Summary
DELLA proteins associate with transcription factors to control plant growth in response to gibberellin 1. Semi-dwarf DELLA mutants with improved harvest index and decreased lodging greatly improved global food security during the “green revolution” in the 1960-70s 2. However, DELLA mutants are pleiotropic and the developmental basis for their effects on plant architecture remains poorly understood. Here, we show that DELLA proteins have genetically separable roles in controlling stem growth and the size of the inflorescence meristem, where flowers initiate. Quantitative 3D image analysis, combined with a genome-wide screen for DELLA-bound loci in the inflorescence tip, revealed that DELLAs limit meristem size in Arabidopsis by directly up-regulating the cell cycle inhibitor KRP2 in the underlying rib meristem, without affecting the canonical WUSCHEL-CLAVATA meristem size regulators3. Mutation of KRP2 in a DELLA semi-dwarf background restored meristem size, but not stem growth, and accelerated flower production. In barley, secondary mutations in the DELLA gain of function mutant Sln1d 4 also uncoupled meristem and inflorescence size from plant height. Our work reveals an unexpected and conserved role for DELLA genes in controlling shoot meristem function and suggests how dissection of pleiotropic DELLA functions could unlock further yield gains in semi-dwarf mutants.
Growth of the stem and of lateral organs, including leaves and flowers, is initiated at the shoot apical meristem (SAM), where a pool of stem cells continuously provides cells to form new tissues3. During reproductive development, the outer cell layers of the SAM (tunica) give rise to floral buds and contribute cells to form the outer tissues of the stem, whereas the inner stem tissues originate from the subapical region of the SAM, called the rib zone (RZ).
Stem growth is promoted by gibberellin, which binds to the GID1 receptor to promote ubiquitin-dependent degradation of DELLA proteins. Mutations that reduce GA levels or disrupt the interaction between DELLAs and GID1 stabilise DELLAs and consequently inhibit stem growth1. Arabidopsis has five DELLA genes with overlapping functions, of which two (GA-INSENSITIVE, GAI, and REPRESSOR of ga1-3, RGA) have a predominant role in regulating stem growth 5 and are transcribed in the inflorescence apex, including the SAM (Supplementary Fig. S1). In accordance with their role in controlling cell proliferation 6, localised activation of a GA-resistant version of the GAI protein 7 using a Cre-loxP recombination system confirmed that most of the inhibitory effect on growth occurred within the apical 2 mm of the inflorescence stem (Supplementary Fig. S1), where cell division is most active 6.
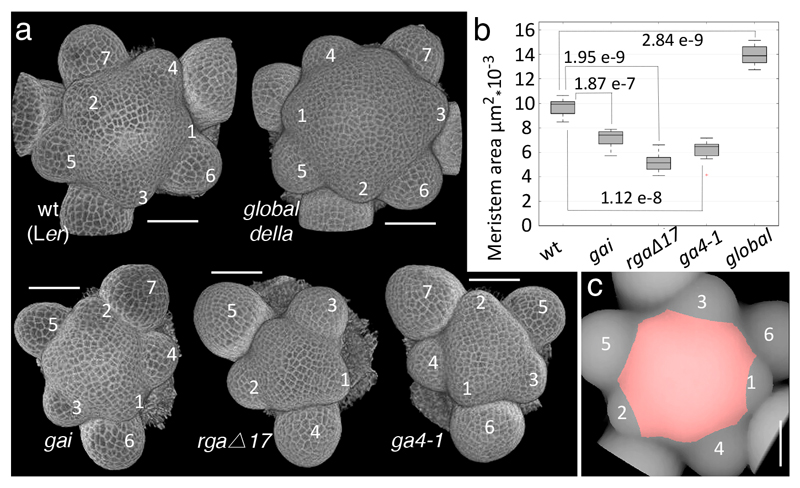
To understand in detail how GAI and RGA control stem growth, we looked for the earliest visible defects in the dwarf gain-of-function gai mutant and in dwarf transgenic plants expressing a GA-resistant version of RGA (RGAp:GFP-rgaΔ17) 8. The CYCB1;1p:GFP 9 reporter showed fewer mitoses in the developing internodes of gai, without obvious differences in cell size and before any visible change in internode length (Supplementary Fig. S2), in line with the role of DELLAs in regulating the supply of new cells for internode growth. In addition, gai and RGAp:GFP-rgaΔ17 appeared to reduce SAM size, including the tunica layers and the RZ (Supplementary Fig. S2). Measurements of projected meristem area confirmed that the SAM was smaller not only in gai and RGAp:GFP-rgaΔ17, but also in the GA-deficient ga4-1 mutant 10; conversely, global della plants, with loss of function of all five Arabidopsis DELLA genes 11, had a notably enlarged SAM (Fig.1a-c). The negative role of DELLA genes in the inflorescence meristem was unexpected, because gibberellin has been shown to antagonise shoot meristem activity in seedlings 12, in contrast to its positive role in the control of root meristem size 9,13. The potentially different roles of DELLA in the vegetative and reproductive SAM might reflect other changes in SAM function, such as activation of the rib meristem during flowering.
Fig. 1. DELLA proteins control inflorescence meristem size.
(a) 3D reconstructions from confocal stacks of shoot apices stained by the modified pseudo-Schiff propidium iodide (mPS-PI) method; rgaΔ17 is short for RGAp:GFP-rgaΔ17 and global della is the quintuple mutant gai-t6, rga-t2, rgl1-1, rgl2-1, and rgl3-411; numbers indicate floral buds emerging from the SAM, in order of age (youngest marked 1). (b) Boxplots showing projected SAM area in genetic backgrounds with altered DELLA function (10 apices each for wt, gai and ga4-1; 8 each for rgaΔ17 and global della); p-values are for equality of means (Student’s t-test). (c) Illustration of how projected SAM areas were measured; in this vertical projection of a wt inflorescence apex, the measured region in red corresponds to the meristem surface determined on a 3D image using landmarks placed on bud boundaries (details in Materials and Methods). Bars: 50 µm.
DELLA proteins interact with transcription factors bound to their target genes, which can be detected by chromatin immunoprecipitation (ChIP) using tagged DELLA 14. To reveal the molecular basis for DELLA control of stem growth and SAM size, we performed ChIP-seq to detect loci bound by RGAp:GFP-rgaΔ17 in inflorescence apices. As internal controls, we used ten genes previously shown to be DELLA-regulated (Supplementary Table S2, Supplementary Fig. S3). Candidate target genes were selected if the region within 3 kb upstream and 1.5 kb downstream of their coding sequences contained ChIP-seq peaks consistently detected in three RGAp:GFP-rgaΔ17 replicates (FDR < 0.001) but not in the negative controls. From these genes, we selected 2327 high-confidence candidates with a peak enrichment at least as high as the internal control with the lowest enrichment (LEAFY) (Supplementary Table S1, Supplementary Fig. S3). In accordance with DELLA roles regulating growth in response to hormonal, developmental and environmental responses 15, the set of high-confidence targets included numerous genes involved in responses to hormones (GA, auxin, brassinosteroid, abscisic acid and cytokinin), floral transition, organ patterning and growth, floral development and cell wall dynamics (Supplementary Table S2). The list was significantly enriched (p-value 1.2 x 10-50, Fisher’s exact test) for genes bound by GFP-RGA in seedlings 14, but the majority of genes was not shared between the two sets, consistent with DELLA function being conditioned by interaction with tissue-specific transcription factors (Fig.2 a). Accordingly, the RGAp:GFP-rgaΔ17 ChIP-seq peaks were significantly enriched for motifs bound by DELLA-interacting transcription factors 14, particularly of the bHLH, bZIP, Dof and SPL families (Supplementary Fig. S3).
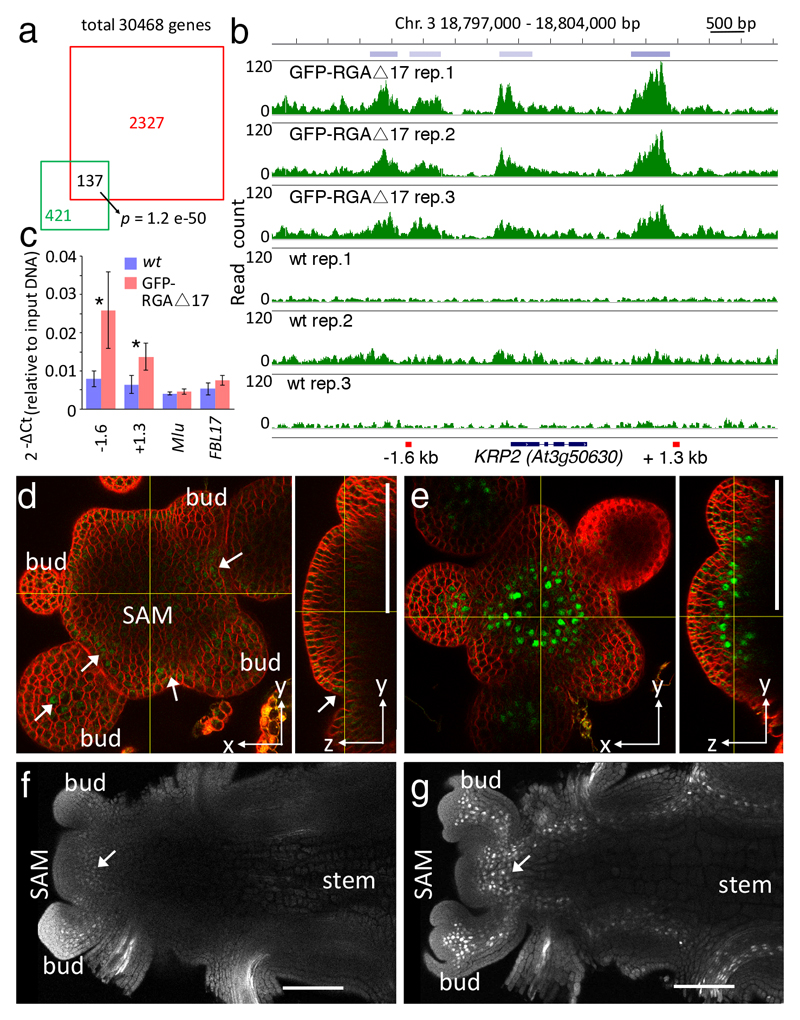
Fig. 2. The cell-cycle inhibitor KRP2 is a direct DELLA target in the shoot apex.
(a) Overlap between RGA target genes identified by ChIP-seq in inflorescence apices (red square) and seedlings 14 (green square); the p-value is for the overlap occurring by chance (Fisher’s exact test, considering a universe of 30,468 genes). (b) Read frequency histograms showing binding of GFP-rgaΔ17 to KRP2 in triplicate ChIP-seq experiments and wild-type controls; top: chromosome position and regions detected as reproducible peaks (blue bars); bottom: black bars and lines indicate KRP2 exons and introns, respectively, and red bars show the regions amplified in the ChIP-qPCR experiment. (c) ChIP-qPCR confirming binding of GFP-rgaΔ17 protein to KRP2; -1.6 kb and +1.3 kb indicate the position of amplified regions relative to the KRP2 coding sequence; negative controls were MU-like transposon (Mlu) and the promoter region of the F-BOX-LIKE 17 gene (FBL17, a representative locus not bound by GFP-rgaΔ17 in the ChIP experiment; Supplementary Fig. S3); bars and lines show means and standard deviation of three biological replicates and asterisks indicate significant difference to the wild type (p<0.05, Student t-test; source data in Supplementary Table S5). (d-g) Confocal images showing KRP2p:KRP2-GFP expression in wild-type (arrows in d, f) and gai (e, g) inflorescence apices, shown as orthogonal views of the inflorescence meristem stained with FM4-64 (d,e) and longitudinal sections of cleared apices (f, g).
The set of high-confidence targets revealed few direct links to the cell cycle machinery (Supplementary Table S2), supporting the idea that DELLA control of cell proliferation is mostly indirect 6. However, the list included three different members (KRP1, 2 and 4) of the Kip-related (KRP) family of CDK inhibitors, which play important roles in plant tissue growth 16,17. To investigate direct links between DELLAs and cell cycle control, we focused on KRP2 because of its higher ChIP-seq peak enrichment relative to KRP1 and KRP4 (Supplementary Table S1) and because KRP2, but not KRP1 or KRP4, was up-regulated in ga1-3 mutant seedlings 13. Binding of GFP-rgaΔ17 to KRP2 was independently confirmed by ChIP-qPCR (Fig. 2b,c). A functional KRP2p:KRP2-GFP fusion (Supplementary Fig. S4) was expressed weakly below the inflorescence and floral meristems and at lateral organ boundaries (Fig. 2d,f), consistent with the repression of growth in boundary regions 18. In gai mutants, KRP2p:KRP2-GFP was up-regulated in the rib zone, in floral pedicels and in developing internodes (Fig. 2e,g). Thus, repression of cell proliferation in the inflorescence apex by DELLAs correlated with increased expression of the cell-cycle inhibitor KRP2.
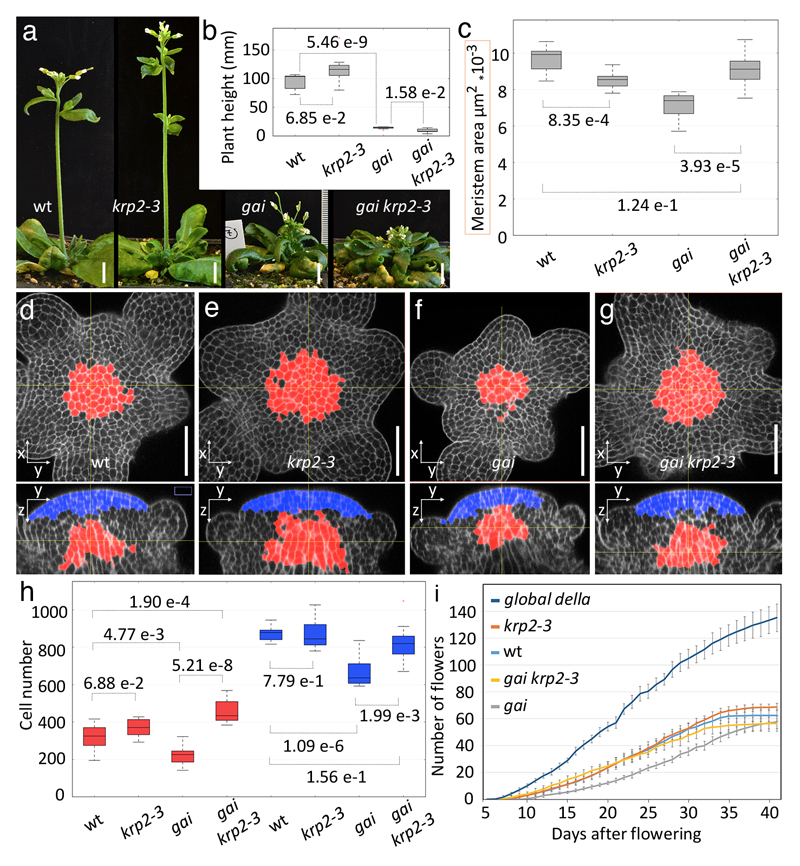
We next tested the functional relevance of KRP2 activation by introducing the krp2-3 loss-of-function allele 19 into genetic backgrounds with enhanced DELLA activity. To detect the effects of KRP2 in different meristem regions and to confirm size differences using a method that is not sensitive to meristem geometry, we also measured the number of cells in the tunica and RZ regions. krp2-3 fully suppressed the reduction in SAM area seen in gai and restored cell numbers not only in the RZ, but also in the overlying tunica layers (Fig.3). However, the more severe defect of RGAp:GFP-rgaΔ17 and ga4-1 meristems was only partially suppressed by krp2-3 (Supplementary Fig. S5), suggesting that RGA restricts SAM size through additional genes; plausible candidates would be KRP1 and KRP4. A more specialised role for RGA in the inflorescence meristem would be in line with its higher expression in the meristem relative to developing buds (Supplementary Fig. S1). Furthermore, the larger meristem of the global della mutant compared to krp2-3 (Figs.1a,b and 3C) and the binding of GFP-rgaΔ17 to known regulators of meristem size, such as CLV1 and cytokinin signalling genes (Supplementary Table S1, Supplementary Fig. S3), suggested that DELLA proteins limit SAM size through additional, KRP2-independent mechanisms. In contrast to the effect on meristem size, krp2-3 did not significantly change stem elongation in wild-type or gai background (Figs. 3 a,b), so activation of KRP2 in developing internodes (Fig.2 g) is not sufficient to explain the inhibition of stem growth by DELLA proteins. In summary, the effect of semi-dwarf DELLA alleles on SAM size is mediated in part by KRP2 and is genetically separable from their effect on stem growth.
Fig. 3. KRP2 mediates DELLA control of meristem size and bud production but not stem elongation.
(a) Representative images of L-er (wild-type), krp2-3, gai and gai krp2-3 plants with 3-5 mature flowers. (b) Boxplots of plant height at the stage shown in (a) (7 plants per genotype). (c) Boxplot of SAM areas measured as in Fig. 1 for wild type, krp2-3, gai and gai krp2-3 (10, 8, 10 and 10 plants, respectively). (d-g) Orthogonal views of confocal image stacks of representative inflorescence apices stained with mPs-PI; within the meristem, defined using landmarks placed on bud boundaries, the RZ (red) was selected as a region within 90 µm of a plane crossing the meristem summit and normal to the main SAM axis, with consistently thicker walls (based on mPS-PI signal) and vertically oriented divisions (based on the orientation of the thinnest wall); the tunica region (blue) was selected within 15 µm of the meristem surface and 30 µm from the plane crossing the summit (details in Supplementary Information). (h) Boxplots of cell numbers in the tunica (blue) and RZ (red) regions for different genotypes (11 plants for krp2-3, 10 for other genotypes). (i) Cumulative number of mature flowers in the main inflorescence of wild-type, krp2-3, gai, gai krp2-3 and global della plants (average and SD for 10 plants per genotype); day 0 was when floral buds were first visible by eye. Bars: 1 cm (a), 50 µm (d-g); p-values in b, c, h are for equality of means (Student’s t-test).
The localised activation of KRP2 suggested that DELLA proteins function in the rib zone to regulate SAM growth. SAM size is controlled by the homeodomain protein WUSCHEL (WUS), which is expressed in the rib zone and moves to the overlying tunica layers to specify stem cells marked by CLAVATA3 (CLV3) expression 20,21. Neither WUS nor CLV3 fulfilled our criteria for high-confidence ChIP-seq targets of GFP-rgaΔ17 (Supplementary Table S1), however, even if these loci are not directly targeted, their function could still be indirectly affected by DELLA proteins. To test this possibility, we compared expression of WUSp:GFP-WUS and CLV3p:GFP in gai and wild-type SAMs. The number of cells expressing either reporter was not significantly different, while the expression level per cell remained the same for CLV3p:GFP and was slightly increased in gai for WUSp:GFP-WUS (Supplementary Fig. S6), indicating that KRP2 activation in gai did not restrict meristem size by interfering with stem cell maintenance. The effect of rib zone-expressed KRP2 on the overlying tunica layers could result from mechanical constraints on tissue growth, as proposed to explain how DELLA function localised specifically in the endodermis limits growth of the whole root meristem 9, in contrast with the hypothesis that plant organ growth is mechanically controlled by the outermost cell layers 22.
Changes in SAM size can affect the number of floral buds formed 23 and disrupt their arrangement around the meristem (phyllotaxis) 24. gai, RGAp:GFP-rgaΔ17 or ga4-1 inflorescence meristens did not show obvious phyllotaxis defects, but the larger meristem of the global della mutant did (Fig.1a). Assuming that successive buds develop at the same speed, we estimated the rate of bud initiation by comparing daily the number of flowers that reached maturity in the main inflorescence of wild-type, krp2-3, gai, gai krp2-3 and global della plants. Reflecting the differences in SAM size, the rate of bud production increased in the global della mutant and decreased in gai in a KRP2-dependent way (Fig.3i). However, increased meristem longevity in gai resulted in a final number of flowers comparable to the wild-type (Fig. 3i). Meristem arrest correlates with seed development 25, although the molecular mechanism remains unknown, so the extended meristem activity in gai may result from the reduced fertility of DELLA gain-of-function mutants 26,27. We conclude that the KRP2-dependent decrease in meristem size in gai plants had a negative effect on meristem function, but the effect on final floral numbers depended also on meristem longevity.
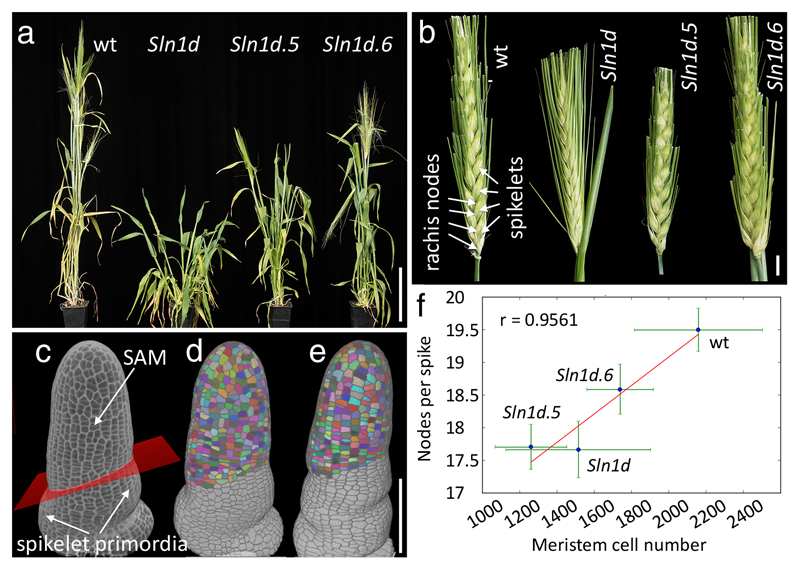
To test whether the role of DELLA in SAM size is conserved in diverse angiosperms, we analyzed mutants for the barley DELLA gene, SLENDER1 (SLN1) 4. The barley inflorescence (spike) has a main axis (rachis) with nodes that support branches (spikelets), each containing a determined number of flowers (florets). 3D imaging of the inflorescence meristem when spikelets were initiated (double ridge stage) 28 revealed that the SAM had significantly fewer cells in the gain-of-function mutant Sln1d 4, compared with the corresponding wild-type (Himalaya) (p = 6.54 e-3, n = 7, Mann-Whitney’s test; Fig. 4f). To test whether control of meristem size is also separable from other DELLA functions in barley, we took advantage of an allelic series of Sln1d 4. The Sln1d.5 and Sln1d.6 alleles, which contain second-site mutations in Sln1d, have similar effects on a subset of the DELLA gain-of-function phenotypes, partially restoring leaf elongation 4 and plant height (Fig. 4a). In spite of these similarities, Sln1d.5 and Sln1d.6 differentially affected meristem size (Fig.4c-f). Furthermore, SAM size correlated with the number of rachis nodes produced by the SAM (Fig. 4b, f). Uncoupling of plant height from SAM size was also shown by the comparable meristems but distinct height of Sln1d and Sln1d.5 (Fig.4a,f). In conclusion, DELLA-induced restriction of meristem size led to reduced inflorescence size in a cereal crop. The smaller inflorescence of Slnd1 is reminiscent of the lower number of flowers seen in the rice semi-dwarf mutant sd1 29, which like ga4-1 affects gibberellin biosynthesis, although it is not known whether sd1 also affects inflorescence meristem size.
Fig. 4. DELLA control of meristem size is conserved and correlates with inflorescence size in barley.
(a) Wild type barley (Himalaya) and della mutants (Sln1d, Sln1d5, Sln1d6) at comparable stages after flowering. (b) Mature spikes of genotypes shown in (a), with alternating spikelets attached to nodes of the main axis (rachis) indicated for the wild type. (c) 3D reconstruction of confocal image of mPS-PI-stained wild-type inflorescence meristem, fixed when the first spikelet primordia were initiated; a plane (red) fitted to landmarks placed on the boundaries of spikelet primordia was used to select SAM cells (details in Supplementary Information). (d, e) 3D reconstructions of segmented images of wild-type (d) and Sln1d (e) apices, with selected SAM cells coloured. (f) Correlation between meristem size (cell numbers) and inflorescence size (rachis nodes) in the wild type and sln1 mutants; points and green lines show average and standard deviation per genotype, the red line shows the linear regression of averages, with correlation coefficient r; for Hymalaya, Sln1d, Sln1d5 and Sln1d6, respectively, n was 18, 15, 18, 18 spikes (for node numbers) and 8, 7, 3, 4 apices (to image cell numbers). Bars: 10 cm (a), 1 cm (b), 100 µm (c-e).
Overall, our results show that DELLA genes have a conserved role in limiting inflorescence meristem size, in line with the role of DELLA in saving resources under environmental stress 15. This function involves a direct link to cell cycle regulation in the rib meristem and is genetically separable from the role of DELLAs in subsequent organ growth. Because the size of the inflorescence meristem limits yield potential in crop plants 23,30, separating the effects of DELLA on stem growth and meristem size could unlock further yield increases in the widely-used semi-dwarf mutants.
Methods
Plant material
Arabidopsis thaliana Landsberg erecta (L-er) accession was used as wild type unless otherwise specified. gai 31, RGAp:GFP-rgaΔ17 26, ga4-1 10, global della 11, krp2-3 19 (backcrossed three times to L-er), jag-1 krp2-317, CYCB1;1p:GFP 9, GAIp:GUS 32, RGAp:GFP-RGA 33, WUSp:GFP-WUS 21, CLV3p:GFP 34, 35Sp:loxGUSlox-GFP 35 and hsp18.2:Cre 36 have been described. 35SploxGUSloxgaiGFP, 35Sp:lox-GFP-lox-GUS and KRP2p:KRP2-GFP were constructed and transformed into Arabidopsis as described in the Supplementary Information. Arabidopsis plants were grown on JIC Arabidopsis Soil Mix (Levington F2 compost with Intercept and 4-mm grit at a 6:1 ratio) at 16°C under continuous light. Dissection of inflorescence apices for imaging, ChIP, and measurement of plant height were performed when the plants had produced the first three mature flowers. The Barley (Hordeum vulgare) Himalaya (wild-type), Slnd1 (M640), Slnd1.5 (TR9) and Slnd1.6 (TR13) mutants 4 were grown on JIC Cereal Mix (containing 1.3kg/m3 PG Mix 14-16-18 and 1kg/m3 Osmocote Mini 16-8-11 2 mg) under long day photoperiod (16 h light/8 h dark) and 20°C/15°C day/night temperatures. Barley inflorescence apices were dissected for imaging at the double ridge stage 37.
Cre-loxP recombination
hsp18.2:Cre, 35Sp:lox-GUS-lox-gai-GFP and hsp18.2:Cre, 35Sp:lox-GUS-lox-GFP plants were grown until they had three self-pollinated flowers. Open flowers were removed and ink marks were placed on the stem. For localised heat-shock, water at 38.5 °C was streamed for 5 minutes onto a 2 mm sponge clamped around the stem. Plants were photographed after 10 days of growth and distances between ink marks were measured with Fiji 38. Similarly treated hsp18.2:Cre, 35Sp:lox-GFP-lox-GUS were stained for GUS 36 30 h after heat shock.
Growth measurements
Arabidopsis stem height was measured from photographs using Fiji 38. To determine the rate of floral initiation, plants were grown at 16 °C under continuous light; starting on the day floral buds became visible, flowers at stage 13 and beyond 39 were counted daily in the main inflorescence of 8 to 10 plants per genotype. The whole experiment was repeated three times with similar results. Rachis nodes in barley inflorescences were counted for the main stem and first tiller in two independent experiments.
Confocal imaging
Dissection, of shoot apices, live imaging and modified pseudo-Schiff propidium iodide (mPS-PI) staining were as described 40,41, before imaging with a Zeiss LSM780 confocal microscope; resolution was 0.42 x 0.42 x 0.50 μm for Arabidopsis and 0.66 x 0.66 x 1.0 µm for barley. GFP was imaged in cleared apices by the ClearSee method 42 before imaging with a Leica SP5 confocal microscope with a 20x/0.75 long-working distance objective (resolution 0.63 x 0.63 x 1 μm). Vibratome sectioning is described in the Supplementary Information.
Image analysis
Custom Python scripts and Fiji macros based on a previous set of scripts 40,41 were used to segment confocal image stacks, measure shoot meristem areas, define the position of cells within the shoot meristem and measure GFP signal within segmented cells. The function of each script is summarised in the Supplementary Information. Supplementary Table S3 lists the scripts used to produce the data for each manuscript figure and Supplementary Table S4 lists the filtering parameters (e.g. to select cells in meristem regions). The annotated source code with detailed instructions and folders containing the confocal images, files produced during image processing and final cell data tables are publicly available (see Data Availability).
Statistics
Data were read and processed in a Python shell using the functions defined and annotated in script /3D_meristem_analysis/python_scripts/statistical_analysis.py (within Supplemental Software, DOI: 10.6084/m9.figshare.4675801), using Scientific Python (http://www.scipy.org) functions for linear regression, Shapiro-Wilk tests for normality, two-tailed Student’s t-tests and two-tailed Mann-Whitney tests. For samples with n less than 100, power analysis was performed with the pwr package in R (http://www.r-project.org/), using function pwr.t2n.test to calculate effect sizes, assuming a significance level of 0.05, power 0.8, and the alternative hypothesis that the mean was smaller for sample 2 than for sample 1. Raw values, descriptive statistics, p-values and effect sizes for the data used in each figure are listed in Supplementary Table S4. In all figures with boxplots, boxes extend from the lower to the upper quartile, with a line marking the median, and whiskers extend to 1.5 times the interquartile range.
ChIP-seq and data analysis
Dissected inflorescence apices of RGAp:GFP-rgaΔ17 and L-er (wt) plants were collected at the same stage used for imaging. Developing siliques and open flowers were removed and the remaining apex was dissected, including 2-5 mm of stem below the SAM. Because DELLA proteins are thought to bind DNA indirectly through interactions with transcription factors, the tissue was fixed with 2% formaldehyde in PBS 1x supplemented with 1 μM Di(N-succinimidyl) glutarate (DSG; Synchem) under vacuum for 20 min to improve protein-protein and protein-DNA crosslinks. Crosslinking was stopped with 100 μM glycine and after two washes with water, the samples were blotted dry and frozen in liquid nitrogen. The subsequent ChIP, generation of ChIP-seq libraries and analysis were performed as described 40. To detect enriched sequence motifs, MEME-ChIP was used in discriminative mode (http://meme-suite.org/tools/meme-chip)43, comparing peak sequences with a ten-fold larger control set of random peaks as described 40. Raw and processed ChIP-seq data have been deposited at NCBI's Gene Expression Omnibus 44 (see Data Availability).
ChIP-PCR
ChIP-qPCR was performed on RGAp:GFP-rgaΔ17 and L-er (wt) dissected inflorescence apices as described 17, using the 2-ΔΔCt method 45 to calculate the relative abundance of immunoprecipitated DNA, compared with controls containing a constant amount of input DNA. The following primers were used for each amplicon: KRP2 -1.6 5’-TGATTGAGTATGCAGCTCGTG-3’ and 5’-AATGCCGTCGTTCTGTATCG-3’; KRP2 +1.3 5’-CCCACGAGGCAAAGATTTTA-3’ and 5’-GTGGAGAAAAAGACTCCAGCTC-3’; MU-like 5’-GATTTACAAGGAATCTGTTGGTGGT-3’ and 5’-CATAACATAGGTTTAGAGCATCTGC-3’; FBL17: 5’- AAGTACAGTGGATCCCAACGTC-3’ and 5’- GCTAATTGAAAGAGCGTGAGGTC-3’.
Data availability
Source data for all boxplots are listed in Supplementary Table 4; original ChIP-PCR data are found in Supplementary Table S5. Raw and processed ChIP-seq data have been deposited at NCBI's Gene Expression Omnibus, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94926, accession GSE94926). The source code for image analysis and detailed instructions are available as Supplementary software (DOI: 10.6084/m9.figshare.4675801). For each image folder listed on Supplementary Tables S3 and S4, the original confocal images, processed images, associated cell data and metadata files data can be downloaded at https://figshare.com (DOI: 10.6084/m9.figshare.4675801).
Supplementary Material
Acknowledgments
We thank Peter Chandler, Lars Østergaard, Tai-Ping Sun, Nick Harberd, Peter Doerner, Venugopala Reddy, Miguel Ángel Pérez Amador and the European Arabidopsis Stock Centre for plasmids and seeds, Grant Calder for advice with confocal microscopy and Bihai Shi for help with vibratome sections. The work was supported by BBSRC grants BB/J007056/1, BB/J004588/1 and BB/M003825/1, and by a grant from the Ministerio de Educación, Cultura y Deporte, Spain (EX-2010-0491) to ASM.
Footnotes
Author contributions
Conceptualisation, A.S.-M., S.Bo. and R.S.; investigation, A.S.-M., S.Be., M.B., S.Bo. and K.S.; software, R.S.; formal analysis and data curation: A.S.-M. and R.S.; writing – original draft, A.S.-M. and R.S.; writing – review & editing, A. S.-M., S.Be., M.B., S.Bo., K.S. and R.S.; funding acquisition, R.S and A. S.-M.; supervision, R.S.
Competing interests
The authors declare no competing financial interests.
References
- 1.Daviere J-M, Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- 2.Pingali PL. Green revolution: impacts, limits, and the path ahead. Proc Natl Acad Sci U S A. 2012;109:12302–12308. doi: 10.1073/pnas.0912953109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holt AL, van Haperen JM, Groot EP, Laux T. Signaling in shoot and flower meristems of Arabidopsis thaliana. Curr Opin Plant Biol. 2014;17:96–102. doi: 10.1016/j.pbi.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Chandler PM, Harding CA. ‘Overgrowth’mutants in barley and wheat: new alleles and phenotypes of the ‘Green Revolution’ Della gene. J Exp Bot. 2013;64:1603–1613. doi: 10.1093/jxb/ert022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King KE, Moritz T, Harberd NP. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;159:767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daviere JM, et al. Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr Biol. 2014;24:1923–1928. doi: 10.1016/j.cub.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Peng J, et al. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997;11:3194–3205. doi: 10.1101/gad.11.23.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dill A, Sun TP. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics. 2001;159:777–785. doi: 10.1093/genetics/159.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubeda-Tomas S, et al. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol. 2009;19:1194–1199. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- 11.Koini MA, et al. High Temperature-Mediated Adaptations in Plant Architecture Require the bHLH Transcription Factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 12.Jasinski S, et al. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr Biol. 2005;15:1560–1565. doi: 10.1016/j.cub.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Achard P, et al. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19:1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- 14.Marín-de la Rosa N, et al. Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins. PLoS Genet. 2015;11:e1005337. doi: 10.1371/journal.pgen.1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 16.De Veylder L, et al. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. The Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiessl K, Muino JM, Sablowski R. Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2830–2835. doi: 10.1073/pnas.1320457111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rast MI, Simon R. The meristem-to-organ boundary: more than an extremity of anything. Curr Opin Genet Dev. 2008;18:287–294. doi: 10.1016/j.gde.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Sanz L, et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell. 2011;23:641–660. doi: 10.1105/tpc.110.080002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 21.Yadav RK, et al. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savaldi-Goldstein S, Peto C, Chory J. The epidermis both drives and restricts plant shoot growth. Nature. 2007;446:199. doi: 10.1038/nature05618. [DOI] [PubMed] [Google Scholar]
- 23.Je BI, et al. Signaling from maize organ primordia via FASCIATED EAR3 regulates stem cell proliferation and yield traits. Nat Genet. 2016;48:785–791. doi: 10.1038/ng.3567. [DOI] [PubMed] [Google Scholar]
- 24.Leyser HO, Furner I. Characterisation of three shoot apical meristem mutants of Arabidopsis thaliana. Development. 1992;116:397–403. [Google Scholar]
- 25.Hensel LL, Nelson MA, Richmond TA, Bleecker AB. The fate of inflorescence meristems is controlled by developing fruits in Arabidopsis. Plant Physiol. 1994;106:863–876. doi: 10.1104/pp.106.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dill A, Jung HS, Sun TP. The DELLA motif is essential for gibberellin-induced degradation of RGA. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14162–14167. doi: 10.1073/pnas.251534098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, et al. Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiol. 2003;131:1270–1282. doi: 10.1104/pp.102.018598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waddington SR, Cartwright PM, Wall PC. A Quantitative Scale of Spike Initial and Pistil Development in Barley and Wheat. Ann Bot. 1983;51:119–130. [Google Scholar]
- 29.Ashikari M, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309:741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 30.Xu C, et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat Genet. 2015;47:784–792. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- 31.Koornneef M, et al. A Gibberellin Insensitive Mutant of Arabidopsis thaliana. Physiol Plant. 1985;65:33–39. doi: 10.1111/j.1399-3054.1985.tb02355.x. [DOI] [Google Scholar]
- 32.Gallego-Giraldo C, et al. Role of the gibberellin receptors GID1 during fruit-set in Arabidopsis. Plant J. 2014;79:1020–1032. doi: 10.1111/tpj.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silverstone AL, et al. Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1565. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 35.Gallois JL, Woodward C, Reddy GV, Sablowski R. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development. 2002;129:3207–3217. doi: 10.1242/dev.129.13.3207. Unsp dev0423. [DOI] [PubMed] [Google Scholar]
- 36.Sieburth LE, Drews GN, Meyerowitz EM. Non-autonomy of AGAMOUS function in flower development: use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development. 1998;125:4303–4312. doi: 10.1242/dev.125.21.4303. [DOI] [PubMed] [Google Scholar]
- 37.Waddington S, Cartwright P, Wall P. A quantitative scale of spike initial and pistil development in barley and wheat. Ann Bot. 1983;51:119–130. [Google Scholar]
- 38.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bencivenga S, Serrano-Mislata A, Bush M, Fox S, Sablowski R. Control of Oriented Tissue Growth through Repression of Organ Boundary Genes Promotes Stem Morphogenesis. Dev Cell. 2016;39:198–208. doi: 10.1016/j.devcel.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serrano-Mislata A, Schiessl K, Sablowski R. Active control of cell size generates spatial detail during plant organogenesis. Curr Biol. 2015;25:2991–2996. doi: 10.1016/j.cub.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurihara D, Mizuta Y, Sato Y, Higashiyama T. ClearSee: a rapid optical clearing reagent for whole-plant fluorescence imaging. Development. 2015;142:4168–4179. doi: 10.1242/dev.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailey TL, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for all boxplots are listed in Supplementary Table 4; original ChIP-PCR data are found in Supplementary Table S5. Raw and processed ChIP-seq data have been deposited at NCBI's Gene Expression Omnibus, (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE94926, accession GSE94926). The source code for image analysis and detailed instructions are available as Supplementary software (DOI: 10.6084/m9.figshare.4675801). For each image folder listed on Supplementary Tables S3 and S4, the original confocal images, processed images, associated cell data and metadata files data can be downloaded at https://figshare.com (DOI: 10.6084/m9.figshare.4675801).