Abstract
Differential Scanning Calorimetry (DSC) has been used in the past to study the thermal unfolding of many different viruses. Here we present the first DSC analysis of rabies virus. We show that non-inactivated, purified rabies virus unfolds cooperatively in two events centered at approximately 62 and 73 °C. Beta-propiolactone (BPL) treatment does not alter significantly viral unfolding behavior, indicating that viral inactivation does not alter protein structure significantly. The first unfolding event was absent in bromelain treated samples, causing an elimination of the G-protein ectodomain, suggesting that this event corresponds to G-protein unfolding. This hypothesis was confirmed by the observation that this first event was shifted to higher temperatures in the presence of three monoclonal, G-protein specific antibodies. We show that dithiothreitol treatment of the virus abolishes the first unfolding event, indicating that the reduction of G-protein disulfide bonds causes dramatic alterations to protein structure. Inactivated virus samples heated up to 70 °C also showed abolished recognition of conformational G-protein specific antibodies by Surface Plasmon Resonance analysis. The sharpness of unfolding transitions and the low standard deviations of the Tm values as derived from multiple analysis offers the possibility of using this analytical tool for efficient monitoring of the vaccine production process and lot to lot consistency.
Keywords: Rabies virus, Differential Scanning Calorimetry, Protein unfolding, Vaccine characterization
Highlights
-
•
Differential Scanning Calorimetry analysis of rabies virus.
-
•
Rabies virus unfolds in two thermal events.
-
•
The first event corresponds to G-protein.
1. Introduction
The Rabies virus (RABV) belongs to the genus Lyssavirus which is part of Rhabdoviridae family [1]. RABV has been known since around 2000 BC [2] and infects a wide number of domestic and wild animal species worldwide. It is spread to people through infected saliva by animal bites or scratches, and remains an important worldwide health problem claiming the lives of an estimated 50,000 people annually [3], [4]. Virions travel quickly along the neuronal pathways to the central nervous system where they cause a fatal encephalomyelitis. It has been shown that the time between infection and fatality ranges between 2 days and 5 years [1]. RABV is an enveloped, negative-stranded RNA virus (NSRV) harboring a single-stranded RNA genome which encodes five proteins, namely the nucleoprotein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), and the viral RNA polymerase (L) [5].
Since the first anti-rabies vaccination conducted by Louis Pasteur and Emile Roux in 1885, a large number of different vaccines have been developed, as reviewed in Wu et al. [6]. Anti-rabies vaccines are also manufactured by Sanofi Pasteur (France). These vaccines correspond to beta-propiolactone (BPL)-inactivated virus particles of the Pitman-Moore strain that were cultured on Vero cells [7]. The analysis of vaccines by novel, state-of-the art approaches has two main purposes, namely to increase the basic knowledge available on the antigen but also to develop analytical protocols that can be of use for production process and lot to lot consistency monitoring.
A characteristic feature of most proteins is their capacity to undergo thermal denaturation in a highly cooperative manner. Unfolding is an endothermic process (heat consumption) and can be monitored by Differential Scanning Calorimetry (DSC) [8]. In a DSC analysis a temperature gradient is applied to the sample and heat changes due to unfolding events are recorded. The analysis of such data provides a number of parameters of which the most relevant are the Tm value (in cases where unfolding occurs in a two-state process, Tm is the temperature at which half the protein is present in the native state and the half in the unfolded state) and the ΔH value representing the enthalpy change upon unfolding [8]. Since the Tm value can be determined with high precision, this parameter is thus a very useful marker of protein conformation.
Although DSC has been used for many decades in life science, over the last decades it has become increasingly popular in both basic and applied science. This is mainly due to important advances at the instrumental level, since latest generation calorimeters are characterised by a very high sensitivity (requiring lower sample amounts) and permit the automated analysis of many samples [9]. DSC was primarily used to study the unfolding of purified proteins in the past [8]. However, viruses are associations of proteins with nucleic acids and in some case lipids. These rather complex associations were found to unfold in a very cooperative manner giving rise in most cases to thermograms with few transitions. For example DSC has been used to study the influenza virus [10], adeno and adeno-asociated viruses [11], [12], [13], Newcastle disease virus [14], tobacco mosaic virus [15], tymoviruses [16], Barley Stripe Mosaic Virus [17], potato virus X [18], human papillomavirus virus-like particles [19] or the polio virus [20]. In addition to these fundamental studies, DSC has also been used for the monitoring of industrial scale vaccine production [21], [22] or the characterization of virus vaccine formulations [23], [24]. However, despite its pharmaceutical relevance, so far no DSC study of the rabies virus has been reported. We show here that the rabies virus unfolding is characterised by two transitions and several approaches indicate that the first transition corresponds to that of the G-protein. Moreover, heat-treated virus in conditions similar to DSC analysis does not recognize conformational G-protein specific antibodies anymore.
2. Materials and methods
2.1. Materials
Viral samples are either non-inactivated or BPL-inactivated bulk lots of strain PM1503 (Pitman-Moore) obtained from the Sanofi Pasteur (Marcy l’étoile, France) production department. BPL-mediated virus inactivation was carried out using BPL-virus ratios of 1 to 4000 (the routine inactivation protocol for industrial scale vaccine production). DSC experiments were conducted in PBS purchased from Eurobio (France). Bromelain was purchased from Sigma-Aldrich (ref. B4882). Purified monoclonal antibodies [D1-25 [25], TJU11-12 [26] and 50AD1 [27]] were obtained from BIOTEM (France). The M protein was recombinantly produced in yeast and purchased from MyBiosource (ref. MBS1194184, San Diego, USA).
2.2. Methods
2.2.1. DSC analysis
Viral samples were dialysed (10 kDa cutoff, Thermo Scientific, ref. 66380) against PBS for 2 times 3 h and an additional overnight dialysis. Experiments were done on an auto-CAP DSC (Microcal, USA) with a scan rate of 200 °C/h (being the same for scan and rescan). A scan rate of 200 °C/h was used to help avoiding aggregation or precipitation of the antibody/virus complex. A lower scan rate (85 °C/h) was tested and shows an identical shape of the thermogram (data not shown). Dialysis buffer was used as a reference. Syringe and cell/valve were rinsed with ultrafiltrated water between each run. One scan only was performed for each sample as DSC analysis required a quite huge amount of virus for one analysis. Sample protein concentrations were in the range of 0.15–1.10 mg/ml (corresponding around 9·1010–7·1011 viral particles). The protein concentration was determined by the Bradford assay. For analysis with DTT, this one was added in the microplate (both in reference cell and sample cell) just before analysis in the calorimeter. There was no alkylation prior to analysis. Due to the irreversibility of the unfolding processes, calorimetric enthalpy values were determined by peak integration using the MicroCal VP capillary automated analysis 2.0 program. Data were normalized using the concentration of the G-protein that accounts to approximately 25% of the total viral protein. G-protein concentration was thus derived by dividing the total protein concentration by four. We are aware that values reported are therefore approximate, but a similar approach, applied to the haemagglutinin protein based normalization of influenza virus data, [10], [20] has permitted to compare experiments from different laboratories.
2.2.2. Bromelain treatment
A modified version of the protocol described in [28] was used. Briefly, to samples dialysed against PBS, pH 6.2, 10% (w/protein weight of virus) bromelain and 0.4% (w/protein weight of virus) l-cysteine were added. Samples were incubated at 35 °C for 3 h prior to the addition of iodoacetamide at 1.56 mg/mg virus to inactivate the protease. Samples were then centrifuged at 112,000×g for 90 min at 10 °C to separate viral particles from the cleaved G protein ectodomain. The pellet was then re-suspended in PBS, pH 7.4 (Eurobio) and then re-dialysed against the same buffer.
2.2.3. SDS-PAGE
Analyses were carried out on a 4–12% (w/v) SDS gradient PAGE gels (XT Criterion gels from Bio-Rad) using MOPS (Bio-Rad) as running buffer. Prior to analysis, samples were denatured by the addition of 9 µl XT Sample Buffer 4× (Bio-Rad) to 25 µl of viral sample. 1.8 µl of XT 20× reducing agent (Bio-Rad) was added prior to an exposure to 100 °C for 5 min. Samples containing 5 µg of viral protein were loaded onto each well. Gels were stained using G250 Coomassie Blue. Densitometry analysis of SDS-PAGE was performed using GS800 densitometer (BioRad). Molecular weight of the major bands and the relative percentage of each band were determined using QuantityOne® software after substraction of the background lane.
2.2.4. Protein identification by mass-spectrometry
Protein containing bands were excised from the SDS-PAGE of inactivated viruses. Samples were washed with 50 mM ammonium bicarbonate (ABC) and subsequently dehydrated by three consecutive additions of acetonitrile. Samples were re-hydrated in 25 mM ABC containing 10 mM DTT and left at 60 °C for 20 min. Alkylation was then performed by incubating the samples with 55 mM iodoacetamide in 25 mM ABC for 30 min at room temperature and in darkness. After removing excess DTT and iodoacetamide, gel pieces were washed in 50 mM ABC and dehydrated with ACN three times before adding 10 ng/μl mass spectrometry grade trypsin (Promega). Digestion was performed at 37 °C for 4 h prior to the addition of 0.5% (v/v) trifluoroacetic acid to stop the reaction. Resulting supernatants are deposited on an anchorchip and analyzed by an UltrafleXtreme MALDI-TOF instrument (Bruker) operated in the reflectron mode. The resulting peptide peak list is then processed by the MASCOT software using the UniProt sequence database to identify the protein. Mascot scores above 89 indicate the reliability of the identification.
2.2.5. Surface Plasmon Resonance (SPR) analysis
Samples have been heated with a ramp temperature mimicking DSC analysis (7–70 °C). Samples (control and heat-treated samples) were then analyzed using Biacore 3000TM instrument (GE Healthcare). Purified monoclonal antibodies (Mab) D1-25, TJU11-12 and 50AD1 were covalently immobilized by an amine coupling mechanism on a CM5 sensor chip (BR-1005-30) following the instructions provided by the manufacturer. Around 12,000 RU of each Mab were immobilized. Samples were diluted in HBS-EP buffer (4 μg/ml total protein) and 10 μl were addressed on the different flow cells at a flow rate of 10 μl/min. The increase of RU over time was followed. The complexes formed on the surface of the sensor chip were dissociated by the injection of 2 times 10 μl of 10 mM glycine, pH 1.5 (BR-1003-54) at a flow rate of 60 μl/min.
3. Results
3.1. The rabies virus unfolds irreversibly in two transitions
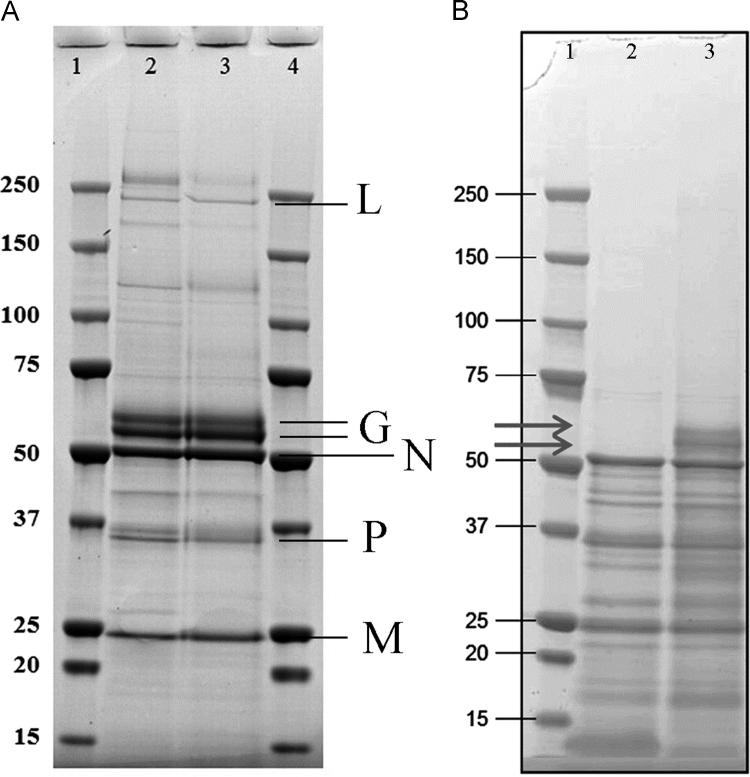
Samples used in this study correspond to bulk material of either non-inactivated or beta-propiolactone-inactivated forms of the rabies virus from the Sanofi Pasteur production line. First experiments were aimed at assessing the impact of viral inactivation on the sample integrity. An SDS-PAGE of non-inactivated and inactivated (1/4000 BPL) samples (Fig. 1A) shows that BPL treatment did not cause any significant changes in the viral protein composition. To identify the individual proteins, bands were excised and the corresponding proteins identified by mass spectrometry based fingerprinting. The five major bands were found to correspond to the viral L-, G-, N-, P- and M-protein. Glycoprotein G of viral strain PM1503 is composed of 524 amino acids with a molecular mass of 58.7 kDa. According to UniProt it is predicted to contain three glycosylation and one palmitoylation sites. As shown in Fig. 1A two bands have been identified by mass spectrometry corresponding to glycoprotein G, that may correspond to different post-translational modification states. SDS-PAGE analysis shows that bulk vaccine corresponds to highly purified virus. The sum of the relative intensity of the different bands corresponding to intact viral proteins L, G, N, P and M is higher than 70%. G protein represents around 25%.
Fig. 1.
Analysis of non-inactivated and BPL-inactivated rabies virus by SDS-PAGE. (A) Assessment of the effect of viral inactivation on the protein profile: lane 2: non-inactivated virus, lane 3: beta-propiolactone (1/4000) inactivated virus. Proteins identified by mass spectrometry fingerprinting are indicated. (B) Protein profile of rabies virus after (lane 2) and prior (lane 3) to bromelain treatment. The arrows indicate glycoprotein G.
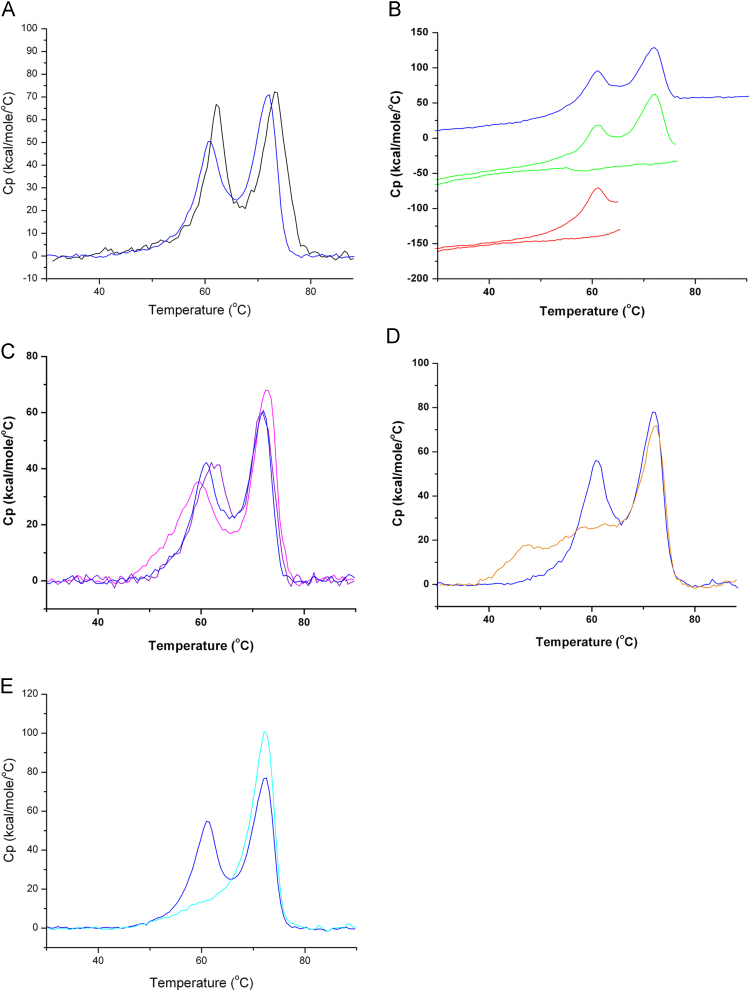
Subsequently, the unfolding behavior of non-inactivated and inactivated rabies virus was assessed by DSC up-screens from 20 to 100 °C. As shown in Fig. 2A the thermograms of both viral forms are similar and unfold in two distinct events. A first event centered at around 61–62 °C was followed by a second event at 71–73 °C (Fig. 2A, Table 1). Calorimetric enthalpy changes of non-inactivated and inactivated samples were also comparable (Table 1). However, the narrower peak shape of the first event of the native sample may suggest a higher cooperativity of unfolding. We thus conclude that viral inactivation has only a minor impact on protein unfolding and have therefore conducted all subsequent analyses with inactivated samples which correspond to the active component of the commercially available anti-rabies vaccine. Repeatability of the method was assessed. Three different vaccine batches (inactivated virus) were analyzed two times independently. Results reported in Supplementary Fig. 1 and Supplementary Table 1 show the good repeatability of the method. To determine the reversibility of unfolding, up-scans to the ends of both transitions (66 and 77 °C) were made, followed by down-scans. In both re-upscans no thermal transitions were observed indicating that both unfolding events are entirely irreversible (Fig. 2B).
Fig. 2.
Analysis of rabies virus by Differential Scanning Calorimetry. (A) Analysis of non-inactivated (black line) and inactivated virus (dark blue line). B) Different DSC up- and re-scans to assess reversibility of unfolding of inactivated rabies virus. Dark Blue line: DSC upscan to 90 °C and rescan; green line: DSC upscan to 77 °C (end of second unfolding event) and rescan, red line: upscan to 66 °C (end of first unfolding event) and rescan. (C) DSC analyses of inactivated rabies virus at different pH values. Dark blue line: pH 7.3; Purple line: pH 5.8, Pink line: pH 6.3. (D) DSC thermogram of inactivated rabies virus in PBS (dark blue line) and PBS supplemented with 5 mM dithiothreitol (orange line). (E) DSC scan of rabies virus prior (dark blue line) and after (cyan line) treatment with bromelain. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Calorimetric parameters of viral unfolding recorded under different experimental conditions and in the presence of different monoclonal antibodies. Data are of the BPL-inactivated virus, except row one showing parameters of the non-inactivated virus.
| Unfolding parameters of rabies virus under different conditions | |||||
|---|---|---|---|---|---|
| Condition | Tm1 (°C) | ΔHcal1 (kcal/mol) | Tm2 (°C) | ΔHcal2 (kcal/mol) | |
| PBS, pH 7.4a | 62.13±0.1 | 455 | 73.04±0.1 | 434 | |
| PBS, pH 7.4b | 61.18±0.1 | 388 | 71.44±0.1 | 447 | |
| PBS, pH 7.3b | 61.40±0.1 | 313 | 71.63±0.1 | 368 | |
| PBS, pH 6.3b | 59.64±0.1 | 365 | 72.43±0.1 | 413 | |
| PBS, pH 5.8b | 62.24±0.1 | 352 | 71.91±0.1 | 362 | |
| PBS, pH 7.4, 5 mM DTTb | Broad transition | 71.78±0.1 | 444 | ||
| PBS, pH 7.4, after bromelain treatmentb | No unfolding event | 71.80±0.1 | 595 | ||
| Tm values determined for equimolar complexes of virusb with different monoclonal antibodies | |||||
| Antibody alone | Tm1 (°C) | Tm2 (°C) | Tm3 (°C) | ||
| D1-25 | 74.90±0.1 | 79.96±0.1 | – | ||
| 50AD1 | 58.88±1.5 | 67.36±0.1 | 75.95±0.1 | ||
| TJU11-12 | 76.28±0.1 | 80.05±0.1 | − | ||
| Virus alone | Tm1 (°C) | Tm2 (°C) | Tm3 (°C) | ||
| PBS, pH 7.4b | 61.18±0.5 | 71.44±0.1 | − | ||
| Antibody/virus complex | Tm1 (°C) | Tm2 (°C) | Tm3 (°C) | ||
| D1-25 | 67.90±0.5 | 73.58±0.1 | 79.94±0.1 | ||
| 50AD1 | 65.35±0.4 | 70.12±0.1 | − | ||
| TJU11-12 | 67.43±0.2 | 73.00±0.1 | 79.72±0.1 | ||
Non-inactivated virus.
BPL-inactivated virus.
3.2. Lowering the pH induces minor changes in the unfolding behavior
Experiments described above were conducted at pH 7.4. There is a body of evidence demonstrating that lowering the pH induces significant structural changes that are particularly pronounced in the G-protein of the virus [28], [29]. To assess how these structural changes influence the viral unfolding behavior, experiments were conducted at pH values of 5.8 and 6.3. As shown in Fig. 2C and Table 1 lowering the pH caused some changes in the Tm of the first event, whereas the corresponding parameter of the second event was relatively conserved. Since the G-protein undergoes conformational changes at lower pH we hypothesized that the first unfolding event may correspond to the unfolding of this protein.
3.3. Large alteration in the unfolding behavior under reducing conditions
We subsequently conducted experiments to establish the influence of reducing conditions on viral unfolding. To this end inactivated samples were dialysed against PBS, pH 7.4 and then 5 mM dithiothreitol was added in the microplate with the sample and incubated at 5 °C for 7 h and 30 min prior to analysis (incubation time was related to the time needed for buffer analysis, 3 runs). Under these conditions disulfide bonds are suppressed. Instead of the first unfolding event of the virus observed under non-reducing conditions, a broad event(s) covering more than 20 °C is seen under reducing conditions (Fig. 2D). As shown in Table 1 the parameters of the second event at approximately 72 °C are very little affected by the reducing conditions. Since the G-protein has several disulfide bonds [30] it appears plausible that the first unfolding event observed under non-reducing conditions is primarily due to its unfolding.
3.4. Bromelain cleavage of G-protein suppresses first unfolding event
To verify this hypothesis we have prepared samples in which the virus was treated with the protease bromelain. It has been reported that such treatment removed the G-protein ectodomain from the viral surface [28]. To verify the efficiency of this treatment samples were analyzed by SDS-PAGE. Fig. 1B shows that bromelain treatment quantitatively removes both G protein bands. The unfolding thermogram of bromelain treated samples (Fig. 2E) is somewhat similar to that of the DTT treated samples and characterized by the absence of the first unfolding event, whereas the second event was very little affected by the protease treatment (Table 1). These data provide further support to the notion that the first unfolding event is primarily due to glycoprotein G unfolding.
3.5. G-protein specific monoclonal antibodies cause an up-shift of the first unfolding event
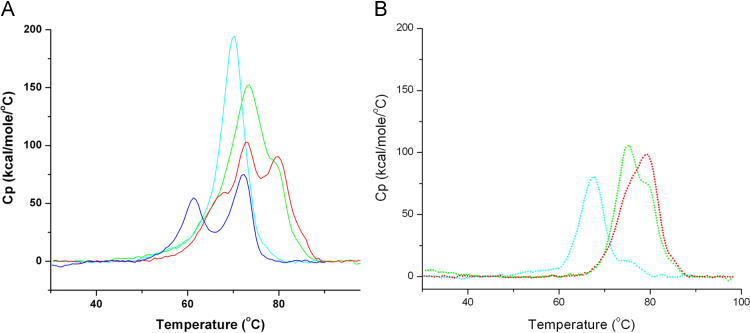
We have then proceeded with the evaluation of the effect of monoclonal G-protein specific antibodies on the viral unfolding. To this end equimolar mixtures of inactivated virus samples (normalized for the G-protein concentration) with the three monoclonal antibodies TJU11-12, D1-25 and 50AD1, were prepared. These antibodies TJU11-12, D1-25 and 50AD1, have been characterized and their binding epitopes at the G-protein were identified (site II, site III and site III, respectively) [25], [26], [27]. The analyses of the virus–antibody mixtures are shown in Fig. 3A and those of the free antibodies in Fig. 3B. All three traces have in common that the first unfolding event at approximately 61 °C has been upshifted to 65–68 °C (Table 1), confirming the notion that it corresponds to the glycoprotein G. In addition, in all samples an event at 70–73 °C was detected which may correspond to the event at 72 °C of the viral sample without Mabs (see also Supplementary Fig. 2).
Fig. 3.
Influence of the binding of glycoprotein G specific antibodies to the BPL-inactivated rabies virus. (A) DSC thermograms of mixtures of inactivated rabies virus with monoclonal antibody. Antibody concentration was adjusted to a 1 : 1 ratio with glycoprotein G. Dark blue line: virus only. Cyan line: in complex with Mab 50AD1, Green line: in complex with Mab D1-25, Red line: in complex with Mab TJU11-12. (B) DSC thermograms of antibodies alone. Cyan dotted line: Mab 50AD1, Green dotted line: Mab D1-25, Red dotted line: Mab TJU11-12. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Inactivated heat-treated virus does not recognize G-protein specific conformational antibodies
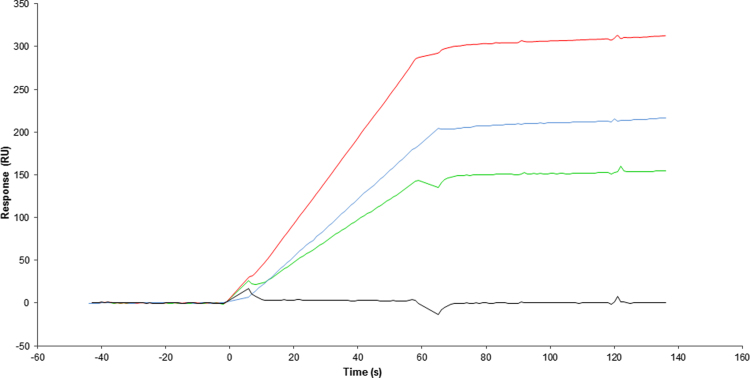
Surface Plasmon Resonance (SPR) experiments were conducted to assess the effect of heat on the structural integrity of the virus. To this end we performed binding assays of the BPL-inactivated virus towards the G-protein specific, conformational Mabs TJU11-12, D1-25 and 50AD1. As shown in Fig. 4, efficient binding was observed in all three cases. When this experiment was repeated with heat-treated virus, using a protocol that mimics heat exposure during a DSC experiment, no viral binding to the three antibodies was observed.
Fig. 4.
SPR analysis of BPL-inactivated non-heated and heated virus with three conformational Mabs. Cyan line: interaction of inactivated non-heated virus with Mab 50AD1, Green line: interaction of inactivated non-heated virus with Mab D1-25, Red line: interaction of inactivated non-heated virus with Mab TJU11-12, Black line: interaction of inactivated heated virus with Mab D1-25 or 50AD1 or TJU11-12. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
These results demonstrate, firstly, that BPL-mediated viral inactivation does not prevent G-protein recognition by specific Mabs and, secondly, show that heat exposure during a DSC experiment causes the irreversible unfolding of the G-protein.
3.7. Analysis of recombinantly produced matrix protein M
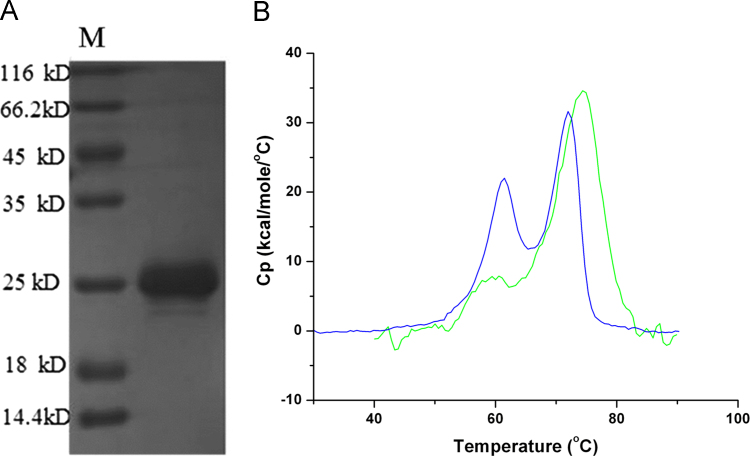
The matrix protein M is a major component of the rabies virus. Recombinantly produced M protein was purchased from MyBiosource (San Diego, USA). An SDS-PAGE analysis (Fig. 5A) shows high purity and confirms the sequence derived molecular weight of 23.3 kDa. The DSC analysis of this protein (Fig. 5B) shows a major unfolding transition at 74 °C with a calorimetric enthalpy change of (3.39±0.1)×102 kcal/mol. The similarities in the Tm and ΔHcal values between the purified M protein and the second event observed for the entire virus may suggest that this second event may be primarily due to the unfolding of the M protein.
Fig. 5.
Comparative analysis of the entire inactivated rabies virus and recombinantly produced purified M protein. (A) SDS-PAGE analysis of purified M protein. (B) DSC analysis of entire inactivated virus (dark blue line) and purified M protein (green line).
4. Discussion
The rabies virus is an assembly of 5 different proteins with RNA. We show here that non-inactivated virus unfolds in two transitions centered at approximately 62 and 72 °C. We show here that BPL mediated virus inactivation does not cause any significant changes in the electrophoretic mobility of viral components (Fig. 1A). Importantly, BPL inactivation does not alter viral unfolding in a significant manner (Fig. 2A). BPL treatment is frequently used for virus inactivation [31]. The chemistry of the BPL reaction with biomolecules has been studied extensively [32] and data indicate that modifications occur primarily at the protein surface [33]. In the case of the influenza vaccine we have demonstrated previously that BPL treatment alters viral adsorption to and fusion with the membrane [34], [35]. The similarity of DSC thermograms of non-inactivated and inactivated rabies virus samples strongly suggests that the chemical modification of viral components by BPL treatment has no major influence on protein structure and unfolding. The standard deviations associated with Tm (Table 1) for the analysis of different sample preparations were remarkably small. This can be attributed, firstly, to the sharpness of both transitions and secondly, to the precision of the instrument. Data thus suggest that the Tm values can be very convenient marker parameters to monitor the industrial vaccine production process.
Changes in the pH caused only minor changes in the viral unfolding behavior. In contrast the exposure to reducing conditions abolished the unfolding event at approximately 61 °C. We show that bromelain treatment removes quantitatively the G-protein. Since the unfolding event at 61 °C is absent from bromelain treated sample, this event corresponds to the unfolding of the G-protein. These observations were confirmed by the up-shift of the first unfolding event upon binding of G-protein specific monoclonal antibodies.
In this context the analysis of the virus under reducing conditions provides interesting insight into G-protein structure. As shown in Fig. 2D the viral thermogram under reducing conditions was characterized by an absence of the first unfolding event. Since the G-protein has 7 potential intra molecular disulfide bonds [30], the DTT treatment of the virus induced their rupture, which in turn had enormous consequences on protein structure and unfolding. As illustrated in Fig. 2D the presence of the reducing agent has little impact on the second event, which is likely to correspond to the cooperative unfolding of the remaining proteins. Viral heat treatment was shown by SPR to prevent G-protein specific Mab recognition. The DSC analysis therefore appears to be a rapid and straightforward tool to assess the structural integrity of the G-protein.
Interestingly, the unfolding parameters of recombinant M-protein are very similar to the second unfolding event observed for the entire virus, suggesting that this latter event may be dominated by M-protein unfolding. However, future studies are necessary to affirm this hypothesis.
In summary, the precision in the determination of the Tm values makes a DSC analysis a very suitable tool to follow the vaccine production process and to assess lot to lot consistency.
Acknowledgments
We thank Fabienne Barriere for her excellent technical assistance in mass spectrometry experiments.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.10.010.
Appendix A. Supplementary material
Supplementary material. Supplementary Fig. (1) DSC thermograms of three different batches of inactivated rabies virus analysed two times independently. (A) Batch no. 1. Pink line: assay no. 1, Purple line: assay no. 2. (B) Batch no. 2. Cyan line: assay no. 1, Green line: assay no. 2. (C) Batch no. 3. Dark blue line: assay no. 1, Dark purple : assay no. 2.
Supplementary material. Supplementary Fig. (2) DSC thermograms of mixture of inactivated rabies virus with monoclonal antibody. Antibody concentration was adjusted to a 1: 1 ratio with glycoprotein G. (A) Dark blue line: virus only. Cyan line: in complex with Mab 50AD1, Cyan dotted line: Mab 50AD1 only. (B) Dark blue line: virus only. Green line: in complex with Mab D1-25, Green dotted line: Mab D1-25 only. (C) Dark blue line: virus only. Red line: in complex with Mab TJU11-12, Red dotted line: Mab TJU11-12 only.
Supplementary material
Supplementary material
References
- 1.Warrell M.J., Warrell D.A. Rabies and other lyssavirus diseases. Lancet. 2004;363:959–969. doi: 10.1016/S0140-6736(04)15792-9. [DOI] [PubMed] [Google Scholar]
- 2.Adamson P.B. The spread of rabies into Europe and the probable origin of this disease in antiquity. J. R. Asiat. Soc. G. B. Irel. 1977;2:140–144. doi: 10.1017/s0035869x00133829. [DOI] [PubMed] [Google Scholar]
- 3.Wunner W.H., Briggs D.J. Rabies in the 21 century. PLoS Negl. Trop. Dis. 2010;4:e591. doi: 10.1371/journal.pntd.0000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigg A.J., Walker P.L. Overview, prevention, and treatment of rabies. Pharmacotherapy. 2009;29:1182–1195. doi: 10.1592/phco.29.10.1182. [DOI] [PubMed] [Google Scholar]
- 5.Finke S., Conzelmann K.K. Replication strategies of rabies virus. Virus Res. 2005;111:120–131. doi: 10.1016/j.virusres.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Wu X., Smith T.G., Rupprecht C.E. From brain passage to cell adaptation: the road of human rabies vaccine development. Expert Rev. Vaccines. 2011;10:1597–1608. doi: 10.1586/erv.11.140. [DOI] [PubMed] [Google Scholar]
- 7.Toovey S. Preventing rabies with the Verorab vaccine: 1985–2005 twenty years of clinical experience. Travel. Med. Infect. Dis. 2007;5:327–348. doi: 10.1016/j.tmaid.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Johnson C.M. Differential scanning calorimetry as a tool for protein folding and stability. Arch. Biochem. Biophys. 2013;531:100–109. doi: 10.1016/j.abb.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Krell T. Microcalorimetry: a response to challenges in modern biotechnology. Microb. Biotechnol. 2008;1:126–136. doi: 10.1111/j.1751-7915.2007.00013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epand R.M., Epand R.F. Thermal denaturation of influenza virus and its relationship to membrane fusion. Biochem. J. 2002;365:841–848. doi: 10.1042/BJ20020290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rayaprolu V., Kruse S., Kant R., Venkatakrishnan B., Movahed N., Brooke D. Comparative analysis of adeno-associated virus capsid stability and dynamics. J. Virol. 2013;87:13150–13160. doi: 10.1128/JVI.01415-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ihnat P.M., Vellekamp G., Obenauer-Kutner L.J., Duan J., Han M.A., Witchey-Lakshmanan L.C. Comparative thermal stabilities of recombinant adenoviruses and hexon protein. Biochim. Biophys. Acta. 2005;1726:138–151. doi: 10.1016/j.bbagen.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Rexroad J., Wiethoff C.M., Green A.P., Kierstead T.D., Scott M.O., Middaugh C.R. Structural stability of adenovirus type 5. J. Pharm. Sci. 2003;92:665–678. doi: 10.1002/jps.10340. [DOI] [PubMed] [Google Scholar]
- 14.Shnyrov V.L., Zhadan G.G., Cobaleda C., Sagrera A., Munoz-Barroso I., Villar E. A differential scanning calorimetric study of Newcastle disease virus: identification of proteins involved in thermal transitions. Arch. Biochem. Biophys. 1997;341:89–97. doi: 10.1006/abbi.1997.9953. [DOI] [PubMed] [Google Scholar]
- 15.Orlov V.N., Kust S.V., Kalmykov P.V., Krivosheev V.P., Dobrov E.N., Drachev V.A. A comparative differential scanning calorimetric study of tobacco mosaic virus and of its coat protein ts mutant. FEBS Lett. 1998;433:307–311. doi: 10.1016/s0014-5793(98)00924-7. [DOI] [PubMed] [Google Scholar]
- 16.Mutombo K., Michels B., Ott H., Cerf R., Witz J. The thermal stability and decapsidation mechanism of tymoviruses: a differential calorimetric study. Biochimie. 1993;75:667–674. doi: 10.1016/0300-9084(93)90097-c. [DOI] [PubMed] [Google Scholar]
- 17.Makarov V.V., Skurat E.V., Semenyuk P.I., Abashkin D.A., Kalinina N.O., Arutyunyan A.M. Structural lability of barley stripe mosaic virus virions. PLoS One. 2013;8:e60942. doi: 10.1371/journal.pone.0060942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemykh M.A., Efimov A.V., Novikov V.K., Orlov V.N., Arutyunyan A.M., Drachev V.A. One more probable structural transition in potato virus X virions and a revised model of the virus coat protein structure. Virology. 2008;373:61–71. doi: 10.1016/j.virol.2007.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Shi L., Sanyal G., Ni A., Luo Z., Doshna S., Wang B. Stabilization of human papillomavirus virus-like particles by non-ionic surfactants. J. Pharm. Sci. 2005;94:1538–1551. doi: 10.1002/jps.20377. [DOI] [PubMed] [Google Scholar]
- 20.Krell T., Manin C., Nicolai M.C., Pierre-Justin C., Berard Y., Brass O. Characterization of different strains of poliovirus and influenza virus by differential scanning calorimetry. Biotechnol. Appl. Biochem. 2005;41:241–246. doi: 10.1042/BA20040113. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X., Wei M., Pan H., Lin Z., Wang K., Weng Z. Robust manufacturing and comprehensive characterization of recombinant hepatitis E virus-like particles in Hecolin((R)) Vaccine. 2014;32:4039–4050. doi: 10.1016/j.vaccine.2014.05.064. [DOI] [PubMed] [Google Scholar]
- 22.Shank-Retzlaff M.L., Zhao Q., Anderson C., Hamm M., High K., Nguyen M. Evaluation of the thermal stability of Gardasil. Hum. Vaccines. 2006;2:147–154. doi: 10.4161/hv.2.4.2989. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan K., Gopinath V.P., Raj G.D. Formulation of newcastle disease virus coupled calcium phosphate nanoparticles: an effective strategy for oculonasal delivery to chicken. Colloids Surf. B: Biointerfaces. 2014;116:9–16. doi: 10.1016/j.colsurfb.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 24.LiCalsi C., Maniaci M.J., Christensen T., Phillips E., Ward G.H., Witham C. A powder formulation of measles vaccine for aerosol delivery. Vaccine. 2001;19:2629–2636. doi: 10.1016/s0264-410x(00)00503-x. [DOI] [PubMed] [Google Scholar]
- 25.Fournier-Caruana J., Poirier B., Haond G., Jallet C., Fuchs F., Tordo N. Inactivated rabies vaccine control and release: use of an ELISA method. Biologicals. 2003;31:9–16. doi: 10.1016/s1045-1056(02)00070-2. [DOI] [PubMed] [Google Scholar]
- 26.Dietzschold B., Gore M., Casali P., Ueki Y., Rupprecht C.E., Notkins A.L. Biological characterization of human monoclonal antibodies to rabies virus. J. Virol. 1990;64:3087–3090. doi: 10.1128/jvi.64.6.3087-3090.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flamand A., Raux H., Gaudin Y., Ruigrok R.W. Mechanisms of rabies virus neutralization. Virology. 1993;194:302–313. doi: 10.1006/viro.1993.1261. [DOI] [PubMed] [Google Scholar]
- 28.Gaudin Y., Tuffereau C., Segretain D., Knossow M., Flamand A. Reversible conformational changes and fusion activity of rabies virus glycoprotein. J. Virol. 1991;65:4853–4859. doi: 10.1128/jvi.65.9.4853-4859.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roche S., Gaudin Y. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology. 2002;297:128–135. doi: 10.1006/viro.2002.1429. [DOI] [PubMed] [Google Scholar]
- 30.Walker P.J., Kongsuwan K. Deduced structural model for animal rhabdovirus glycoproteins. J. Gen. Virol. 1999;80:1211–1220. doi: 10.1099/0022-1317-80-5-1211. [DOI] [PubMed] [Google Scholar]
- 31.Suomela H. Inactivation of viruses in blood and plasma products. Transfus. Med. Rev. 1993;7:42–57. doi: 10.1016/s0887-7963(93)70032-2. [DOI] [PubMed] [Google Scholar]
- 32.Uittenbogaard J.P., Zomer B., Hoogerhout P., Metz B. Reactions of beta-propiolactone with nucleobase analogues, nucleosides, and peptides: implications for the inactivation of viruses. J. Biol. Chem. 2011;286:36198–36214. doi: 10.1074/jbc.M111.279232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.She Y.M., Cheng K., Farnsworth A., Li X., Cyr T.D. Surface modifications of influenza proteins upon virus inactivation by beta-propiolactone. Proteomics. 2013;13:3537–3547. doi: 10.1002/pmic.201300096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desbat B., Lancelot E., Krell T., Nicolai M.C., Vogel F., Chevalier M. Effect of the beta-propiolactone treatment on the adsorption and fusion of influenza A/Brisbane/59/2007 and A/New Caledonia/20/1999 virus H1N1 on a dimyristoylphosphatidylcholine/ganglioside GM3 mixed phospholipids monolayer at the air-water interface. Langmuir: ACS J. Surf. Colloids. 2011;27:13675–13683. doi: 10.1021/la2027175. [DOI] [PubMed] [Google Scholar]
- 35.Bonnafous P., Nicolai M.C., Taveau J.C., Chevalier M., Barriere F., Medina J. Treatment of influenza virus with beta-propiolactone alters viral membrane fusion. Biochim. Biophys. Acta. 2014;1838:355–363. doi: 10.1016/j.bbamem.2013.09.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material. Supplementary Fig. (1) DSC thermograms of three different batches of inactivated rabies virus analysed two times independently. (A) Batch no. 1. Pink line: assay no. 1, Purple line: assay no. 2. (B) Batch no. 2. Cyan line: assay no. 1, Green line: assay no. 2. (C) Batch no. 3. Dark blue line: assay no. 1, Dark purple : assay no. 2.
Supplementary material. Supplementary Fig. (2) DSC thermograms of mixture of inactivated rabies virus with monoclonal antibody. Antibody concentration was adjusted to a 1: 1 ratio with glycoprotein G. (A) Dark blue line: virus only. Cyan line: in complex with Mab 50AD1, Cyan dotted line: Mab 50AD1 only. (B) Dark blue line: virus only. Green line: in complex with Mab D1-25, Green dotted line: Mab D1-25 only. (C) Dark blue line: virus only. Red line: in complex with Mab TJU11-12, Red dotted line: Mab TJU11-12 only.
Supplementary material
Supplementary material