Abstract
Several somatic ribosome defects have recently been discovered in cancer, yet their oncogenic mechanisms remain poorly understood. Here we investigated the pathogenic role of the recurrent R98S mutation in ribosomal protein L10 (RPL10-R98S) found in T-ALL. The JAK-STAT signaling pathway is a critical controller of cellular proliferation and survival. A proteome screen revealed overexpression of several Jak-Stat signaling proteins in engineered RPL10-R98S mouse lymphoid cells, which we confirmed in hematopoietic cells from transgenic Rpl10-R98S mice and T-ALL xenograft samples. RPL10-R98S expressing cells displayed JAK-STAT pathway hyper-activation upon cytokine stimulation, as well as increased sensitivity to clinically used JAK-STAT inhibitors like pimozide. A mutually exclusive mutation pattern between RPL10-R98S and JAK-STAT mutations in T-ALL patients further suggests that RPL10-R98S functionally mimics JAK-STAT activation. Mechanistically, besides transcriptional changes, RPL10-R98S caused reduction of apparent programmed ribosomal frameshifting at several ribosomal frameshift signals in mouse and human Jak-Stat genes, as well as decreased Jak1 degradation. Of further medical interest, RPL10-R98S cells showed reduced proteasome activity and enhanced sensitivity to clinical proteasome inhibitors. Collectively, we describe modulation of the JAK-STAT cascade as a novel cancer-promoting activity of a ribosomal mutation, and expand the relevance of this cascade in leukemia.
Keywords: T-cell leukemia, ribosomal protein L10/uL16, JAK-STAT signaling, proteasome
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy caused by mutations in developing T-cells. Despite treatment improvements, T-ALL remains lethal in 20% of pediatric and 40% of adult patients. Moreover, treatment regimens comprise intensive chemotherapy associated with severe toxicities.1 Ectopic expression of transcription factors, inactivation of the tumor suppressors CDKN2A/CDKN2B, and hyper-activation of the NOTCH1 cascade are hallmarks of T-ALL. Other factors that are often altered include PHF6, PTEN and the JAK-STAT signaling pathway, with the latter being hyper-activated in 20-30% of patients due to aberrations in IL7RA, JAK1, JAK3 and/or STAT5 proteins.2–6 Recent genomics studies have revealed that ~20% of T-ALL patients also carry somatic defects in ribosomal protein (RP) genes RPL5, RPL10, RPL22 or RPL11.6–8 Importantly, somatic ribosomal protein defects are not confined to T-ALL. RPS15 is altered in 10-20% of aggressive chronic lymphocytic leukemias and RPL5 shows heterozygous deletions or mutations in 11-34% of multiple myeloma, glioblastoma, melanoma and breast cancer samples.9–12 Furthermore, loss of one allele of Rpl22 or Rpl11 accelerates lymphoma in mice,7,13 ribosomal protein genes act as haploinsufficient tumor suppressors in zebrafish,14 and patients with congenital lesions in ribosomal proteins or ribosome assembly factors (ribosomopathies) have elevated cancer risks.15,16 This body of evidence supports that ribosomal deregulation is a general route contributing to cancer development.
These findings raise important questions regarding the molecular mechanisms and cellular pathways by which ribosomal defects promote cancer. Inactivation of Rpl22 leads to endoplasmic reticulum (ER) stress, followed by NF-κB activation and induction of stemness factor Lin28B.7 Many ribosomal proteins have roles outside the ribosome and alteration of these may also contribute to carcinogenesis. For example, RPS15 mutations abrogate its ability to activate the TP53 tumor suppressor,12 RPL5 and RPL11 have extra-ribosomal roles in regulating TP53 as well as the oncogene MYC, and inactivation of Rpl11 in mice compromises Trp53 function and increases Myc protein levels.13,17 It remains unclear whether these extra-ribosomal mechanisms can fully explain the oncogenic action of ribosomal protein defects and whether alterations of ribosome function are also relevant. Moreover, the cancer promoting mechanisms exploited by the T-ALL associated RPL10 mutations are currently unexplored.
In our T-ALL patient cohorts, the arginine to serine mutation at position 98 (R98S) in RPL10 (also known as uL16)18 was by far the most recurrent ribosomal defect, detected in 7.9% of pediatric patients.6 The R98 residue of RPL10 closely approaches the catalytic core of the ribosome and RPL10 R98S impairs ribosome assembly, translational fidelity, and cell proliferation in yeast models.19,20 Here, we investigated the role of the RPL10 R98S induced defects in T-ALL pathogenesis. Two independent RPL10 R98S mouse hematopoietic cell models as well as T-ALL patient-derived samples showed upregulation of components of the oncogenic JAK-STAT signaling cascade, hyper-activation of the cascade upon cytokine stimulation, and enhanced JAK-STAT inhibitor sensitivity. We propose that altered transcription, protein degradation and, to a lesser extent, levels of apparent programmed -1 ribosomal frameshifting may function as novel mechanisms dysregulating this cascade in T-ALL. Our results link a ribosomal mutation to the JAK-STAT pathway, adding to the small but rapidly expanding repertoire of oncogenic mechanisms exploited by ribosomal defects in cancer. Moreover, we identify the JAK-STAT cascade as an attractive therapeutic target in RPL10 R98S positive T-ALL.
Methods
Patient samples
Mutational status for RPL10 and JAK-STAT genes was analyzed in 195 pediatric T-ALL cases (age<16). These patient samples and determination of their mutational status has been described previously.2,6 This study was approved by the ethics committees of the institutes involved and informed consent was obtained from the participants. Samples and clinical data were stored in accordance with the declaration of Helsinki.
Plasmids
RPL10 wild type (WT) and R98S encoding constructs have been described previously.6 A short hairpin RNA sequence (AACCGACGATCCTATTGTCATC) targeting mouse Rpl10 was cloned into a mir30 cassette and introduced into the pMSCV-Neo vector.
Dual luciferase reporter plasmids were constructed to monitor programmed -1 ribosomal frameshifting. Two overlapping synthetic oligonucleotides or a gene fragment (IDT, sequences in Table S1) containing the -1 PRF signal and homology with Firefly and Renilla were inserted into the reporter plasmid, pJD175f, using an In-Fusion HD Cloning kit (Clonetech). The plasmid was linearized through a double digestion with BamH1 and Sal1.
Cell culture
The RPL10 gene is located on the X chromosome and RPL10 R98S positive leukemia cells only express mutant RPL10.6 To mimic this, Ba/F3 cells (DSMZ) were transduced with retroviral vectors encoding WT or R98S RPL10 cDNAs6 and endogenously expressed Rpl10 was knocked down with the Rpl10 targeting shRNA construct described above. Liquid cultures were established from single cell colonies grown in Clonacell-TCS medium (Stemcell technologies) followed by selection of cultures with ≥90% knock-down of endogenous Rpl10 as determined by qPCR. Expression of RPL10 R98S and knock-down of endogenous Rpl10 were confirmed by Sanger sequencing of cDNA (Figure S1). Cells derived from 3 monoclonal RPL10 WT or R98S expressing Ba/F3 cultures were used for all the experiments.
Lineage negative cells were extracted and cultured from transgenic MX1-Cre Rpl10cKI R98S mice and from Rpl10cKI R98S controls as described in supplementary methods and Figure S2.
Inhibitor proliferation experiments
Ba/F3 cells were seeded in triplicate into 96-well plates (1x104cells/well) and treated with indicated compound concentrations or vehicle (DMSO). Proliferation was assessed using the ATPlite 1step assay (PerkinElmer). To calculate relative proliferation, luminescence values at the start of the drug incubation were subtracted from the values after 24 hours of drug incubation and normalized to the DMSO control.
For T-ALL xenograft experiments, viably frozen xenograft cells were thawed and incubated overnight in xenograft MEMα medium containing 10% FBS6450 (Stemcell Technologies), 10% human AB+ serum (Institut de Biotechnologies Jacques BOY), Pen/Strep, Glutamine, 10ng/mL hIL7 (R&D systems), 50ng/ml hSCF and 20ng/ml hFLT3L (Stemcell Technologies) and 20nM insulin (Sigma). The next day, cells were passed through Lymphoprep (Stemcell Technologies) and plated in triplicate in 96-well plates previously coated with 10µg/ml anti-Fc (ab1927, Abcam) followed by overnight coating with 2µg/ml mDL4-Fc peptide (VIB PSF). Plating was done in drug containing xenograft medium at 1x105cells/well. After 72 hrs, relative cell numbers were measured using the ATPlite 1step assay (PerkinElmer) and apoptosis was determined by flow cytometry analysis of Annexin V expression.
Cytokine starvation/stimulation experiments and effects of drugs
Ba/F3 cells were starved of serum and interleukin 3 (IL3) for two hours, followed by stimulation with 1ng/ml IL3 or 5ng/ml interferon gamma (IFNy). Xenograft T-ALL samples were freshly collected from NSG mice, starved in CTS OpTmizer T Cell Expansion SFM (Gibco) for 2.5 hours, drug or DMSO was added for 30 minutes, followed by 30 minutes stimulation with 10ng/ml hIL7. Cells were lysed in lysis buffer (Cell Signaling Technology) with addition of 5mM Na3VO4 and protease inhibitors (Complete, Roche) and processed for immunoblotting.
Quantitative proteomics
Cells derived from 3 monoclonal Ba/F3 cultures expressing WT or R98S RPL10 were lysed in lysis buffer (Cell Signaling Technology) with addition of 5mM Na3VO4 and protease inhibitors (Complete, Roche). Twenty µg of protein as determined by the Bradford method were processed for quantitative proteomics as described in the supplementary methods. Data are available via ProteomeXchange with identifier PXD005995. The entire list of identified proteins was ranked according to log2 fold changes and used as input for GSEA against the MSigDB C2 KEGG gene sets.21,21,22 Only GSEA results with a FDR q-value <0.25 were considered.
Cycloheximide chase assay
Cells were treated with 10mg/mL cycloheximide for the indicated times followed by lysis and immunoblotting.
Proteasome activity assay
Ba/F3 cells were seeded in triplicate into 96-well plates (4500 cells/well). Proteasome activity was tested using Proteasome-Glo 3-substrate Cell-Based Assay (Promega).
Programmed ribosomal frameshifting (PRF) assays
Enrichment analysis was performed on human genes predicted to contain -1 PRF signals and extracted from the PRF database.23 G:profiler software24 in combination with GO (Biological Process) and KEGG databases were used to determine pathways enriched for predicted -1 PRF signals and significance (Fisher’s one-tailed test).
Dual luciferase assays (Promega) were performed as previously described and as detailed in the supplementary methods.25–27 Briefly, Hek293T cells or Ba/F3 cells were transfected with plasmids harboring the in-frame control, the out-of-frame control, or a -1 PRF signal between the upstream renilla and downstream firefly luciferase open reading frames (Figure S3A). Tested -1 PRF signals are reported in Table S10.
Statistical testing
Throughout this study, statistical significance was defined as P < 0.05. Statistical testing methods are described in figure and table legends.
Additional methods are available in the supplementary methods section.
Results
Four percent of proteins show significantly altered expression levels in RPL10 R98S cells
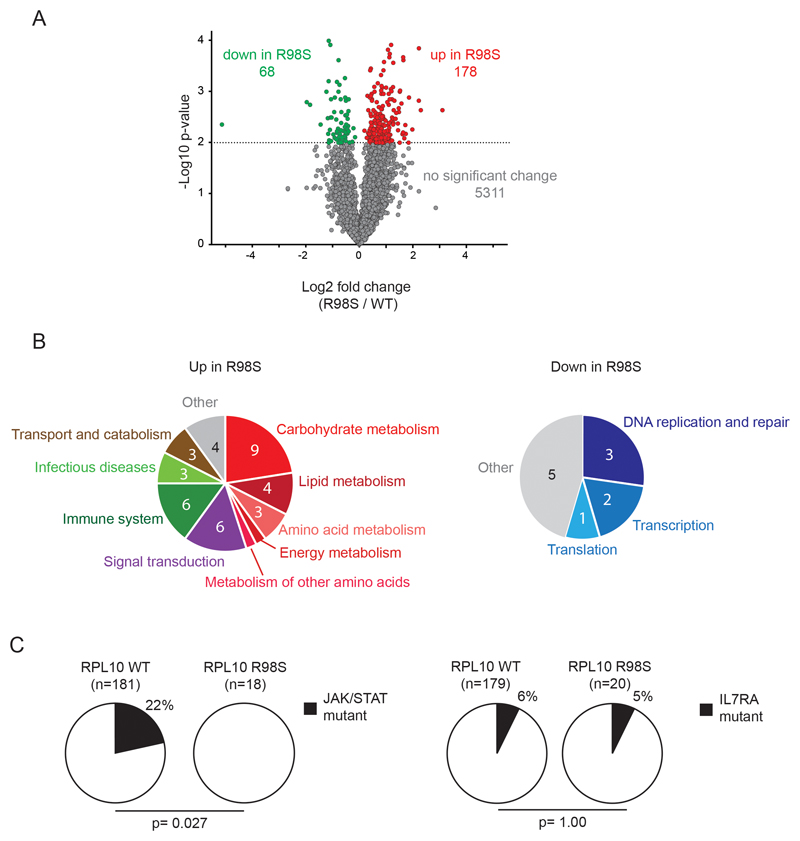
To gain insights into the mechanisms by which the RPL10 R98S mutation contributes to T-ALL development, we screened for proteins that are differentially expressed between RPL10 wild type (WT) and RPL10 R98S (R98S) expressing cells. These experiments were conducted in the mouse pro-B Ba/F3 cell line, a well-established hematopoietic model for oncogenic studies. A label-free quantitative proteomics approach was used to compare the abundance of the 5557 most highly expressed proteins, and unsupervised hierarchical clustering showed that WT and R98S samples grouped into 2 independent clusters (Figure S4). Whereas 96% (5311/5557) of proteins did not change significantly between WT and R98S samples, 3% (178) were significantly upregulated in the R98S samples, and 1% (68) were significantly downregulated (Figure 1A, Tables S4-S5). Gene Set Enrichment Analysis (GSEA) revealed that many of the overexpressed proteins in the R98S cells fit into pathways of cell metabolism, signaling, and function of the immune system. In contrast, proteins related to essential cellular processes such as DNA replication and repair, transcription and translation, were downregulated in the mutant cells (Figure 1B; Tables S6-S7). RPL10 R98S cells thus showed altered expression of 4% of proteins, with differential expression of several known cancer-associated processes.
Figure 1. Mass spectrometry screen for proteins and pathways showing differential expression between RPL10 WT and R98S cells.
A) Volcano plot of the quantitative proteomics data comparing RPL10 WT and R98S samples. The dashed line illustrates the cut-off for significance (p<0.01; T-test). Red dots represent the 178 proteins that are significantly upregulated in the R98S samples, green dots correspond to the 68 proteins that are significantly downregulated. B) Pathways/processes that are significantly up- (left) or down- (right) regulated in R98S samples. The numbers in the pie chart represent the number of significant pathways corresponding to each process that are up or down in the cells. The individual pathways are listed in Tables S6-S7. C) Pie charts reporting mutation frequencies in JAK and STAT genes (left) or in IL7RA (right) detected in RPL10 WT versus R98S mutant pediatric T-ALL patients. The P-value was calculated using a 2-tailed Fisher’s test.
RPL10 R98S cells express elevated levels of Jak-Stat signaling mediators
Analysis of the GSEA results of the proteomic data revealed an enrichment of JAK-STAT pathway members among the upregulated proteins in the RPL10 R98S cells (Figure S4B, Table S6+S8). The JAK-STAT cascade is well established in the pathogenesis of T-ALL, with 20-30% of patients carrying mutations in the IL7RA, JAK and/or STAT genes that hyper-activate the cascade.2,4,28 Analysis of the occurrence of these defects in a cohort of 195 pediatric T-ALL cases2,6 revealed JAK, or STAT gene mutations in 22% (39/181) of RPL10 WT cases as compared to 0% (0/18) in RPL10 R98 cases (p=0.027; Figure 1C). In contrast, mutations in IL7RA showed the same frequency in RPL10 WT and R98S cases. The mutually exclusive mutation pattern for RPL10 R98S mutations and JAK-STAT lesions as well as the GSEA enrichment hinted that RPL10 R98S may modulate the JAK-STAT cascade in human T-ALL and this pathway was therefore selected for further follow-up in this study.
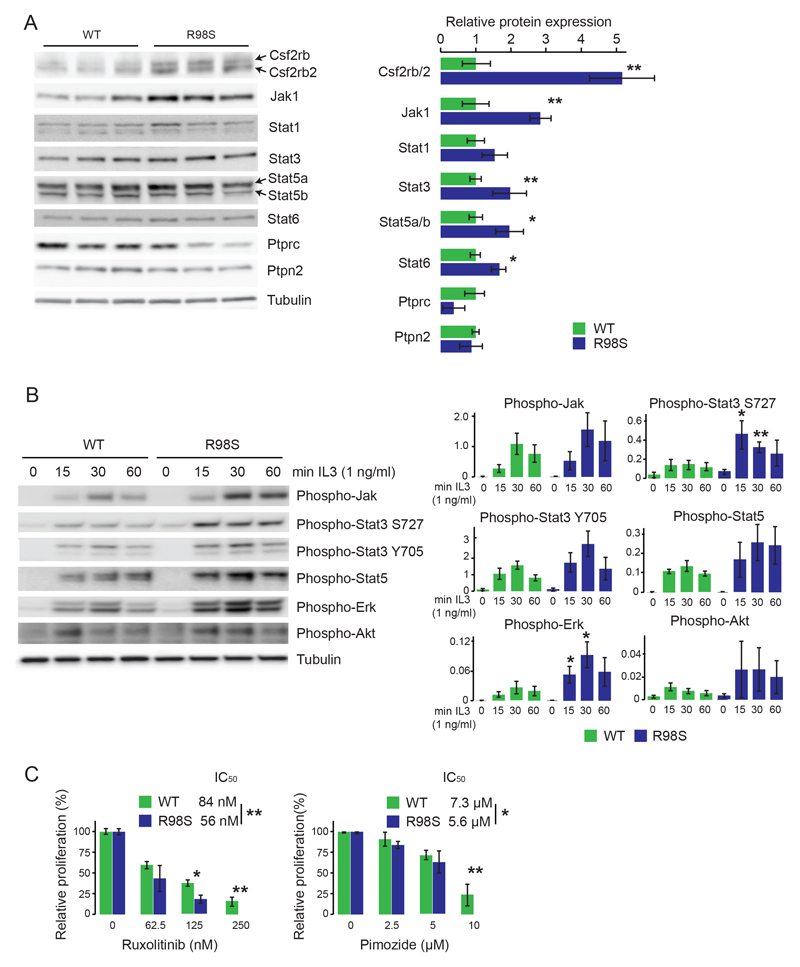
Three Jak kinases (Jak1, Jak3 and Tyk2) were detected in the mass spectrometry screen, of which Jak1 showed a significant 1.9-fold upregulation in the R98S cells (Figure S4C, Table S4). Of the five Stat proteins that were detectable, only Stat2 was unchanged. All others trended towards upregulation in the R98S cells, with Stat6 attaining a statistically significant increase. The mass spectrometric data also revealed a significant 2- to 3-fold upregulation of the Csf2rb and Csf2rb2 proteins. Csf2rb corresponds to the common beta chain of the receptor for IL3, IL5 and GM-CSF, whereas Csf2rb2 is an IL3-receptor specific beta subunit in mouse.29 IL3, IL5 and GM-CSF signaling are all mediated via JAK-STATs. JAK-STAT signaling is tightly controlled via negative regulators including Pias proteins, Socs proteins and phosphatases such as Ptprc and Ptpn2.30 Only Ptprc and Ptpn2 were detectable among the mass spectrometry data, and Ptprc was significantly downregulated in the R98S cells (Figure S4C, Table S5). Immunoblot analysis of Jak-Stat cascade proteins confirmed upregulation of Csf2rb and Csf2rb2, Jak1, Stat1, Stat3, Stat5a, Stat5b, Stat6, downregulation of Ptprc and no change for Ptpn2 in R98S cells (Figure 2A). As no consistent changes were detected for Tyk2 and Jak3 by immunoblot, these were not considered in further analyses. Whereas the protein changes detected for Csf2rb/2, Stat5a and Ptprc were associated with corresponding changes on mRNA level, the transcripts of the other genes were unchanged in R98S cells (Figure S5).
Figure 2. RPL10 R98S Ba/F3 cells express altered levels of Jak-Stat signaling components, are hyper-reactive towards cytokine stimulation and are sensitized to JAK-STAT inhibitors.
A) Left: immunoblot validation of differential expression of Jak-Stat pathway genes in three independent clones of RPL10 R98S versus WT expressing cells. The figure shows a representative blot of 3 independent experiments. Right: quantification of the immunoblot validations. Signal of the JAK-STAT pathway proteins was normalized for loading. The quantification represents the average +/- standard deviation of a representative experiment comparing 3 independent RPL10 WT versus 3 independent R98S cell clones. B) Left: immunoblot analysis of protein phosphorylation of Jak-Stat pathway genes in RPL10 R98S versus WT expressing cells at different time points after addition of 1 ng/ml of IL3. The figure shows a representative blot of 6 independent experiments. Right: phospho-levels were quantified by dividing the phospho-signal through the signal of the loading control on the blot. The quantification represents the average +/- standard deviation of a representative experiment comparing 3 biologically independent RPL10 WT versus 3 independent R98S cell clones. P-values were calculated using a T-test. C) Relative proliferation of RPL10 WT and R98S cells treated with the JAK1/JAK2 inhibitor ruxolitnib and the STAT5 inhibitor pimozide measured using the ATPlite luminescence assay (Perking Elmer). The panel shows the average +/- standard deviation of a representative experiment comparing 4 biologically independent RPL10 WT versus 4 independent R98S cell clones. P-values were calculated using a T-test *p<0.05, **p<0.01.
RPL10 R98S expressing cells are hyper-reactive to cytokine stimulation and sensitized to JAK-STAT inhibitors
Cancer associated hyper-activation of the JAK-STAT signaling cascade is associated with constitutive phosphorylation of its components. Under basal conditions, phosphorylation of the cascade was variable in the Ba/F3 cells, likely due to uncontrollable amounts of residual IL3 in the culture medium. However, phosphorylation of Jak and Stat proteins trended towards higher levels in the R98S cells upon starvation and cytokine depletion. Moreover, consistently stronger and more prolonged phosphorylation of Jak, Stat3 and Stat5, as well as of Erk and Akt was observed in the R98S expressing cells upon cytokine depletion and restimulation with IL3 (Figure 2B), while general tyrosine phosphorylation was not increased (Figure S6). Jak proteins also showed increased phosphorylation upon stimulation with IFNγ, and Stat3 phosphorylation tended to be prolonged in the mutant cells (Figure S7). R98S expressing Ba/F3 cells were more sensitive to the JAK1/JAK2 inhibitor ruxolitinib and the STAT5 inhibitor pimozide, but not to the JAK3 inhibitor tofacitinib, consistent with observed upregulation of Jak1 but not Jak3. R98S cells were also not sensitized to the MEK inhibitor CI-1040, suggesting that R98S cells rely mainly on Stat5 and not Erk activation for proliferation (Figure 2C + S8). The enhanced Jak-Stat activation upon cytokine stimulation and increased sensitivity to ruxolitinib and pimozide in R98S cells indicates that the observed overexpression of Jak-Stat components is of biological relevance.
RPL10 R98S T-ALL patient samples have elevated expression of the JAK-STAT cascade, are hyper-reactive to cytokine stimulation and sensitized to JAK-STAT inhibitors
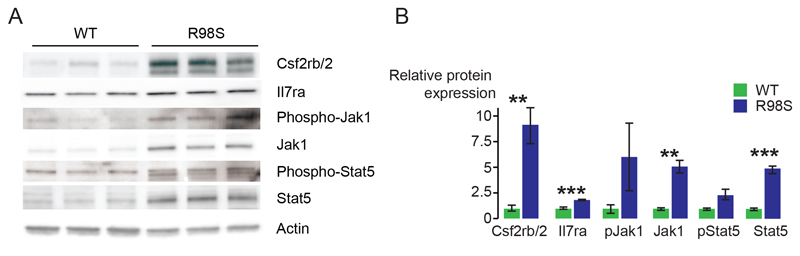
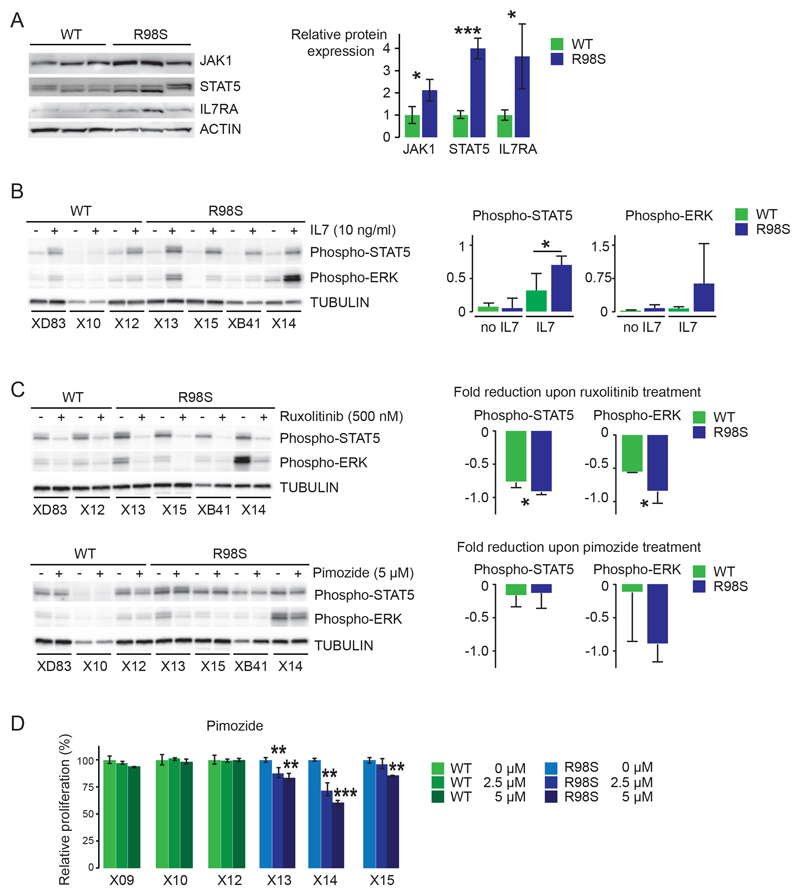
To determine whether RPL10 R98S also causes upregulation of the Jak-Stat cascade in other cellular models, we confirmed elevated expression and/or phosphorylation of Csf2rb/2, Jak1 and Stat5 in mouse hematopoietic cells derived from a conditional Rpl10 R98S knock-in mouse model (Figure 3). Furthermore, immunoblotting of protein lysates from xenografted T-ALL patient samples confirmed elevated expression of JAK1 (2.1-fold) and STAT5 (4-fold) (Figure 4A) in RPL10 R98S mutant T-ALL samples. Downregulation of PTPRC was also confirmed in these samples (Figure S9). None of these patient samples contained any IL7R-JAK-STAT mutations (Table S9). The IL3 receptor was not expressed in the T-ALL samples analyzed here. Instead, the IL7 receptor is important for normal T-cell development and in T-ALL.31 This receptor is not expressed in the Ba/F3 cell model, but Il7ra chain expression was upregulated in the cells of the Rpl10 R98S knock-in model (Figure 3). Also in the RPL10 R98S T-ALL samples, IL7RA chain expression was 3.7-fold upregulated as compared to RPL10 WT samples (Figure 4A). Upon stimulation with IL7, phosphorylation of STAT5 and ERK was consistently induced to higher levels in RPL10 R98S positive T-ALL samples as compared to RPL10 WT samples (Figure 4B). These phosphorylations were inhibited by ruxolitinib and pimozide, with a tendency towards stronger inhibition of phosphorylation in the R98S mutant xenografts (Figure 4C). Upon in vitro culture for 72 hrs, cell numbers of RPL10 R98S mutant xenograft cultures were reduced by pimozide, but this was not associated with apoptosis induction (Figure 4D + S10). Due to technical limitations, effects of ruxolitinib could not be analyzed in this in vitro culture system.
Figure 3. RPL10 R98S mouse knock-in hematopoietic cells display increased expression and phosphorylation of Jak-Stat signaling components.
A) Representative immunoblots of expression and phosphorylation of Jak-Stat proteins in hematopoietic cells derived from Rpl10cKI R98S (labeled as WT in the figure) and Mx1-Cre Rpl10cKI R98S (labeled as R98S in the figure) mice. The blots show data from cells derived from 3 biologically independent RPL10 WT versus 3 independent R98S mice. Cells were cultured in cytokine-rich Methocult GF M3534 media containing stem cell factor (SCF), IL3 and IL6. B) Quantification representing the average +/- standard deviation of the experiment shown in A). P-values were calculated using a T-test.
Figure 4. RPL10 R98S T-ALL patient samples have elevated expression and activation of the JAK-STAT cascade and are sensitized to JAK-STAT inhibitors.
A) Left: immunoblot analysis of JAK-STAT pathway protein expression in 3 RPL10 WT and 3 RPL10 R98S mutant human T-ALL xenograft samples. Only those components of the JAK-STAT pathway for which significant changes were observed are represented. Right: quantification of the immunoblots. The bars indicate the average +/- standard deviation of 3 independent RPL10 WT T-ALL patient samples versus 3 RPL10 R98S positive patient samples. P-values were calculated using a T-test. B) Left: Immunoblot analysis of STAT5 and ERK phosphorylation in RPL10 WT (XD83, X10, X12) versus R98S mutant (X13, X15, XB41, X14) T-ALL xenograft samples in unstimulated conditions or after 30 mins of stimulation with 10 ng/ml IL7. Right: quantification of the immunoblots. The bars indicate the average +/- standard deviation of 3 independent RPL10 WT T-ALL patient samples versus 4 RPL10 R98S positive patient samples that were analyzed. P-values were calculated using a T-test. C) Left: Immunoblot analysis of STAT5 and ERK phosphorylation in RPL10 WT versus R98S mutant T-ALL xenograft samples treated for 30 minutes with DMSO, ruxolitinib (500 nM) or pimozide (5 µM) followed by addition of 10 ng/ml IL7 for 30 mins. Right: quantification of the immunoblots. The bars represent the fold reduction in phospho-protein in the drug treated versus DMSO condition and show the average +/- standard deviation of all analyzed RPL10 WT T-ALL patient samples versus R98S positive patient samples. P-values were calculated using a T-test. D) Relative proliferation of 3 RPL10 WT (X09, X10 and X12) and 3 RPL10 R98S mutant (X13, X14 and X15) in vitro cultured T-ALL xenograft samples treated with the STAT5 inhibitor pimozide. Read out was done using the ATPlite assay (Perkin Elmer). The panel shows the average +/- standard deviation of three technical replicates per xenograft sample. P-values were calculated using a T-test. Due to limited sample availability, all experiment in this figure could only be performed once.*p<0.05, **p<0.01, ***p<0.001.
RPL10 R98S decreases apparent programmed -1 ribosomal frameshifting on several JAK-STAT pathway mRNAs
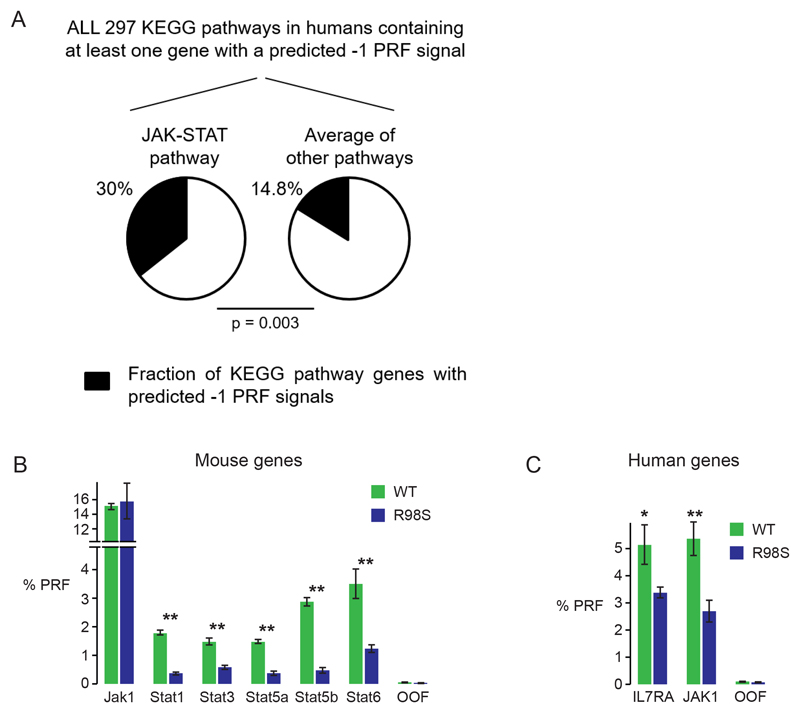
We next investigated cellular mechanisms that could be contributing to the specific JAK-STAT overexpression. We have previously shown in yeast models that the R98S mutation can alter apparent programmed -1 ribosomal frameshifting (-1 PRF) rates.19 In this process, cis-acting mRNA elements (-1 PRF signals) direct translating ribosomes to slip on an mRNA by one base in the 5’ direction, thus establishing a new reading frame. In mammalian cells, such -1 PRF events direct translating ribosomes towards premature termination codons, resulting in destabilization of the -1 PRF signal-containing mRNA. -1 PRF thus serves as a mechanism to fine-tune gene expression, and in silico algorithms predict that approximately 10% of human genes contain -1 PRF signals.23,32 We have recently shown that several cytokine receptors, including the IL7RA chain, contain functional -1 PRF signals.33 To test whether the RPL10 R98S associated differences in JAK-STAT protein expression might be influenced by alterations in -1 PRF rates, the JAK-STAT pathway was first screened in silico for predicted -1 PRF signals. Enrichment analysis revealed that, within the 10% of human genes predicted to contain -1 PRF signals, the JAK-STAT pathway was enriched for such predicted signals using both GO (p=0.00056) and KEGG (p=0.0031) databases. Specifically, 30% of genes in the JAK-STAT pathway are predicted to contain -1 PRF signals compared to the 14.8% average in other pathways (Figure 5A). Several human and mouse JAK-STAT pathway members for which RPL10 R98S associated differential protein levels were observed harbored such predicted -1 PRF signals (Table S10). We validated the frameshifting-promoting activity of these signals using dual luciferase reporter constructs (Figure S3A).25 Efficient rates of apparent -1 PRF promoted by several mouse and human sequences were confirmed in human Hek293T cells (Figure S3B), as well as in mouse Ba/F3 cells (Figure 5B-C). Interestingly, frameshifting levels induced by the signals in the mouse Stat genes were three to six times reduced in R98S cells (Figure 5B). In contrast, high levels (15%) of apparent frameshifting induced by the mouse Jak1 -1 PRF signal were unaffected by the R98S mutation in the Ba/F3 cells (Figure 5B). A 1.5-fold and 2-fold decrease in frameshifting rates induced by human -1 PRF signals in IL7RA and JAK1 mRNAs were observed in cells expressing the R98S mutation (Figure 5C). In summary, we have identified apparent functional -1 PRF signals in several mouse and human JAK-STAT genes, the frameshifting levels on some of which were influenced by the RPL10 R98S mutation.
Figure 5. Jak-Stat pathway genes contain functional apparent -1 PRF signals, the frameshifting levels on some of which are influenced by the RPL10 R98S mutation.
A) Results of enrichment analysis within all human genes containing predicted -1 PRF signals and extracted from the PRF database23. Analysis was performed using the G:profiler24 software and KEGG databases, and statistical significance was calculated using Fisher’s one-tailed test. B) Results from dual luciferase reporter assays (explained in Figure S3A) testing apparent -1 PRF levels on computationally predicted -1 PRF signals in the indicated mouse genes. The out-of-frame (OOF) is a negative control. Assays were performed in Ba/F3 cells expressing RPL10 WT or R98S. The bars indicate the average +/- standard error of at least 5 biologically independent measurements. C) Percentages of -1 PRF on human IL7RA and JAK1 mRNAs as determined by dual luciferase reporter assays performed in WT versus R98S Ba/F3 cells. Plots show the average +/- standard error of at least 5 biologically independent measurements.
RPL10 R98S cells show altered proteasome activity
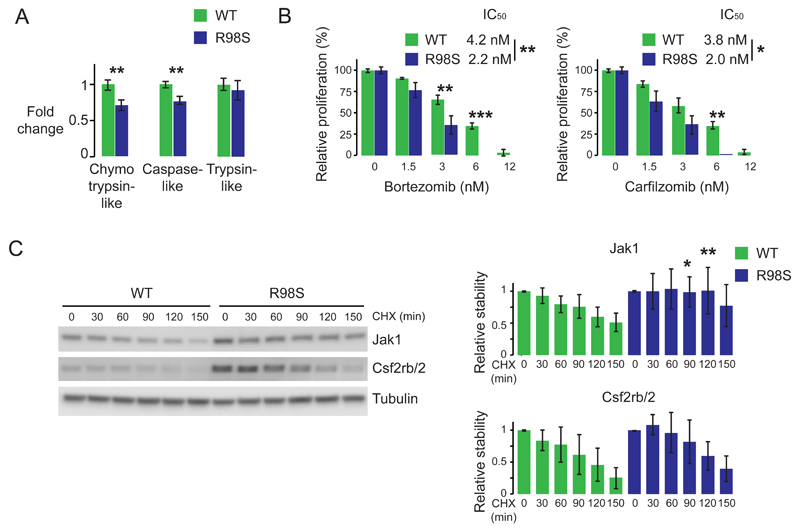
Altered frameshifting rates can only partially explain the upregulation of the Jak-Stat cascade, as there was one -1 PRF signal (Jak1) in which frameshifting levels were unaffected by RPL10 R98S, despite a change at the protein level. Additionally, our computational tools could not identify -1 PRF signals in the mRNAs encoding some of the other proteins in which changes at the protein level were observed. The altered levels of mouse Stat proteins are also likely not entirely explained by altered -1 PRF alone. The transcriptional changes detected for Csfr2b/2, Stat5a and Ptprc (Figure S5) might partially explain the detected protein changes, and we investigated additional potential regulation at the translational or post-translational level. Polysomal RNA-seq analysis in the Ba/F3 cell model did not reveal any significantly altered translation efficiency of Csf2rb/2, Jak or Stat mRNAs (Table S11). Interestingly, mass spectrometry data and RNA-seq revealed up- and downregulation of several proteasomal proteins in R98S cells (p<0.05), including upregulation of Psmb10, a catalytic subunit specific for the immunoproteasome (Table S12). Upregulation of Psmb10 and Psmb9, another immunoproteasome specific catalytic subunit, was confirmed on immunoblots from both available mouse hematopoietic cell models (Figure S11). In light of this, proteasome activities were assayed. R98S Ba/F3 cells displayed 28% and 23% decreases in chymotrypsin-like and caspase-like activity of the proteasome (Figure 6A). Consistent with these observations, R98S cells were more sensitive to the proteasome inhibitors bortezomib and carfilzomib (Figure 6B). Interestingly, this altered proteasomal activity was not accompanied by differences in total protein polyubiquitination (Figure S12). However, chase experiments after treatment with the translational inhibitor cycloheximide revealed increased stability of Jak1, but not Csf2rb/2, in R98S cells (Figure 6C). Stat protein stability remained unchanged within the 150 minutes of the CHX-chase (Figure S13). Altogether, our data indicate that RPL10 R98S cells display alterations in proteasome composition and activity, which may explain the increased stability of particular proteins such as Jak1 in these cells.
Figure 6. RPL10 R98S expressing cells show altered proteasome expression and activity and enhanced stability of Jak1.
A) Chymotrypsin-like, caspase-like and trypsin-like proteasomal activity of Ba/F3 cells expressing either WT or R98S RPL10. Plots show the average +/- standard deviation of three independent experiments comparing 3 biologically independent RPL10 WT versus 3 independent R98S cell clones. B) Relative proliferation of RPL10 WT and R98S cells treated with the indicated proteasome inhibitors measured using the ATPlite luminescence assay (Perking Elmer). The panel shows the average +/- standard deviation of a representative experiment comparing 3 biologically independent RPL10 WT versus 3 independent R98S cell clones. C) Left: immunoblots illustrating stability of Jak1 and Csf2rb/2 proteins as assessed by cycloheximide chase assays. Right: quantification of the immunoblots representing the average +/- standard deviation of three independent experiments comparing 3 biologically independent RPL10 WT versus 3 independent R98S cell clones. P-values were calculated using a T-test *p<0.05, **p<0.01, ***p<0.001. CHX: cycloheximide
Discussion
The link between somatic mutations in ribosomal proteins and cancer is a recent finding, and insights into the molecular mechanisms through which these defects promote cancer are only beginning to emerge.34 The current study illuminates how the recurrent ribosomal RPL10 R98S mutation may promote T-ALL. While the upregulation of the JAK-STAT cascade identified in our proteome screening was the focus of this study, it is reasonable to assume that some of the other detected changes induced by the RPL10 R98S mutation, for example metabolic reprogramming and reduced DNA repair may also contribute to oncogenesis. Upregulation of the oncogenic JAK-STAT cascade was confirmed in an independent hematopoietic cell model derived from an Rpl10 R98S conditional knock-in mouse, and in T-ALL patient samples. Whereas the IL7RA, JAK and STAT mutations in T-ALL increase cell proliferation and survival through constitutive phosphorylation of the cascade without cytokine stimulation, strong phosphorylation in the absence of cytokines was not observed in R98S cells. These findings suggest that R98S induced upregulation of the cascade is insufficient to drive constitutive activation, but that it rather provides the cells with an advantage upon cytokine exposure. RPL10 R98S cells have the capacity to react with a stronger response, which may lead to their clonal selection at the expense of WT cells over time. Interestingly, there was the recent observation of elevated STAT3 phosphorylation in leukocytes from Shwachman-Diamond Syndrome (SDS) patients, a cancer-predisposing ribosomopathy caused by mutations in the SBDS gene.35 RPL10 and SBDS are functionally linked and the R98S mutation yields similar biogenesis impairments as the SBDS defects.19,36 Altered JAK-STAT regulation may thus also be a shared feature of the RPL10 R98S and SBDS lesions.
We explored the molecular mechanism by which the RPL10 R98S mutation may drive selective upregulation of the JAK-STAT cascade. These effects may, at least partially, be mediated via reduced levels of -1 PRF on several JAK-STAT mRNAs. Initially described in viruses, it is becoming clear that this process is also relevant in mammalian cells.33 Only a limited set of predicted mammalian -1 PRF signals have been experimentally validated, including signals in several cytokine receptors such as IL7RA.33 We show the functionality of several additional predicted -1 PRF signals in mammalian cells, and that the JAK-STAT signaling cascade, downstream of cytokine receptors, is enriched for such signals. The dual luciferase data are consistent with -1 PRF. However, because of absent additional confirmatory data, e.g. identification of the trans-frame peptides by LC-MS/MS, we choose to err on the side of caution, and refer to this as apparent -1 PRF. Levels of -1 PRF can be regulated by trans-acting proteins and miRNAs.33,37,38 It is plausible that these -1 PRF signals function in fine-tuning and controlling immune responses, opening the possibility for oncogenic factors such as RPL10 R98S to deregulate this control mechanism.
Given the relatively low rates of -1 PRF promoted by Jak-Stat signals, it is clear that the altered frameshifting levels detected here cannot fully explain the observed protein level changes, suggesting contribution by additional mechanisms. We propose that R98S associated decreases in -1 PRF levels, combined with changes in the degradation of particular proteins and potential other mechanisms such as transcriptional regulation, leads to an oncogenic program (Figure 7). The specificity of the frameshifting alterations can be explained by the presence of unique -1 PRF signals in Jak-Stat genes. However, the specificity of the transcriptional and degradation phenotype is less clear. We show altered levels of specific catalytic components of the immunoproteasome. This finding, together with the altered proteasome activity in R98S cells, may indicate expression of a distinct type of proteasome, and is consistent with previously described ‘mixed type’ proteasomes containing constitutive as well as immuno-subunits.39,40,40 Different proteasome varieties show quantitative differences in cleavage efficiency of particular epitopes, which might provide a certain degree of protein specificity.41,42 This specificity is probably not restricted to JAK-STAT proteins, possibly explaining why R98S cells are sensitized to proteasome inhibitors despite potential further JAK-STAT stabilization by these drugs.
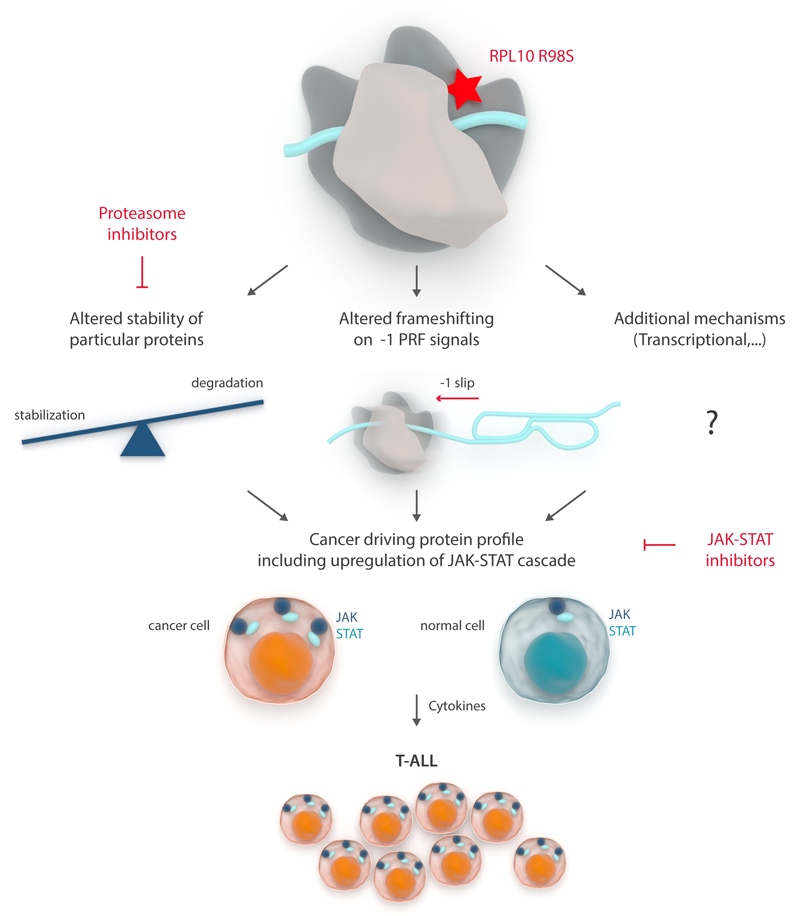
Figure 7. Model of RPL10 R98S promoted JAK-STAT upregulation and T-ALL pathogenesis.
Expression of RPL10 R98S alters protein stability and -1 PRF rates. These and potentially other mechanisms such as altered transcription drive a cancer promoting cellular protein profile, including overexpression of the JAK-STAT cascade. Particular conditions, such as cytokine stimulation, provide mutant cells with a competitive advantage, causing clonal selection and T-ALL development. Attractive targets for which clinically used drugs are available, some of which RPL10-R98S cells are more sensitive to, are indicated.
The JAK-STAT cascade is an established oncogenic driver with activating mutations in its signaling components in 20-30% of T-ALL patients. In our cohort, RPL10 R98S mutations were mutually exclusive with JAK and STAT mutations, with no difference in the incidence of IL7RA mutations in RPL10 WT versus R98S samples. This might reflect the fact that IL7RA mutations also activate the PI3K and MAPK cascade.
Our findings support that the RPL10 R98S mutation modulates JAK-STAT signaling in T-ALL. In addition to known JAK-STAT stimulating mutations and deregulation by DNM2 and ZEB2 lesions, this contributes another mechanism for activation of the pathway and further expands the relevance of this cascade in leukemia.43,44 Our results might also be therapeutically relevant. JAK-STAT cascade inhibitors ruxolitinib and pimozide are FDA approved drugs for myelofibrosis and schizophrenia/psychotic disorders respectively. A phase II trial of ruxolitinib in B-ALL has started (NCT02723994) and pre-clinical data on its effect in JAK mutant driven T-ALL tumors are available.45 Pre-clinical studies have indicated the potential of pimozide in AML.46,47 In light of the observation that cells harboring the RPL10 R98S mutation are sensitized towards ruxolitinib and pimozide, RPL10 R98S patients might benefit from these drugs. Additionally, we observed R98S induced changes in cellular proteasome expression and activity, rendering these cells more sensitive to clinically used proteasome inhibitors. This study thus provides the basis for the design of future therapy for RPL10 R98S defective cancer.
Supplementary Material
Acknowledgements
We thank the EPFL proteomics core facility and the VIB Nucleomics core facility for their excellent support on mass spectrometry and RNA sequencing respectively. We thank Somersault1824 for preparing Figure 7. TG is funded by the fellowship “Emmanuel van der Schueren” Kom op tegen kanker. SOS was supported by a long-term EMBO Fellowship (ALTF 1263-2014), the European Commission and Marie Curie Actions (LTFCOFUND2013, GA-2013-609409) and by an EHA junior research grant. SV is SB PhD fellow at FWO (n°1S49817N). KRK is supported by the Lady Tata Memorial Trust International Award for research in Leukaemia. This research was funded by an ERC starting grant (n°334946), FWO funding (G067015N, G084013N, and 1509814N) and a Stichting Tegen Kanker grant (grant n° 2012-176) to KDK. This work was partially supported by funding to JDD by the National Institutes of Health (R01HL119439, and R01GM117177).
Footnotes
Author’s Contributions
T.G., J.D.D., and K.D.K. designed the research. T.G., S.V., S.O.S., K.R.K. and S.B. developed methodology. T.G., S.V., S.O.S., K.R.K., Y.K., E.D., Z.F., J.O.D.B., and J.V. performed the research. T.G., S.V., S.O.S., K.R.K., Y.K., L.F., C.V., K.V., and K.D.K. analyzed data. J.P.M., C.S., E.G., C.J.H., and J.C. provided administrative, technical, and material support. T.G., S.V., S.O.S., L.F., C.J.H., J.P.M., J.C., J.D.D., and K.D.K. wrote and edited the paper. J.D.D. and K.D.K. supervised the study.
Disclosure of Conflicts of Interest
The authors disclose no potential conflicts of interest.
References
- 1.Pui C-H, Robison LL, Look AT. Acute lymphoblastic leukaemia. The Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Vicente C, Schwab C, Broux M, Geerdens E, Degryse S, Demeyer S, et al. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica. 2015;100:1301–1310. doi: 10.3324/haematol.2015.130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canté-Barrett K, Spijkers-Hagelstein JAP, Buijs-Gladdines JGCAM, Uitdehaag JCM, Smits WK, van der Zwet J, et al. MEK and PI3K-AKT inhibitors synergistically block activated IL7 receptor signaling in T-cell acute lymphoblastic leukemia. Leukemia. 2016;30:1832–1843. doi: 10.1038/leu.2016.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girardi T, Vicente C, Cools J, De Keersmaecker K. The genetics and molecular biology of T-ALL. Blood. 2017;129:1113–1123. doi: 10.1182/blood-2016-10-706465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2012;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao S, Lee S-Y, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z, et al. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120:3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fancello L, Kampen KR, Hofman IJF, Verbeeck J, De Keersmaecker K. The ribosomal protein gene RPL5 is a haploinsufficient tumor suppressor in multiple cancer types. Oncotarget. 2017;8:14462–14478. doi: 10.18632/oncotarget.14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofman IJF, Van Duin M, De Bruyne E, Fancello L, Mulligan G, Geerdens E, et al. RPL5 on 1p22.1 is recurrently deleted in multiple myeloma and its expression is linked to bortezomib response. Leukemia. 2017 doi: 10.1038/leu.2016.370. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ljungstrom V, Cortese D, Young E, Pandzic T, Mansouri L, Plevova K, et al. Whole-exome sequencing in relapsing chronic lymphocytic leukemia: clinical impact of recurrent RPS15 mutations. Blood. 2015;127:1007–1016. doi: 10.1182/blood-2015-10-674572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morgado-Palacin L, Varetti G, Llanos S, Gómez-López G, Martinez D, Serrano M. Partial Loss of Rpl11 in Adult Mice Recapitulates Diamond-Blackfan Anemia and Promotes Lymphomagenesis. Cell Rep. 2015;13:712–722. doi: 10.1016/j.celrep.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, et al. Many Ribosomal Protein Genes Are Cancer Genes in Zebrafish. PLoS Biol. 2004;2:e139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Keersmaecker K, Sulima SO, Dinman JD. Ribosomopathies and the paradox of cellular hypo-to hyperproliferation. Blood. 2015;125:1377–1382. doi: 10.1182/blood-2014-10-569616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Liao W-J, Liao J-M, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ban N, Beckmann R, Cate JHD, Dinman JD, Dragon F, Ellis SR, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulima SO, Patchett S, Advani VM, De Keersmaecker K, Johnson AW, Dinman JD. Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc Natl Acad Sci USA. 2014;111:5640–5645. doi: 10.1073/pnas.1400247111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sulima SO, Gülay SP, Anjos M, Patchett S, Meskauskas A, Johnson AW, et al. Eukaryotic rpL10 drives ribosomal rotation. Nucleic Acids Res. 2013;42:2049–2063. doi: 10.1093/nar/gkt1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 23.Belew AT, Hepler NL, Jacobs JL, Dinman JD. PRFdb: a database of computationally predicted eukaryotic programmed -1 ribosomal frameshift signals. BMC Genomics. 2008;9:339. doi: 10.1186/1471-2164-9-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–9. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs JL, Dinman JD. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 2004;32:e160. doi: 10.1093/nar/gnh157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hara T, Miyajima A. Two distinct functional high affinity receptors for mouse interleukin-3 (IL-3) EMBO J. 1992;11:1875–1884. doi: 10.1002/j.1460-2075.1992.tb05239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vainchenker W, Constantinescu SN. JAK/STAT signaling in hematological malignancies. Oncogene. 2012;32:2601–2613. doi: 10.1038/onc.2012.347. [DOI] [PubMed] [Google Scholar]
- 31.Ribeiro D, Melão A, Barata JT. IL-7R-mediated signaling in T-cell acute lymphoblastic leukemia. Adv Biol Regul. 2012;53:211–222. doi: 10.1016/j.jbior.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Advani VM, Dinman JD. Reprogramming the genetic code: The emerging role of ribosomal frameshifting in regulating cellular gene expression. Bioessays. 2015;38:21–26. doi: 10.1002/bies.201500131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belew AT, Meskauskas A, Musalgaonkar S, Advani VM, Sulima SO, Kasprzak WK, et al. Ribosomal frameshifting in the CCR5 mRNA is regulated by miRNAs and the NMD pathway. Nature. 2014;512:265–269. doi: 10.1038/nature13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bezzerri V, Vella A, Calcaterra E, Finotti A, Gasparello J, Gambari R, et al. New insights into the Shwachman-Diamond Syndrome-related haematological disorder: hyper-activation of mTOR and STAT3 in leukocytes. Sci Rep. 2016;6:33165. doi: 10.1038/srep33165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weis F, Giudice E, Churcher M, Jin L, Hilcenko C, Wong CC, et al. Mechanism of eIF6 release from the nascent 60S ribosomal subunit. Nat Struct Mol Biol. 2015;22:914–919. doi: 10.1038/nsmb.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anzalone AV, Lin AJ, Zairis S, Rabadan R, Cornish VW. Reprogramming eukaryotic translation with ligand-responsive synthetic RNA switches. Nat Methods. 2016;13:453. doi: 10.1038/nmeth.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Treffers EE, Napthine S, Tas A, Zhu L, Sun Z, et al. Transactivation of programmed ribosomal frameshifting by a viral protein. Proceedings of the National Academy of Sciences. 2014;111:E2172. doi: 10.1073/pnas.1321930111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klare N, Seeger M, Janek K, Jungblut PR, Dahlmann B. Intermediate-type 20 S proteasomes in HeLa cells: ‘asymmetric’ subunit composition, diversity and adaptation. J Mol Biol. 2007;373:1–10. doi: 10.1016/j.jmb.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 40.Dahlmann B, Ruppert T, Kuehn L, Merforth S, Kloetzel PM. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J Mol Biol. 2000;303:643–653. doi: 10.1006/jmbi.2000.4185. [DOI] [PubMed] [Google Scholar]
- 41.Huber EM, Basler M, Schwab R, Heinemeyer W, Kirk CJ, Groettrup M, et al. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell. 2012;148:727–738. doi: 10.1016/j.cell.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 42.Mishto M, Liepe J, Textoris-Taube K, Keller C, Henklein P, Weberruß M, et al. Proteasome isoforms exhibit only quantitative differences in cleavage and epitope generation. Eur J Immunol. 2014;44:3508–3521. doi: 10.1002/eji.201444902. [DOI] [PubMed] [Google Scholar]
- 43.Tremblay CS, Brown FC, Collett M, Saw J, Chiu SK, Sonderegger SE, et al. Loss-of-function mutations of Dynamin 2 promote T-ALL by enhancing IL-7 signalling. Leukemia. 2016;30:1993–2001. doi: 10.1038/leu.2016.100. [DOI] [PubMed] [Google Scholar]
- 44.Goossens S, Radaelli E, Blanchet O, Durinck K, Van der Meulen J, Peirs S, et al. ZEB2 drives immature T-cell lymphoblastic leukaemia development via enhanced tumour-initiating potential and IL-7 receptor signalling. Nat Commun. 2015;6:5794. doi: 10.1038/ncomms6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Degryse S, de Bock CE, Cox L, Demeyer S, Gielen O, Mentens N, et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood. 2014;124:3092–3100. doi: 10.1182/blood-2014-04-566687. [DOI] [PubMed] [Google Scholar]
- 46.Schafranek L, Nievergall E, Powell JA, Hiwase DK, Leclercq T, Hughes TP, et al. Sustained inhibition of STAT5, but not JAK2, is essential for TKI-induced cell death in chronic myeloid leukemia. Leukemia. 2014;29:76–85. doi: 10.1038/leu.2014.156. [DOI] [PubMed] [Google Scholar]
- 47.Nelson EA, Walker SR, Xiang M, Weisberg E, Bar-Natan M, Barrett R, et al. The STAT5 Inhibitor Pimozide Displays Efficacy in Models of Acute Myelogenous Leukemia Driven by FLT3 Mutations. Genes Cancer. 2012;3:503–511. doi: 10.1177/1947601912466555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.