Abstract
More and more evidences suggested that the flow of genetic information can be spatially and temporally regulated by non-coding RNAs (ncRNAs), such as microRNAs (miRNAs). Although biogenesis and function of miRNAs have been well detailed, elucidation of the dynamic interplays between miRNAs and mRNAs have just begun. Here, we highlighted that the miRNA–mRNA interactions which could take place in different cellular locations. During dynamic interactions, miRNA binding sites included not only 3′UTRs, but also 5′UTRs and CDSs. Under different physiological or pathological conditions, miRNAs could switch from translational inhibition to activation. Dynamic miRNA–mRNA paradigms which suggested a novel tip of the iceberg beneath the gene regulation network will provide clues for function studies of other ncRNAs.
Keywords: MiRNA, RNA interaction, RNA traffic, Mitochondria, Gene regulation
Highlights
-
•
miRNA–mRNA interactions which could take place in different cellular locations.
-
•
miRNA binding sites included not only 3′UTRs, but also 5′UTRs and CDSs.
-
•
miRNAs could switch from translational inhibition to activation.
1. Introduction
Eukaryotic genomes are nearly entirely transcribed, generating huge number of non-coding RNAs (ncRNAs) [1], [2], [3], [4]. There are more than 100 unique classes of functional ncRNAs so far [5], [6]. As important molecules, ncRNAs could function at different levels such as DNA transcription, RNA processing, protein translation and so on [7], [8]. Among these ncRNAs, a distinctly different type named microRNAs (miRNAs), have attracted a great deal of global attention.
MiRNAs are small, single-stranded and regulatory RNA molecules, playing important roles in cells [9], [10], [11]. It is currently widely recognized that miRNAs are transcribed in the nucleus, being exported to cytoplasm as a post-transcriptional negative regulator. Firstly, most miRNAs are independently transcribed or generated from introns of host-genes [12], [13]. After being transcribed, they are usually processed from primary-miRNAs (pri-miRNAs) into precursor-miRNA (pre-miRNA) in the nucleus [14], [15], [16]. After exported to the cytoplasm, pre-miRNAs are usually processed to mature, ~22 nt in length [9], [10], [11]. Finally, the mature miRNAs are incorporated into the RNA-induced silencing complex (RISC) for function [9], [10], [11]. Within RISC, Argonaute (AGO) proteins (AGO1–AGO4 in mammals) are core components that are directly associated with miRNAs [9], [10], [11]. And miRNA–mRNA duplex is required as miRNA seed region (nucleotides 2–7) interacts with the 3′UTR of target mRNA with perfect base-pair sequences [17], [18]. Owing to short base-pairs within miRNA-3′UTR duplex, one miRNA can bind to several even hundreds target mRNAs or a single mRNA can be targeted by many different miRNAs [17], [18]. Finally, target mRNAs are usually degraded or translationally repressed [9], [10], [11].
As a major regulator of gene expression at post-transcriptional level in the cytoplasm, expression of miRNA is physiologically or pathologically specific, leading to spatial and temporal specificity of mRNA expression [10]. Although biogenesis and functional patterns of miRNAs have been well documented, dynamic interactions between miRNAs and mRNAs are usually neglected. As crucial relationships, their interactions with different subcellular locations, distinct binding sites and cellular circumstances would shed flashlight for their biological function studies and provide paradigms for comprehending regulation and function of other ncRNAs.
2. MiRNAs acting in the nucleus, mitochondria and exosomes
It is generally recognized that mature miRNAs function in the cytoplasm. Increasing evidences suggest that mature miRNAs are found in the nucleus, mitochondria and exosomes, suggesting non-canonical roles of miRNA [19], [20], [21], [22], [23], [24], [25], [26] (Fig. 1A). Here, we summarized some miRNAs with unexpected cellular locations in mammalians (Table. 1). Systematic studies of sub-cellular distribution of mature miRNAs reveal that miRNAs can shuttle from cytoplasm to nucleus in different mammals [27], [28], [29], [30], [31], [32] (Table 1). Mature miRNAs discovered in the nucleus suggested that miRNAs can function in transcriptional silencing or activation, post-transcriptional silencing and alternative splicing [19]. Recent studies of RNAs in exosomes indicate that miRNAs can be transferred by exosomes, regulating inter-cellular communication [26], [33], [34], [35], [36] (Table 1). However, weather or/and how miRNAs interact with mRNAs in the nucleus and exosomes are still elusive. In this article, we emphasize on dynamic miRNA–mRNA interactions with cytoplasmic and mitochondria locations in the followed paragraphs.
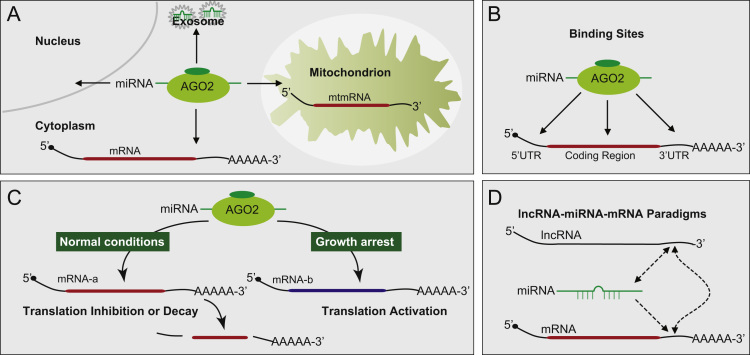
Fig. 1.
Dynamic miRNA–mRNA paradigms. (A) miRNAs acting in different cellular locations. The cytoplasm, mitochondrion, nucleus and exosomes are indicated with drawings. Cellular location of miRNA–AGO2 complex are indicated with arrows. mtmRNA: mitochondrion mRNA. (B) Distinct binding sites of miRNAs. MiRNA targets are indicated with arrows pointed 5′UTR, coding region and 3′UTR of mRNA. (C) Dual faces of miRNA. During normal conditions, miRNA–AGO2 complex mediates target mRNAs for translational repression or decay. As cells are arrested, the translation of target mRNAs is activated. (D) Possible lncRNA-miRNA–mRNA interactions. MiRNA, mRNA and lncRNA constitute complex interactions. Among these interactions, miRNA can interact with both mRNA and lncRNA. Meanwhile, lncRNA can interact with miRNA and mRNA. RNA–RNA interactions are indicated with dotted line with arrows.
Table 1.
Examples of miRNAs with unexpected cellular locations. miRNAs (column 1) with different cellular locations (column 2), functional description (column 3) and the corresponding references (column 4). Rno: Rattus norvegicus. Mmu: Mus musculus. Hsa: Homo sapiens.
| miRNAs | Localizations | Functions | References |
|---|---|---|---|
| Hsa-mir-29b, Hsa-mir-320, Hsa-mir-32, Hsa-mir-148a, Hsa-mir-148b, Hsa-mir-1285, Hsa-mir-29c, Hsa-mir-1, Hsa-mir-652, Hsa-mir-15b, Hsa-mir-135b | Nucleus cytoplasm | Transcriptional regulation and alterantive splicing | [27], [28], [29] |
| Rno-mir-206, Rno-mir-351, Rno-mir-494, Rno-mir-664, Rno-mir-1, Rno-let-7a, Rno-mir-21, Rno-mir-199a-3p, Rno-mir-125b-5p | [30], [31] | ||
| Mmu-mir-709, Mmu-mir-805, Mmu-mir-690, Mmu-mir-122, Mmu-mir-30e | [32] | ||
| Hsa-mir-365, Hsa-mir-1, Hsa-mir-181c, Hsa-mir-720, let-7 familly, Hsa-mir-133a, Hsa-mir-206, Hsa-mir-195, Hsa-mir-181a, Hsa-mir-181b | Mitochondria cytoplasm | Metabolism, development, apoptosis | [23], [42], [43] |
| Mmu-miR-1, let-7, Mmu-miR-15, Mmu-miR-181, Mmu-miR-16, Mmu-miR-375, Mmu-miR-17, Mmu-miR-18 | Exosome cytoplasm | Cellular communication | [26] |
| let-7b, Hsa-miR-150, Hsa-miR-27b, Hsa-miR-29b, Hsa-miR-29c, Hsa-miR-335, Hsa-miR-379, Hsa-miR-433, Hsa-miR-454, Hsa-miR-483-3p, Hsa-miR-584, Hsa-miR-621, Hsa-miR-652, Hsa-miR-760, Hsa-miR-888, Hsa-mir-292, Hsa-mir-103, Hsa-mir-15b, Hsa-mir-17, Hsa-mir-199a, Hsa-mir-20a, Hsa-mir-210 | [34], [35] |
As a central regulator of energy metabolism, the inter-organellar crosstalk is essential for the coordination or rapidly changed situations [37], [38]. Previous studies indicate that many nuclear encoded proteins and RNAs can be imported to mitochondria for vitally cellular processes [39], [40]. Through cellular fractionation, together with deep sequencing, microarray and qRT-PCR methods, many miRNAs in different mammals which are mitochondria located are systematically identified [23], [24], [41] (Table 1). More importantly, AGO2 protein which is directly associated with miRNAs is also found to be located in mitochondria by immunoblotting and confocal microscopy [22]. New evidences suggest that some miRNAs can regulate the function of mitochondria, even function in mitochondria [22], [23], [24], [41], [42], [43], [44].
Although there are lots of reports about miRNAs can interact with mRNAs that related to mitochondria function, the interactions occuring in the mitochondria are rare [25], [45]. Up to now, miR-181c and miR-1 are two miRNAs that can interact with mitochondria encoded mRNAs. Both miRNAs are nuclear encoded, after being processed to maturation in the cytoplasm, they can be imported to the mitochondria. When miR-181c is over-expressed, one of its target mRNA mt-COX1 (cytochrome c oxidase subunit 1) is reduced, leading to cardiac dysfunction in rats [42], [44]. As for miR-1, it is specifically expressed during myogenesis. Increasing levels of miR-1 represses mRNA translation in the cytoplasm, while the translation of mt-ND1 and mt-COX1 in the mitochondria is stimulated unexpectedly [43]. These evidences provided new clues for functional studies of miRNA–mRNA interactions in mitochondria or other cellular localizations, suggesting highly coordinated inter-organellar crosstalk during rapidly physiological or pathological changes.
3. MiRNAs targeting 5′UTRs and CDSs
Beside different cellular localizations, miRNAs can target different regions of mRNAs during dynamic miRNA–mRNA interactions, changing the fates of different target mRNAs. Although it is well established that the mRNA 3′UTRs are usually targeted by miRNAs, un-conventional sites in the 5′UTRs or coding regions have also been reported [21], [46], [47] (Fig. 1B).
In vitro experiment, artificial lin-4-like miRNA can interact with the 5′UTR of lin-28-like mRNA which is consistent with let-7a:lin-41 pairs, resulting in repression of translation of target mRNAs [47], [48]. Further functional studies of miR-103a-3p, miR-122 and miR-10a whose target sites are within 5′UTR regions confirmed that miRNAs can efficiently target 5′UTR sites in the cells [49], [50], [51]. MiRNAs which can target both 3′UTRs and 5′UTRs is found [48]. This kind of miRNA is sequences dependent when targeting both UTRs of mRNA. The function of endogenous miRNAs interacted with both UTRs such as miR-34a:AXIN2, miR-605:SEC24D pairs are also validated in vivo [48]. As both UTRs are targeted by miRNAs, the fold of target mRNA levels change is only modest, while the fold of corresponding protein levels change is significant [52].
Besides UTRs, CDSs can also be targeted by miRNAs. Through genome-wide screen, highly conserved target sequences within CDSs are found in the human genome [53]. Interestingly, conserved CDSs target sites are also widely observed in the Drosophila [54]. When miRNAs binding sites are in the CDSs of mRNAs, the translation of target mRNAs are usually inhibited [55]. As the target sites of miR-199a are cloned in the CDS of the luciferase, increasing levels of miR-199a can reduce its activity in vitro [55]. In vivo, let-7 can target the CDS of Dicer, forming a negative feedback loop [53]. Meanwhile, miR-148 targets the CDS of DNA methyltransferase 3b (Dnmt3b) is also confirmed in human cells [56]. As CDSs contain rich regulatory sequences, the miRNA-CDS patterns may influence gene architecture, alternative splicing and alternative poly-adenylation, tuning protein abundance with more flexible time-scale and magnitude [46].
A new algorithm (MinoTar-miRNA ORF Targets) is used to estimate preferentially conserved miRNA-target sites, suggesting the number of CDSs and 5′UTRs targets may exceed 3′UTRs in mammalians [54]. Systematic analysis of miRNA target sites located within 3′UTR and CDS between species indicate that CDS target sites are also functional [55], [57]. MiRNAs whose center nucleotides 4–14 or 5–15 pair can target mRNAs are reported. They have extensive target sites, including UTRs and CDSs [58]. Crosslinking and immunoprecipitation (CLIP) experiments provide genome-wide miRNA target sites, indicating CDS target sites are frequent as 3′UTRs [59], [60], [61]. All these suggest that miRNA target sites within UTRs and CDSs are possibly functional. However, individual miRNA show different preferences in targeting the CDSs or the UTRs. RIP-Chip experiments in H4 cells suggest that miR-107 prefers to target the CDSs while miR-320 and miR-124 tends to bind the 3′UTRs [62], [63]. So, it is still challenging to study the function and mechanism of non-canonical miRNA target sites in the cells.
4. MiRNA-mediated activation of translation
Most (although not all) of miRNA-mRNA interactions showed negative regulation at the post-transcriptional level [52]. As dynamic interactions, dual function of miRNAs has been found under different cellular circumstances. On normal conditions, miRNA-mRNA interactions mediated target mRNAs for translational repression or decay. When cell growth was arrested, miRNA-AGO2 complex activated the translation of target mRNAs (Fig. 1C).
MiR-369 was the firstly reported miRNA that had double faces in regulating target mRNAs [64], [65]. When HEK293 cells are grown with serum, target mRNAs (such as TNF-α) whose 3′ UTR contained the AU-rich elements (AREs) are subject to be translational inhibited or decayed, mediated by miR-369. But when the growth of cells are serum-starved, miR-369 unexpectedly activated the translation of target mRNAs [64], [65]. The mode of translational repression or activation requires the FXR1 protein (fragile X mental retardation-related protein 1) whose presence can activate translation of proteins [64], [65]. Further studies of let-7 and the synthetic miRNA miRcxcr4 suggest that dual roles of miRNPs are common during different cellular conditions [64], [65]. However, when the same mRNA TNF-α is targeted within the 3′UTR (perhaps not the same binding site as miR-369) by miR-16, its translation is unexpectedly inhibited under changed cellular conditions [66].
Besides different physiological or pathological conditions, miRNAs can also translationally activate target mRNAs via binding different regions of target mRNAs or by distinctly cellular localizations. In the HCV (hepatitis C virus) life cycle, miR-122 can positively activate the translation of HCV by targeting its 5′UTR in the cytoplasm [49]. During the early phase of liver regeneration, increased miR-21 can accelerate the translation of cyclin D1 by interacting with its 3′UTR in the cytoplasm [67]. During cellular stress, miR-10a can stimulate the translation of ribosomal proteins by binding their 5′UTRs in the cytoplasm [50]. Contrast to miR-181c, miR-1 can activate the translation of mt-COX1 in the mitochondria by binding its 3′UTR in the absence of GW182 which is a functional partner of AGO2 [42], [43].
Under different cellular conditions, miRNA–mRNA interactions with different binding sites or/and cellular localizations make the interactions more complicated than we can anticipate. So, how many positive and novel miRNA–mRNA interactions in the cytoplasm and mitochondria are there still needs further studies.
5. Conclusions and perspectives
MiRNAs with different cellular locations and binding sites suggest possibly novel function in gene expression. At the post-transcriptional level, beyond translation repression or mRNA decay, miRNA can also active protein translation under different cellular circumstances.
In addition to mRNAs, lncRNAs can be the target of miRNAs, constituting complex circuits of gene regulatory network [68] (Fig. 1D). The ceRNA (competing endogenous RNA) hypothesis suggests that lncRNAs sharing miRNA response elements could keep corresponding mRNAs from miRNAs mediated repression [69]. However, this hypothesis is challenged by quantitative study of abundance of miRNA and its binding site in vivo, suggesting this hypothesis is unlikely to cause significant effects on gene regulation through miRNA antagonist [70]. So, how mRNAs and lncRNAs harbored the same miRNA binding sites, communicating to each other still need further studies. Using novel experimental technology such as miR-CLIP would reveal more dynamic miRNA–mRNA, miRNA–lncRNA interactions under different physiological and pathological conditions [71]. Systematic and integrative analysis of miRNAs, lncRNAs, and mRNAs may unveil their regulatory relationships with different times and spaces [72]. Further studies of miRNA–mRNA interactions with distinct cellular locations and binding sites under differently physiological or pathological conditions will provide more discoveries before we can anticipate. And this will shed more lights on functional studies of other ncRNAs.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Acknowledgment
This work was supported by Grants from The First Affiliated Hospital of Xinxiang Medical University (xyyfy2014BS-006).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.10.011.
Contributor Information
Wen-Juan Ni, Email: 290361612@qq.com.
Xiao-Min Leng, Email: m13781936681@163.com.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Amaral P.P., Dinger M.E., Mercer T.R., Mattick J.S. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., Bell I., Cheung E., Drenkow J., Dumais E., Patel S., Helt G., Ganesh M., Ghosh S., Piccolboni A., Sementchenko V., Tammana H., Gingeras T.R. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G.K., Khatun J., Williams B.A., Zaleski C., Rozowsky J., Roder M., Kokocinski F., Abdelhamid R.F., Alioto T., Antoshechkin I., Baer M.T., Bar N.S., Batut P., Bell K., Bell I., Chakrabortty S., Chen X., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Falconnet E., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M.J., Gao H., Gonzalez D., Gordon A., Gunawardena H., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Luo O.J., Park E., Persaud K., Preall J.B., Ribeca P., Risk B., Robyr D., Sammeth M., Schaffer L., See L.H., Shahab A., Skancke J., Suzuki A.M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Wrobel J., Yu Y., Ruan X., Hayashizaki Y., Harrow J., Gerstein M., Hubbard T., Reymond A., Antonarakis S.E., Hannon G., Giddings M.C., Ruan Y., Wold B., Carninci P., Guigo R., Gingeras T.R. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 5.Robinson V.L. Rethinking the central dogma: noncoding RNAs are biologically relevant. Urol. Oncol. 2009;27:304–306. doi: 10.1016/j.urolonc.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Morris K.V., Mattick J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014;15:423–437. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cech T.R., Steitz J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Patil V.S., Zhou R., Rana T.M. Gene regulation by non-coding RNAs. Crit. Rev. Biochem. Mol. Biol. 2014;49:16–32. doi: 10.3109/10409238.2013.844092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 11.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez A., Griffiths-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohler A., Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 15.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim V.N. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell. Biol. 2004;14:156–159. doi: 10.1016/j.tcb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Roberts T.C. The microRNA biology of the mammalian nucleus. Mol. Ther. Nucleic Acids. 2014;3:e188. doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang H., Zhang J., Zen K., Zhang C.Y., Chen X. Nuclear microRNAs and their unconventional role in regulating non-coding RNAs. Protein Cell. 2013;4:325–330. doi: 10.1007/s13238-013-3001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cipolla G.A. A non-canonical landscape of the microRNA system. Front. Genet. 2014;5:337. doi: 10.3389/fgene.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandiera S., Ruberg S., Girard M., Cagnard N., Hanein S., Chretien D., Munnich A., Lyonnet S., Henrion-Caude A. Nuclear outsourcing of RNA interference components to human mitochondria. Plos One. 2011;6:e20746. doi: 10.1371/journal.pone.0020746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrey E., Saint-Auret G., Bonnamy B., Damas D., Boyer O., Gidrol X. Pre-microRNA and mature microRNA in human mitochondria. Plos One. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sripada L., Tomar D., Prajapati P., Singh R., Singh A.K., Singh R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: detailed analysis of mitochondrial associated miRNA. Plos One. 2012;7:e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duarte F.V., Palmeira C.M., Rolo A.P. The role of microRNAs in mitochondria: small players acting wide. Genes. 2014;5:865–886. doi: 10.3390/genes5040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 27.Hwang H.W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 28.Kim D.H., Saetrom P., Snove O., Jr., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao J.Y., Ma L.M., Guo Y.H., Zhang Y.C., Zhou H., Shao P., Chen Y.Q., Qu L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3’ trailers. Plos One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Politz J.C., Hogan E.M., Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15:1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Politz J.C., Zhang F., Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. USA. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang R., Li L., Zhu D., Hou D., Cao T., Gu H., Zhang J., Chen J., Zhang C.Y., Zen K. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res. 2012;22:504–515. doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Squadrito M.L., Baer C., Burdet F., Maderna C., Gilfillan G.D., Lyle R., Ibberson M., De Palma M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell. Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 34.Michael A., Bajracharya S.D., Yuen P.S., Zhou H., Star R.A., Illei G.G., Alevizos I. Exosomes from human saliva as a source of microRNA biomarkers. Oral. Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray W.D., French K.M., Ghosh-Choudhary S., Maxwell J.T., Brown M.E., Platt M.O., Searles C.D., Davis M.E. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 2015;116:255–263. doi: 10.1161/CIRCRESAHA.116.304360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Li S., Li L., Li M., Guo C., Yao J., Mi S. Exosome and exosomal microRNA: trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ashwell M., Work T.S. The biogenesis of mitochondria. Annu. Rev. Biochem. 1970;39:251–290. doi: 10.1146/annurev.bi.39.070170.001343. [DOI] [PubMed] [Google Scholar]
- 38.Attardi G., Schatz G. Biogenesis of mitochondria. Annu. Rev. Cell Biol. 1988;4:289–333. doi: 10.1146/annurev.cb.04.110188.001445. [DOI] [PubMed] [Google Scholar]
- 39.Tarassov I., Kamenski P., Kolesnikova O., Karicheva O., Martin R.P., Krasheninnikov I.A., Entelis N. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell. Cycle. 2007;6:2473–2477. doi: 10.4161/cc.6.20.4783. [DOI] [PubMed] [Google Scholar]
- 40.Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 41.Kren B.T., Wong P.Y., Sarver A., Zhang X., Zeng Y., Steer C.J. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S., Bedja D., Campbell N., Dunkerly B., Chenna V., Maitra A., Steenbergen C. miR-181c regulates the mitochondrial genome, bioenergetics, and propensity for heart failure in vivo. Plos One. 2014;9:e96820. doi: 10.1371/journal.pone.0096820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y., Huang J., Zhao X., Zhou J., Yan Y., Zhang H., Guo P., Sun H., Guo L., Zhang Y., Fu X.D. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das S., Ferlito M., Kent O.A., Fox-Talbot K., Wang R., Liu D., Raghavachari N., Yang Y., Wheelan S.J., Murphy E., Steenbergen C. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P., Jiao J., Gao G., Prabhakar B.S. Control of mitochondrial activity by miRNAs. J. Cell. Biochem. 2012;113:1104–1110. doi: 10.1002/jcb.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brummer A., Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays. 2014;36:617–626. doi: 10.1002/bies.201300104. [DOI] [PubMed] [Google Scholar]
- 47.Lytle J.R., Yario T.A., Steitz J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee I., Ajay S.S., Yook J.I., Kim H.S., Hong S.H., Kim N.H., Dhanasekaran S.M., Chinnaiyan A.M., Athey B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts A.P., Lewis A.P., Jopling C.L. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Orom U.A., Nielsen F.C., Lund A.H. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 51.Zhou H., Rigoutsos I. MiR-103a-3p targets the 5′ UTR of GPRC5A in pancreatic cells. RNA. 2014;20:1431–1439. doi: 10.1261/rna.045757.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forman J.J., Legesse-Miller A., Coller H.A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. USA. 2008;105:14879–14884. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schnall-Levin M., Zhao Y., Perrimon N., Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc. Natl. Acad. Sci. USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hausser J., Syed A.P., Bilen B., Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duursma A.M., Kedde M., Schrier M., le Sage C., Agami R. miR-148 targets human DNMT3b protein coding region. RNA. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu G., Zhang R., Xu J., Wu C.I., Lu X. Functional conservation of both CDS- and 3′-UTR-located microRNA binding sites between species. Mol. Biol. Evol. 2015;32:623–628. doi: 10.1093/molbev/msu323. [DOI] [PubMed] [Google Scholar]
- 58.Shin C., Nam J.W., Farh K.K., Chiang H.R., Shkumatava A., Bartel D.P. Expanding the microRNA targeting code: functional sites with centered pairing. Mol. Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chi S.W., Zang J.B., Mele A., Darnell R.B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jr., Jungkamp A.C., Munschauer M., Ulrich A., Wardle G.S., Dewell S., Zavolan M., Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erhard F., Dolken L., Jaskiewicz L., Zimmer R. PARma: identification of microRNA target sites in AGO-PAR-CLIP data. Genome Biol. 2013;14:R79. doi: 10.1186/gb-2013-14-7-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelson P.T., Wang W.X., Mao G., Wilfred B.R., Xie K., Jennings M.H., Gao Z., Wang X. Specific sequence determinants of miR-15/107 microRNA gene group targets. Nucleic Acids Res. 2011;39:8163–8172. doi: 10.1093/nar/gkr532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang W.X., Wilfred B.R., Xie K., Jennings M.H., Hu Y.H., Stromberg A.J., Nelson P.T. Individual microRNAs (miRNAs) display distinct mRNA targeting “rules”. BRNA Biol. 2010;7:373–380. doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 65.Buchan J.R., Parker R. Molecular biology. The two faces of miRNA. Science. 2007;318:1877–1878. doi: 10.1126/science.1152623. [DOI] [PubMed] [Google Scholar]
- 66.Jing Q., Huang S., Guth S., Zarubin T., Motoyama A., Chen J., Di Padova F., Lin S.C., Gram H., Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 67.Ng R., Song G., Roll G.R., Frandsen N.M., Willenbring H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J. Clin. Investig. 2012;122:1097–1108. doi: 10.1172/JCI46039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yoon J.H., Abdelmohsen K., Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imig J., Brunschweiger A., Brummer A., Guennewig B., Mittal N., Kishore S., Tsikrika P., Gerber A.P., Zavolan M., Hall J. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction. Nat. Chem. Biol. 2015;11:107–114. doi: 10.1038/nchembio.1713. [DOI] [PubMed] [Google Scholar]
- 72.Guo L., Zhao Y., Yang S., Zhang H., Chen F. An integrated analysis of miRNA, lncRNA, and mRNA expression profiles. Biomed. Res. Int. 2014;2014:345605. doi: 10.1155/2014/345605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material