Abstract
P. aegyptiaca is one of the most destructive root parasitic plants worldwide, causing serious damage to many crop species. Under natural conditions P. aegyptiaca seeds must be conditioned and then stimulated by host root exudates before germinating. However, preliminary experiments indicated that TIS108 (a triazole-type inhibitor of strigolactone) and fluridone (FL, an inhibitor of carotenoid-biosynthesis) both stimulated the germination of P. aegyptiaca seeds without a water preconditioning step (i.e. unconditioned seeds). The objective of this study was to use deep RNA sequencing to learn more about the mechanisms by which TIS108 and FL stimulate the germination of unconditioned P. aegyptiaca seeds. Deep RNA sequencing was performed to compare the mechanisms of germination in the following treatments: (i) unconditioned P. aegyptiaca seeds with no other treatment, (ii) unconditioned seeds treated with 100 mg/L TIS108, (iii) unconditioned seeds treated with 100 mg/L FL + 100 mg/L GA3, (iv) conditioned seeds treated with sterile water, and (v) conditioned seeds treated with 0.03 mg/L GR24. The de novo assembled transcriptome was used to analyze transcriptional dynamics during seed germination. The key gene categories involved in germination were also identified. The results showed that only 119 differentially expressed genes were identified in the conditioned treatment vs TIS108 treatment. This indicated that the vast majority of conditions for germination were met during the conditioning stage. Abscisic acid (ABA) and gibberellic acid (GA) played important roles during P. aegyptiaca germination. The common pathway of TIS108, FL+GA3, and GR24 in stimulating P. aegyptiaca germination was the simultaneous reduction in ABA concentrations and increase GA concentrations. These results could potentially aid the identification of more compounds that are capable of stimulating P. aegyptiaca germination. Some potential target sites of TIS108 were also identified in our transcriptome data. The results of this experiment suggest that TIS108 and FL+GA3 could be used to control P. aegyptiaca through suicidal germination.
1. Introduction
Approximately 3500 to 4000 species of angiosperms lost their autotrophic lifestyle during evolution. These plants, which include broomrape (Orobanche and Phelipanche spp.), witchweed (Striga spp.), and dodder (Cuscuta spp.), now directly invade and parasitize other plants[1]. Severe infestations by these species can cause complete yield loss[2]. The Parasitic Plant Genome Project has sequenced the transcripts of three root-parasitic species (P. aegyptiaca, S. hermonthica and Triphysaria versicolor) at key life stages from seed conditioning through anthesis. The information gained from this project offers greater potential for increasing understanding not only about population dynamics, parasite virulence, and host resistance mechanisms but also about target sites for herbicide action and other novel control strategies[2].
Broomrapes are holoparasites, devoid of chlorophyll, and found largely in the Mediterranean and warm-temperate areas of Europe, North Africa, the Middle East, and northern China[3]. Broomrape species spend most of their life cycle underground. After germination, they form a haustorium which attaches to the roots and vascular tissues of host plants, facilitating the exchange of water, nutrients, hormones, toxins, and almost anything else able to travel through vascular connections[4]. Broomrape species affect legumes and to a lesser extent, a range of crops in Apiaceae, Asteraceae, Cucurbitaceae and Solanaceae. Crop losses range between 20 and 70% in many countries and regions[3]. Numerous physical, chemical, and biological approaches have been explored against broomrape, including soil solarization, organic amendment, and trap crops[5–8]. However, these control methods are generally ineffective and uneconomical [9,10]. Broomrape releases numerous small and long-living seeds. Reports indicate that topsoil may contain up to 4 million broomrape seeds m−2[4]. The seeds remain viable for up to 20 years in soil in the absence of a suitable host[4]. Broomrape seeds require chemical stimulants secreted by host roots to germinate, and the seedlings must contact the host root within a few days to survive [11]. It has been suggested that synthetic or natural chemicals could be used to reduce seedbanks by stimulating seed germination in the absence of suitable hosts. This control method (i.e., suicidal germination) is an attractive means of keeping seedbanks below a certain threshold. Many research projects are currently trying to identify natural or synthetic chemicals that can induce suicidal germination of broomrape.
Several types of chemical compounds [e.g., dihydrosorgoleone, glucosinolate derivatives, and strigolactones (SLs)], have been identified as chemical signals or germination stimulants for Striga and Orobanche [12,13]. Among these germination stimulants, SLs at concentrations of 10−7 to 10−15 mol/L result in the highest germination rates [1,5]. Treatment with a synthetic analogue of SL, GR24 (10−7 mol/L), resulted in P. aegyptiaca germination rates > 80%. However, P. aegyptiaca seeds must be conditioned for 4 to 7 d in a humid environment before GR24 can stimulate high germination rates. During conditioning, gibberellin (GA) is synthesized and plays an important role in subsequent germination[11]. Fluridone (FL), a carotenoid biosynthesis inhibitor, shortens the conditioning period required for O. minor seeds to germinate after stimulation by strigol. Fluridone also has the capacity to inhibit SL production and exudation in crops[14,15]. Gibberellic acid (GA3) and brassinolide influence the seed conditioning and germination of Orobanche and Phelipanche spp.[16]. A triazole-type SL-biosynthesis inhibitor, TIS108, can reduce the level of 2′-epi-5-deoxystrigol in rice. It has been hypothesized that TIS108 could potentially be applied to reduce the germination of root parasitic weeds [17].
In a preliminary experiment, it was observed that TIS108 and FL + GA3 both stimulated the rapid and high germination of P. aegyptiaca seeds without a water preconditioning period (i.e. hereafter referred to as unconditioned seeds). In contrast, GR24 required a water preconditioning period to stimulate P. aegyptiaca germination. The objective of this study was to better understand the seed germination mechanisms by comparing the transcriptome profiles of P. aegyptiaca seeds treated with TIS108-, FL+GA3-, and GR24.
2. Materials and methods
2.1. Plant material
Mature seeds of P. aegyptiaca were collected from a processing tomato field in 2016 at Junhu, Xinjiang Uyghur Autonomous Region, China. These seeds were stored at 4°C.
2.2. Seed germination tests
An improved culture method was used to compare the effects of various stimulants on the germination of unconditioned P. aegyptiaca seeds [14,18]. The P. aegyptiaca seeds were disinfected with 75% ethanol for 2 min and 1% sodium hypochlorite for 20 min [19]. The seeds were then rinsed five times with sterile water. Petri dishes were lined with two layers of filter paper and then autoclaved. Three glass fiber discs were laid on top of the filter paper. The sterilized unconditioned seeds were then spread evenly on the discs. A 100 μL aliquot of 1 of 14 treatment solutions was then applied to each disc in the petri dish. The treatment solutions included four combinations of FL + GA3 (100 mg/L FL + 1000, 100, 10, or 1 mg/L GA3). The 10 additional treatments included sterile water (negative control), GA3 (10 mg/L), abscisic acid (ABA, 1 mg/L), acetone (4 g/L), FL (100 mg/L), TIS108 (1000, 100, 10, and 1 mg/L) and GR24 (0.03 mg/L). The dishes were incubated in the dark at 25°C for seven days. In one additional treatment (positive control), conditioned seeds (sterile water for 7 d) were treated with 0.03 mg/L GR24 and then incubated for 5 d. All the seeds were then examined with a microscope to determine germination rates.
2.3. Transcriptome analysis
2.3.1. Sample preparation
P. aegyptiaca seeds were surface sterilized and then spread evenly on a double layer of autoclaved filter paper in petri dishes (15 cm diam). The seeds in some dishes received no other treatment (unconditioned treatment). The remaining dishes were incubated in the dark at 25°C after treatment with 100 mg/L TIS108 (4 d incubation), 100 mg/L FL + 100 mg/L GA3 (4 d incubation), sterile water (7 d incubation), or sterile water (7 d incubation) followed by 0.03 mg/L GR24 (2 d incubation). These treatments will be respectively referred to as the TIS108, FL+GA3, conditioned, and GR24 treatments in the remainder of the paper. The unconditioned sample treated with water was the negative control and the conditioned sample treated with GR24 was the positive control. After incubation, the seeds were transferred from the glass fiber disks onto a piece of aluminum foil on a clean lab bench. The seeds were wrapped inside the foil, frozen in liquid N, and then stored at -80°C until RNA extraction.
2.3.2. RNA extraction, RNA-seq library preparation, and sequencing
The total RNA of each sample was isolated using Trizol Reagent (Invitrogen, Gaithersburg, MD, USA) according to the manufacturer’s recommendations. High-quality RNA, with a 28S:18S ratio of more than 1.5 and a 260/280 absorbance ratio between 1.8 and 2.2, was used for library construction and sequencing. 3 μg RNA of each sample was used for the RNA sample preparations. Sequencing libraries were obtained using a NEBNext®Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) according to the manufacturer’s recommendations. The index codes were added to attribute sequences to each sample. In short, mRNA was purified from the total RNA by using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized by random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Subsequently, Second strand cDNA synthesis was performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends by exonuclease/polymerase activities. After adenylation of the DNA fragments 3′ ends, NEBNext Adaptors with hairpin loop structures were ligated to prepare for hybridization. In order to select cDNA fragments of 150~200 bp length, the library fragments were purified using AMPure XP system (Beckman Coulter, Beverly, USA). 3 μL of USER Enzyme (NEB, USA) was then used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. PCR was then performed using Phusion High-Fidelity DNA polymerase, universal PCR primers and Index (X) Primer. Finally, the PCR products were purified (AMPure XP system) and the library quality was evaluated on the Agilent Bioanalyzer 2100 system. The Illumina sequencing platform was Hiseq X ten. The RNA library construction and sequencing were performed at Biomarker Technologies Co. Ltd., Beijing, China.
2.3.3. Preprocessing of illumina reads and de novo transcriptome assembly
Raw data (raw reads) of fastq format were first processed through in-house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapters, reads containing poly-N, and low quality reads (i.e., reads where the Q-scores were <20 and the ratio of bases was >20%) from the raw data. The Q20, Q30, GC-content, and sequence duplication level of clean data were calculated simultaneously. High-quality clean data were used for downstream analyses. Transcriptome assembly was accomplished by Trinity [20].
2.3.4. Functional annotation and differential expression analysis
Gene function was annotated using the following databases: non-redundant (NR) (National Center for Biotechnology Information NR protein sequences); Protein family (Pfam); EuKaryotic Orthologous Groups (KOG)/Clusters of Orthologous Groups of proteins (COG)/evolutionary genealogy of genes: Non-supervised Orthologous Groups (eggNOG); Swiss-Prot (A manually annotated and reviewed protein sequence database); Kyoto Encyclopedia of Genes and Genomes (KEGG); and Gene Ontology (GO). Bowtie was used to compare the sequenced reads with the unigene library. Gene expression levels were estimated by RSEM [21]. The clean data were mapped back to the assembled transcriptome and the read count for each gene was obtained from the mapping results. The FPKM value was used to express the corresponding unigene abundance. DESeq was used to identify the differentially expressed genes. The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling false discovery rate. Genes with an adjusted P value < 0.05 were assigned as differentially expressed. The False Discovery Rate (FDR) was used to determine the threshold of the P value in multiple tests. The cutoff thresholds for significance of expression were FDR < 0.01 and fold change ≥ 1.
2.3.5. Endogenous hormone level analysis and validation of related genes
Endogenous hormone levels were determined using the frozen seed samples from each treatment in Section 2.3.1. The seed samples (1 g) were separately ground in liquid N using a mortar and pestle. The concentrations of ABA and GA were quantified using appropriate enzyme-linked immunosorbent assay kits (Chengling, Beijing, China) according to manufacturer’s instructions [11]. The ABA and GA concentrations are reported as the mean of three biological replicates. The RNA samples for RNA-seq were also used for real-time qRT-PCR validation. First-strand cDNA was synthesized using a Takara PrimeScriptTM RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Dalian, China). Six plant-hormone-related genes, two SL perception-related genes, and two genes related to P450s were selected for validation of RNA-seq by qRT-PCR. PaTubulin1 was used as an internal control [11,22]. The qRT-PCR was conducted using SYBR GreenER™ qPCR SuperMix Universal (Invitrogen, Gaithersburg, MD, USA). Thermal cycle conditions for PCR were as follows: 94°C for 3 min followed by 40 cycles at 94°C for 15 s and 59°C for 30 s. The expression level of the genes was calculated as the means of three biological replicates. Specific primers were designed using Primer 5.0 and are listed in S1 Table. All data were analyzed using one-way ANOVA (IBM SPSS Statistics 19.0, Armonk, NY, USA).
2.4. Accession numbers
RNA-seq data were submitted to NCBI under BioProject accession number PRJNA388245. The quality filtered and trimmed short read data set was deposited in the NCBI Sequence Read Archive (SRA) under accession numbers: SRR5680424, SRR5680425, SRR5680426, SRR5680427, SRR5680428, SRR5680429, SRR5680430, SRR5680431, SRR5680432, SRR5680433, SRR5680434, SRR5680435, SRR5680436, SRR5680437 and SRR5680438. The assembled transcripts can be accessed from NCBI Transcriptome Shotgun Assembly Sequence (TSA) Database under the accession number GFQM00000000.
3. Results
3.1. Seed germination rates
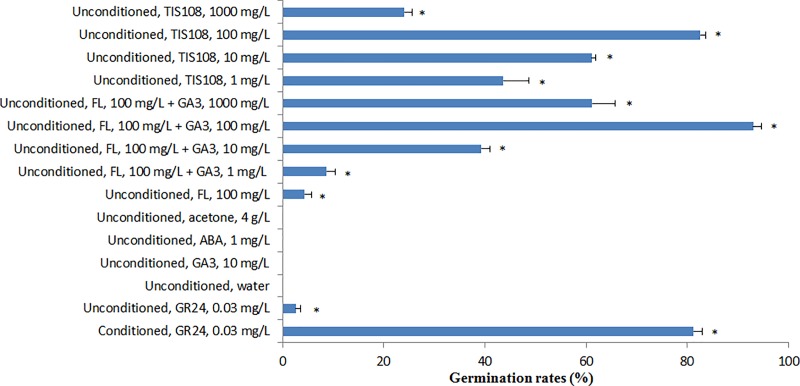
The germination rates of unconditioned P. aegyptiaca seeds were 0% when treated with water, GA3, ABA and acetone alone (Fig 1). The germination rate of unconditioned seeds treated with GR24 was 3%, whereas the germination rate of conditioned seeds treated with GR24 was 81%. Application of FL (100 mg/L) alone stimulated P. aegyptiaca germination; however the germination rate was only 4%. In comparison, FL + GA3 increased P. aegyptiaca germination significantly, with germination rates as high as 93% in the 100 mg/L FL+100 mg/L GA3 treatment. The SL biosynthesis inhibitor, TIS108, also promoted P. aegyptiaca germination, with germination rates reaching 83% in the 100 mg/L TIS108 treatment. More importantly, the FL + GA3 treatment and the TIS108 treatment both stimulated P. aegyptiaca germination without a water preconditioning period (Fig 2). It should be noted that there was acetone (as the solvent) in the solutions containing FL and TIS108. A preliminary experiment indicated that the acetone concentrations (0.04 to 40.00 g/L) in these solutions did not stimulate P. aegyptiaca germination. We chose to present the 4 g/L acetone treatment in Fig 1 because this was the acetone concentration in the treatments with the highest germination rates (FL 100 mg/L + GA3 100 mg/L and TIS108 100 mg/L).
Fig 1. Germination rates of unconditioned P. aegyptiaca seeds as affected by water, gibberellic acid (GA3), GR24, acetone, fluridone (FL), and TIS108.
Germination rates were determined after incubation in the dark at 25°C for 7 d. Conditioned seeds treated with GR24 and unconditioned seeds treated with water were used as positive and negative controls, respectively. Mean values ± standard deviations are from measurements on three independent germination assays. An asterisk (*) indicates that the values are significantly different than those in the water treatment according to Fisher’s protected LSD test (P <0.05).
Fig 2. P. aegyptiaca germination.
(a) Germinated and ungerminated seeds. Seeds were considered to have germinated when the radical length was more than half the seed diameter. (b) and (c) Germination of unconditioned P. aegyptiaca seeds after treatment with FL + GA3 and TIS108, respectively.
3.2. RNA-seq and de novo assembly of the P. aegyptiaca transcriptome
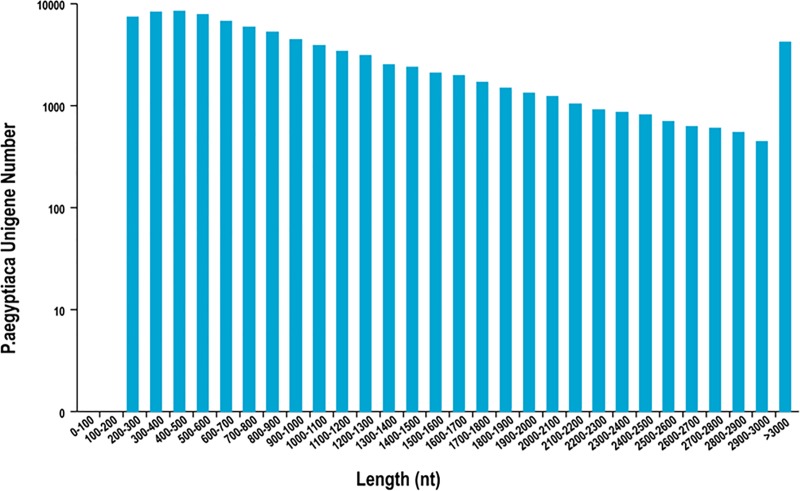
To learn more about the germination mechanism, RNA-seq was performed using mRNA extracted from P. aegyptiaca seeds in five treatments: (i) unconditioned seeds, (ii) conditioned seeds, (iii) unconditioned seeds treated with TIS108, (iv) unconditioned seeds treated with FL+GA3, and (v) conditioned seeds treated with GR24. There were three biological replications of each treatment. After filtering, 429,090,579 pair-end reads of clean data were obtained. The percentage of Q30 bases in each sample was more than 94.90%, and the GC percentage ranged from 45.76% to 48.91% (S2 Table). Clean reads were de novo assembled using the Trinity. By combining unigenes from all 15 samples, 89,434 unigenes were assembled. The average N50 length of the unigenes was 1105.26 bp. The major length of the unigenes ranged from 300 to 2000 bp (Fig 3 and S3 Table). The clean reads for each sample was mapped to the library of assembled transcripts and unigenes. Mapped reads were used for subsequent analysis (Table 1).
Fig 3. Unigene length distribution of P. aegyptiaca seeds.
Table 1. RNA-seq data and assembly results of P. aegyptiaca seeds as affected by different treatments.
The number after each treatment name represents the replication number.
| Samples | Clean Reads | Mapped Reads | Mapped Ratio |
|---|---|---|---|
| Unconditioned-1 | 21,312,030 | 13,682,131 | 64.20% |
| Conditioned, DI-water-1 | 23,760,435 | 15,338,029 | 64.55% |
| Unconditioned, FL+GA3-1 | 22,111,556 | 14,592,535 | 66.00% |
| Unconditioned, TIS108-1 | 25,793,060 | 16,708,666 | 64.78% |
| Conditioned, GR24-1 | 24,005,284 | 15,977,457 | 66.56% |
| Unconditioned-2 | 25,209,866 | 16,019,275 | 63.54% |
| Conditioned, DI-water-2 | 24,515,750 | 15,991,440 | 65.23% |
| Unconditioned, FL+GA3-2 | 25,539,216 | 16,587,180 | 64.95% |
| Unconditioned, TIS108-2 | 24,391,846 | 15,675,998 | 64.27% |
| Conditioned, GR24-2 | 21,836,698 | 14,021,636 | 64.21% |
| Unconditioned-3 | 22,107,194 | 14,123,756 | 63.89% |
| Conditioned, DI-water-3 | 21,551,978 | 13,684,833 | 63.50% |
| Unconditioned, FL+GA3-3 | 22,089,391 | 14,149,684 | 64.06% |
| Unconditioned, TIS108-3 | 21,954,428 | 14,215,757 | 64.75% |
| Conditioned, GR24-3 | 21,646,354 | 13,696,284 | 63.27% |
3.3. Functional annotation of P. aegyptiaca unigenes
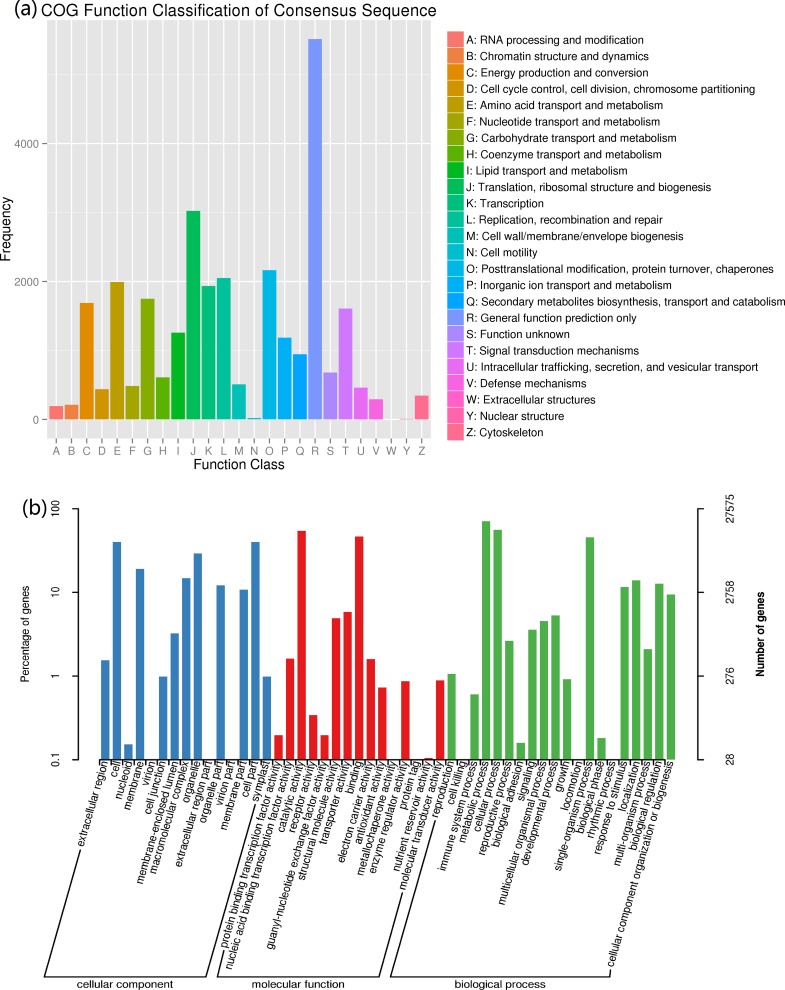
Functional annotation of P. aegyptiaca unigenes was conducted using BLAST software with an E-value ≤ 10−5. Among 89,434 non-redundant unigenes, there were 21,995 hits in COG, 27,575 hits in GO, 21,763 hits in KEGG, 33,118 hits in KOG, 27,364 hits in Swiss-Prot, 47,886 hits in eggNOG, and 52,917 hits in NR. The predicted amino acid sequences of the unigenes were subsequently compared with the Pfam database at an E-value ≤ 10−10. The HMMER software package was used to annotate the unigenes (S4 Table). The unigenes were classified into 25 groups based on COG function classification. The unigenes were mainly related to transcription and translation, including (i) translation, ribosomal structure, and biogenesis (3025); (ii) replication, recombination, and repair (2050); and (iii) post-translational modification, protein turnover, and chaperones (2164) (Fig 4A). Many unigenes were also related to transport and metabolism, especially the transport and metabolism of carbohydrates (1749), amino acids (1992), and lipids (1258). Some unigenes were involved in signal transduction mechanisms (1608), and cell cycle control, cell division, and chromosome partitioning (440) (Fig 4A).
Fig 4. Functional classification of all unigenes in the COG and GO databases.
(a) COG functional classification of the unigenes. (b) GO functional classification of the unigenes.
According to GO function classification, the majority of unigenes under cellular component participate in cell (11,101), membrane (5302), macromolecular complex (5302), organelle (8042), organelle part (3348), membrane part (2961), and cell part (11,101). Molecular functions of these unigenes were mostly clustered in catalytic activity (15,042), structural molecule activity (1356), transporter activity (1607), and binding (12,843). The biological process categories were mainly metabolic process (19,482), cellular process (15,442), single-organism process (12,552), stimulus response (3222), localization (3845), and biological regulation (3507) (Fig 4B).
3.4. Comparative analysis of differential expression
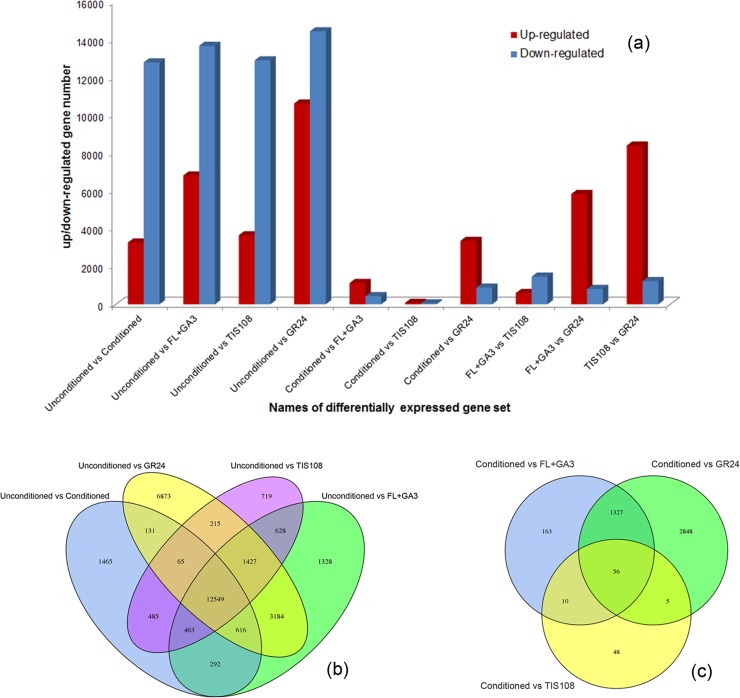
Spatial analysis of differentially expressed unigenes (DEGs) was also performed to determine the degree of overlap between the five different treatments during seed germination. DESeq software was used to perform differential expression analysis between pairs of treatments to obtain a set of DEGs[23]. Compared with the unconditioned treatment, the number of up-regulated genes was 3268, 6814, 3650, and 10,620 in the conditioned, FL+GA3, TIS108, and GR24 treatments, respectively. The number of down-regulated genes was 12,798, 13,673, 12,901, and 14,440 in the conditioned, FL+GA3, TIS108, and GR24 treatments, respectively. Compared with conditioned samples, FL+GA3 contained 1,127 up-regulated and 429 down-regulated unigenes, TIS108 exhibited 74 up-regulated and 45 down-regulated unigenes, and GR24 showed 3356 up-regulated and 880 down-regulated unigenes. Compared with FL+GA3, TIS108 contained 596 up-regulated and 1464 down-regulated unigenes and GR24 had 5837 up-regulated and 818 down-regulated unigenes. Compared with TIS108, GR24 exhibited 8,394 up-regulated and 1,227 down-regulated unigenes (Fig 5A and S5 Table).
Fig 5. Statistical analysis of DEGs in P. aegyptiaca seeds as influenced by conditioning and germination stimulants.
(a) Statistical analysis of up/down regulated unigenes in unconditioned, conditioned, FL+GA3-, TIS108-, and GR24 treated seeds. (b) Venn diagram of the DEGs in unconditioned vs conditioned, unconditioned vs GR24, unconditioned vs TIS108 and unconditioned vs FL+GA3. (c) Venn diagram of DEGs in conditioned vs GR24, conditioned vs TIS108, and conditioned vs FL+GA3.
Different process genes were also compared using a Venn diagram. There were 16,066 DEGs in unconditioned vs unconditioned; 20,487 DEGs in unconditioned versus FL+GA3; 16,551 DEGs in unconditioned vs. TIS108; 25,060 DEGs in unconditioned vs GR24; 1,556 DEGs in conditioned vs FL+GA3; 119 DEGs in conditioned vs TIS108, and 4,236 DEGs in conditioned vs GR24 (Fig 5B and 5C). There were 12,549 DEGs among unconditioned vs conditioned, unconditioned vs FL+GA3, unconditioned vs TIS108, and unconditioned vs GR24 (Fig 5B). These results indicated that 12,549 genes were essential during germination of P. aegyptiaca seeds. There were 56 DEGs among conditioned versus FL+GA3, conditioned versus TIS108, and conditioned versus GR24 (Fig 5C). This suggested that these 56 genes may be key genes for P. aegyptiaca germination.
3.5. Pathways of DEGs based on KEGG database
KOBAS 2.0 software was used to test the significance (P <0.05) of the enrichment of DEGs in KEGG pathways[24]. The total number of DEGs annotated in the databases were as follows: 14,226 DEGs in unconditioned vs conditioned; 18,320 DEGs in unconditioned vs FL+GA3; 14,992 DEGs in unconditioned vs TIS108; 21,551 DEGs in unconditioned vs GR24; 1332 DEGs in conditioned vs FL+GA3; 79 DEGs in conditioned vs TIS108; 3476 DEGs in conditioned versus GR24; 1876 DEGs in FL+GA3 vs TIS108; 4887 DEGs FL+GA3 vs GR24; and 7503 DEGs in TIS108 vs GR24 (S6 Table). The KEGG annotation indicated that 128 pathways in unconditioned vs conditioned; 127 pathways in unconditioned vs FL+GA3, unconditioned vs TIS108, and unconditioned vs GR24; 99 pathways in conditioned versus FL+GA3, 20 pathways in conditioned vs TIS108, 123 pathways in conditioned versus GR24, 110 pathways in FL+GA3 vs TIS108, 121 pathways in FL+GA3 vs GR24, and 125 pathways in TIS108 vs GR24 were enriched. The pathways of unconditioned vs conditioned, unconditioned vs FL+GA3, unconditioned vs TIS108, and unconditioned vs GR24 were very similar. Only 20 pathways were significantly altered in conditioned vs TIS108 based on the KEGG database. These results indicated that majority of requirements for P. aegyptiaca germination were met after conditioning. Pathways related to metabolism (alpha-linolenic acid metabolism, pyrimidine metabolism, starch and sucrose metabolism, carbon metabolism, purine metabolism, and glutathione metabolism) and biosynthesis (aminoacyl-tRNA biosynthesis and sesquiterpenoid and triterpenoid biosynthesis) covered almost half of the DEGs. Many of the DEGs were involved in energy production processes such as oxidative phosphorylation, pentose phosphate pathway, and glycolysis/gluconeogenesis. Other processes included plant hormone signal transduction, plant–pathogen interaction, and ubiquitin-mediated proteolysis.
3.6. Role of GA in P. aegyptiaca germination
The KEGG annotation indicated six pathways related to GA biosynthesis in P. aegyptiaca seeds (S1A and S1B Fig). This suggested that GA plays an important role in P. aegyptiaca germination. Pairwise comparison (unconditioned vs conditioned, unconditioned vs FL+GA3, unconditioned vs TIS108, unconditioned vs GR24, conditioned vs FL+GA3, and conditioned vs GR24) resulted in the identification of five DEGs related to GA biosynthesis in P. aegyptiaca. The expressions of all five DEGs were significantly greater in the conditioned, GR24, TIS108, and FL+GA3 treatments than in the unconditioned treatment (Table 2). These results indicated that the five DEGs play important roles in the conditioning stage and germination of P. aegyptiaca.
Table 2. Key DEGs related to the endogenous hormones under unconditioned, conditioned, GR24, TIS108 and FL+GA3 treatments.
| Gene ID | Gene name | Gene expressions (FPKM) | Annotation in KEGG | ||||
|---|---|---|---|---|---|---|---|
| Unconditioned | Conditioned | GR24 | TIS108 | FL+GA3 | |||
| c176949.graph_c0 | CPS | 0b | 0.8283a | 0.1006b | 1.0013a | 0.1211b | GA biosynthesis |
| c213846.graph_c0 | KS | 2.1824c | 8.0045a | 3.4058c | 7.0654ab | 6.1675b | GA biosynthesis |
| c212148.graph_c0 | KO | 20.5620d | 39.1807c | 60.6104a | 40.1073bc | 48.4856b | GA biosynthesis |
| c199689.graph_c0 | Ga20ox1-D | 0.6047e | 17.3994a | 7.6034c | 13.5352b | 5.1710d | GA biosynthesis |
| c190116.graph_c0 | GA3ox2 | 0.0211d | 0.3660d | 5.1775a | 1.6195c | 3.1649b | GA biosynthesis |
| c206041.graph_c0 | PSY | 3.9156a | 0.5036b | 0.0397b | 0.1961b | 0.1250b | Carotene biosynthesis |
| c208625.graph_c0 | PDS | 2.1621a | 0.2367b | 0.1057b | 0.3714b | 0.4898b | Carotene biosynthesis |
| c157830.graph_c0 | crtISO | 4.4044e | 9.7842d | 13.4980b | 11.0922c | 15.8902a | Carotene biosynthesis |
| c206406.graph_c0 | D27 | 1.2269c | 3.8825a | 2.6401b | 2.4140b | 2.6363b | SL biosynthesis |
| c214275.graph_c0 | NCED3 | 11.5488b | 12.3965b | 3.7011c | 28.3315a | 11.5441b | ABA biosynthesis |
| c186185.graph_c0 | NCED2 | 67.4027a | 3.0454bc | 1.2129c | 9.9701b | 3.0231bc | ABA biosynthesis |
| c196600.graph_c0 | CYP707A1 | 0.1752d | 1.9444bc | 4.0840a | 3.3379ab | 1.3514cd | ABA catabolism |
| c220152.graph_c0 | ACO | 41.1838b | 2.4058c | 62.4597a | 2.7025c | 32.0591b | Ethylene biosynthesis |
| c178284.graph_c0 | ACO1 | 9.3564a | 0b | 0b | 0b | 0.1150b | Ethylene biosynthesis |
| c172189.graph_c0 | ACO5-like | 0b | 0.4073b | 4.5449a | 0.4551b | 5.2078a | Ethylene biosynthesis |
| c217397.graph_c0 | CYP90B1 | 0.494812b | 1.169578b | 4.223323a | 1.191781b | 3.699762a | Brassinosteroid biosynthesis |
| c203876.graph_c1 | CYP90C19 | 0.179022d | 1.496152d | 17.70538a | 3.866753c | 14.95575b | Brassinosteroid biosynthesis |
| c91829.graph_c0 | CYP734A1 | 0.305962b | 1.361314b | 6.643272a | 2.313917b | 6.136895a | Brassinosteroid biosynthesis |
| c208011.graph_c0 | CYP72A13 | 10.00686a | 0.785742c | 4.046461b | 0c | 0.063386c | Brassinosteroid biosynthesis |
CPS: ent-copalyl diphosphate synthase; KS: ent-kaurene synthase; KO: ent-kaurene oxidase; Ga20ox1-D: gibberellin 20-oxidase 1-D; GA3ox2: gibberellin biosynthesis-related protein GA3ox2; PSY: phytoene synthase; PDS: phytoene desaturase; crtISO: prolycopene isomerase; D27: beta-carotene isomerase D27; NCED3: 9-cis-epoxycarotenoid dioxygenase 3; NCED2: 9-cis-epoxycarotenoid dioxygenase 2; CYP707A1: ABA 8'-hydroxylase CYP707A1; ACO: ACC oxidase; ACO1: 1-aminocyclopropane-1-carboxylate oxidase 1; ACO5-like:1-aminocyclopropane-1-carboxylate oxidase 5-like; CYP90B1: cytochrome P450 90B1; CYP90C19: cytochrome P450 90C19; CYP734A1: cytochrome P450 734A1; CYP72A13: Cytochrome P450 72A13; Means within a line followed by the same letter does not differ significantly according to Fisher’s protected LSD test (P<0.05).
3.7. Role of ABA in P. aegyptiaca germination
The KEGG annotation indicated seven pathways related to ABA biosynthesis in P. aegyptiaca seeds (S1C Fig). This suggested that ABA, in addition to GA, also plays an important role in P. aegyptiaca germination. Pairwise comparison of the treatments (unconditioned vs conditioned, unconditioned vs FL+GA3, unconditioned vs TIS108, unconditioned vs GR24, FL+GA3 vs TIS108, FL+GA3 vs GR24, and TIS108 vs GR24) resulted in the identification of three DEGs related to GA biosynthesis in P. aegyptiaca (Table 2). This suggested that these three DEGs also participate in P. aegyptiaca germination. The expression of CYP707A1 was significantly greater, but that of NCED2 was significantly less, in the conditioned, GR24, TIS108, and FL+GA3 treatments compared with the unconditioned treatment. These results indicated that these genes are indispensable for P. aegyptiaca germination. The TIS108 treatment significantly increased NCED3 expression compared with the unconditioned treatment, whereas the GR24 treatment had the opposite effect.
3.8. Other important genes related to germination
The KEGG pathway analysis revealed three DEGs (PSY, PDS, and crtISO) related to carotenoid biosynthesis, one DEG (D27) related to strigolactone biosynthesis, and three DEGs (ACO, ACO1, and ACO5-like) related to ethylene biosynthesis (Table 2 and S1D Fig). The expression of PSY and PSD was significantly less but the expression of crtISO and D27 was significantly greater in the GR24, TIS108, and FL+GA3 treatments than in the unconditioned treatment (Table 2). The expression of ACO was significantly less in the TIS108, and FL+GA3 treatments but significantly greater in the GR24 treatment compared with the conditioned treatment. The expression of ACO1 was significantly less, but the expression of ACO5-like was significantly greater in the conditioned, GR24, TIS108, and FL+GA3 treatments than in the unconditioned treatment. These results suggest that these genes are closely related to P. aegyptiaca germination. The KEGG pathway analysis also revealed some DEGs related to SL biosynthesis and signaling (Table 3) as well as to brassinosteroid (BR) biosynthesis (Table 2). When the expression of a gene was significantly less in the TIS108 treatment than in the other treatments, then such gene may be a potential target site of TIS108. Based on this theory, some potential target sites of TIS108 were obtained from the transcriptome data (Table 3).
Table 3. Expression of genes related to strigolactone and potential target site(s) of TIS108.
| Gene ID | Gene name | Gene expressions (FPKM) | Annotation in KEGG | ||||
|---|---|---|---|---|---|---|---|
| Unconditioned | Conditioned | GR24 | TIS108 | FL+GA3 | |||
| c192595.graph_c0 | MAX2 | 26.8149b | 57.5734a | 39.5196b | 53.4968a | 30.1277b | SL biosynthesis |
| c221871.graph_c0 | KAI2 | 0.5237c | 1.5407c | 10.1703b | 1.1781c | 15.4844a | SL biosynthesis |
| c212844.graph_c1 | KAI2-like | 4.0092a | 3.6226ab | 1.3980d | 2.7338bc | 2.1810cd | SL biosynthesis |
| c211198.graph_c0 | D14-like | 2.6704c | 7.4137bc | 32.6106a | 11.1315b | 35.4002a | SL biosynthesis |
| c225931.graph_c0 | DAD2 | 0.0824b | 1.4485a | 0.7500ab | 0.8229ab | 1.6309a | SL biosynthesis |
| c210504.graph_c0 | NADPH-cytochrome P450 reductase-like | 49.9144bc | 54.1220b | 73.8767a | 41.7903c | 57.7514b | potential target sites of TIS108 |
| c202864.graph_c0 | Cytochrome P450 704C1-like | 30.5226a | 29.7833a | 28.7952a | 16.8037b | 20.1252b | potential target sites of TIS108 |
| c185935.graph_c0 | Cytochrome P450 CYP736A12-like | 4.4690a | 2.1528bc | 3.2614ab | 1.6387c | 2.2211bc | potential target sites of TIS108 |
| c224503.graph_c0 | Cytochrome P450 CYP81Q2 | 2.2118a | 0.0665bc | 1.0619b | 0c | 0.0655bc | potential target sites of TIS108 |
| c187679.graph_c0 | Cytochrome P450 71D13-like | 10.4351b | 21.2041b | 82.9333a | 2.7094b | 16.7248b | potential target sites of TIS108 |
| c208011.graph_c0 | Cytochrome P450 72A13 | 10.0069a | 0.7857c | 4.0465b | 0c | 0.0634c | potential target sites of TIS108 |
| c209299.graph_c0 | Cytochrome P450 84A1-like | 3.2949b | 3.7305b | 20.0044a | 1.6184b | 19.2370a | potential target sites of TIS108 |
| c145815.graph_c0 | Cytochrome P450 714C2-like | 0.4289b | 0.4271b | 2.9095a | 0.1194b | 0.3632b | potential target sites of TIS108 |
| c219495.graph_c0 | Cytochrome P450 76A1-like | 86.7924a | 64.9125ab | 67.4746ab | 17.5377c | 39.6643bc | potential target sites of TIS108 |
Means within a line followed by the same letter do not differ significantly according to Fisher’s protected LSD test (P<0.05).
3.9. Endogenous hormone concentrations and validation of related genes by qRT-PCR
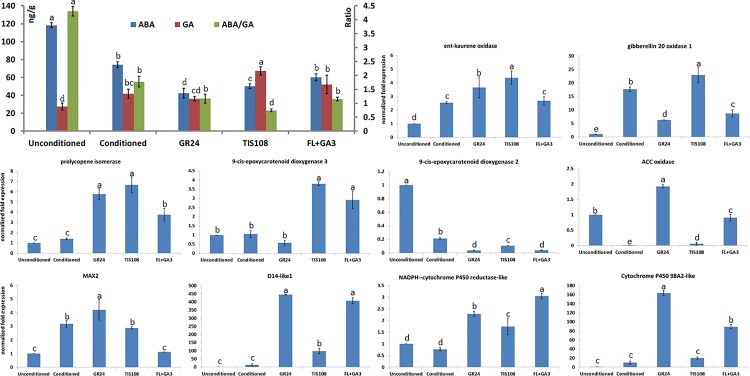
Endogenous ABA and GA concentrations were measured to learn more about the role of these hormones in P. aegyptiaca germination. The ABA concentrations were significantly less, but the GA was significantly greater in the conditioned, GR24, TIS108, and FL+GA3 treatments compared with the unconditioned treatment. The ABA:GA ratio steadily decreased during germination. This suggests that P. aegyptiaca can germinate only when the ABA:GA ratio declines to a certain value (Fig 6). The expression of ten representative genes was confirmed by qRT-PCR. The expression patterns of these genes were consistent with their transcriptional expression models (Fig 6).
Fig 6. Endogenous ABA and GA concentrations and verification of the expression patterns of ten genes by qRT-PCR.
The error bars indicate standard deviation. Different lowercase letters indicate significant differences among the treatments according to Fisher’s protected LSD test (P<0.05).
4. Discussion
In this study, transcriptome sequencing was used to learn more about the germination mechanisms of P. aegyptiaca as affected by TIS108, FL+GA3, and GR24. By comparing treatments, several key genes and pathways were identified that are associated with P. aegyptiaca germination. Most of the DEGs were related to protein, DNA, RNA, energy biosynthesis, and metabolism. Only a few DEGs were involved in hormone biosynthesis; however, these DEGs are indispensable for seed germination.
4.1. Energy requirements for P. aegyptiaca germination
Physiological processes during germination require considerable energy. Lacking mineral absorption systems and photosynthetic apparatus, seeds depend on reserves such as starch, protein, and lipids to provide the necessary energy for germination[25–27]. Degradation products of energy reserves are constantly fed into glycolysis, followed by ATP synthesis through the TCA cycle, and mitochondrial electron transport [28]. In a previous study involving P. aegyptiaca, the adenylate energy charge (AEC) in GR24-treated seeds reached a maximum (0.9) after 1 d of conditioning and then remained constant for the next 9 d [29]. In the present study, two genes related to the glycolysis pathway (PFK and PK) were obtained in the transcriptome data of P. aegyptiaca. The expressions of PFK and PK were significantly down-regulated in the conditioned samples compared with unconditioned samples. These results indicated that most of the energy required for seed germination was generated during conditioning (S7 Table). During the early stage of seed germination, the energy for seed germination is mainly provided by glycolysis; however, during late germination, the energy provided by anaerobic respiration cannot meet the requirements of germinating seeds. At this point, the TCA cycle provides considerable energy under oxygen enrichment [30]. In the present study, the pyruvate dehydrogenase (PDHA) sequence, which is important in the TCA cycle, was obtained in the transriptome data. Compared with the unconditioned samples, PDHA expression was significantly up-regulated in the GR24, TIS108, and FL+GA3 treatments, suggesting that the energy required for later stages of germination is mainly provided by the TCA cycle.
4.2. Activation of endogenous hormone-related genes
Throughout the life cycle of plants, GAs regulate various developmental processes including seed development and seed germination[31]. The major bioactive GAs includes GA1, GA3, GA4, and GA7[32]. Previous studies showed that GA synthesis occurs during conditioning [33]. In this study, the following DEGs were annotated to the GA biosynthesis pathway: CPS, KS, KO, and Ga20ox1-D. Genes related to GA1, GA3, GA4, and GA7 syntheses were all up-regulated significantly in the conditioned, GR24, TIS108, and FL+GA3 treatments compared with the unconditioned treatment (Table 2). Previous studies showed that applying GA biosynthesis inhibitors during seed conditioning can suppress the germination of Striga hermonthica, P. ramose, and P. aegyptiaca in response to GR24[34–36]. Taken together, these results show that GA synthesis is an essential step during seed germination. This assumption is also supported by the increases in endogenous GA concentrations that were observed during P. aegyptiaca germination (Fig 5). The expression of Ga20ox1-D increased significantly in the order unconditioned < GR24 < conditioned (Table 2). This suggests that when GR24 is applied, the most important thing is not the synthesis of GA, but the degradation of ABA.
The hormones ABA and GA play antagonistic roles in regulating seed germination, and the concentrations of GA and ABA are negatively correlated in germinating seeds [37,38]. The GA is closely related to the promotion of seed germination. However, ABA inhibits this process. The antagonistic relationship and ratio of these two hormones regulate processes ranging from embryogenesis to seed germination [39]. Previous studies showed that exogenous ABA can inhibit P. ramosa germination [36]. The results of the present study also showed two significantly down-regulated DEGs associated with ABA biosynthesis and two up-regulated DEGs associated with ABA catabolism (Table 2). These results indicated that ABA is closely related to P. aegyptiaca germination. This idea was also supported by the decreases in endogenous ABA that were observed during P. aegyptiaca germination (Fig 5).
Previous studies showed that ethylene also plays an important role in seed germination, and this hormone neutralizes a number of negative functions of ABA during germination[40–44]. The interaction between GA and ethylene is complicated, as shown by their negative and positive reciprocal effects [45]. Three genes (ACO, ACO1, and ACO5-like) related to ethylene-directed synthesis precursors were observed in our data. However, the treatments in our study had inconsistent effects on these ethylene-related genes, increasing the expression of some of these genes and reducing the expression of others (Table 2). Therefore, further studies should be conducted to determine the role of ethylene in germination of P. aegyptiaca seeds.
4.3. Comparison of the three hormones involved in P. aegyptiaca germination
Previous studies showed that P. aegyptiaca seeds need to be conditioned at 20 to 26°C for several days in the dark before germination[11,46]. Researchers have proposed the following functions of the conditioning stage: (i) formation or activation of receptors for germination stimulants, (ii) leaching of germination inhibitors, and (iii) biosynthesis of plant hormones which may play important roles in germination [14,33,47]. Only 119 DEGs and 20 pathways were obtained in the transcriptome data of conditioned vs TIS08 (Fig 4). Seed germination rate tests showed that the TIS108 treatment but not the conditioned treatment can stimulate P. aegyptiaca germination (Fig 1). We suggest that the conditioned stage is important for seed germination, providing most, but not all of the conditions required for P. aegyptiaca germination.
Fluridone [i.e., 1-methyl-3-phenyl-5-[3-trifluoromethy1-(phenyl)]-4-(lH)-pyridinone] is an inhibitor of phytoene desaturase, which converts phytoene to phytofluene in the carotenoid biosynthesis pathway [48,49]. Carotenoids correspond to major precursors of ABA in plants; therefore FL also blocks ABA biosynthesis [50]. The transcriptome data in this study also showed this relationship between carotene biosynthesis and ABA biosynthesis (Table 2). The application of FL alone stimulated P. aegyptiaca germination to a small degree (Fig 1), indicating that a reduction in ABA increased P. aegyptiaca germination. The germination rate of P. aegyptiaca was significantly increased by the combined application of FL and GA. This suggested that P. aegyptiaca germination was caused by increasing GA3 concentration and decreasing ABA concentration. This phenomenon was also supported by transcriptome data (Table 2).
The synthetic analog of SL, GR24, can stimulate P. aegyptiaca germination [11,51]. Studies have shown that GR24 promotes the degradation of ABA and reduces its concentration in P. ramosa [29,52]. Other studies indicate that ABA and ethylene play important roles in P. aegyptiaca germination. It is also known that GR24 application dramatically changes ABA concentrations and the expression of ethylene-associated genes[11]. Analysis of the transcriptome data in this study showed that the expression of genes related to ABA biosynthesis and ethylene biosynthesis varied significantly among the five treatments (Table 2). The ABA concentrations were significantly less in the conditioned and GR24 samples than in the unconditioned samples (Fig 5). The ABA catabolic gene CYP707A1 is a key component in the germination of P. ramosa seed treated with GR24 [29]. In the present study, PaCYP707A1 was also obtained from the transcriptome data. The PaCYP707A1 expression was significantly up-regulated in the conditioned, TIS108, FL+GA3, and GR24 samples compared with the unconditioned samples (Table 2). This indicated the importance of PaCYP707A1 activation during the conditioning stage.
Researchers previously reported that TIS108, a triazole-type SL-biosynthesis inhibitor, reduced SL concentrations in Arabidopsis and rice [17,53]. Those authors suggested that the germination of root parasitic weeds could be reduced by treating the host plants with TIS108. The results of this study, however, indicated that TIS108 stimulated P. aegyptiaca germination rates to as high as 82.60% (Fig 1). This suggested that TIS108 can be used to control P. aegyptiaca by inducing suicidal germination. Previous studies showed that the target site(s) of TIS108 may be P450 (MAX1) homologues, and that TIS108 may affect some P450s involved in either the GA or BR biosynthesis pathways[17,53]. Our analysis of the transcriptome data suggested another potential target site (CYP72A13) in the BR biosynthesis pathway (Table 2).
4.4. Transcriptome annotation of P. aegyptiaca seeds
Only 62.63% of the filtered unigenes were annotated in our transcriptome data. These annotation results were similar to many other de novo-assembled transcriptomes[11,54–56]. The results showed that 18.53% of the unigenes were annotated to Sesamum indicum, 5.01% to Erythranthe guttata, and 4.91% to Rhizopus delemar (S2 Fig). A total of 119 DEGs were obtained when we compared conditioned with TIS108 samples. Only 79 of these DEGs were annotated, suggesting that 40 genes involved in germination may not be annotated and were possibly missed. These missing genes should be further studied in the future.
5. Conclusions
Transcriptome approaches were used to study the effects of TIS108, FL+GA3, and GR24 on P. aegyptiaca germination. Many DEGs related to seed germination were obtained. Some key genes associated with germination were detected. These genes were annotated as “hormone-associated”. The expression patterns of some important genes and the roles of hormones during seed germination were further verified by qRT-PCR analysis and endogenous hormone content analysis. The results showed that all three compounds (i.e., TIS108, FL+GA3, and GR24) stimulated P. aegyptiaca germination without a water preconditioning period. The common effect of all three compounds was that they reduced ABA concentrations in the seeds and increased GA concentrations. However, the target sites for these compounds differ. The results of this study provide information that is helpful for identifying other compounds that can stimulate P. aegyptiaca germination. We suggest that the important characteristic of these compounds is that they ultimately inhibit ABA synthesis or promote ABA degradation. This study also suggests that TIS108 or FL+GA3 could be used as an economical and effective means of controlling P. aegyptiaca by promoting suicidal germination.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(a) Pathway of gibberellic acid biosynthesis in unconditioned vs TIS108. (b) Pathway of gibberellic acid biosynthesis in unconditioned vs GR24. (c) Pathway of abscisic acid biosynthesis in unconditioned vs conditioned. (d) Pathway of ethylene biosynthesis in unconditioned vs FL+GA3.
(DOCX)
(DOCX)
Data Availability
RNA-seq data were submitted to NCBI under BioProject accession number PRJNA388245. The quality filtered and trimmed short read data set was deposited to the NCBI Sequence Read Archive (SRA) under accession numbers: SRR5680424, SRR5680425, SRR5680426, SRR5680427, SRR5680428, SRR5680429, SRR5680430, SRR5680431, SRR5680432, SRR5680433, SRR5680434, SRR5680435, SRR5680436, SRR5680437 and SRR5680438. The assembled transcripts can be accessed from NCBI Transcriptome Shotgun Assembly Sequence (TSA) Database under the accession number GFQM00000000.
Funding Statement
This work was supported by the National Natural Science Foundation of China (No. 31460467).
References
- 1.Xie X, Yoneyama K, Yoneyama K. The strigolactone story. Annu Rev Phytopathol. 2010; 48: 93–117. doi: 10.1146/annurev-phyto-073009-114453 [DOI] [PubMed] [Google Scholar]
- 2.Westwood JH, Depamphilis CW, Das M, Fernández-Aparicio M, Honaas LA, Timko MP, et al. The Parasitic Plant Genome Project: New tools for understanding the biology of Orobanche and Striga. Weed Sci. 2012; 60:295–306. [Google Scholar]
- 3.Parker C. Observations on the current status of Orobanche and Striga problems worldwide. Pest Manag Sci. 2009; 65:453–459. doi: 10.1002/ps.1713 [DOI] [PubMed] [Google Scholar]
- 4.Joel DM, Hershenhorn J, Eizenberg H, Aly R, Ejeta G, Rich PJ, et al. Biology and management of weedy root parasites. Hortic Rev. 2007; 8:267–350. [Google Scholar]
- 5.Bouwmeester HJ, Matusova R, Zhongkui S, Beale MH. Secondary metabolite signalling in host-parasitic plant interactions. Curr Opin Plant Biol. 2003; 6:358–364. [DOI] [PubMed] [Google Scholar]
- 6.Hershenhorn J, Eizenberg H, Dor E, Kapulnik Y, Goldwasser Y. Phelipanche aegyptiaca management in tomato. Weed Res. 2009; 49 (s1):34–47. [Google Scholar]
- 7.Mauro RP, Monaco AL, Lombardo S, Restuccia A, Mauromicale G. Eradication of Orobanche/Phelipanche spp. seedbank by soil solarization and organic supplementation. Sci Hortic. 2015; 193:62–68. [Google Scholar]
- 8.Pérez-de-Luque A, Eizenberg H, Grenz JH, Sillero JC, Ávila C, Sauerborn J, et al. Broomrape management in faba bean. Field Crops Res. 2010; 115:319–328. [Google Scholar]
- 9.Joel DM. The long-term approach to parasitic weeds control: manipulation of specific developmental mechanisms of the parasite. Crop Prot. 2000; 19:753–758. [Google Scholar]
- 10.Goldwasser Y, Kleifeld Y. Recent approaches to Orobanche management. Weed Biol Manag. 2004; 6:439–466. [Google Scholar]
- 11.Yao Z, Tian F, Cao X, Xu Y, Chen M, Xiang B, et al. Global transcriptomic analysis reveals the mechanism of Phelipanche aegyptiaca seed germination. Int J Mol Sci. 2016; 17:1139–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang M, Netzly DH, Butler LG, Lynn DG. Chemical regulation of distance. Characterization of the first natural host germination stimulant for Striga asiatica. J Am Chem Soc. 1986; 108:7858–7860. doi: 10.1021/ja00284a074 [DOI] [PubMed] [Google Scholar]
- 13.Auger B, Pouvreau JB, Pouponneau K, Yoneyama K, Montiel G, Bizec BL, et al. Germination stimulants of Phelipanche ramosa in the rhizosphere of Brassica napus are derived from the glucosinolate pathway. Mol Plant Microbe Interact. 2012; 25:993–1004. doi: 10.1094/MPMI-01-12-0006-R [DOI] [PubMed] [Google Scholar]
- 14.Chae SH, Yoneyama K, Takeuchi Y, Joel DM. Fluridone and norflurazon, carotenoid-biosynthesis inhibitors, promote seed conditioning and germination of the holoparasite Orobanche minor. Physiol Plant. 2004; 120:328–337 doi: 10.1111/j.0031-9317.2004.0243.x [DOI] [PubMed] [Google Scholar]
- 15.Matusova R., Rani K, Verstappen FW, Franssen MC, Beale MH, Bouwmeester HJ. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 2005; 139: 920–934. doi: 10.1104/pp.105.061382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi Y, Omigawa Y, Ogasawara M, Yoneyama K, Konnai M, Worsham D. Effects of brassinosteroids on conditioning and germination of clover broomrape (Orobanche minor) seeds. Plant Growth Regul. 1995; 16:153–160. [Google Scholar]
- 17.Ito S, Umehara M, Hanada A, Kitahata N, Hayase H, Yamaguchi S, et al. Effects of triazole derivatives on strigolactone levels and growth retardation in rice. PLoS ONE. 2011; 6(7):e21723 doi: 10.1371/journal.pone.0021723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto Y, Ueyama T. Production of (+)-5-deoxystrigol by Lotus japonicus root culture. Phytochemistry. 2008; 69:212–217. doi: 10.1016/j.phytochem.2007.06.011 [DOI] [PubMed] [Google Scholar]
- 19.Mangnus EM, Stommen PLA, Zwanenburg B. A standardized bioassay for evaluation of potential germination stimulants for seeds of parasitic weeds. J Plant Growth Regul. 1992; 11:91–98. [Google Scholar]
- 20.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011; 29: 644–652. doi: 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12: 323–339. doi: 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.González-Verdejo CI, Die JV, Nadal S, Jiménez-Marín A, Moreno MT, Román B. Selection of housekeeping genes for normalization by real-time RT-PCR: analysis of Or-MYB1 gene expression in Orobanche ramosa development. Anal Biochem. 2008; 379:38–43. [DOI] [PubMed] [Google Scholar]
- 23.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BMG, et al. EBSeq: An empirical bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013; 29:1035–1043. doi: 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011; 39: W316–W322. doi: 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bewley JD. Seed germination and dormancy. Plant Cell.1997; 9:1055–1066. doi: 10.1105/tpc.9.7.1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheoran IS, Olson DJ, Ross AR, Sawhney VK. Proteome analysis of embryo and endosperm from germinating tomato seeds. Proteomics. 2005; 5:3752–3764. doi: 10.1002/pmic.200401209 [DOI] [PubMed] [Google Scholar]
- 27.Pritchard SL, Charlton WL, Baker A, Graham IA. Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 2002; 31:639–647. [DOI] [PubMed] [Google Scholar]
- 28.Weitbrecht K, Müller K, Leubner-Metzger G. First off the mark: early seed germination. J Exp Bot. 2011; 62:3289–3309. doi: 10.1093/jxb/err030 [DOI] [PubMed] [Google Scholar]
- 29.Lechat MM, Pouvreau JB, Péron T, Gauthier M, Montiel G, Véronési C, et al. PrCYP707A1, an ABA catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J Exp Bot. 2012; 63: 5311–5322. doi: 10.1093/jxb/ers189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang P, Li X, Wang X, Chen H, Chen F, Shen S. Proteomic analysis of rice (Oryza sativa) seeds during germination. Proteomics. 2007; 7:3358–3368. doi: 10.1002/pmic.200700207 [DOI] [PubMed] [Google Scholar]
- 31.Sun TP, Gubler F. Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol. 2004; 55:197–223. doi: 10.1146/annurev.arplant.55.031903.141753 [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008; 59: 225–251. doi: 10.1146/annurev.arplant.59.032607.092804 [DOI] [PubMed] [Google Scholar]
- 33.Joel DM, Back A, Kleifeld Y, Gepstein S. Seed conditioning and its role in Orobanche seed germination: Inhibition by paclobutrazol. In K Wegmann, LJ Musselman, eds, Progress in Orobanche Research, Proceeding of the International Workshop on Orobanche Research. Obermachtal, Germany. 1991. pp 147–156.
- 34.Song WJ, Zhou WJ, Jin ZL, Cao DD, Joel DM, Takeuchi Y, et al. Germination response of Orobanche seeds subjected to conditioning temperature, water potential and growth regulator treatments. Weed Res. 2005; 45: 467–476. [Google Scholar]
- 35.Uematsu K, Nakajima M, Yamaguchi I, Yoneyama K, Fukui Y. Role of cAMP in gibberellin promotion of seed germination in Orobanche minor Smith. J Plant Growth Regul. 2007; 26: 245–254. [Google Scholar]
- 36.Zehhar N, Ingouff M, Bouya D, Fer A. Possible involvement of gibberellins and ethylene in Orobanche ramosa germination. Weed Res. 2002; 42:464–469. [Google Scholar]
- 37.Batge SL, Ross JJ, Reid JB. Abscisic acid levels in seeds of the gibberellin-deficient mutant lh-2 of pea (Pisum sativum). Physiol Plant. 1999; 105: 485–490. [Google Scholar]
- 38.White CN, Proebsting WM, Hedden P, Rivin CJ. Gibberellins and seed development in maize. I. Evidence that gibberellin/abscisic acid balance governs germination versus maturation pathways. Plant Physiol. 2000; 122: 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Razem FA, Baron K, Hill RD. Turning on gibberellin and abscisic acid signaling. Curr Opin Plant Biol. 2006; 9: 454–459. doi: 10.1016/j.pbi.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 40.Linkies A, Muller K, Morris K, Tureckova V, Wenk M, Cadman CSC, et al. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell. 2009; 21:3803–3822. doi: 10.1105/tpc.109.070201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graeber K, Linkies A, Müller K, Wunchova A, Rott A, Leubner-Metzger G. Cross-species approaches to seed dormancy and germination: conservation and biodiversity of ABA-regulated mechanisms and the brassicaceae DOG1 genes. Plant Mol Biol. 2010; 73: 67–87. doi: 10.1007/s11103-009-9583-x [DOI] [PubMed] [Google Scholar]
- 42.Kucera B, Cohn MA, Leubner-Metzger G. Plant hormone interactions during seed dormancy release and germination. Seed Sci Res. 2005; 15:281–307. [Google Scholar]
- 43.Matilla AJ, Matilla-Vázquez MA. Involvement of ethylene in seed physiology. Plant Sci. 2008; 175:87–97. [Google Scholar]
- 44.Morris K, Linkies A, Müller K, Oracz K, Wang X, Lynn JR, et al. Regulation of seed germination in the close Arabidopsis relative Lepidium sativum: a global tissue-specific transcript analysis. Plant Physiol. 2011; 155: 1851–1870. doi: 10.1104/pp.110.169706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss D, Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007; 144:1240–1246. doi: 10.1104/pp.107.100370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joel DM, Bar H, Mayer AM, Plakhine D, Ziadne H, Westwood JH, et al. Seed ultrastructure and water absorption pathway of the root-parasitic plant Phelipanche aegyptiaca (Orobanchaceae). Ann Bot. 2012; 109: 181–195. doi: 10.1093/aob/mcr261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bar-Nun N, Mayer AM. Composition of and changes in storage compounds in Orobanche aegyptiaca seeds during preconditioning. Isr J Plant Sci. 2002; 50:277–279. [Google Scholar]
- 48.Bartels PG, Watson CW. Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Sci. 1978; 26:198–203. [Google Scholar]
- 49.Fong F, Schiff JA. Blue-light-induced absorbance changes associated with carotenoids in Euglena. Planta.1979; 146:119–127. doi: 10.1007/BF00388221 [DOI] [PubMed] [Google Scholar]
- 50.Quatrano RS, Bartels D, Ho TD, Pages M. New insights into ABA-mediated processes. Plant Cell. 1997; 9:470–475. [Google Scholar]
- 51.Malik H, Rutjes FPJT, Zwanenburg B. A new efficient synthesis of GR24 and dimethyl A-ring analogues, germinating agents for seeds of the parasitic weeds Striga and Orobanche spp. Tetrahedron. 2010; 66:7198–7203. [Google Scholar]
- 52.Lechat MM, Brun G, Montiel G, Véronési C, Simier P, Thoiron S, et al. Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel. J Exp Bot. 2015; 66:3129–3140. doi: 10.1093/jxb/erv119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito S, Umehara M, Hanada A, Yamaguchi S, Asami T. Effects of strigolactone-biosynthesis inhibitor TIS108 on Arabidopsis. Plant Signal Behav. 2013; 8(5):e24193 doi: 10.4161/psb.24193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S, Li W, Wu Y, Chen C, Lei J. De novo transcriptome assembly in Chili Pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids. PloS ONE. 2013, 8(1):e48156 doi: 10.1371/journal.pone.0048156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakasugi K, Crowhurst RN, Bally J, Wood CC, Hellens RP, Waterhouse PM. De novo transcriptome sequence assembly and analysis of RNA silencing genes of Nicotiana benthamiana. PloS ONE. 2013; 8(3):e59534 doi: 10.1371/journal.pone.0059534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ranjan A, Ichihashi Y, Farhi M, Zumstein K, Townsley B, David-Schwartz R, et al. De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol. 2014; 166: 1186–1199. doi: 10.1104/pp.113.234864 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(a) Pathway of gibberellic acid biosynthesis in unconditioned vs TIS108. (b) Pathway of gibberellic acid biosynthesis in unconditioned vs GR24. (c) Pathway of abscisic acid biosynthesis in unconditioned vs conditioned. (d) Pathway of ethylene biosynthesis in unconditioned vs FL+GA3.
(DOCX)
(DOCX)
Data Availability Statement
RNA-seq data were submitted to NCBI under BioProject accession number PRJNA388245. The quality filtered and trimmed short read data set was deposited to the NCBI Sequence Read Archive (SRA) under accession numbers: SRR5680424, SRR5680425, SRR5680426, SRR5680427, SRR5680428, SRR5680429, SRR5680430, SRR5680431, SRR5680432, SRR5680433, SRR5680434, SRR5680435, SRR5680436, SRR5680437 and SRR5680438. The assembled transcripts can be accessed from NCBI Transcriptome Shotgun Assembly Sequence (TSA) Database under the accession number GFQM00000000.