Abstract
pH-induced conformational changes in dengue virus (DENV) are critical to its ability to infect host cells. The envelope protein heterodimers that make up the viral envelope shift from a dimer to a trimer conformation at low-pH during membrane fusion. Previous studies have suggested that the ionization of histidine residues at low-pH is central to this pH-induced conformational change. We sought out to use molecular modeling with structure-based pKa prediction to provide a quantitative basis for the role of histidines in pH-induced conformational changes and identify which histidine residues were primarily responsible for this transition. We combined existing crystallographic and cryo-electron microscopy data to construct templates of the dimer and trimer conformations for the mature and immature virus. We then generated homology models for the four DENV serotypes and carried out structure-based pKa prediction using Rosetta. Our results showed that the pKa values of a subset of conserved histidines in DENV successfully capture the thermodynamics necessary to drive pH-induced conformational changes during fusion. Here, we identified the structural determinants underlying these pKa values and compare our findings with previous experimental results.
Keywords: pKa shift, Viral fusion, Histidine, Flavivirus
Highlights
-
•
Structure-based pKa prediction for histidines in dengue virus was carried out.
-
•
Conserved histidine residues drive pH-dependent conformation changes.
-
•
The mature dimer form of the envelop protein is destabilized at low-pH.
1. Introduction
Dengue virus (DENV) is an RNA virus that is transmitted into human hosts through the bite of an infected female Aedes mosquito. DENV belongs to the Flaviviridae family, which includes many mosquito- and arthropod-borne human viruses, including yellow fever, Japanese encephalitis, and West Nile virus. In addition to dengue fever, infection can cause serious complications such as dengue hemorrhagic fever and dengue shock syndrome. DENV affects >200 million people worldwide, and currently there are no licensed vaccines or effective antiviral drugs for treatment of the disease.
DENV is found as one of four serotypes (DENV-1, DENV-2, DENV-3, and DENV-4), and its genome encodes for 10 proteins, including envelope (E) and precursor membrane (PrM) proteins, which constitute the outer layer of the virus. X-ray crystallography studies [1] have shown that the structure of the soluble part of E consists of three domains and includes the fusion loop, which is critical for fusion of the host and viral membranes and putative receptor-binding region. The membrane-associated region of E includes an α-helical segment known as the “stem” region, which has also been shown to be critical for membrane fusion [2], [3], along with two transmembrane helices that anchor E to the viral membrane. The PrM protein consists of a globular domain that “caps” the fusion loop of E to prevent premature fusion [4], a linker region containing a furin cleavage motif, and an α-helical membrane-associated region, including two transmembrane helices. During viral maturation, the globular domain of PrM (termed Pr) in the immature virus is cleaved, releasing Pr and resulting in the mature virus.
Like other flaviviruses, DENV goes through a number of different pH conditions and conformational states during its life-cycle. Viral assembly takes place in the high-pH environment (~7.2) of the endoplasmic reticulum (ER) of the host cell. There, heterodimers of E and PrM (E-PrM) aggregate to form a rough surface capsid composed of 60 trimer spikes of E-PrM. During subsequent transport and processing through the trans-Golgi network (TGN), a decrease in pH (from roughly 7 to ~6) induces a series of conformational rearrangements that results in the formation of the mature capsid [5]. Structural studies based on cryo-electron microscopy (cryo-EM) have shown that, initially, the capsid surface evolves from a “rough” [4] form (~600-Å radius) to a “smooth” (~500-Å radius) form [6] due to a change in the E-PrM oligomerization state. The immature rough capsid, formed by 60 spike-like trimers, converts to a smooth, non-infective form composed of 90 dimers.
After Pr cleavage, E becomes the only protein exposed on the viral surface, making it a critical target for dengue vaccine development. During invasion of a new host cell, E fulfills two roles: (1) it is involved in host-receptor binding and endocytosis and (2) once within the endosome, it is responsible for viral and host membrane fusion. Viral fusion is initiated by acidic conditions in the endosome. The pH drop from 7.0 to <6.0 results in a dramatic change in the oligomeric arrangement of E on the viral envelope from the dimeric state to a trimeric state [7], a behavior that is consistent with experimental observations from in vitro studies on the related flavivirus tick-borne encephalitis virus [8], [9]. Such studies have shown that E associates as dimers at neutral pH, whereas a drop in pH leads to dimer dissociation, followed rapidly by irreversible trimerization. Further studies have shown that trimerization is possible even with recombinant E proteins that lack the stem region [10] and domain III [11], underscoring an inherent proclivity for trimerization of the E protein at low-pH.
Despite extensive biochemical and structural knowledge on the conformational states, the dynamics and structural mechanisms underlying the pH-driven transitions during flavivirus maturation and fusion remain unclear. Some studies have suggested that the generalized ionization of multiple histidines acts to destabilize the pre-fusion conformation and drive the system towards fusion, showing that no single histidine residue is essential for viral fusion [12], [13]. Other studies have suggested that selective ionization of key conserved histidines drives the necessary conformational transitions, such as fusion loop exposure or dimer dissociation [14], [15]. Additional evidence of the importance of selective ionization over generalized destabilization of the pre-fusion conformation comes from in vitro studies that showing that only a decrease in pH, and not an increase in temperature or other denaturing condition, results in fusion [9], [16].
The pH-induced ionization of a given histidine residue is a function of the solution's pH and its pKa as determined by its local environment [17], [18]. In solution, histidine has a pKa of 6.3, but within a protein, this value can vary widely from 3 to 9, depending on the degree of burial and polar and ionic interactions with neighboring residues [19]. We proposed to elucidate the mechanisms of pH-induced conformational changes in DENV using a structure-based approach to determine conformation-specific pKa values for conserved histidine residues. Through this effort, we aimed to identify the role of specific histidine residues in the flavivirus maturation and fusion and quantify the thermodynamic basis for the role of histidines in pH-induced conformational changes in DENV.
We used existing structural data in conjunction with molecular modeling in the Rosetta software suite [20], [21] to generate templates for three conformational states: immature-dimer, mature-dimer, and postfusion trimer. We used these templates to construct homology models for representative strains of all four DENV serotypes. We then used in silico structure-based pKa prediction in Rosetta [22] to determine the pKa of histidine residues in each conformation and calculated the pH-dependent change in stability for each conformation based on the thermodynamic framework developed by Isom et al. [23]. The Rosetta pKa algorithm has been extensively tested on a benchmark set of 264 residues across 34 proteins and predicted the pKa of ionizable residues to within 0.5 pH units in over half the cases, and within 1.5 pH units in over 90% of the cases [22]. However, because pKa shifts are exquisitely sensitive to a complex array of factors including electrostatics, protein conformational fluctuations, and solvent thermodynamics, accurate pKa predictions remain extremely challenging. Our goal in using computational pKa prediction is to provide some quantitative thermodynamic basis for largely qualitative observations about the structural mechanisms underlying pH-induced conformational changes in DENV.
Our results showed that the pKa values of conserved histidine residues within E and PrM are sufficient to explain the pH-induced oligomeric and conformational transitions in both the immature and mature forms of the virus. We identified the histidine residues responsible for driving the pH-induced conformational shifts and how the local environment around these residues “tunes” their pKa values. Finally, we explored the implications of our thermodynamic model for pH-induced conformational changes within the context of a generalized mechanism for membrane fusion in flaviviruses.
2. Materials and methods
2.1. Generating template structures
We used the homology modeling program NEST [30], included in our Protein Structure Prediction Pipeline [31] (PPSP), to generate three-dimensional (3-D) models for the dengue proteins E, PrM, and M. Two types of data inputs are required to produce these models: (1) a template file, usually an experimentally determined structure obtained from the Protein Data Bank [32] (PDB), and (2) a pair-wise alignment between each target sequence and the template structure. We generated models using different templates to account for the conformers of E associated with high, neutral and low-pH values and also to assess the variability of different parts of the structure. The alignments were obtained using the BLAST program. Coordinates of the protein systems were derived from the experimental structures of E and PrM as shown in Supplemental Table S1. To generate missing fragments, produce complete structures for each of the four stages of DENV, and perform analyses of the final structures, we resorted to the following molecular modeling programs: the PyMOL Molecular Graphics System (Accelrys, San Diego, CA) and ECEPPAK.
After the generation of template structures using the automated homology modeling methods of NEST and PSPP, we carried out manual refinement of the structures with the following three criteria: (1) the highest resolution template would be used to define the proper atomic contacts between residue side chains whenever possible, (2) we allowed for minimal adjustments to be made to relieve atomic overlaps caused by the inclusion of residues or atoms not resolved in the experimental structures, and (3) we allowed for minimal adjustments to accommodate overlaps caused by quaternary contacts between E-PrM heterodimers.
We made refinements to the template structures manually by altering relevant dihedral angles, followed by local optimization with a simple contact potential. To generate missing fragments necessary to produce contiguous structures for each of the three conformational states, we used modeling tools in the following software: PyMol (Schrodinger), Discovery Studio (Accelrys), and ECEPPAK [33], [34]. We used a combination of molecular modeling and structural alignments to construct a hybrid template that contained the initial structural template merged with the modeled missing region. These hybrid templates were then manually refined as stated above followed by all-atom minimization using Rosetta (see below).
2.2. Dengue envelope homology modeling pipeline
Once the template structures for the immature dimer, mature dimer, and postfusion trimer structures were completed, we developed a high-throughput homology modeling pipeline that can rapidly generate structure for all four configurations from an input E and PrM sequence. This pipeline was written in Python and used the Pyrosetta [20] interface for the Rosetta molecular modeling suite [21]. First, we used clustalW to align the input query sequence to the template structure sequence. We then threaded the query sequence into the template structure by mutating any mismatched residues to that of the query sequence. We then used the rotamer packing functionality to pack all side chains in the structure. We set the packing parameters to include all original sidechain rotamers into the rotamer library (-include_current) and used an expanded rotamer set that included extra rotamers for the X1 and X2 angles (-ex1 -ex2). After side chain packing, the structure was minimized using Davidon–Fletcher–Powell minimization, allowing all backbone and side chain torsion angles to move. We used the standard full-atom score function [35] modified for soft repulsive forces (“soft_rep”) to carry out both packing and minimization.
DENV-3 has a two-residue deletion in the E protein corresponding to residues 156 and 157 in DENV-1, DENV-2, and DENV-4. We made the deletion in each of the template structures and then reformed the loop surrounding the deletion (defined from residue 152 to 161) using Rosetta loop modeling [25]. This allowed the DENV-3 structure templates to be the appropriate length for threading of DENV-3 sequences.
We used a single representative sequence for each DENV serotype. DENV-1 was the Western Pacific strain, DENV-2 was the New Guinea C strain, DENV-3 was S221/03 strain C, and DENV-4 was the SG/06K2270DK1/2005 strain. Sequences for each strain were downloaded from GenBank and separated into E and PrM sequences as inputs for the homology modeling pipeline.
2.3. Structure-based pKa prediction
We carried out pKa prediction using Rosetta-pKa using a previously described protocol [22]. Briefly, we first carried out pKa prediction for each conserved histidine residue in the E and PrM sequence for each serotype. In order to better accommodate conformational variations in the protein structure that result from side-chain ionization, we opted to use the enhanced side-chain sampling option which allows for the packing of all side chains within 8 Å of the selected histidine residue. We set the packing parameters to include all original side chain rotamers (-include current) as well as include extra rotamers for X1, X2, and X3 angles (-ex1 -ex2 -ex3) to maximize the available sampling of neighboring side chain conformations. We used the default score function for Rosetta-pKa to carry out packing and pKa prediction.
Rosetta pKa simulates the titration of a single ionizable residue within the context of a high-resolution protein structure using a side-chain packing algorithm that includes both protonated and de-protonated forms of all amino acids. The pH at which the free energy of the protonated state is equivalent to the free energy of the unprotonated state is reported as the pKa. Rosetta pKa [22] uses a score function that approximate the free energy of folding for a given protein structure and includes terms for Van der Waals potential (), implicit solvation model (), electrostatic potential (), hydrogen bonds (), amino acid pairwise potential (), intrinsic side-chain conformation energies (), protonation potential (), and reference energies or each amino acid () that are summed up to represent the free energy of the unfolded state (Eq. (1)).
| (1) |
Rosetta pKa is a stochastic method that produces a narrow range of predicted pKa values for each histidine residue. We calculated 10 pKa values for each histidine within a structure and selected the median pKa value as the representative prediction that histidine residue. Since the dimer and trimer configurations are arranged in symmetrical fashion, there are two and three symmetrically related histidines within a single dimer or trimer (hereafter referred to as “oligomerically related” residues). We used the least shifted pKa value among the oligomerically related histidines (the lowest magnitude shift from the solution pKa of 6.3) as the representative pKa for that histidine residue position. Our rationale for this was that if multiple conformations are available to a given histidine, it would tend to adopt the conformation with the lowest free energy, which is the conformation with the least shifted pKa. Finally, we set upper and lower boundaries for the pKa prediction at 3.0 and 9.0 based on a survey of documented pKa values for histidines [19].
2.4. Structure analysis and pH-dependent free energy profile
We calculated the change in free energy of folding as a function of pH using the thermodynamic framework outlined previously [23] and adapted for use in this system. Briefly, we calculated the pKa values of all conserved histidine residues in the system for the three conformational states: immature dimer, mature dimer, and postfusion trimer. We then used these pKa values with Eq. (4) (see Section 3) to generated a curve that reflects the change in free energy as a function of pH. We reported the pH-dependent free energy profile for each of three conformational states for DENV-1 through DENV-4.
Analysis of the structural models was carried out using Schrodinger's PyMol software. SASA was calculated in PyMol using the get_area function using a probe with a radius of 1.8 Å. We calculated the SASA for all conserved histidine residues for all four conformational states over all four DENV serotypes. We reported the SASA value for each conformational state that reflects the average across all oligomerically related histidine residues across all four serotypes.
3. Results
3.1. pH dependence of DENV conformations
DENV has two major forms: an immature form and a mature form, defined by cleavage of the globular domain of Pr from PrM. After viral assembly, the immature virus is found in the dimer state in the low-pH of the TGN, where the viral surface is studded with a regular arrangement of Pr domains. The fully mature virus is found in the dimer state, after both the cleavage of Pr and release into the neutral pH of the extracellular environment. During subsequent host cell infection, the low-pH of the late endosome results in a conformational change to the postfusion trimer state, which precedes membrane fusion and infection. Because the major titratable amino acid at the pH range throughout the maturation and infection is histidine, it is theorized to play a central role in driving the pH-induced conformational changes.
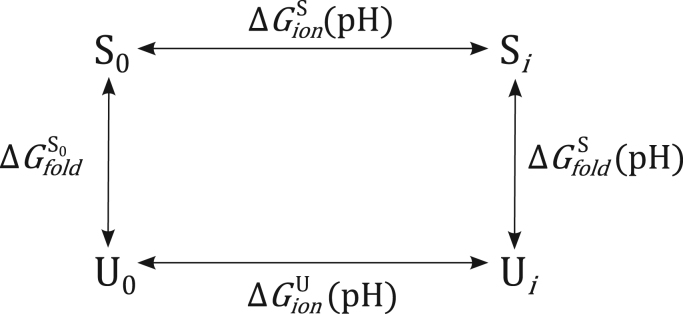
We constructed a thermodynamic cycle based on studies carried out by Isom et al. [23] (Fig. 1), which allowed us to evaluate the thermodynamic contribution of each histidine residue to the stability of a conformational state of the virus (immature dimer, mature dimer, and postfusion trimer). This thermodynamic cycle is analogous to the change in folding free energy associated with the substitution of a mutant amino acid in place of a wild-type amino acid. In this case, the “wild-type” amino acid is a fixed-charge unprotonated histidine (denoted by subscript 0) and the “mutant” amino acid is a pH-sensitive, ionizable histidine (denoted by subscript i). The cycle captures the unfolded state (U) and a particular folded state (S). Through this cycle, we can distinguish between pH-dependent and pH-independent paths for any folded conformational state of the virus and calculate the pH-dependent change in folding energy of a given conformational state and compare these folding energies between different conformations.
Fig. 1.
Thermodynamic cycle of pH dependence of DENV E-PrM. Thermodynamic cycle of the pH dependence of the unfolded state (U) and a folded state (S) of dengue virus in the fixed uncharged (subscript 0) state and ionizable (subscript i) state. The folded state (S) can refer to any folded conformation, including the immature dimer, mature dimer, and postfusion trimer states.
Based on this thermodynamic cycle, Eq. (2) describes the folding free energy of a folded state, S, at a given pH,, as the sum of free energy of the folded state with fixed uncharged histidine residues, (hereafter referred to as the intrinsic folding energy), and the ionization energies of the folded and unfolded states at that pH, and , respectively. Isom et al. derived the contribution of a single ionizable group to stability as a function of the pH and the pKa of that group in the folded state () and unfolded state () [23]. We extended this approach to capture the sum total of j ionizable groups in the protein (Eq. (3)). Finally, we expressed the pH-dependent contribution to the folding free energy of state S at a given pH,, as the difference between the folding free energy at that pH and the intrinsic folding energy (Eq. (4)), which, when combined with Eq. (3), is represented as a function of the pKa values of the ionizable groups in the folded state and unfolded state (Eq. 5). In Eqs. (2), (3), (4), (5), R is the gas constant and T is temperature.
| (2) |
| (3) |
| (4) |
| (5) |
This approach makes two critical assumptions. First, the total contribution of pH to the stability of the protein is a sum of the individual contributions of each ionizable group. Second, the pKa of a residue in the unfolded state is equivalent to its ideal pKa (for histidine, pKa=6.3), based on a theoretical model of the unfolded state in which any ionizable residue would be solvent exposed and, thus, have no major pKa shifts.
The free energy difference between two conformational states, such as the mature dimer and postfusion trimer, includes both pH-independent and pH-dependent components. By isolating the pH-dependent component of free energy of folding, we are comparing the change in pH-dependent stability within each state, but we cannot determine the overall relative free energy difference between each state. For example, we can show that one state becomes less stable and another state becomes more stable at one pH compared to another, but we cannot determine at what pH one state becomes favored over the other, because folding energy () is not explicitly accounted for.
We used homology modeling and structure-based pKa calculations to determine the pKa of each histidine residue in each conformational state. Using the thermodynamic framework above, we can calculate the contribution of each histidine in the dengue E and PrM proteins to the pH-dependent component of folding free energy for the immature dimer, mature dimer, and postfusion trimer states. We focused the analysis on histidine residues because they are theorized to be the primary drivers of pH-induced conformational change in DENV. As such, we modeled not the overall change in pH-dependent stability, but the contribution of histidine residues to pH-dependent stability.
3.2. Structural modeling of DENV envelope proteins
The structure of DENV envelope proteins are highly complex with heterodimers of E and PrM proteins deeply intertwined with each other, each containing transmembrane regions. This topology combined with the mostly low to moderate resolution structural data available through cryo-EM makes modeling of dengue viral proteins challenging using standard methods. We developed a custom protocol for modeling dengue structures from sequence by manually constructing low-resolution templates for three conformational states (immature dimer, mature dimer, and postfusion trimer) followed by high-resolution structural refinement using the Rosetta molecular modeling package.
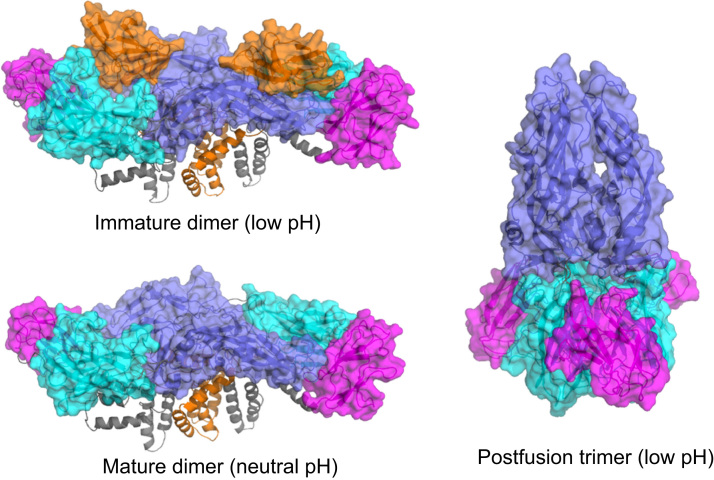
Supplemental Table S1 shows the structures used as the basis for constructing each of the four templates. With one exception, all structures were from DENV-2. Fig. 2 shows the template structures for each of the three conformations. For the immature dimer form, we started from a low-resolution structure of the mature dimer [4] followed by the addition of the pre-cleaved PrM region from a previously studied immature trimer structure [24] and a superposition of the high-resolution X-ray structure of the non-membrane region of the E protein for the immature dimer. For the mature dimer, we started with the EM structure of the mature dimer [14], and for the postfusion trimer we started with the EM structure of the trimer “spike” structure [7]. We did not have any structural data for the membrane regions of either E or PrM proteins in the postfusion trimer, and we omitted those regions entirely.
Fig. 2.
Template structures for DENV E-PrM conformational states. Template structures for the trimer (right) and dimer configurations for mature (bottom left) and immature (top left) DENV. Domains I, II and III of E are shown in cyan, purple, and magenta and PrM is shown in orange. The soluble portion of PrM and E are shown as surfaces, the peri- and trans-membrane helices of PrM and E are shown as cartoons.
For each template structure, the sequence corresponding to each of the four serotypes was threaded into the template, and the structure was optimized using a high-resolution structure refinement protocol using PyRosetta [20], [21]. Briefly, the side chain conformations were repacked iteratively while the protein backbone conformations were re-sampled and minimized. For DENV-3, which has a two-residue deletion in the E protein, Rosetta loop modeling [25] was used to refine the shortened loop.
Fig. 2 shows each of the three template structures and highlights the position and orientation of the E, Pr, and M regions. At the low-pH of the TGN, the immature dimer form dominates, studded with the as-of-yet uncleaved Pr domains. The mature dimer form results from the cleavage and subsequent release of Pr. Finally, during host cell invasion, the low-pH of the endosomal compartment triggers a conformational change of the mature virus particle to form a tightly packed postfusion trimer. In addition to differences in the overall oligomeric states of E-PrM, there are numerous internal conformational rearrangements within these domains that distinguish the three conformational states from each other.
3.3. Structure-based pKa calculations
We calculated the pKa values of each histidine in the E and PrM proteins for all four DENV serotypes in three structural states: (1) immature dimer, (2) mature dimer, and (3) postfusion trimer. We used Rosetta-pKa [22], a structure-based method for pKa prediction, which uses side-chain sampling around the local environment surrounding a given histidine to predict pKa shifts. Although we calculated pKa values for all histidines, we focused our analysis on a subset of histidine residues that are conserved across all DENV serotypes to determine their potential role in driving pH-induced conformational changes in DENV. Overall, conserved histidines make up 9 of 11 histidines present in all four serotypes of E, and 2 of 10 histidines present in all four serotypes of DENV.
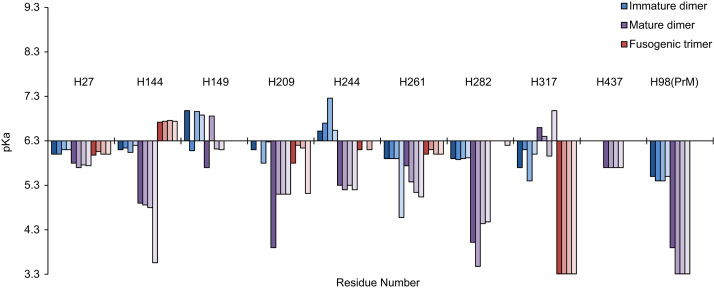
Fig. 3 shows the pKa values for each of the conserved histidine residues in the three conformational states as determined by our Rosetta-pKa protocol. The results show that the pKa values for the conserved histidines were relatively stable across the four serotypes despite the fact that they were generated from structures based on homology modeling, suggesting that the local environments around these histidines is largely conserved across serotypes. A comparison between mature dimer and postfusion trimer states revealed significant changes in the pKa values across all four serotypes that reflect systematic changes in these local environments. Likewise, a comparison between mature and immature dimer forms of the virus also showed consistent differences in the pKa values, demonstrating that the structural differences between the mature and immature forms of the virus contribute to an altered ionization environment for these conserved histidine residues.
Fig. 3.
Structure-based pKa predictions for DENV E-PrM. Calculated pKa values for conserved histidine residues in the immature dimer (blue), mature dimer (purple), and postfusion trimer (red) states using Rosetta-pKa. The ideal pKa value for histidine was set to 6.3, and the maximal allowable pKa range was set between 3.3 and 9.3. Each residue has four pKa values to reflect the pKa value calculated for DENV-1 through DENV-4, respectively.
A number of conserved histidines showed significant pKa shifts in the three conformations studied. In the immature dimer form, only H98 on PrM showed a significantly downshifted pKa. In contrast, the mature dimer showed a large number of conserved histidines, including H27, H144, H209, H244, H261, and H282 on the E protein, that all showed a significant down-shift in pKa. In the postfusion trimer, with one exception (H317), all conserved histidines showed minor pKa shifts. These pKa values were in qualitative agreement with experimental data that showed that low-pH conditions favor the dimer conformation in the immature virus and the trimer conformation in the mature virus.
3.4. pH-dependent effects on conformational stability
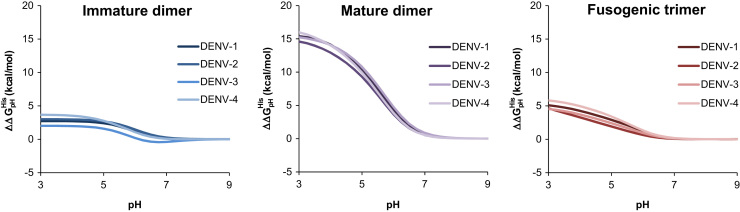
We next sought to analyze these pKa shifts in the context of the thermodynamic cycle shown in Fig. 1 to quantify the degree to which the above histidine pKa values contribute to the overall pH-dependent stability of the three conformational states. Fig. 4 shows the contribution of histidine residues to the pH-dependent folding energy, , for the immature dimer, mature dimer and postfusion trimer conformations across all four DENV serotypes.
Fig. 4.
Changes in folding free energy as a function of pH. Contribution of histidine to changes in folding energy as a function of pH () based on the predicted pKa values for conserved histidines for the immature dimer (blue), mature dimer (purple), and postfusion trimer (red) for DENV-1 through DENV-4.
The pH-dependent folding energy captures the change in folding energy as a function of pH. In the immature dimer, the folding energy is largely insensitive to changes in pH. In contrast, the mature dimer becomes significantly less stable as the pH decreases, when compared with the postfusion trimer, for all four serotypes. These results suggest that the sum total contribution of conserved histidines in the dengue E protein are responsible for a strong thermodynamic destabilization of the dimer configuration at low-pH in the mature virion.
The calculated values of pH-dependent stability are determined by the pKa values for conserved histidines in the dengue E and PrM proteins as based on structural models of each of the three major conformational states. This leads to two main conclusions: (1) changes in the local environment of the conserved histidines observed in our structural models are sufficient to explain the pH dependence that characterizes the mature dimer to postfusion trimer transition and (2) these local environments are largely conserved across all four DENV serotypes and potentially in all members of the flavivirus family.
3.5. Individual residue contributions
Our analysis allowed us to evaluate the relative contribution of each histidine residue and identify those residues that are primarily responsible for providing pH-dependent stabilization of the immature dimer, mature dimer, and postfusion trimer conformations. Table 1 shows the change in the pKa of conserved histidines for two different conformational transitions: (1) the immature dimer-to-mature dimer transition that results from the cleavage of Pr and (2) the mature dimer-to-postfusion trimer transition that occurs in the late endosome. A positive ΔpKa indicates an increase in pKa of that histidine residue, indicating that the residue stabilizes the conformational change at low-pH.
Table 1.
Changes in predicted pKa values of conserved histidines in DENV.
| Residue | Immature dimer → mature dimer | Mature dimer → fusogenic trimer |
|---|---|---|
| ΔpKa (s.d.) | ΔpKa (s.d.) | |
| H27 | −0.3 (0.1) | +0.3 (0.1) |
| H144 | −1.6 (0.7) | +2.2 (0.6) |
| H149 | −0.5 (0.9) | +0.1 (0.5) |
| H209 | −1.3 (0.6) | +1.0 (0.8) |
| H244 | −1.5 (0.3) | +1.0 (0.2) |
| H261 | −0.2 (0.5) | +0.7 (0.3) |
| H282 | −1.8 (0.4) | +2.2 (0.5) |
| H317 | +0.7 (0.3) | -3.2 (0.4) |
| H437 | −0.6 (0.0) | +0.6 (0.0) |
| H98 (PrM) | −2.0 (0.3) | +2.8 (0.3) |
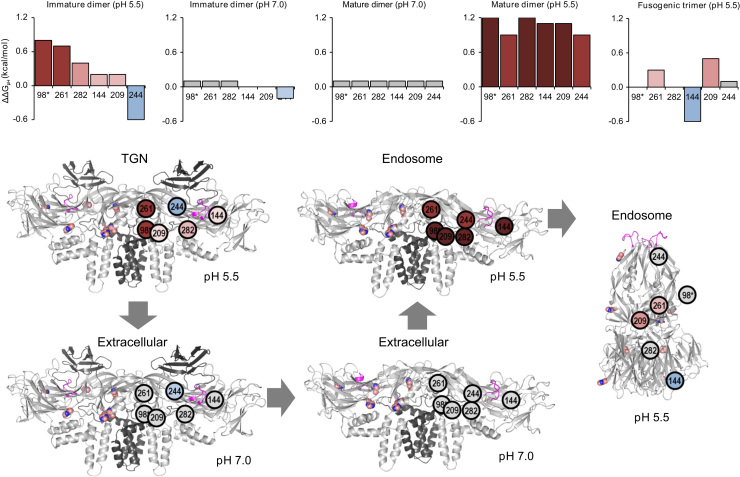
The change in pKa associated with the immature dimer-to-mature dimer transition reflects the effects of Pr association with E: Residues H144, H209, H244, and H282 all showed significant shifts. Likewise, the change in pKa associated with the mature dimer-to-postfusion trimer transition reflects the change in the local environment surrounding histidine residues in E as a result of trimerization. In particular, H144, H209, H244, H261, and H282 in E, and H98 in PrM showed substantial pKa shifts during trimerization. To more easily visualize the contribution of the pKa shifts of these residues to protein stability in the context of viral maturation and infection, we calculated the pH-dependent contribution to the folding free energy (based on Eq. (4)) in the immature dimer, mature dimer, and postfusion trimer conformations at the appropriate environmental pH for various stages of the virus life-cycle (Fig. 5).
Fig. 5.
Individual residue contributions to protein stability as a function of pH. Individual residue contributions to protein stability at low-pH and neutral pH for the immature dimer, mature dimer, and postfusion trimer shown as by bar graphs (top) and graphical representation (bottom). The various stages and respective environmental pH of the viral life cycle are shown as viral maturation [in the trans-Golgi network (TGN)], viral release into the extracellular environment, and host cell invasion via the endosome. Colors correspond to the energetic contribution to stability with red as destabilizing, gray as neutral, and blue as stabilizing. Structures of E are colored dark gray (PrM), magenta (fusion loops), and salmon (conserved histidine residues).
During viral maturation, in the low-pH of the TGN, where the virus is found as an immature dimer, H261, H282, and H98 (PrM) are destabilizing, whereas H244 is strongly stabilizing. As the virus is released into the neutral extracellular environment, the low-pH-induced destabilization caused by these residues is removed, along with the stabilizing effect of H244. Likewise, in the mature dimer, with Pr removed, the ionizability of these conserved histidine residues has no effect on stability. During host cell infection, the low endosomal pH leads to strong pH-induced destabilization of all six histidine residues [H144, H209, H244, H261, H282, H98 and (PrM)]. Finally, at low-pH, the postfusion trimer conformation largely relieves this destabilizing effect.
These results, based on conformation-specific pKa predictions, show two distinct trends with respect to individual residue contributions to stability at low-pH. The first trend is a trimerization effect, where the dimer form seems “primed” to become destabilized at low-pH in both the mature and immature forms of the virus. As the pH decreases, the ionizability of these conserved histidine residues – particularly H98 (PrM), H261, H282, and H209 – becomes increasingly destabilizing. The second trend is the chaperone effect, whereby the presence of Pr bound to the E protein acts as a “chaperone,” stabilizing or reversing the low-pH-induced destabilization of conserved histidine residues, particularly H244, H144, and H209. The trimerization effect explains the increased relative stability of the trimer form over the dimer form at low-pH (Fig. 4), whereas the chaperone effect accounts for the lack of pH sensitivity of the immature dimer compared with the mature dimer (also in Fig. 4).
3.6. Structural mechanisms of pH-induced conformational changes
The pKa shifts calculated using Rosetta are a function of the local environment around a given histidine for a particular DENV protein structure in terms of both the (1) specific arrangement of polar and charged residues within that local environment, and (2) more general features, such as its overall level of solvent exposure and hydrophobicity. We explored the local environment around a subset of these conserved histidines that were determined to be most responsible for the pH-dependent conformational shifts in DENV to try to identify the structural mechanisms guiding the pKa shifts.
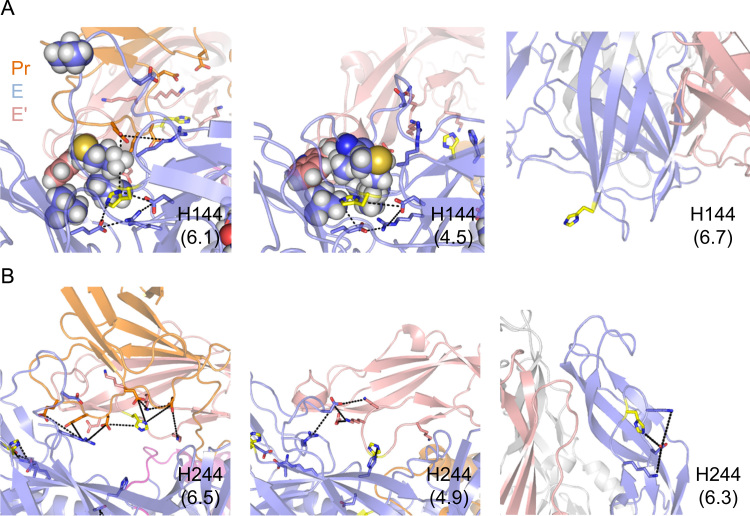
H144 of the E protein is found adjacent to the fusion loop in domain II. In the immature dimer, H144 interacts with E60 of PrM and has a pKa of 6.1 (Fig. 6A, left). In the mature dimer form, Pr cleavage removes E60, and H144 moves into a hydrophobic pocket formed by M1, I4, V151, and V321 – along with W101 of the fusion loop – and its pKa decreases to 4.5 (Fig. 6A, middle). In the postfusion trimer, a significant rearrangement results in H144 becoming almost entirely solvent-exposed while maintaining its interaction with D42 and a pKa of 6.7 (Fig. 6A, right). The low pKa value of H144 in the mature dimer form and its subsequent increase in the trimer is consistent with the trimerization effect that favors the postfusion trimer over the mature dimer at low-pH. Likewise, direct interaction with E60 of Pr provides a mechanism for the chaperone effect, whereby the Pr–E interactions act to increase the stability of the immature dimer at low-pH.
Fig. 6.
Local environment of H144 and H244 in DENV E-PrM. The local environment around conserved residues H144 (A) and H244 (B), in the immature dimer (left), mature dimer (middle), and postfusion trimer (right). Median pKa values are shown in parentheses. Pr is shown in orange, and E is shown in slate and magenta. Salt bridges between positive and negatively charged residues are shown as dotted lines. Hydrophobic residue side-chains are shown as spheres.
H244 is found on domain II at the Pr–E interface in the immature form of DENV. In the immature dimer, H244 has a median pKa of 6.5 and forms salt bridges with D63 and D65 of Pr (Fig. 6B, left). In the mature dimer, with Pr removed, there is a significant rearrangement of H244, which moves inwards toward the E protein core and forms a hydrogen bond with H27 (Fig. 6B, middle) and adopts a pKa of 4.9. In the postfusion trimer, D244 has a median pKa of 6.3, is almost completely solvent-exposed, and forms an interaction with D249 (Fig. 6B, right). Like H144, H282 plays a role in both the trimerization effect, where burial and interaction with the ionizable H27 leads to destabilization of the dimer at low-pH, as well as in the chaperone effect, where intermolecular salt bridges with Pr lead to stabilization of the immature dimer at low-pH.
H282 of the E protein is found in domain I, at the interface between the stem helices of E that are thought to be critical for membrane fusion in the postfusion form of the protein. In the immature dimer, H282 interacts with the conserved K284, D417, and R106 of PrM, resulting in a subsequent decrease in the median pKa to 5.9 (Supplemental Fig. S1, left). In the mature dimer, after Pr cleavage, there is a shift in the location of H282, which maintains its contact with E26 but forms additional contacts with conserved hydrophobic residues L191, I414, and I415, which further decreases its median pKa to 4.1 (Supplemental Fig. S1, middle). Finally, in the postfusion trimer, a dramatic rearrangement leads to the release of H282 from this hydrophobic residue cluster, creates a new contact with E368, increases its solvent accessibility, and has a median pKa of 6.3 (Supplemental Fig. S1, right). Like H144, the increase in pKa of H282 in the postfusion trimer state is consistent with the low-pH-induced transition to the trimer form.
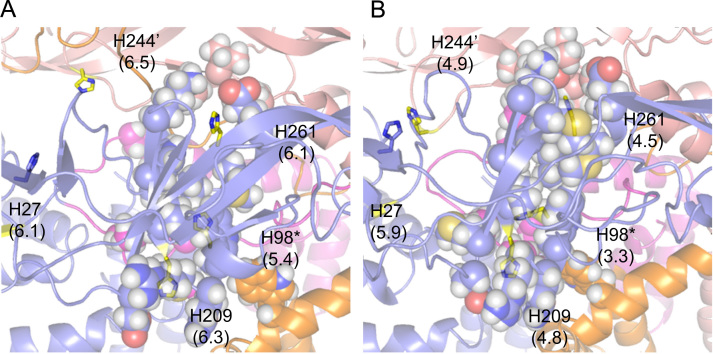
Finally, H98 (PrM) and the neighboring H261 and H209 of E are found at the interface of the linker that connects Pr, M, and E. In the immature dimer, H98 (PrM), H261, and H209 are largely buried and loosely packed along the interior of E (Fig. 7A) and have slightly downshifted pKa values of 5.4, 6.1, and 6.3. In the mature dimer form, conformational changes induced by the release of Pr lead to a tight packing of H98, H261, and H209 along the hydrophobic core of the E protein (Fig. 7B), which is made up of a number of highly conserved residues, including A263, A267, W206, W212, and L103. In this arrangement, these three histidine residues are not only buried within the core but are located near each other and have significantly downshifted median pKa values of 3.3, 4.5, and 4.8. Finally, in the postfusion trimer, H261 and H209 are fully or partially solvent exposed and have median pKa values of 6.2 and 6.0, respectively; whereas H98 (PrM), which is not modeled in the structure, is assumed to be either solvent-exposed or disordered and, thus, has a pKa of 6.3. The tight packing of these three residues, H98 (PrM), H261, and H209, within the hydrophobic core of E in the mature dimer, followed by the subsequent release toward solvent in the fusogenic trimer, contributes significantly to the destabilization of the mature dimer at low-pH.
Fig. 7.
Local environment of H98 (PrM), H209, and H261 in DENV E-PrM. The local environment around conserved residues H98 (PrM), H209, and H261, as well as neighboring H27 and H244 in the immature (left) and mature (right) dimer. Median pKa values are shown in parentheses. The coloring and format is identical to Fig. 6.
3.7. Solvent accessibility of conserved histidine residues in DENV
Solvent accessibility is a critical feature of the local residue environment that determines its pKa: The more solvent-exposed a residue is, the more likely it is to have a pKa comparable to the solution state or an ideal pKa value [19]. We calculated the SASA for each of the conserved histidine residues for each conformational state for all four DENV serotypes. Table 2 shows the median values from these calculations and Supplemental Fig. S2 shows the SASA in the structures of the immature dimer, mature dimer, and postfusion trimer.
Table 2.
Solvent-accessible surface area for conserved histidine residues.
| Residue | Immature dimer (Å2) | Mature dimer (Å2) | Fusogenic trimer (Å2) |
|---|---|---|---|
| H27 | 49 | 19 | 43 |
| H144 | 10 | 2 | 185 |
| H149 | 110 | 110 | 148 |
| H209 | 36 | 21 | 38 |
| H244 | 75 | 43 | 90 |
| H261 | 0 | 6 | 78 |
| H282 | 0 | 26 | 3 |
| H317 | 38 | 25 | 3 |
| H437 | 143 | 100 | 148 |
| H98 (PrM) | 10 | 2 | 153 |
| Total | 471 | 354 | 889 |
Overall, there are large systematic shifts in the degree to which conserved histidine residues are solvent-accessible during the different stages of the viral life cycle. In the immature dimer, the total SASA for conserved histidine residues is 471 Å2. The transition to the mature dimer leads to a decrease to 354 Å2, whereas the transition to the postfusion trimer leads to a large increase in the SASA to 889 Å2. An illustration of the SASA in the context of the protein structures is shown in Supplementary Fig. S2. These results show a >2-fold increase in the solvent exposure of conserved histidine residues between the dimer and trimer in the mature virus. Since solvent exposure of these residues is largely a consequence of the general fold and topology of the E protein in these conformational states, as opposed to the individual atomic-level interactions of these histidine residues, it underscores a general feature of the dimer conformation: Buried histidine residues “prime” this conformation to be unstable at low-pH.
3.8. Conservation of structural mechanisms underlying pKa
We sought to determine how conserved the electrostatic and hydrophobic interactions between these histidine residues were within both DENV sequences and the entire flavivirus family. We carried out a multiple sequence alignment of 23 representative flavivirus sequences (shown in Supplementary Table 2). Supplementary Fig. 3 shows the multiple sequence alignment of the flavivirus sequences for both the PrM and E proteins and highlights conserved histidine residues, the fusion loop motif in E, and the furin cleavage site in PrM. In addition to the conserved histidine residues, it also highlights the amino acids identified above as being responsible for altering the local environment and shifting the pKa for H98 in PrM and H282 and H144 in E. The figure also shows the consensus sequence for DENV seqeunces and all flaviviruses and identifies conserved residues in both cases (denoted by either the residue letter in cases of identity or by a + in cases of similarity).
The results of the sequence alignment illustrate that flavivirus sequences show significant diversity, with only 20% of the sequence being identical and with 42% similarity across the entire family. In contrast, the residues forming interactions with the conserved histidine residues described above show significantly higher rates of conservation in the flavivirus family. H282 interacts with E26, D417, and R106 (PrM) in the immature dimer and hydrophobic residues I414 and I415 in the mature dimer. All these residues are either identical or highly similar in all flaviviruses. H144 interacts with R9, D42, E368, and E60 (PrM) in the dimer state for both the immature and mature forms of the virus, whereas H244 interacts with D63 and D65 of Pr. These residues were completely conserved among all sequences we analyzed. Likewise, in the dimer form, H98 (PrM) and H261 are buried in a hydrophobic core formed by residues A263, A267, W206, W212, and L103. Again, this hydrophobic region is conserved across the flavivirus family. These results show that the pKa shifts observed for these residues are the result of highly conserved interactions within their local environment – interactions that change systematically during the maturation and fusion.
4. Discussion
In the present study, we sought to use structure-based calculations of pKa values for conserved histidine residues to elucidate the thermodynamic basis for pH-induced conformational changes in the immature and mature forms of the DENV envelope. We used existing low-to-moderate resolution crystallography and cryo-EM structural data to generate template models for three conformational states: immature dimer, mature dimer, and postfusion trimer. We then generated high-resolution homology models for all four DENV serotypes and carried out structure-based pKa prediction using Rosetta-pKa for each histidine residue in the context of all three conformational states. We integrated the pKa values into a thermodynamic framework developed by Isom et al. to calculate pH-dependent changes in stability of the immature dimer, mature dimer, and postfusion trimer for a pH range of 3.0–9.0. We showed that stability of the immature dimer conformation are only weakly sensitive to pH. In contrast, the dimer state of the mature form of the virus is highly sensitive to pH and exhibits strong destabilization at a low-pH.
We identified two pH-dependent effects on the conformational landscape of the dengue E protein. First and foremost was that the trimerization effect, in which the dimer form – and particularly the mature dimer – showed large pH-dependent instability at low-pH. Second was the chaperone effect, by which Pr–E interactions in the immature virion mitigated the destabilizing effects of low-pH in the immature dimer. Our pKa analysis identified several histidine residues – including H98 in PrM, along with H144, H209, H244, H261, and H282 – as responsible for the trimerization effect, destabilizing the mature dimer at low-pH, while a subset of these residues, H144, H244, and H209, was responsible for the chaperone effect stabilizing the immature dimer at low-pH.
Since the predicted pKa shifts of histidine residues are a function of the local environment, we explored the protein structure around these residues to identify the mechanisms that underlie these pKa shifts. We showed that the trimerization effect is caused by H98 (PrM), H209, and H261, and was largely the result of the formation of a tightly packed hydrophobic core in E in the mature dimer that closely integrated these three ionizable residues within it. Furthermore, we showed that there was a systematic increase in the degree of solvent accessibility of a number of additional histidine residues, including H144, H244, and H261, between the mature dimer and postfusion trimer forms. This agrees with an experimental study that found that the degree of burial within a protein was a key determinant of histidine pKa values [19].
These findings are corroborated in previous studies of flaviviruses. Zhang et al. observed that the E–M interface includes H98 (PrM), H209, and H261, packed within a conserved hydrophobic region between E and M, and postulated that low-pH would destabilize this E–M interface [14]. Mutagenesis studies of histidine residues homologous to H209 and H261, and to a greater extent, H144 and H244 in the related West Nile virus led to a significant reduction in infection [13]. A similar study carried out in tick-borne encephalitis virus found that a single mutation at H317 and a double mutation at H244/H282 showed decreased viral fusion [12]. Furthermore, both studies showed that no single histidine residue was absolutely essential for infection, supporting the finding that the aggregate effect of multiple histidines is responsible for the pH-dependent conformation changes in DENV. Finally, previous molecular dynamics studies have highlighted the role of electrostatics in destabilizing the mature dimer conformation at low-pH [26], [27], and the stabilizing role of solvation energy on the trimer conformation [28].
We showed that the chaperone effect driven by H144, H244, and H209 was the result of conformational changes between the immature and mature dimer form, largely due to the release of Pr. H144 and H244; and that they form a number of stabilizing intermolecular interactions with Pr. All three residues show significant degrees of burial into neighboring hydrophobic pockets after the removal of Pr. H144 becomes buried in a hydrophobic pocket that includes W101 of the fusion loop and a shifted V151. H244 moves deeper inside the E–E interface and forms a direct interaction with H27. H209 forms a part of a hydrophobic core of E that is more tightly packed in the mature dimer than in the immature dimer.
Previous studies of the chaperone effect of Pr largely agree with our findings. In vitro studies have found that the presence of Pr stabilizes the dimer form of E at low-pH and dissociates from E at neutral pH [6]. In a cryo-EM study, Li et al. observed that the Pr–E interface is formed by salt bridges, including the conserved D63 in Pr with H244 in E [4]. Zhang et al. postulated that an increase in pH leads to the loss of the H244-D63 salt bridge followed by unbinding of E [14], which was later supported by a study that showed that H244A mutation leads to a loss of E–Pr interactions in vitro [29]. Finally, results from comprehensive mutagenesis of E suggested that H144, along with H149 and H317 function as a switch that triggers the exposure of the fusion loop in the mature dimer [15].
5. Conclusion
This study represents an attempt to integrate atomic-scale models of the dengue envelope protein with biophysics and computational biology to identify structural mechanisms that underlie key aspects of viral maturation and fusion. Rapid advances continue to be made in our understanding of the structural biology of flaviviruses, and as additional information on intermediate structures becomes available, they can be used to more clearly define the thermodynamics and pH dependence of the flavivirus life-cycle.
Competing interests
The authors declare that they have no competing interests.
Author's contributions
SC and DRR performed the template construction, homology modeling, structural refinement, and pKa calculations. SC carried out the data analysis. SC and AW wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We would like to acknowledge Dr. Krisha Kilambi for his assistance in selecting the appropriate sampling options in the Rosetta-pKa algorithm. Support for this research was provided by the Military Infectious Diseases Research Project, (Grant no. MIDRP Z0019_14_TC) the United States (US) Army Medical Research and Materiel Command (Fort Detrick, Maryland), as part of the U.S. Army’s Network Science Initiative, and the US Department of Defense (DoD) High-Performance Computing Modernization Program. The opinions and assertions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the US Army or the US DoD. This paper has been approved for public release with unlimited distribution.
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi:10.1016/j.bbrep.2015.10.014.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Modis Y., Ogata S., Clements D., Harrison S.C. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA. 2003;100(12):6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison S.L., Stiasny K., Stadler K., Mandl C.W., Heinz F.X. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 1999;73(7):5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stiasny K., Kiermayr S., Bernhart A., Heinz F.X. The membrane-proximal "stem" region increases the stability of the flavivirus E protein postfusion trimer and modulates its structure. J. Virol. 2013;87(17):9933–9938. doi: 10.1128/JVI.01283-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., Lok S.M., Yu I.M., Zhang Y., Kuhn R.J., Chen J., Rossmann M.G. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319(5871):1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 5.Mukhopadhyay S., Kuhn R.J., Rossmann M.G. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005;3(1):13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 6.Yu I.M., Zhang W., Holdaway H.A., Li L., Kostyuchenko V.A., Chipman P.R., Kuhn R.J., Rossmann M.G., Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319(5871):1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 7.Modis Y., Ogata S., Clements D., Harrison S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 8.Allison S.L., Schalich J., Stiasny K., Mandl C.W., Kunz C., Heinz F.X. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995;69(2):695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stiasny K., Allison S.L., Mandl C.W., Heinz F.X. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 2001;75(16):7392–7398. doi: 10.1128/JVI.75.16.7392-7398.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stiasny K., Allison S.L., Schalich J., Heinz F.X. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 2002;76(8):3784–3790. doi: 10.1128/JVI.76.8.3784-3790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-San Martin C., Sosa H., Kielian M. A stable prefusion intermediate of the alphavirus fusion protein reveals critical features of class II membrane fusion. Cell Host Microbe. 2008;4(6):600–608. doi: 10.1016/j.chom.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritz R., Stiasny K., Heinz F.X. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 2008;183(2):353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson S., Poddar S., Lin T.Y., Pierson T.C. Protonation of individual histidine residues is not required for the pH-dependent entry of west nile virus: evaluation of the "histidine switch" hypothesis. J. Virol. 2009;83(23):12631–12635. doi: 10.1128/JVI.01072-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Ge P., Yu X., Brannan J.M., Bi G., Zhang Q., Schein S., Zhou Z.H. Cryo-EM structure of the mature dengue virus at 3.5-A resolution. Nat. Struct. Mol. Biol. 2013;20(1):105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Christian E.A., Kahle K.M., Mattia K., Puffer B.A., Pfaff J.M., Miller A., Paes C., Davidson E., Doranz B.J. Atomic-level functional model of dengue virus Envelope protein infectivity. Proc. Natl. Acad. Sci. USA. 2013;110(46):18662–18667. doi: 10.1073/pnas.1310962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stiasny K., Kossl C., Lepault J., Rey F.A., Heinz F.X. Characterization of a structural intermediate of flavivirus membrane fusion. PLOS Pathog. 2007;3(2):e20. doi: 10.1371/journal.ppat.0030020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi M.D., Hedberg A., McIntosh L.P. Complete measurement of the pKa values of the carboxyl and imidazole groups in Bacillus circulans xylanase. Protein Sci. 1997;6(12):2667–2670. doi: 10.1002/pro.5560061224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagi M., Nakazawa T. Determination of pKa values of individual histidine residues in proteins using mass spectrometry. Anal. Chem. 2008;80(17):6481–6487. doi: 10.1021/ac8009643. [DOI] [PubMed] [Google Scholar]
- 19.Edgcomb S.P., Murphy K.P. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins. 2002;49(1):1–6. doi: 10.1002/prot.10177. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhury S., Lyskov S., Gray J.J. PyRosetta: a script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics. 2010;26(5):689–691. doi: 10.1093/bioinformatics/btq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leaver-Fay A., Tyka M., Lewis S.M., Lange O.F., Thompson J., Jacak R., Kaufman K., Renfrew P.D., Smith C.A., Sheffler W. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilambi K.P., Gray J.J. Rapid calculation of protein pKa values using Rosetta. Biophys. J. 2012;103(3):587–595. doi: 10.1016/j.bpj.2012.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isom D.G., Castaneda C.A., Cannon B.R., Garcia-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc. Natl. Acad. Sci. USA. 2011;108(13):5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostyuchenko V.A., Zhang Q., Tan J.L., Ng T.S., Lok S.M. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 2013;87(13):7700–7707. doi: 10.1128/JVI.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohl C.A., Strauss C.E., Chivian D., Baker D. Modeling structurally variable regions in homologous proteins with rosetta. Proteins. 2004;55(3):656–677. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]
- 26.Dubey K.D., Chaubey A.K., Ojha R.P. Role of pH on dimeric interactions for DENV envelope protein: an insight from molecular dynamics study. Biochim. Biophys. Acta. 2011;1814(12):1796–1801. doi: 10.1016/j.bbapap.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Prakash M.K., Barducci A., Parrinello M. Probing the mechanism of pH-induced large-scale conformational changes in dengue virus envelope protein using atomistic simulations. Biophys. J. 2010;99(2):588–594. doi: 10.1016/j.bpj.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubey K.D., Chaubey A.K., Ojha R.P. Stability of trimeric DENV envelope protein at low and neutral pH: an insight from MD study. Biochim. Biophys. Acta. 2013;1834(1):53–64. doi: 10.1016/j.bbapap.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 29.Zheng A., Umashankar M., Kielian M. In vitro and in vivo studies identify important features of dengue virus pr-E protein interactions. PLOS Pathog. 2010;6(10):e1001157. doi: 10.1371/journal.ppat.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrey D., Xiang X., Tang C.L., Xie L., Gimpelev M., Mitors T., Soto C.S., Goldsmith-Fischman S., Kernytsky A., Schlessinger A. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins: Struct. Function Genetics. 2003;3(6):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 31.Lee M.S., Bondugula R., Desai V., Zavaljevski N., Yeh I.C., Wallqvist A., Reifman J. PSPP: a protein structure prediction pipeline for computing clusters. PLOS One. 2009;4(7):0006254. doi: 10.1371/journal.pone.0006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemethy G., Gibson K.D., Palmer K.A., Yoon C.N., Paterlini G., Zagari A., Rumsey S., Scheraga H.A. Energy parameters in polypeptides. 10. Improved geometrical parameters and nonbonded interactions for use in the ECEPP/3 algorithm, with application to proline-containing peptides. J. Phys. Chem. 1992;96:6472–6484. [Google Scholar]
- 34.Ripoll D.R., Liwo A., Czaplewski C. The ECEPP package for conformational analysis of polypeptides. TASK Q. 1999;3:313–331. [Google Scholar]
- 35.Simons K.T., Kooperberg C., Huang E., Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268(1):209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material