light, intermediate and heavy forms of dimethylation as metabolic modifications. Peptide candidates were filtered using DTASelect 2.0 [19]. Quantitative analysis of the entire proteome was performed by Census integrated into IP2 pipeline [20]. Mtb-GMPS and Msm-GMPS ratios were calculated using Qual Browser from Thermo XCalibur 2.2 (Thermo Fisher Scientific) and Skyline [21].
4. Results
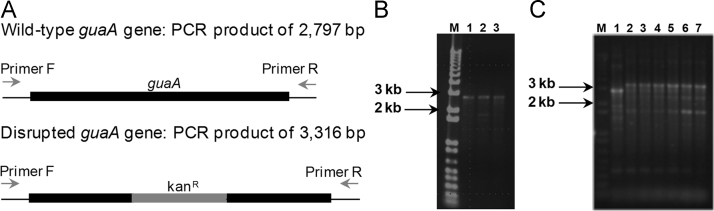
GMPS activity was shown to be essential for M. tuberculosis growth in previous high density mutagenesis studies [7], [8]. In order to directly evaluate GMPS essentiality, a genetic strategy using the pPR27xylE vector was employed to disrupt the functional guaA gene from the chromosome of M. tuberculosis H37Rv. The pPR27xylE vector contains the counter-selective sacB gene, the reporter gene xylE, and a mycobacterial thermosensitive origin of replication, which enable the selection of mutants in M. tuberculosis [9]. The genomic environment of guaA gene and the regions used to construct the suicide delivery vector are shown in Fig. 1. The pPR27xylE construction containing the guaA gene disrupted by a selection marker was introduced into the M. tuberculosis H37Rv strain; six recombinant yellow colonies were selected. In the next step, all the possible DCO white colonies obtained on plates, either containing guanine or not, were inoculated on liquid media, their genomic DNAs extracted and PCRs carried out using gene-specific primers to determine whether the wild-type or the disrupted guaA gene was present. Of the 17 potential DCOs screened, all were wild-type for the guaA gene either from plates containing guanine or not (Fig. 2A and B). This result is an indication that the guaA gene is essential for M. tuberculosis growth in the experimental conditions employed.
Fig. 2.
The guaA gene is essential for M. tuberculosis growth. A. Expected PCR product sizes using gene-specific screening primers forward (F) and reverse (R) for strains containing guaA gene either wild-type or disrupted. B. Agarose gel electrophoresis of PCR products from possible double crossover (DCO) strains which were transformed with pPR27xylE::guaA Kan. Among 17 screened clones, 2 representative clones are shown. M – molecular marker 1 kb plus DNA Ladder (Invitrogen), PCRs were carried out with: Lane 1 – M. tuberculosis H37Rv genomic DNA, Lanes 2 and 3 – possible DCO genomic DNA. C. Agarose gel electrophoresis of PCR products from complemented strains with M. smegmatis guaA gene. M – molecular marker 1 kb plus DNA Ladder (Invitrogen), PCRs were carried out with: Lane 1 – M. tuberculosis H37Rv genomic DNA, Lanes 2–7 – genomic DNA from complemented strains.
In order to confirm guaA essentiality in M. tuberculosis, a strain carrying a copy of the ortholog guaA gene from M. smegmatis mc2155 expressing from its native promoter using a mycobacteriophage Ms6-based integrating vector was constructed [10]. This strain was transformed with the pPR27xylE construction to obtain the complemented strain, and six clones were screened for DCOs by PCR. Of the six screened clones, all contained the guaA disrupted allele (Fig. 2C). This result confirmed the guaA essentiality in M. tuberculosis, since the DCO event only occurred when an extra copy of the gene was present.
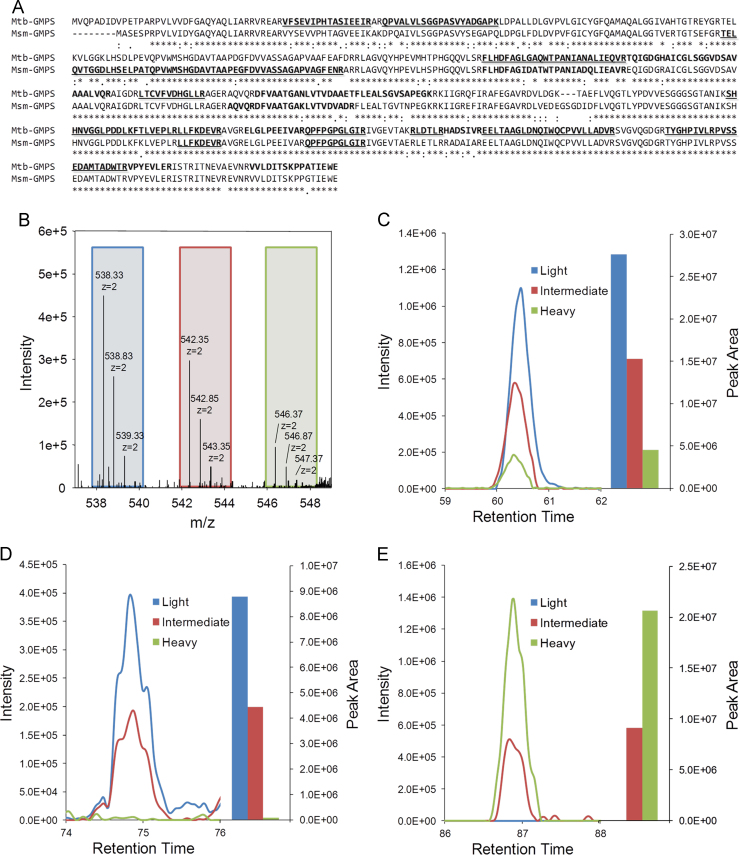
Protein level validation of the guaA essentiality observed by PCR (Fig. 2) was approached by shotgun proteomic analysis. Mtb- and Msm-GMPS were identified in wild-type and complemented cellular extracts of M. tuberculosis, respectively. Among 1666 proteins detected using reversed phase peptide separation, we were able to identify Mtb-GMPS (P9WMS7, Rv3396c) and Msm-GMPS with 45.7% and 19.4% of sequence coverage, respectively (Fig. 3A). The uniqueness of these peptides was confirmed by a quantitative MudPIT analysis, where peptides from wild-type and complemented cellular extracts were labeled with Light and Heavy forms of dimethylation, respectively. As an internal control, a mixture of equal proportions of wild-type and complemented cellular extracts was labeled with Intermediate form of dimethylation. These analyses increased the sequence coverage of Mtb- and Msm-GMPS to 51.2% and 23.0%, respectively (Fig. 3A).
Fig. 3.
Identification of GMPS by LC–MS/MS. (A) GMPS amino acid sequences alignment from M. tuberculosis (Mtb-GMPS) and M. smegmatis (Msm-GMPS); sequences identified by label-free LC–MS/MS are in bold; those identified by MudPIT are underlined. (B) Mass spectrum of precursor ions of triply charged peptide LLFKDEVR; Light (left box), Intermediate (middle box) and Heavy (right box). (C–E) Extracted ion chromatograms (XICs) and peak areas of the Light (blue), Intermediate (red) and Heavy (green) forms of the peptides: (C) LLFKDEVR, doubly charged, common to Mtb-GMPS and Msm-GMPS; (D) QPVALVLSGGPASVYADGAPK, triply charged, unique to Mtb-GMPS; (E) TELQVTGGDLHSELPATQPVWMSHGDAVTAAPEGFDVVASSAGAPVAGFENR, quadruply charged, unique to Msm-GMPS.
The expression levels of Mtb- and Msm-GMPS in wild-type and complemented cellular extracts were measured by the relative abundance displayed by peptides common in both proteins. Fig. 3B shows the precursor ions of doubly charged peptide LLFKDEVR, shared by both proteins. The extracted ion chromatograms (XICs) and the respective peak areas of Light, Intermediate and Heavy forms of the peptide are shown in Fig. 3C. Using all common peptides identified in MudPIT technical triplicates, an average ratio of Mtb-GMPS (Light)/Msm-GMPS (Heavy) was calculated. To account for experimental inaccuracies, a global normalization factor calculated by Census, using all peptide ratios from the entire M. tuberculosis proteome, was used (data not shown), giving a corrected ratio value for Mtb-GMPS/Msm-GMPS of 3.41.
The quantitative method employed here also allowed the identification of peptides unique to either Mtb- or Msm-GMPS. Fig. 3D shows XICs of the Light, Intermediate and Heavy forms of a Mtb-GMPS unique, triply charged peptide, QPVALVLSGGPASVYADGAPK. Peak areas were calculated for each isotopic label; Light channel presented a peak area of 8.8×106 whereas the peak area detected in the Heavy channel was not identified as the Heavy labeled peptide, being only at noise level (7.6×104), as expected. The Intermediate peak area was 4.4×106, an approximate average between Light and Heavy areas.
In contrast, a quadruply charged Msm-GMPS unique peptide, TELQVTGGDLHSELPATQPVWMSHGDAVTAAPEGFDVVASSAGAPVAGFENR, displayed detectable XIC only in the Heavy form of the peptide (2.1×107). While a Light peak area was undetectable, the Intermediate form was 9.1×106, an approximate half of the Heavy peak area (Fig. 3E).
The Intermediate XIC peaks of unique and common peptides further confirm the reliability of the quantitative data. The peak area of Intermediate XICs is an experimental measurement of the average intensity of Light and Heavy peaks. Indeed, the comparison of calculated Light and Heavy average areas to the experimentally obtained Intermediate area of all Mtb-GMPS/Msm-GMPS common peptides showed an error of less than 1% (data not shown).
5. Discussion
In this report, we evaluate the essentiality of guaA by gene replacement in M. tuberculosis H37Rv strain. GMP synthesis from inosine 5′-monophosphate requires both IMPDH and GMPS, which are important enzymes from the purine biosynthesis pathway. These genes are present as independent loci in M. tuberculosis genomic DNA while in some bacteria they are located in the same operon. The disruption of guaA and guaB genes have been previously reported in several bacterial pathogens, resulting in attenuated strains which, in some cases, were also auxotrophic for guanine [4], [5], [6]. In contrast, the results presented here showed that a M. tuberculosis strain containing a disrupted guaA gene could not be isolated, which is an indication that this gene is essential for growth. Despite the presence of hypoxantine-guanine phosphoribosyltransferase enzyme in M. tuberculosis, an auxotrophic strain for guanine could not be obtained by deleting guaA gene. The guaA gene could only be disrupted at its native locus if a functional copy is supplied elsewhere. Msm-GMPS complemented the function of the correspondent Mtb enzyme, once the lethal phenotype of M. tuberculosis guaA disruption was avoided by the expression of M. smegmatis guaA.
In order to confirm guaA essentiality at protein level, we performed label-free and dimethyl labeling quantitative LC–MS/MS analyses of wild-type and complemented cellular extracts. Although Mtb- and Msm-GMPS proteins share 85.69% amino acid sequence identity, we were able to evaluate the origin of unique and common peptides from both proteins. Through these quantitative proteomic experiments, we confirmed that the complemented strain contained only Msm-GMPS. Further evidence of the origin of the unique and common peptides was obtained using equal proportion of wild-type and complemented cellular extracts. Peptide peak areas were expected as an average of the intensity of those found in the wild-type and complemented samples, as shown in Fig. 3C–E.
In addition, the method employed here allows the relative quantification of GMPS in the wild-type and in the complemented strains. Using peptide sequences common to both Mtb- and Msm-GMPS, we verified an approximate 3.4-fold difference in the expression levels of guaA gene in the wild-type strain compared to the complemented strain. The reduced sequence coverage of Msm-GMPS compared to Mtb-GMPS obtained by reversed phase and MudPIT analyses also corroborates with the difference in expression levels. The insertion of the ortholog guaA gene in a different position on the mycobacterial genomic DNA, and the expression of this gene under the natural M. smegmatis promoter in the complemented strain could explain the different expression levels seen here.
Multiple sequence alignment of polypeptide chains of GMPS enzymes showed that M. tuberculosis has high identity to prokaryotic proteins (47.39% with Escherichia coli) while it has a relatively low identity to the human enzyme (15.50%) [3]. Based on sequence alignment of primary sequences and tertiary structures, the main differences between the monomeric eukaryotic and dimeric prokaryotic GMPSs rely on several large insertions in C-terminal region of eukaryotic protein, which form an additional domain that eliminates the need for dimerization [3], [22]. These differences could be further explored to the development of specific Mtb-GMPS inhibitors. The low identity between human and mycobacterial GMPS enzymes, and its essentiality in M. tuberculosis make guaA-encoded enzyme a potential drug target. The biochemical characterization of GMPS from M. tuberculosis may help future efforts towards the development of inhibitors based on particular features of the mycobacterial enzyme [3].
In conclusion, we showed that the disruption of the guaA gene resulted in M. tuberculosis lethality. The complementation with a rescue copy of this gene is required to avoid the lethal phenotype, which was confirmed at protein level via quantitative LC–MS/MS. These results validate GMPS as a molecular target for drug design against Mtb, and GMPS inhibitors might prove to be useful for future development of new drugs to treat human TB.
Conflict of interest
The authors declare that there is no conflict of interest in this work.
Acknowledgments
We acknowledge Dr. Mary Jackson and Dr. Brigitte Gicquel for providing the pNIP40/b and pPR27xylE plasmids. D.S.S. (CNPq, 304051/1975-06) and L.A.B. (CNPq, 520182/99-5) are Research Career Awardees of the National Council for Scientific and Technological Development (CNPq, Brazil). A.D.V. acknowledges the fellowship by Coordination for the Improvement of Higher Education Personnel (CAPES, Brazil) (PNPD, 10/2013), V.R.J. by FAPERGS-CAPES (DOCFIX, 05/2013), and P.E. and A.F.M.P. by CNPq (163507/2014-7, 304156/2014-0). This research has been supported by funds by Decit/SCTIE/MS-MCT-CNPq-FNDCT-CAPES to National Institute of Science and Technology on Tuberculosis (INCT-TB), CNPq (441720/2014-5), National Center for Research Resources (5P41RR011823), BNDES (14.2.0914.1), and National Institute of General Medical Sciences (8P41GM103533). D.S.S. and L.A.B. acknowledge financial support by FAPERGS-CNPq-PRONEX-2009.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.10.005.
Contributor Information
Anne Drumond Villela, Email: anne.villela@pucrs.br.
Diógenes Santiago Santos, Email: diogenes@pucrs.br.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.WHO. WHO Global Tuberculosis Report, 2014.
- 2.Ducati R.G., Breda A., Basso L.A., Santos D.S. Purine salvage pathway in Mycobacterium tuberculosis. Curr. Med. Chem. 2011;18:1258–1275. doi: 10.2174/092986711795029627. [DOI] [PubMed] [Google Scholar]
- 3.Franco T.M., Rostirolla D.C., Ducati R.G., Lorenzini D.M., Basso L.A., Santos D.S. Biochemical characterization of recombinant guaA-encoded guanosine monophosphate synthetase (EC 6.3.5.2) from Mycobacterium tuberculosis H37Rv strain. Arch. Biochem. Biophys. 2012;517:1–11. doi: 10.1016/j.abb.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Santiago A.E., Cole L.E., Franco A., Vogel S.N., Levine M.M., Barry E.M. Characterization of rationally attenuated Francisella tularensis vaccine strains that harbor deletions in the guaA and guaB genes. Vaccine. 2009;27:2426–2436. doi: 10.1016/j.vaccine.2009.02.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noriega F.R., Losonsky G., Lauderbaugh C., Liao F.M., Wang J.Y., Levine M.M. Engineered deltaguaB-A deltavirG Shigella flexneri 2a strain CVD 1205: construction, safety, immunogenicity, and potential efficacy as a mucosal vaccine. Infect. Immun. 1996;64:3055–3061. doi: 10.1128/iai.64.8.3055-3061.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jewett M.W., Lawrence K.A., Bestor A., Byram R., Gherardini F., Rosa P.A. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J. Bacteriol. 2009;191:6231–6241. doi: 10.1128/JB.00450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassetti C.M., Boyd D.H., Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 8.Griffin J.E., Gawronski J.D., Dejesus M.A., Ioerger T.R., Akerley B.J., Sassetti C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelicic V., Jackson M., Reyrat J.M., Jacobs W.R., Jr., Gicquel B., Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freitas-Vieira A., Anes E., Moniz-Pereira J. The site-specific recombination locus of mycobacteriophage Ms6 determines DNA integration at the tRNA(Ala) gene of Mycobacterium spp. Microbiology. 1998;144:3397–3406. doi: 10.1099/00221287-144-12-3397. [DOI] [PubMed] [Google Scholar]
- 11.Wessel D., Flügge U.I. A method for quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 12.Klammer A.A., MacCoss M.J. Effects of modified digestion schemes on the identification of proteins from complex mixtures. J. Proteome Res. 2006;5:695–700. doi: 10.1021/pr050315j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boersema P.J., Raijmakers R., Lemeer S., Mohammed S., Heck A.J.R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 14.Wolters D.A., Washburn M.P., Yates J.R., III An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 15.Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open source tandem mass spectrometry sequence database search tool. Proteomics. 2012;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho P.C., Fischer J.S., Yates J.R., 3rd, Barbosa V.C. PatternLab: from mass spectra to label-free differential shotgun proteomics. Curr. Protoc. Bioinform. 2012 doi: 10.1002/0471250953.bi1319s40. Chapter 13:Unit 13.19. [DOI] [PubMed] [Google Scholar]
- 17.McDonald W.H., Tabb D.L., Sadygov R.G., MacCoss M.J., Venable J. MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 18.Xu T., Venable J.D., Park S.K., Conciorva D., Lu B. ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cell. Proteomics. 2006;5:S174. [Google Scholar]
- 19.Tabb D.L., McDonald W.H., Yates J.R., III DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S.K.R., Aslanian A., McClatchy D.B., Han X., Shah H., Singh M. Census 2: isobaric labeling data analysis. Bioinformatics. 2014;30:2208–2209. doi: 10.1093/bioinformatics/btu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tesmer J.J., Klem T.J., Deras M.L., Davisson V.J., Smith J.L. The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families. Nat. Struct. Biol. 1996;3:74–86. doi: 10.1038/nsb0196-74. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material