Abstract
Gene transfer agents (GTAs) are shaped like bacteriophage particles but have many properties that distinguish them from bacteriophages. GTAs play a role in horizontal gene transfer in nature and thus affect the evolution of prokaryotic genomes. In the course of studies on the extracellular production of designed RNAs using the marine bacterium Rhodovulum sulfidophilum, we found that this bacterium produces a GTA-like particle. The particle contains DNA fragments of 4.5 kb, which consist of randomly fragmented genomic DNA from the bacterium. This 4.5-kb DNA production was prevented while quorum sensing was inhibited. Direct observation of the particle by transmission electron microscopy revealed that the particle resembles a tailed phage and has a head diameter of about 40 nm and a tail length of about 60 nm. We also identified the structural genes for the GTA in the genome. Translated amino acid sequences and gene positions are closely related to those of the genes that encode the Rhodobacter capsulatus GTA. This is the first report of a GTA-like particle from the genus Rhodovulum. However, gene transfer activity of this particle has not yet been confirmed. The differences between this particle and other GTAs are discussed.
Keywords: Artificial RNAs, Extracellular nucleic acids, Gene transfer, Genome sequence, Rhodobacter capsulatus
Highlights
-
•
Rhodovulum sulfidophilum produces the GTA-like particle.
-
•
The GTA-like particle contains 4.5 kbp DNA.
-
•
The GTA-like particle was directly observed by TEM.
-
•
The structural genes for the particle were identified in the host genome.
1. Introduction
Gene transfer agents (GTAs) are shaped like bacteriophage particles but are different from bacteriophages in many ways [1], [2], [3], [4]. The GTA particles do not have their own genomic DNA coding their structure; instead, they contain relatively short DNA fragments uniform in size, randomly cut from the host genome by an unknown cleavage mechanism. The amount of DNA in a single particle is too small to code for all of the particle. The DNA fragments have been thought to be used for genetic exchange between host cells by a mechanism similar to phage-mediated generalized transduction. Although the structural genes of GTAs are present in the host bacterial genome (as for the prophage genes of lysogen), the GTAs are not inducible by mitomycin C [5] and do not form plaques [6]. The expression of GTA genes studied to date is controlled by cellular regulatory systems such as those involved in signaling and quorum sensing [2], [3]. GTAs were first discovered in Rhodobacter capsulatus (basionym Rhodopseudomonas capsulata) in 1974 [5]. Clusters of genes homologous to the GTA structural genes of Rba. capsulatus have been found in many other species of bacteria by genome sequencing projects [7].
The marine phototrophic bacterium Rhodovulum sulfidophilum, which is closely related to the GTA producer Rba. capsulatus, is known to produce extracellular nucleic acids in soluble form in culture medium [8], [9], [10], [11]. Some other bacteria related to this bacterium have also been reported to produce extracellular nucleic acids (reviewed by Kikuchi [11]). Previously, we demonstrated that the extracellular nucleic acids of Rdv. sulfidophilum are cellular DNAs and RNAs [8], [9]. Although the production mechanism of the extracellular nucleic acids is not known, using this bacterial property, we developed a new method for extracellular production of artificially designed, functional, soluble RNAs (RNA aptamers and short hairpin RNAs) in the culture medium [11], [12], [13], [14], [15]. Extracellular production of artificial RNAs was achieved by introduction into Rdv. sulfidophilum cells of an engineered plasmid containing an artificial gene for RNA expression. For completion and improvement of this method, it is necessary to elucidate the mechanism of extracellular nucleic acid production by the bacterium. During the course of studies of this mechanism, we found that Rdv. sulfidophilum produces a GTA-like particle; this report describes the discovery of the genes for the GTA-like particle and the detection of the particles.
2. Materials and methods
2.1. Bacterial strain and growth conditions
The purple phototrophic marine α-proteobacterium Rhodovulum sulfidophilum DSM 1374 was used throughout this study. Rhodovulum sulfidophilum was grown anaerobically at 30 °C in 1.5-mL tubes filled with PYS medium [16] containing 2% (wt/vol) NaCl under incandescent illumination (about 3000 lux). Aerobic growth of Rdv. sulfidophilum was achieved by shaking a 10-mL culture in a 50-mL centrifuge tube at 150 rpm. When inhibition of possible quorum sensing was required, 20 mM α-cyclodextrin was added to the culture. Cell growth was evaluated by measuring the turbidity of the culture medium at 600 nm.
2.2. Preparation of extracellular soluble DNA
Cells were removed from culture medium by centrifugation, and the nucleic acid fraction of the supernatant was precipitated with isopropanol. The precipitate was dried and solubilized with water. This preparation was directly subjected to 0.5% agarose electrophoresis. Because this preparation was not treated with protein-denaturation chemicals such as SDS or phenol–chloroform, the bands on the gel were thought to be free soluble nucleic acids present in the culture medium.
2.3. Preparation of putative GTA fraction from the culture medium of Rdv. sulfidophilum
The putative GTA fraction was prepared essentially as described [17]. A culture of Rdv. sulfidophilum DSM 1374 (150 mL) was centrifuged and the supernatant was collected. To remove bacterial cells completely, the solution was filtered using a 0.22-μm-pore filter. Polyethylene glycol and NaCl were added to the filtrate to concentrations of 10% and 1 M, respectively, and the solution was incubated at 4 °C for 16 h. After centrifugation, the precipitate was suspended in 1.3 mL of SM buffer [100 mM NaCl, 8 mM MgSO4, 50 mM Tris–HCl (pH 7.5), 0.002% (w/v) gelatin]. To remove possible free nucleic acids from the solution it was treated with DNase I and RNase A, and then with CHCl3. The putative GTA particle fraction was harvested by CsCl gradient ultracentrifugation. The band was collected from the gradient and the virions were concentrated by ultrafiltration using Microcon YM-100 centrifugal filter devices (Amicon).
2.4. Identification in the Rdv. sulfidophilum genome of genes homologous to the GTA genes of Rba. capsulatus
Masuda et al. reported the whole-genome sequence of Rdv. sulfidophilum DSM 1374 [18]. The whole-genome sequence and the genes encoding the GTA of Rba. capsulatus were obtained from NCBI (accession number NC_014034). Hereafter, we refer to the GTA from Rba. capsulatus as “RcGTA”. Based on the translated nucleotide sequences of the RcGTA genes, we searched for possible GTA genes in the genome of Rdv. sulfidophilum using BLASTp.
2.5. Transmission electron microscopy of Rdv. sulfidophilum GTA particles
The GTA fraction concentrated by ultrafiltration (Section 2.3) was negatively stained with 2% (wt/vol) phosphotungstic acid (pH 7.0). Transmission electron microscopy using a JEM-1200EX instrument (80 kV) was performed to visualize the particles.
2.6. DNase I treatment of 4.5-kb DNAs in extracellular soluble nucleic acid preparations or particles
DNase I was purchased from Promega. The concentration of DNase I in the mixture was 5 U/mL in 50 mM phosphate buffer (pH 7.5). The mixture was incubated at 37 °C for 3 h. When using the putative GTA preparation as a substrate, the ultrafiltered virion preparations (Section 2.3) were used directly.
2.7. PCR analysis of the GTA-like particle
To test whether the particle contains random pieces of the producing cell's genome, PCR analysis was performed using specific primers for genes from the host genome. The genes were nifH, soxA, pufQ, and regA, which are spread throughout the genome of Rdv. sulfidophilum. PCR was performed in a total volume of 15 µL containing 0.5 µM each primer, 0.2 mM each deoxyribonucleotide triphosphate, 1×GoTaq® Green Flexi Buffer (Promega), 1.5 mM MgCl2, 0.75 U GoTaq® DNA polymerase (Promega), and 10 ng of 4.5-kb DNA. The PCR reaction cycle consisted of a 2-min denaturation at 94 °C, followed by 30 cycles of 1 min at 95 °C, 1 min at 55 °C, and 1 min at 72 °C. The nucleotide sequences of primers for PCR analysis were: for nifH, 5ʹ-CGTGATGAAGATCGGCTACA-3ʹ and 5ʹ-TGTAGATTTCCTGGGCCTTG-3′; for soxA, 5ʹ-AGGGCCGCGAGATCTACTAT-3ʹ and 5ʹ-CATTGAGCCTGGCATTCTTC-3ʹ; for pufQ, 5ʹ-ACCAGACTTCCGACGTTCAC-3ʹ and 5ʹ-AAAATCATGGGCGTGATGA-3ʹ; and for regA, 5ʹ-GACCCGTCGCTATTGATTGT-3ʹ and 5ʹ-TCCTCAAGCCTGAGATCGAC-3ʹ. The samples from PCR were analyzed by 2% agarose gel electrophoresis.
3. Results and discussion
3.1. Detection of 4.5-kb-long DNA fragment in the extracellular soluble DNA preparation of Rdv. sulfidophilum
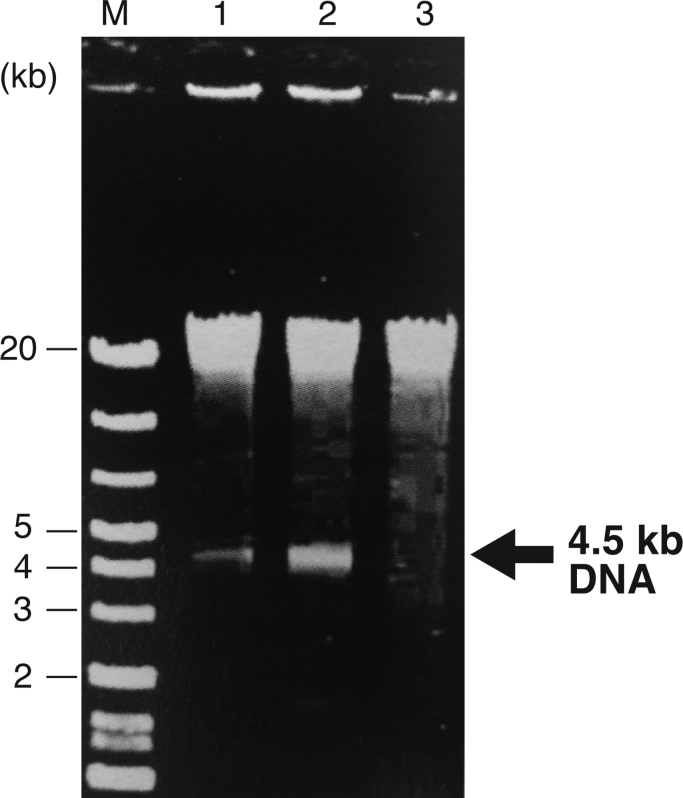
During the course of our extracellular DNA production study, we noticed that the extracellular soluble DNA preparation from Rdv. sulfidophilum DSM 1374 contained approximately 4.5-kb-long DNA. Fig. 1 shows a discrete 4.5-kb-long band (marked with an arrow) when the extracellular soluble DNA preparation from a stationary phase culture was analyzed by agarose gel electrophoresis. This band was not produced when the quorum sensing inhibitor α-cyclodextrin was added into the culture (Fig. 1, lane 3). Quorum sensing is a mechanism by which bacteria can monitor their population density through extracellularly produced signal molecules. In many Gram-negative bacteria, N-acyl homoserine lactones (AHLs) are signaling compounds involved in quorum sensing [19]. α-Cyclodextrin is known to form inclusion complexes with AHLs and shows an inhibitory effect on quorum sensing [20]. To eliminate the possibility that α-cyclodextrin had only a nutritional effect and did not inhibit quorum sensing, Rdv. sulfidophilum was cultivated in medium containing an equivalent amount (in glucose units) of glucose. The extracellular DNA from this culture contained the 4.5-kb DNA (Fig. 1, lane 2). Although the quorum sensing system(s) in this bacterium are unknown, these results suggest that the generation of the extracellular 4.5-kb DNA is regulated by AHL-mediated quorum sensing.
Fig. 1.
Detection of 4.5-kb DNA in the extracellular soluble DNA fraction of Rhodovulum sulfidophilum. Extracellular soluble DNA fractions from the stationary phase of Rdv. sulfidophilum cultures were electrophoresed on 0.5% agarose gels and stained by ethidium bromide. Lane 1, DNA from PYS medium; lane 2, DNA from glucose containing medium; and lane 3, DNA from α-cyclodextrin containing medium. M, size marker. DNA sizes (2–20 kb) are shown to the left of the panel. The 4.5-kb DNA bands are indicated with an arrow. See text for full description.
GTA-like genes are widely conserved in α-proteobacteria [21]. The GTA of Rba. capsulatus (RcGTA) was discovered first and has been studied most extensively [1], [2], [3], [4], [5], [6], [7], [21]. The RcGTA particle packages 4.5-kb DNA randomly produced from the Rba. capsulatus genome by an unknown cleavage mechanism. Although the 4.5-kb DNAs seen in Fig. 1 were detected in the culture medium in soluble form, rather than in a particle, the results concerning the response of the uniform-sized DNA fragment production to a quorum-sensing inhibitor led us to speculate that Rdv. sulfidophilum could produce a GTA.
3.2. Identification of GTA-like structural genes in the Rdv. sulfidophilum genome
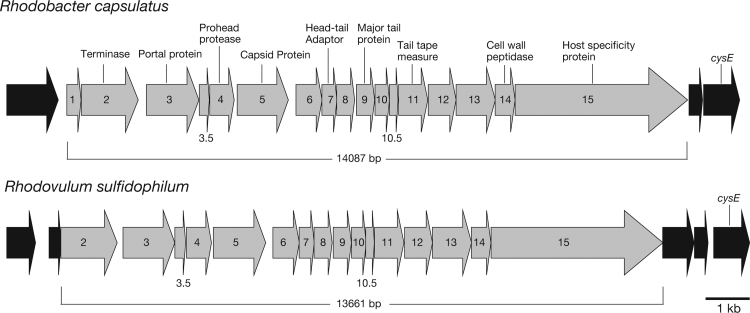
Using next-generation DNA sequencing technology, we have analyzed the whole genome sequence of Rdv. sulfidophilum DSM 1374 (unpublished) and DSM 2351 [22]. Masuda et al. also published the whole-genome sequence of Rdv. sulfidophilum DSM 1374 [18]. In these sequences, we found a cluster (a copy in the genome) of genes homologous to the RcGTA structural genes (Fig. 2). A position of the cluster in the chromosome is 1,923,465–1,937,125. The RcGTA gene cluster consists of 15 genes (ORFs), orfg1–orfg15, and is thought to be transcribed as an operon [7]. As Fig. 2 shows, nine of these ORFs (orfgs 2, 3, 4, 5, 7, 9, 11, 14, and 15) encode identified proteins, but the functions of the other ORFs have not been determined [23], [24]. The genome of Rdv. sulfidophilum DSM 1374 contains homologs of orfg2–orfg15 of Rba. capsulatus, but does not have a homolog of orfg1, although an ORF is present at the corresponding position (Fig. 2). The amino acid sequence identities of orfg2–orfg15 between two strains were calculated by BLASTp to be from 51.4% to 73.7% (Table 1). The presence of the RcGTA-like gene cluster as well as the presence of the 4.5-kb DNA fragment in extracellular DNA preparations motivated us to search for GTA particles in Rdv. sulfidophilum culture.
Fig. 2.
Organization of the GTA structural gene cluster of Rhodobacter capsulatus and the GTA-like gene cluster of Rhodovulum sulfidophilum. The arrows show the approximate scale of the open reading frames (ORFs). A 1-kb scale bar is shown. The numbers, 2–15, in the genes of Rdv. sulfidophilum are homologs of the same numbered genes of Rba. capsulatus. The predicted functions of the ORFs are shown for Rba. capsulatus. GTA structural genes and other host-associated genes are shaded in gray and black, respectively. See text for full description.
Table 1.
Amino acid identities between RcGTA and RsGTA predicted proteins.
| orfg no. | RcGTA CDS locus tag | RsGTA CDS locus tag | % Identity | E value |
|---|---|---|---|---|
| 2 | RCAP_rcc01683 | TY12_RS09170 | 72.3 | 0 |
| 3 | RCAP_rcc01684 | TY12_RS09175 | 71.0 | 0 |
| 3.5 | RCAP_rcc01685 | TY12_RS09180 | 51.4 | 1.00E−25 |
| 4 | RCAP_rcc01686 | TY12_RS09185 | 67.0 | 3.00E−74 |
| 5 | RCAP_rcc01687 | TY12_RS09190 | 71.7 | 0 |
| 6 | RCAP_rcc01688 | TY12_RS09195 | 54.0 | 1.00E−66 |
| 7 | RCAP_rcc01689 | TY12_RS09200 | 54.0 | 5.00E−35 |
| 8 | RCAP_rcc01690 | TY12_RS09205 | 60.5 | 4.00E−45 |
| 9 | RCAP_rcc01691 | TY12_RS09210 | 73.7 | 7.00E−78 |
| 10 | RCAP_rcc01692 | TY12_RS09215 | 64.1 | 4.00E−29 |
| 10.5 | RCAP_rcc01693 | TY12_RS09220 | 63.3 | 3.00E−24 |
| 11 | RCAP_rcc01694 | TY12_RS09225 | 59.3 | 3.00E−82 |
| 12 | RCAP_rcc01695 | TY12_RS09230 | 69.1 | 2.00E−109 |
| 13 | RCAP_rcc01696 | TY12_RS09235 | 57.8 | 4.00E−120 |
| 14 | RCAP_rcc01697 | TY12_RS09240 | 70.1 | 9.00E−72 |
| 15 | RCAP_rcc01698 | TY12_RS09245 | 60.4 | 0 |
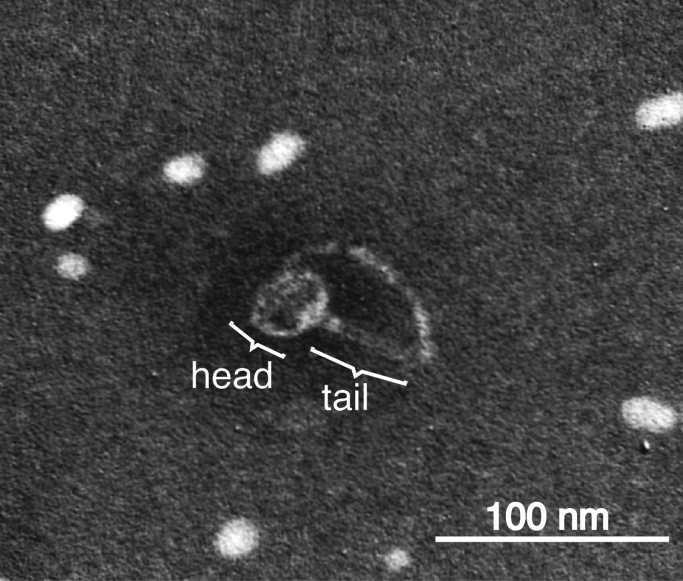
3.3. Direct observation of the GTA-like particle of Rdv. sulfidophilum
To obtain direct evidence for GTA-like particle production by Rdv. sulfidophilum, we tried electron microscopic analysis of the bacterial culture. We observed GTA-like particles in culture of Rdv. sulfidophilum by transmission electron microscopy (Fig. 3). The particle has a head diameter of about 40 nm and a tail length of about 60 nm. The particle is very similar to RcGTA, but a little larger (the head and tail of RcGTA were reported to be 30 nm and 50 nm, respectively [25]). The sizes of almost all the GTA ORFs of Rdv. sulfidophilum (orfg2–orfg15) are very similar to those of the homologous ORFs of Rba. capsulatus [18]. However, the amino acid sequence identity between the ORFs from these two strains is about 64% on average. The highest homology, observed in orfg9, is only 73.7%. In both strains, orfg9 is thought to code the major tail protein consisting of 137 amino acids. Therefore, the difference in GTA particle sizes between these two strains may be owing to differences in the amino acid compositions of the molecules, but not to the sizes (numbers of amino acid residues) of the component polypeptides.
Fig. 3.
Transmission electron micrograph of the GTA-like particle of Rhodovulum. sulfidophilum. The preparation was negatively stained with 2% (wt/vol) phosphotungstic acid (pH 7.0). Bar=100 nm.
3.4. The GTA-like particle of Rdv. sulfidophilum packages 4.5-kb DNA fragments
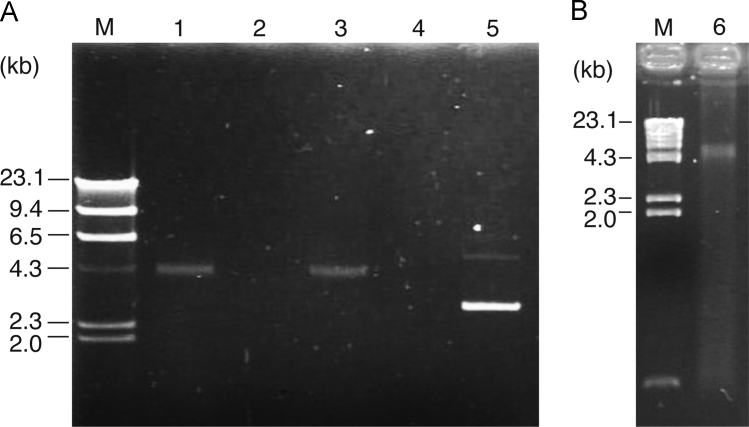
GTAs reported to date package random pieces of the producing cell's genome [1], [2], [3], [4]. To test whether the GTA-like particle of Rdv. sulfidophilum contains DNA fragments, we analyzed the GTA fraction prepared from the culture of Rdv. sulfidophilum using gel electrophoresis. First, we could detect a DNA fragment of 4.5 kb after phenolization of this GTA fraction (Fig. 4A, lane 1). We did not detect any bands other than the 4.5-kb DNA in this lane, indicating that the particle contains DNA of 4.5 kb only. The band was easily lost on DNase I treatment (Fig. 4A, lane 2), indicating that this band was DNA. However, if the treatment with DNase I was performed before phenolization of the GTA fraction, the DNA fragments almost completely remained (Fig. 4A, lane 3). When the GTA fraction itself (without any treatment) was directly subjected to gel electrophoresis, no discrete DNA band of 4.5 kb was detected, although a broad DNA band was observed (Fig. 4B, lane 6). This smear band was thought to be the DNAs from particles broken in the gel during the electrophoresis run. These results indicate that the DNA fragment of 4.5 kb was resistant to DNase I when it was packaged in the particle. It should be emphasized that the DNA fragment prepared from the particle preparation by phenolization showed a clear discrete band of 4.5 kb only, without any smeared or broad bands (Fig. 4A, lane 1). This reveals that the GTA-like particles of Rdv. sulfidophilum package DNAs that are uniform in size, and that the particles may truly be GTA like.
Fig. 4.
Analysis of the DNA from the Rhodovulum sulfidophilum GTA-like particle. (A) The DNA preparation from the GTA-like particle or the particle itself was treated with DNase I and analyzed by 0.8% agarose gel electrophoresis. Lane 1, the DNA from the particle fraction purified by phenolization; lane 2, the same sample as in lane 1 but treated with DNase I; lane 3, the particle was first treated with DNase I, then the DNA sample was prepared by phenolization; lanes 4 and 5, DNase I treated and untreated plasmid DNA (pGEM-3Z), respectively, as a positive control for DNase I activity. (B) The particle fraction was directly subjected to 1% agarose gel electrophoresis (lane 6). M, size markers (phage lambda DNA treated with HindIII). Sizes of markers (in kb) are indicated on the left-hand side of the panel.
3.5. The 4.5-kb DNA fragments in the GTA-like particle may originate from whole-genome DNA
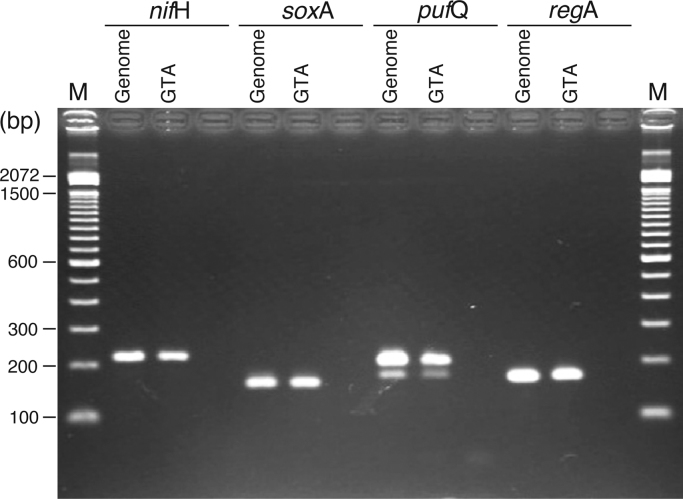
The RcGTA has been reported to constitutively package random pieces of the producing cell’s genome [1], [2], [3], [4], [7]. To test whether our GTA-like particles contained random fragments originating from the host cell's genome, the GTA-like particle fractions prepared were directly probed for the genes nifH, soxA, pufQ, and regA of Rdv. sulfidophilum. These genes were chosen because of their widespread locations on the genome of this organism [18]. Using specific primers for these genes, specific sequences from all four genes could be amplified from the DNA from the GTA-like particle by PCR (Fig. 5). To confirm that these bands (Fig. 5) were not false signals, we performed another experiment as a negative control using a possible GTA fraction from the ΔctrA mutant of Rdv. sulfidophilum as template for the PCR experiment. No DNA was amplified by PCR using this template (data not shown). CtrA is a cell-cycle regulator required for RcGTA production [1]. We also isolated a ctrA mutant of Rdv. sulfidophilum and confirmed that the mutant lost the ability to produce the GTA-like particle (these results will be reported elsewhere). Our results strongly indicate that the 4.5-kb fragments in the particle are random pieces of the cell's genome and that the GTA-like particle has very similar characteristics to RcGTA.
Fig. 5.
The DNAs in the particle are random fragments from the host genome. Using primers specific for the genes nifH, soxA, pufQ, and regA (known to have widespread location on the genome of Rhodovulum sulfidophilum[18]), PCRs were performed with the GTA-like particle fraction as a template. The PCR products analyzed by 2% agarose gel electrophoresis are shown. “Genome” (for control) and “GTA” indicate the templates used for the PCRs. Exactly the same sized bands were amplified from “Genome” and “GTA” DNA by each primer set. M, size marker, 100-bp DNA ladder (Invitrogen). See text for full description.
3.6. Gene transfer activity of the GTA-like particle
We tested the gene transfer ability of the GTA-like particle of Rdv. sulfidophilum. Using the same assay method as for RcGTA and the kanamycin-resistance gene as a marker gene [26], we have not yet obtained reproducible and reliable results showing that our GTA-like particle has gene transfer activity. It might be necessary to develop a new method to detect the gene transfer activity of this GTA-like particle. Brimacombe et al. recently reported that the receptor for RcGTA is a capsular polysaccharide and that the polysaccharide synthesis gene clusters were discovered in the Rba. capsulatus genome [3]. They also reported that mutants deficient in this polysaccharide synthesis showed very low recipient ability from the RcGTA. We could not identify genes homologous to the polysaccharide synthesis genes of Rba. capsulatus in the genome of Rdv. sulfidophilum [18]. However, as mentioned above, our GTA-like particle’s structural genes are very similar to the genes that encode RcGTA (Fig. 2). If our GTA-like particle also requires this type of polysaccharide as receptor, it is possible that the particle is not for self-genetic exchange but acts as a gene carrier to other species. Studies of the gene transfer ability of the Rdv. sulfidophilum GTA-like particle are in progress.
3.7. Views on the formation of 4.5-kb DNA fragments from the cell’s genome
The GTA or GTA-like particle contains 4.5-kb DNAs, uniform in size, from the host genome. This is a typical point that distinguishes GTAs from bacteriophages. To make the evolutionary relationship between GTAs and bacteriophages clear, it is important to know the mechanism of production of the 4.5-kb DNA found in the GTA. In Section 3.1 of the present paper, we mentioned the detection of a soluble, extracellular 4.5-kb DNA fragment in the culture medium (Fig. 1). This finding suggests an insight into the mechanism of formation of the 4.5-kb fragments. As described in Section 2 (Section 2.2), the extracellular soluble DNA was prepared from the culture supernatant by simple isopropanol precipitation. This precipitated preparation was directly subjected to gel electrophoresis without any pretreatment such as phenolization. However, the discrete 4.5-kb DNA fragment band could not be detected when the GTA-like particles were directly subjected to gel electrophoresis (Fig. 4B, lane 6). Therefore, the 4.5-kb DNA fragments were present in soluble form in the culture supernatant. It is thus plausible that the fragmentation of the genomic DNA into 4.5-kb-long segments occurs concomitantly with GTA particle assembly. There is, however, also a possibility that the GTA particles, once formed, decompose during cultivation, or that the fragmentation of the genomic DNA is independent of the particle assembly.
4. Conclusions
The phototrophic marine bacterium Rdv. sulfidophilum produces GTA-like particles that are very similar to RcGTA. Although the gene transfer activity of this GTA-like particle is still not clear, the presence of the structural genes for the particle in the host genome and electron microscopic observation of the particle are enough evidence to establish the presence of the GTA-like particle in Rdv. sulfidophilum.
Acknowledgments
We thank Mrs. Chinatsu Yonekawa for technical assistance. This work was supported in part by a Grant-in-Aid for Science Research to Y. Kikuchi [No. 25252017] from the Japan Society for the Promotion of Science, and by the Institute for Fermentation, Osaka, to S. Umekage.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.11.002.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Lang A.S., Beatty J.T. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. USA. 2000;97:859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaefer A.L., Taylor T.A., Beatty J.T., Greenberg E.P. Long-chain acyl-homoserine lactone quorum-sensing regulation of Rhodobacter capsulatus gene transfer agent production. J. Bacteriol. 2002;184:6515–6521. doi: 10.1128/JB.184.23.6515-6521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brimacombe C.A., Stevens A., Jun D., Mercer R., Lang A.S., Beatty J.T. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA) Mol. Microbiol. 2013;87:802–817. doi: 10.1111/mmi.12132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang A.S., Beatty J.T. Gene transfer agents and defective bacteriophages as sources of extracellular prokaryotic DNA. In: Kikuchi Y., Rykova E.Y., editors. Extracellular Nucleic Acids. Springer-Verlag; Heidelberg: 2010. pp. 15–24. [Google Scholar]
- 5.Marrs B. Genetic recombination in Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA. 1974;71:971–973. doi: 10.1073/pnas.71.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solioz M., Marrs B. The gene transfer agent of Rhodopseudomonas capsulata. Purification and characterization of its nucleic acid. Arch. Biochem. Biophys. 1977;181:300–307. doi: 10.1016/0003-9861(77)90508-2. [DOI] [PubMed] [Google Scholar]
- 7.Lang A.S., Beatty J.T. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. doi: 10.1016/j.tim.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Ando T., Suzuki H., Nishimura S., Tanaka T., Hiraishi A., Kikuchi Y. Characterization of extracellular RNAs produced by the marine photosynthetic bacterium Rhodovulum sulfidophilum. J. Biochem. 2006;139:805–811. doi: 10.1093/jb/mvj091. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H., Daimon M., Awano T., Umekage S., Tanaka T., Kikuchi Y. Characterization of extracellular DNA production and flocculation of the marine photosynthetic bacterium Rhodovulum sulfidophilum. Appl. Microbiol. Biotechnol. 2009;84:349–356. doi: 10.1007/s00253-009-2031-7. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H., Umekage S., Tanaka T., Kikuchi Y. Extracellular tRNAs of the marine photosynthetic bacterium Rhodovulum sulfidophilum are not aminoacylated. Biosci. Biotechnol. Biochem. 2009;73:425–427. doi: 10.1271/bbb.80465. [DOI] [PubMed] [Google Scholar]
- 11.Kikuchi Y. Extracellular nucleic acids of the marine phototrophic bacterium Rhodovulum sulfidophilum and related bacteria: physiology and biotechnology. In: Kikuchi Y., Rykova E.Y., editors. Extracellular Nucleic Acids. Springer-Verlag; Heidelberg: 2010. pp. 55–67. [Google Scholar]
- 12.Suzuki H., Ando T., Umekage S., Tanaka T., Kikuchi Y. Extracellular production of an RNA aptamer by ribonuclease-free marine bacteria harboring engineered plasmids: a proposal for industrial RNA drug production. Appl. Environ. Microbiol. 2010;76:786–793. doi: 10.1128/AEM.01971-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikuchi Y., Suzuki H., Umekage S. Produktion definierter RNAs im Kulturüberstand von Bakterien. LABORWELT. 2010;11:6–7. [Google Scholar]
- 14.Suzuki H., Umekage S., Tanaka T., Kikuchi Y. Artificial RNA aptamer production by the marine bacterium Rhodovulum sulfidophilum: improvement of the aptamer yield using a mutated transcriptional promoter. J. Biosci. Bioeng. 2011;112:458–461. doi: 10.1016/j.jbiosc.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 15.Nagao N., Suzuki H., Numano R., Umekage S., Kikuchi Y. Short hairpin RNAs of designed sequences can be extracellularly produced by the marine bacterium Rhodovulum sulfidophilum. J. Gen. Appl. Microbiol. 2014;60:222–226. doi: 10.2323/jgam.60.222. [DOI] [PubMed] [Google Scholar]
- 16.Nagashima K.V., Hiraishi A., Shimada K., Matsuura K. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J. Mol. Evol. 1997;45:131–136. doi: 10.1007/pl00006212. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey S.B., Stanton T.B., Jensen N.S., Zuerner R.L. Purification and characterization of VSH-1, a generalized transducing bacteriophage of Serpulina hyodysenteriae. J. Bacteriol. 1997;179:323–329. doi: 10.1128/jb.179.2.323-329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masuda S., Hori K., Maruyama F., Ren S., Sugimoto S., Yamamoto N., Mori H., Yamada T., Sato S., Tabata S., Ohta H., Kurokawa K. Whole-genome sequence of the purple photosynthetic bacterium Rhodovulum sulfidophilum Strain W4. Genome Announc. 2013;1 doi: 10.1128/genomeA.00577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitehead N.A., Barnard A.M.L., Slater H., Simpson N.J.L., Salmond G.P.C. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 20.Morohoshi T., Tokita K., Ito S., Saito Y., Maeda S., Kato N., Ikeda T. Inhibition of quorum sensing in gram-negative bacteria by alkylamine-modified cyclodextrins. J. Biosci. Bioeng. 2013;116:175–179. doi: 10.1016/j.jbiosc.2013.01.022. [DOI] [PubMed] [Google Scholar]
- 21.Lang A.S., Zhaxybayeva O., Beatty J.T. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 2012;10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagao N., Hirose Y., Misawa N., Ohtsubo Y., Umekage S., Kikuchi Y. Complete genome sequence of Rhodovulum sulfidophilum DSM 2351, an extracellular nucleic acid-producing bacterium. Genome Announc. 2015;3 doi: 10.1128/genomeA.00388-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fogg P.C., Westbye A.B., Beatty J.T. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One. 2012;7:e43772. doi: 10.1371/journal.pone.0043772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung M.M., Florizone S.M., Taylor T.A., Lang A.S., Beatty J.T. The gene transfer agent of Rhodobacter capsulatus. In: Hallenbeck P.C., editor. Recent Advances in Phototrophic Prokaryotes. Springer-Verlag; New York: 2010. pp. 253–264. [Google Scholar]
- 25.Yen H.C., Hu N.T., Marrs B.L. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 1979;131:157–168. doi: 10.1016/0022-2836(79)90071-8. [DOI] [PubMed] [Google Scholar]
- 26.Solioz M., Yen H.C., Marris B. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J. Bacteriol. 1975;123:651–657. doi: 10.1128/jb.123.2.651-657.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material