Abstract
We investigated the safety and efficacy of liver-directed gene therapy using lentiviral vectors in a large animal model of hemophilia B, and evaluated the risk of insertional mutagenesis in tumor-prone mouse models. We show that gene therapy using lentiviral vectors targeting expression of a canine factor IX transgene to hepatocytes was well-tolerated and provided stable long-term production of coagulation factor IX in dogs with hemophilia B. By exploiting three different mouse models designed to amplify the consequences of insertional mutagenesis, we show that no genotoxicity was detected with these lentiviral vectors. Our findings suggest that lentiviral vectors may be an attractive candidate for gene therapy targeted to the liver and may be useful for the treatment of hemophilia.
Introduction
Hemophilia is a monogenic X-linked disease caused by deficiency of coagulation factor VIII (hemophilia A) or factor IX (hemophilia B) (1). Bleeding is the hallmark of hemophilia, either spontaneous or post-traumatic, which can be fatal if left untreated. According to residual factor activity, hemophilia is classified as severe (<1%), moderate (1-5%) or mild (6-30%). Patients with moderate or mild hemophilia have occasional to rare spontaneous hemorrhages, thus rescuing factor activity at ≥1% of normal substantially benefits the clinical phenotype of severely affected patients (2).
Prophylactic or on-demand replacement therapy with recombinant products is the current standard of care for hemophilia in high-income countries and has improved the quality of life and life expectancy of patients with severe hemophilia (3). Nevertheless, this treatment has high costs and entails discomfort for patients and has the risk of inducing neutralizing anti-factor antibodies, which complicates further treatment (4). Moreover, about 80% of people affected by hemophilia live with no or unsatisfactory treatment, mainly in developing countries (5). Gene therapy could help to address these needs by establishing long-term endogenous production of the clotting factor at therapeutic levels after a single treatment (2, 6, 7).
Recently, factor IX activity at 1-7% of normal has been reported long-term in adult patients with severe hemophilia B after administration of a single dose of an adeno-associated viral (AAV) vector targeting expression of human factor IX cDNA to hepatocytes (8). These results establish the therapeutic potential of liver-directed gene therapy in humans and offer the prospect of a definitive treatment for hemophilia. However, there are still important hurdles to overcome before this gene therapy can be applied to the majority of severely affected patients (9). In particular, pre-existing neutralizing antibodies to AAV following natural exposure to the wild-type virus may inhibit gene transfer with AAV vectors. In addition, AAV-specific cellular immune responses to the transduced hepatocytes may curtail long-term transgene expression and require transient immune suppression to allow clearance of AAV-derived antigens (8). HIV-derived lentiviral vectors may complement the therapeutic reach of AAV vectors because of the low prevalence of HIV infection in humans and the vector’s capacity to accommodate larger gene inserts. Moreover, the efficient integration of lentiviral vectors into the genome of target cells may eventually make these vectors better suited for treatment of pediatric patients, in which hepatocyte turnover is high and episomal vectors may be progressively lost (10).
We have developed a lentiviral vector platform that achieves stable and robust transgene expression in the mouse liver and induces transgene-specific immune tolerance upon systemic administration (11–15). This lentiviral vector stringently targets transgene expression to hepatocytes through transcriptional and microRNA (miR)-mediated regulation. We and others have shown the efficacy of this vector in establishing correction of hemophilia B and A in mouse models and of hyperbilirubinemia in rats (12, 16, 17).
Although encouraging, these results were obtained in rodents and it is crucial to assess the feasibility and safety of scaling-up lentiviral gene therapy in large animal models. In addition, whereas liver gene transfer by lentiviral vectors appeared to be safe in treated mice, concerns remain regarding the risk of insertional mutagenesis. We and others recently reported safe and efficacious clinical testing of lentiviral vectors for ex vivo gene therapy with hematopoietic stem cells (18–20). The safe outcome of these trials to date supports the predictions about vector safety developed in our preclinical tumor-prone mouse models, where the consequences of insertional mutagenesis are amplified in a model species that otherwise limits detection of low incidence, vector-induced oncogenesis (21, 22).
Here, we investigated liver-directed gene therapy using lentiviral vectors in dogs with hemophilia B and tested the potential for genotoxicity in mouse models prone to develop hepatocellular carinoma.
Results
Lentiviral vectors efficiently transduce and regulate transgene expression in canine cells
We generated three lentiviral vectors with Self-Inactivating (SIN) Long Terminal Repeats (LTR) expressing cDNA transgenes for canine factor IX (cFIX) under the control of an internal synthetic hepatocyte-specific promoter (Enhanced Transthyretin, ET) and carrying 4 tandem repeats of miR-142 target sequences (142T, Fig. 1A). The lentiviral vectors contained the wildtype, codon-usage optimized, or codon-usage optimized hyper-functional cFIX carrying the R338L mutation associated with human thrombophilia (cFIX, co-cFIX, co-cFIXR338L, respectively) (14, 23). All lentiviral vectors were pseudotyped with the vesicular stomatitis virus glycoprotein G (VSV-G).
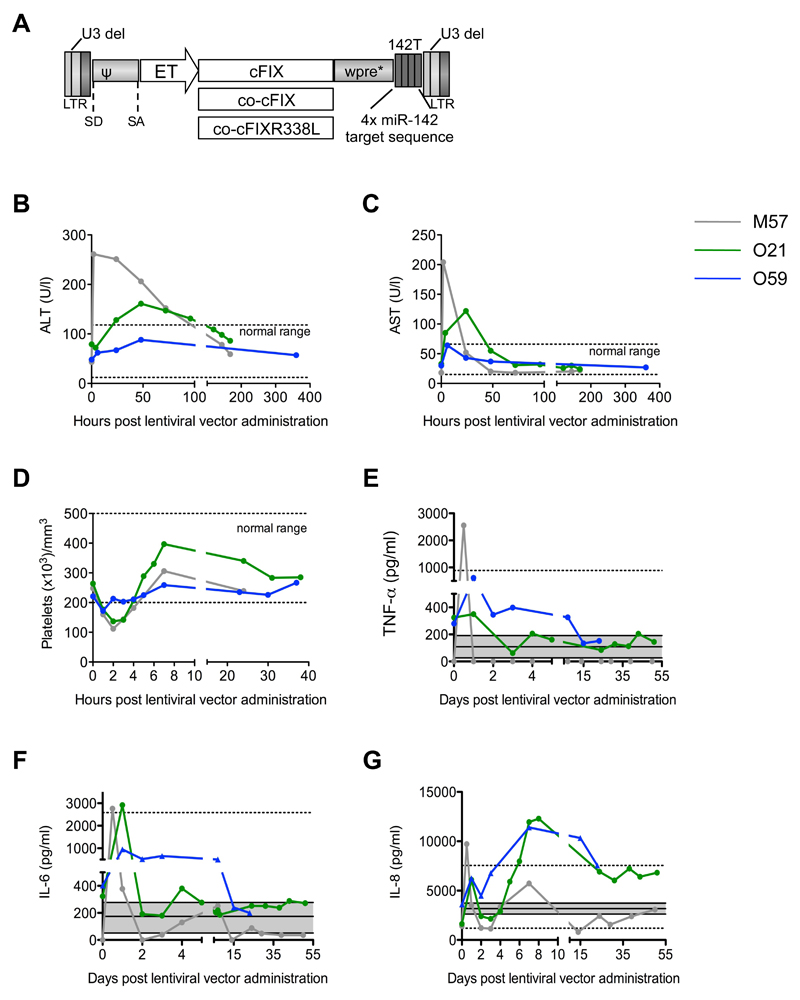
Figure 1. Intraportal administration of lentiviral vectors to dogs with hemophilia B.
(A) Schematic representation of the third-generation Self Inactivating (SIN) lentiviral vectors (proviral form) used in this work [U3 del: deletion of the promoter/enhancer of the HIV Long Terminal Repeats (LTR) (43)]. SD: splicing donor site. SA: splicing acceptor site. ψ: packaging signal. Wpre*: mutated woodchuck hepatitis virus post-transcriptional regulatory element (44). 142T: miR-142 target sequence made of 4 tandem copies of a sequence perfectly complementary to miR-142. Hepatocyte-specific Enhanced Transthyretin (ET) promoter composed of synthetic hepatocyte-specific enhancers and transthyretin promoter (45). The wildtype, codon-usage optimized, and codon-usage optimized and hyper-functional cDNAs of canine factor IX (cFIX, co-cFIX, co-cFIXR338L) were used as transgenes (14). Serum concentrations of alanine aminotransferase, ALT (B) and aspartate aminotransferase, AST (C), platelet counts (D), and serum concentrations of TNF-α (E), IL-6 (F) and IL-8 (G) were measured in blood samples collected at the indicated time points after lentiviral vector administration to dogs M57 (grey line), O21 (green line), and O59 (blue line). Baseline values are shown as “time 0”. (B-D) The normal range is shown (dashed lines). (E-G) The mean ± standard deviation (grey area) and range (dashed lines) of the serum concentrations of each cytokine measured in samples collected from 11 control untreated dogs are shown. Note that the lowest range for TNF-α and IL-6 is 0. Dog O59 was administered corticosteroids and anti-histamine drugs before lentiviral vector infusion to reduce inflammation.
We observed 2-fold to 5-fold reduction in lentiviral vector titer after incubation with pooled and individual dog sera, possibly mediated by complement (fig. S1A) (24). We selected for infusion those dogs whose serum showed the lowest neutralizing potential against lentiviral vectors. To verify lentiviral vector transduction and promoter activity in canine hepatocytes, we transduced primary human and canine hepatocytes ex vivo at increasing multiplicity of infection (MOI) with lentiviral vectors expressing GFP driven by the ubiquitously expressed phosphoglycerate kinase (PGK) promoter or the hepatocyte-specific Enhanced Transthyretin (ET) promoter. We observed high levels of transgene expression in hepatocytes of both species with both promoters (fig. S1B). We assessed miR expression in DH82 cells, a cell line derived from canine macrophages and found miR-142 to be expressed at high levels (fig. S1C). We then transduced DH82 cells with reporter lentiviral vectors encoding GFP with or without 142T, and showed ≥100-fold downregulation of GFP expression (fig. S1D). These data indicate that the regulatory elements of our lentiviral vectors are functional in canine cells.
Intraportal lentiviral vector administration is well tolerated in dogs with hemophilia B
We produced three large-scale batches of lentiviral vectors according to a manufacturing process previously developed for clinical-grade lentiviral vectors (19, 20). Quality assessment of the vector batches (2009/D2, 2011/D13-15 and 2012/DG) is summarized in table S1. The process yielded 1.1-4.5x1010 transducing units (TU), corresponding to 864 - 3,151 μg HIV Gag p24 equivalents (p24) of viral particles in 160-230 ml saline for infusion. Lentiviral vector infectivity was 0.63-4.4x104 transducting units (TU)/ng p24. The lentiviral vector batches had low endotoxin content, were sterile and free of replication-competent lentiviral vectors. The 3 batches differed for the cFIX transgene: they were either wildtype, co-cFIX, or co-cFIXR338L. Each lentiviral vector batch was infused into one male dog with hemophilia B by portal vein administration (Table 1).
Table 1. Gene therapy dose-response in treated dogs with hemophilia B.
| DOG | M57 (Hemil) | O21 (Valentine) | O59 (Enzo) |
|---|---|---|---|
| Age at treatment | 8 months | 21 months | 21 months |
| Weight at treatment | 20 kg | 22 kg | 18 kg |
| Transgene | wt cFIX | co-cFIX | co-cFIXR338L |
| TU/kg | 5.7x108 | 2.3x109 | 1.1x109 |
| μg p24/kg | 44 | 47 | 174 |
| FU (days) | 1831 | 900 | 637 |
| WBCT (min) | 20.31 ± 0.91 range: 14.5-32 |
17.36 ± 0.66 range: 13.5-22.5 |
15.73 ± 0.5 range: 11-19.5 |
| cFIX activity (% normal) | 0.08 ± 0.01 range: 0.01-0.25 |
1.05 ± 0.12 range: 0.3-1.7 |
1.18 ± 0.08 range: 0.7-1.9 |
| cFIX antigen (% normal) | 0.05 ± 0.004 range: 0.01-0.09 |
0.6 ± 0.06 range: 0.2-0.85 |
0.16 ± 0.005 range: 0.14-0.2 |
The table shows the age and weight at treatment of three dogs with hemophilia B (M57, O21, O59), the infused dose of lentiviral vector (transducing units, TU and physical particles) per weight, the follow up (FU) time in days, the Whole Blood Clotting Time (WBCT) in minutes (min), and the canine factor IX (cFIX) activity (determined by activated partial thromboplastin time, aPTT). Also shown are the cFIX antigen (determined by ELISA), the type of transgene contained in the infused lentiviral vector: wildtype (wt), codon-usage optimized, or codon-usage optimized hyperfunctional cFIX cDNAs (cFIX, co-cFIX, co-cFIXR338L; see also Fig. 1). When possible, results are presented as mean±standard error of the mean of values over time (range of determined values is also shown). The values of WBCT, cFIX activity and antigen are considered valid only if measured 27 days after the last canine plasma transfusion (to ensure washout of exogenous cFIX).
The lentiviral vector infusion was well tolerated by the first dog (M57) except for a transient rise in body temperature (1° C above baseline). The second dog (O21) experienced acute hypotension during the infusion, attributed to an anaphylactoid reaction to an unknown component of the vector batch. This event was successfully managed by immediate administration of an antihistamine drug (Benadryl 1 mg/kg, i.v.) and corticosteroid (Dexamethasone 25 mg/kg i.v.). Lentiviral vector infusion was subsequently completed upon blood pressure recovery. We observed a transient rise in body temperature. Based on these events, the third dog (O59) was pre-treated with corticosteroid (Prednisone, 1 mg/kg oral) and anti-histamine drugs (Benadryl 1 mg/kg i.m., Famotidine 0.5 mg/kg i.m.) the day before, the morning before surgery, and just prior to vector infusion (Dexamethasone 0.2 mg/kg i.v., Benadryl and Famotidine as above). Using this regimen, there was no change in blood pressure, the infusion was uneventful and body temperature did not increase.
In M57 and O21, serum concentrations of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) increased slightly above the normal range for the first few days post-infusion, indicating minor, self-limiting hepatocellular toxicity (Fig. 1B,C). In the third dog, both ALT and AST remained in the normal range throughout the follow-up, suggesting that anti-histamine and anti-inflammatory treatment prevented acute hepatotoxicity due to lentiviral vector infusion. In all three dogs, platelet counts fell slightly below the normal range for 2-3 days after lentiviral vector administration (Fig. 1D). This drop may be due to consumption at sites of surgical bleeding and to the large amount of fluid infused (vector vehicle plus normal canine plasma administered as a source of factor IX on the day of surgery and the following day), as also suggested by the concurrent transient drop in hematocrit (fig. S2A). Plasma concentrations of fibrinogen and thrombin/anti-thrombin complex (TAT) increased in the first few days after lentiviral vector administration, with the least evident changes in O59 (fig. S2B,C). The fibrin degradation product D-dimer increased only slightly above pre-treatment values in the first 2 dogs, and did not change in the third dog (fig. S2D). These data were consistent with the induction of an inflammatory response and activation of the clotting system upon abdominal surgery and intraportal lentiviral vector administration. These responses were self-limiting and effectively prevented by a one-day anti-inflammatory pre-treatment in the third dog. Indeed, in all treated dogs, we found an increase in tumor necrosis factor-α (TNF-α), interleukin (IL)-6 and IL-8 serum concentrations after lentiviral vector administration, which declined rapidly over the following hours or days (Fig. 1E-G). These increases were less pronounced in the dogs receiving anti-histamine and anti-inflammatory treatment. Other cytokines tested (IL-2, IL-4, IL-7, IL-10, IL-15, transforming growth factor-β, interferon-γ) were found not to be significantly different from control untreated animals (n=11). All of the other blood chemistry parameters tested and cell counts were in the normal range throughout the follow-up, with minor sporadic fluctuations (tables S2-4)
We found very low concentrations of p24 in the serum at the first sampling, which was three hours after lentiviral vector administration (approximately 0.26% of the infused dose), indicating a rapid clearance of vector particles from the circulation (fig. S2E, F). We did not find detectable p24 in oral, nasal, lachrymal, genital and rectal swabs taken from day 1 to 8 after lentiviral vector administration, except for a borderline signal in the nasal secretion of O21 at day 1 (table S5). Overall, these data suggest that administration of lentiviral vectors to dogs by intraportal delivery is well tolerated provided that anti-inflammatory and anti-histamine treatment is given before infusion.
Lentiviral vector gene therapy provides stable improvement in clotting time and clinical benefit in dogs with hemophilia B
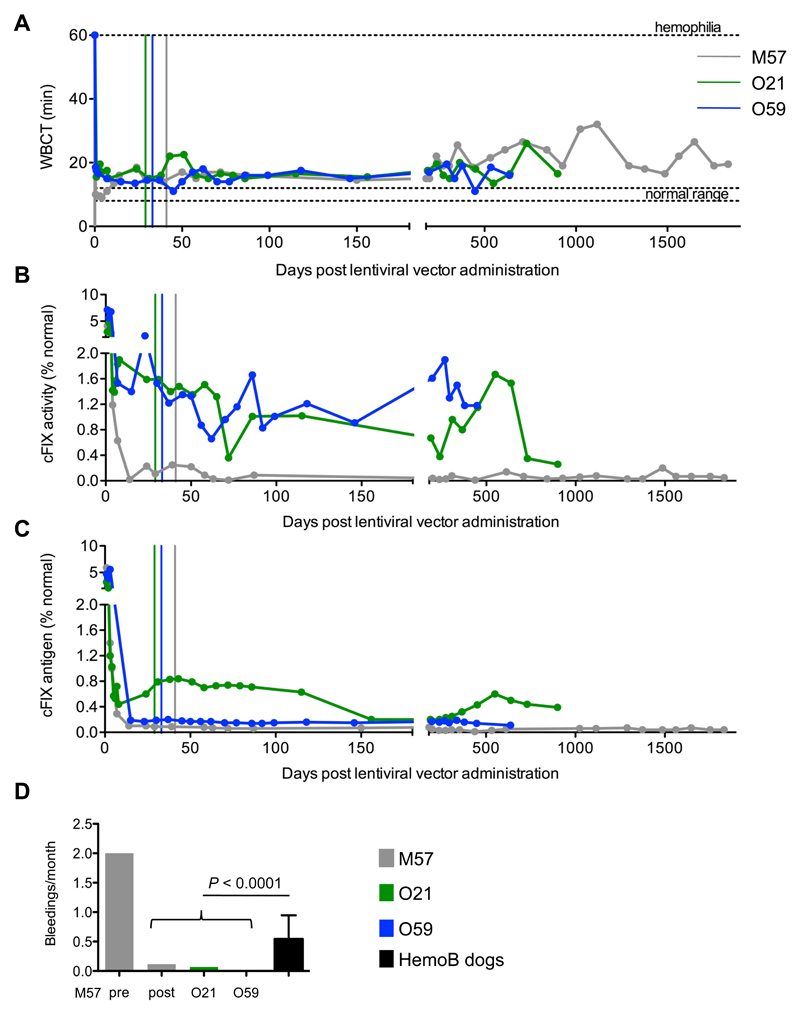
We measured the whole blood clotting time (WBCT), cFIX activity (by activated partial thromboplastin time, aPTT) and cFIX antigen in blood or plasma samples collected from treated dogs at routine intervals after lentiviral vector administration (Fig. 2A-C). The three dogs were followed up and were alive and well at 5, 2.5 and 1.75 years after lentiviral vector infusion for M57, O21 and O59, respectively. The WBCT was shortened and remained stable, albeit without reaching normal levels. The average WBCT over the follow-up time was approximately 20, 17 and 15.7 minutes for M57, O21, O59, respectively (Table 1). In M57, cFIX activity and antigen levels averaged 0.08% and 0.05% of normal, respectively (Table 1). Although the reconstituted activity was low, this dog experienced only 7 spontaneous bleedings in the 5 years following gene therapy (out of 27 expected from the bleeding frequency in the colony), whereas it had experienced 6 bleedings in the 3 months before gene therapy (Fig. 2D). In dog O21, cFIX activity ranged between 0.3-1.7% and antigen levels were 0.2-0.9% of normal. This dog experienced only 2 bleedings over the last 2.5 years. Since this dog received a 4-fold higher lentiviral vector dose and showed approximately 10-14 fold higher amounts of cFIX antigen and activity compared to dog M57, transgene codon-usage optimization may have increased cFIX expression by 3-fold as compared with the wildtype transgene, in line with our data obtained in mice (14). In dog O59, cFIX activity ranged between 0.6-1.9% but cFIX antigen levels was only 0.1-0.2% of normal (Table 1). Whereas the low antigen levels reflect the lower lentiviral vector dose administered compared to O21, the measured activity likely reflects the 5-10 fold increased activity conferred by the R338L mutation (14). This dog has not experienced any spontaneous bleeding to date. There is a marked difference in the monthly bleeding frequency between treated and untreated dogs from the same colony (Fig. 2D, P < 0.0001; table S6). Thromboelastogram values were within or close to the normal range in all treated dogs, with a shorter time to clot in O21 and O59 dogs (table S7). Anti-factor IX inhibitory antibodies tested negative in all treated dogs (table S8). A liver biopsy was taken from M57 and O21 at 16 and 12 months after lentiviral vector administration, respectively, and was scored normal by pathology (fig. S3). We found approximately 0.9 and 2.4 lentiviral vector DNA copies for every 1,000 diploid genomes, respectively (fig. S4). This finding confirms the presence of lentiviral vector in the dog liver in amounts consistent with the measured factor IX output. We did not find detectable lentiviral vector DNA in blood and sperm samples obtained from the treated dogs (fig. S4). These data show the long-term persistence of lentiviral vector-transduced cells in canine liver, stable reconstitution of factor IX activity up to 1% of normal and amelioration of the clinical phenotype in 3 treated dogs affected by hemophilia B.
Figure 2. Lentiviral vector-mediated gene therapy targeted to liver provides stable improvement in clotting time in dogs with hemophilia B.
Whole blood clotting time, WBCT (A) measured in blood samples, canine factor IX activity (cFIX) (B) and cFIX antigen (C) measured by activated partial thromboplastin time, aPTT (B) or ELISA (C) in plasma samples collected at the indicated times after lentiviral vector administration from dogs M57 (grey line), O21 (green line), O59 (blue line) 0. The colored vertical lines indicate 27 days after the last normal plasma transfusion of the dogs at which time exogenous canine factor IX had been washed-out. (D) Frequency of spontaneous bleedings (bleeding events/month of observation) in the treated dogs after gene therapy. For M57, the frequency of spontaneous bleeding before gene therapy is shown. The mean ± SD bleeding frequency of 10 untreated dogs with hemophilia B in the colony is shown (black bar) (46). P < 0.0001 (2-sample test for equality of proportions; see also table S6).
Treated mice did not show evidence of genotoxicity after lentiviral vector integration in liver
The normal blood chemistry and the stability of factor IX expression in the long-term follow up of the treated dogs suggests a low risk for the development of neoplasia from transduced hepatocytes. To better investigate the risk of oncogenesis, we turned to mice and analyzed the safety of lentiviral integration into the liver in multiple settings of escalating stringency. In our prior studies of lentiviral vector gene therapy in mice with hemophilia B, we did not observe macroscopic liver lesions at necropsy in the treated mice (12, 14, 15). Here, we analyzed the integration site distribution in the liver of treated mice and scored for the potential enrichment of integration sites at specific genomic loci over time. Such analyses could reveal a selective growth advantage conferred on hepatocytes by lentiviral vector integration close to cancer genes before the development of overt neoplasia.
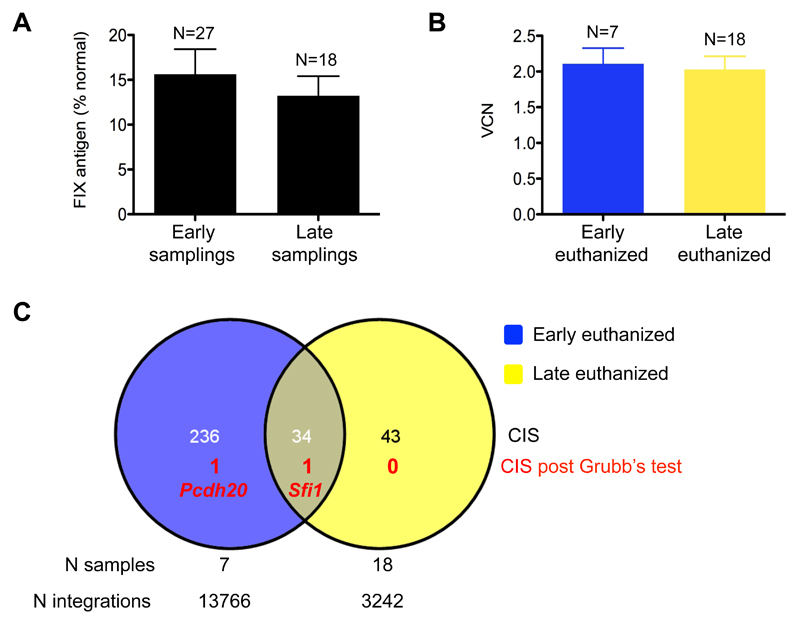
Twenty-seven adult mice with hemophilia B were administered 2.5-5x1010 transducing units/kg therapeutic lentiviral vector (which carries Self-Inactivating long terminal repeats and an internal ET promoter, SIN.ET, expressing human or canine wildtype factor IX, see also Fig. 1A) in 5 different experiments (table S9). As expected, plasma factor IX was at 10-15% of normal levels and did not significantly change between sampling performed “early” (<3 months) or “late” (6-12 months) after lentiviral vector administration (Fig. 3A) (12, 14). Nine mice were euthanized “early” and 18 were euthanized “late” to measure lentiviral vector content and integration site distribution in the liver. There was no significant difference in the average vector copies per diploid genome (Vector Copy Number, VCN) between the early and late time points (Fig. 3B). We then deep sequenced integration sites by linear-amplification mediated-PCR followed by 454 pyrosequencing or Illumina sequencing and mapped a total of 17,008 unique integration sites onto the murine genome (table S9-10). We applied standard criteria to identify genomic regions recurrently hit by lentiviral vector integrations with a frequency significantly higher than random, and defined them as Common Insertion Sites (CIS) (25). We identified 270 and 77 CIS in the datasets of integration sites retrieved from early or late euthanized mice, respectively, each CIS being identified by the most targeted gene within the genomic region (table S11). The higher number of CIS from the “early” integration site dataset is likely to be the consequence of the higher number of integration sites retrieved. Nearly half of the “late” CIS were also found among the “early” dataset (Fig. 3C), suggesting that they are unlikely to be the result of in vivo selection of clones carrying integration at those genes. In contrast, lentiviral vector CIS often result from intrinsic biases of viral integration that preferentially target gene-dense genomic regions (26). We thus used the genome-wide Grubb’s test for outliers to exclude all the CIS occurring within a larger genomic cluster of lentiviral vector integrations, likely representing integration biases, as previously described (19, 20). None of the “late” CIS passed the Grubb’s test, except for the Sfi1 gene, which was also found among the “early” CIS. Overall, this analysis did not find evidence of in vivo selection of mouse hepatocytes carrying SIN.ET integrations.
Fig. 3. No evidence of genotoxicity after lentiviral vector integration into liver of mice.
(A) Factor IX antigen measured by ELISA in the plasma collected from mice “early” (<3 months) or “later” (6-12 months) after lentiviral vector administration. P = 0.391, Student’s t test. (B) Vector copy number (VCN) in liver DNA collected from mice euthanized early or later after lentiviral vector administration. P = 0.806, Student’s t test. (A, B) Data are mean±standard error of the mean (SEM). (C) Venn diagram representing Common Insertion Sites (CIS) identified in liver DNA of mice euthanized early or later after lentiviral vector administration. The overlap is calculated considering the gene associated with each CIS; the number of CIS that passed the Grubb’s test is shown along with the gene name. The number of samples analyzed and integration sites retrieved are indicated for the two data sets.
No evidence of genotoxicity after SIN lentiviral vector administration to tumor-prone mice
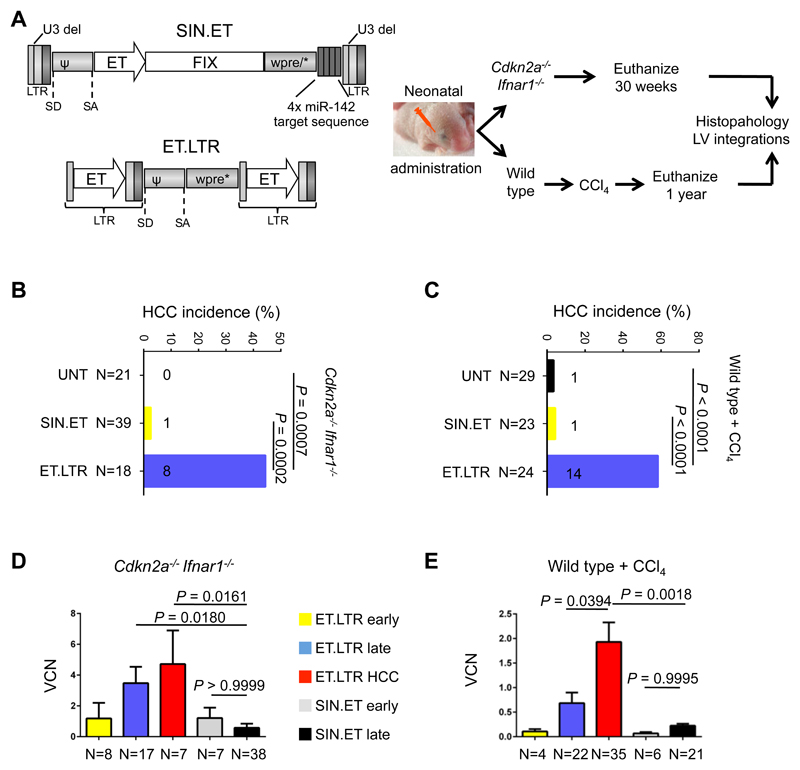
Mice can only be administered a limited dose of vector and followed-up for a short time given their short lifespan compared to humans. For this reason, insertions leading to gain or loss of function in cancer genes that have a delayed effect on hepatocyte proliferation and/or selection may have escaped detection in the previous analysis (Figure 3). In order to increase the sensitivity of our analysis, we administered the SIN.ET lentiviral vector to newborn tumor-prone mice or wildtype mice given a tumor-promoting regimen (27). We used a previously described genotoxic lentiviral vector carrying transcriptionally active Long Terminal Repeats containing the ET promoter as a positive control for genotoxicity (ET.LTR lentiviral vector; Fig. 4A). Matched doses of ET.LTR or SIN.ET lentiviral vector (2.5x1010 transducing units/kg) were administered to newborn Cdkn2a-/-Ifnar1-/- mice or newborn wildtype mice that were then given a regimen of CCl4; 125 new experimental mice and 29 historical controls (27) were analyzed (table S12). Cdkn2a-/-Ifnar1-/- mice were euthanized if they displayed signs of illness or at 30 weeks of age. Wildtype mice given the CCl4 regimen were euthanized at 1 year of age. By visual inspection at necropsy and histopathological analysis of serial liver sections from multiple lobes, we detected two microscopic hepatocellular carcinomas (HCC) in 62 tumor-prone or CCl4-treated wildtype mice administered SIN.ET, an incidence overlapping with that observed in nontransduced mice. On the contrary, ET.LTR induced a significantly higher incidence of HCC (P = 0.0007 for Cdkn2a-/-Ifnar1-/-, P < 0.0001 for CCl4-treated wildtype mice), most of which were visible at necropsy (Fig. 4B-C and fig. S5A-D). We measured the vector copy number in non-tumoral mouse liver and in the HCCs collected from the experimental groups at the end of the experiment, and from 4 cohorts of mice (Cdkn2a-/-Ifnar1-/- or wildtype) euthanized 2 weeks after neonatal administration of SIN.ET or ET.LTR lentiviral vectors. The SIN.ET and ET.LTR vector copy number were comparable in mice euthanized at 2 weeks of age, indicating comparable in vivo transduction. However, the ET.LTR vector copy number was higher in livers harvested at the end of the experiment and showed an even greater increase in HCCs, indicating expansion of transduced hepatocytes (P = 0.0394, Fig. 4D, E). This increased vector copy number over time was not observed in SIN.ET-treated mice. Moreover, we did not detect any significant change in circulating factor IX over time in either Cdkn2a-/-Ifnar1-/- mice or wildtype mice transduced with SIN.ET, confirming the stability of gene transfer after administration to neonatal mice (fig. S5E, F). These data suggest that there was not transformation or expansion of SIN.ET-transduced hepatocytes even in sensitive mouse models administered lentiviral vectors as neonates.
Figure 4. SIN.ET lentiviral vectors do not induce hepatocellular carcinoma in tumor-prone mice.
(A) Experimental outline of the in vivo biosafety study in mice. (Left) Schematic representations of the lentiviral vectors used U3 del: deletion of the promoter/enhancer of the HIV Long Terminal Repeats (LTR) (43). SD: splicing donor site. SA: splicing acceptor site. ψ: packaging signal. Wpre*: mutated woodchuck hepatitis virus post-transcriptional regulatory element (44). 142T: miR-142 target sequence made of 4 tandem copies of a sequence perfectly complementary to miR-142. Hepatocyte-specific Enhanced Transthyretin (ET) promoter composed of synthetic hepatocyte-specific enhancers and transthyretin promoter (45). Either SIN.ET (gene therapy lentiviral vector with Self Inactivating Long Terminal Repeats and an internal Enhanced Transthyretin promoter) or ET.LTR (oncogenic lentiviral vector with transcriptionally active Long Terminal Repeats containing the Enhanced Transthyretin promoter) were administered at matched doses to newborn Cdkn2a-/-Ifnar1-/- tumor-prone mice or wildtype mice resulting in four experimental groups. Wildtype mice then were given a CCl4-based tumor promoting regimen. Mice were euthanized at the indicated time points or earlier if sick. Necropsy was performed and samples were collected for DNA extraction (for determination of vector copy number and the retrieval of integration sites ) and for histopathology analysis. (B, C) Shown is the incidence of hepatocellular carcinoma (HCC) in Cdkn2a-/-Ifnar1-/- mice (B) or wildtype mice (C) transduced with the two different lentiviral vectors (SIN.ET or ET.LTR) or untransduced (UNT). Untransduced mice include historical controls (n=20 Cdkn2a-/-Ifnar1-/- and n=9 wildtype mice) (27). P values were calculated by two-tailed Fisher’s exact test. Numbers on the histograms indicate the number of mice that developed HCC. (D, E) Vector copy number in liver DNA from Cdkn2a-/-Ifnar1-/- mice (D) or wildtype mice (E) collected two weeks after lentiviral vector administration (early) or at necropsy (late). Data are mean±SEM. P values were calculated by One-Way ANOVA and Bonferroni’s multiple correction test. All vector copy numbers were measured in non-tumoral liver tissue except for ET.LTR-induced HCCs.
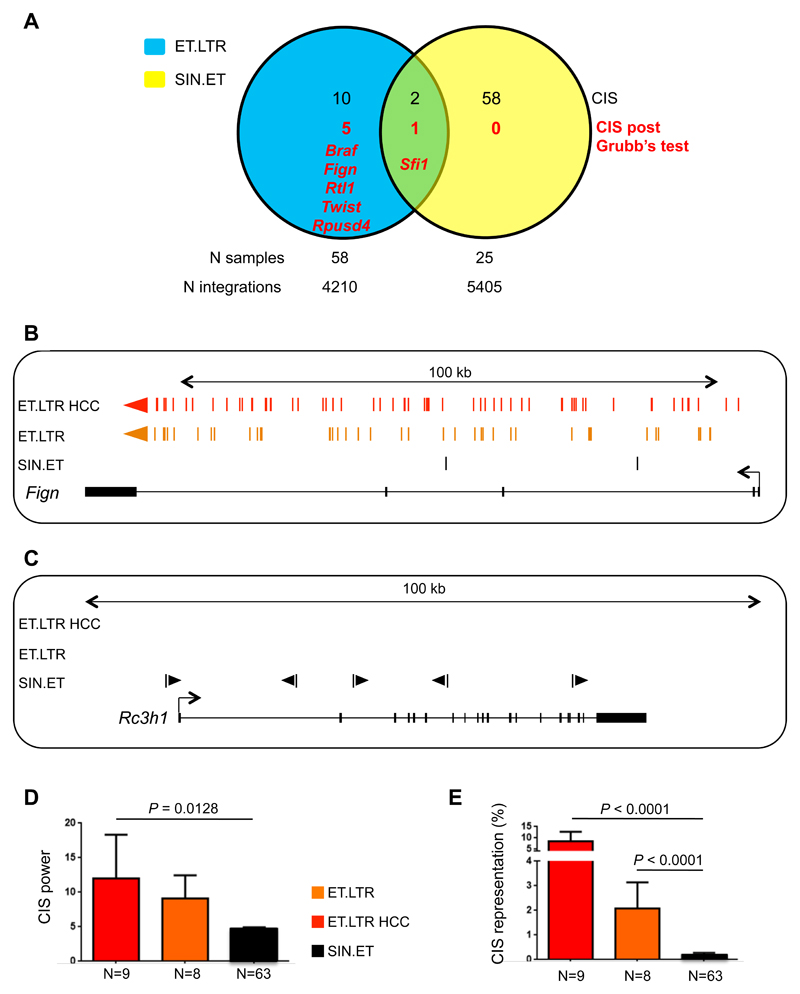
We then retrieved lentiviral vector integrations from 83 liver samples (by linear-amplification mediated-PCR and 454 pyrosequencing) and identified 9,615 unique integration sites (table S13). We identified 60 and 12 CIS in the datasets of integration sites from SIN.ET- and ET.LTR-treated mice, respectively (table S13). There was almost no overlap between the SIN.ET and ET.LTR CIS (Fig. 5A), and the latter CIS were also different from the CIS identified in SIN.ET-transduced mice with hemophilia reported in Figure 3C (fig. S6A). Moreover, when we applied the Grubb’s test, the only significant CIS of SIN.ET was Sfi1, already found in the integration sites of mice with hemophilia euthanized early and late after lentiviral vector administration, and thus most likely due to an intrinsic lentiviral vector integration bias (table S14). Conversely, 5 of the ET.LTR CIS passed the Grubb’s test for outliers (table S14), among which were found the genes Braf, Rtl1 and Fign and three previously validated liver oncogenes (27). ET.LTR integration sites within these CIS were clustered in narrow regions and almost always in the same orientation of transcription as the targeted gene, consistent with a previously described mechanism of insertional mutagenesis. This mechanism involves transcription from the inserted active LTR and splicing into the oncogene, leading to up-regulation of expression of a truncated or full-length oncogenic transcript and in vivo expansion of cells harboring that integration site (Fig. 5B, fig. S6B, C) (27). These data confirm the genotoxic features of the positive control lentiviral vector and the sensitivity of the mice to insertional oncogenesis. In contrast, integration sites for SIN.ET CIS had a lower integration density without orientation bias (Fig 5C, fig. S6D). Additionally, we found that the power and density of SIN.ET CIS, two measures of the extent of enrichment over random occurrence, were significantly lower (P = 0.0128, P < 0.0001 respectively) as compared to those of ET.LTR (Fig. 5D and figure S6E). We also compared the relative sequence counts of all the integration sites in a CIS as a surrogate readout of the relative abundance of cell clones harboring integration at that CIS within sampled liver tissues of that experimental group (Fig. 5E). The relative sequence counts of SIN.ET CIS were significantly lower than those of ET.LTR CIS retrieved from non-tumoral or tumoral samples (Fig. 5E, P < 0.0001). These data further indicate that SIN.ET CIS were not the result of in vivo clonal selection or expansion. Accordingly, the analysis of molecular pathways enriched in the CIS datasets showed that ET.LTR-targeted genes frequently act in pathways associated with cancer and transformation, whereas SIN.ET-targeted genes did not (fig. S6F). Overall, we did not detect evidence of SIN.ET-induced genotoxicity, even after investigation of early molecular readouts of transformation in highly permissive tumor-prone mice or mice in which tumors were chemically promoted.
Fig. 5. Integration sites analysis does not reveal genotoxicity of SIN.ET lentiviral vectors in tumor-prone mice.
(A) Venn diagram representing Common Insertion Sites (CIS) identified in liver DNA of SIN.ET-transduced and ET.LTR-transduced mice. The overlap is calculated considering the gene associated with each CIS. The number of CIS that passed the Grubb’s test is shown with the gene name (red). The number of samples analyzed and the total number of integration sites are indicated for the two data sets. (B, C) Schematic drawing of two representative CIS of ET.LTR (B) and SIN.ET (C) lentiviral vectors. Each colored bar represents an integration site (red: from ET.LTR-induced HCCs; orange: from non-tumoral liver of mice transduced with ET.LTR; black: from liver of mice transduced with SIN.ET). Colored arrows indicate the orientation of the integration site. The gene within the region is represented below, with black boxes indicating exons and arrows indicating the transcription orientation. The span of the outlined genomic region is indicated on top. (D) Common insertion sites (CIS) power, calculated as the number of different integration sites targeting each CIS. (E) CIS representation, calculated as % of sequencing reads from all integration sites comprised within a CIS over the total number of reads within an experimental data set. (D, E) Data are mean±SEM. P values were calculated by One-Way ANOVA and Bonferroni’s multiple correction test. For all the analyses, integration sites from the two mouse models were merged.
Discussion
Here, we show that lentiviral vector-mediated gene therapy in the liver of dogs with hemophilia B is feasible and provides stable long-term factor IX activity up to 1% of normal activity with therapeutic benefit. The treatment was associated with manageable self-limiting acute toxicity without any detectable long-term toxicity or development of anti-transgene immune responses.
We produced 3 large-scale batches of lentiviral vectors for in vivo administration and analyzed them using a panel of tests for identity, potency and purity, according to methods and specifications previously used for clinical trials (20). Upon portal vein administration of these lentiviral vectors carrying a factor IX transgene to dogs with hemophilia B, we observed mild transient fever and hepatocellular toxicity accompanied by a transient rise in circulating TNF-α, IL-6, IL-8, fibrinogen and thrombin/anti-thrombin complex. This acute inflammatory response may be a consequence of abdominal surgery and type-I interferon release by hepatosplenic plasmacytoid dendritic cells, triggered by nucleic acids contaminating or associated with the infused vector particles, as has been observed in previous mouse studies (28). This response was alleviated in one dog pretreated with anti-inflammatory and anti-histamine drugs one day before lentiviral vector infusion, a mild regimen that could also be translated to the clinical setting. In one dog, we observed a hypotensive anaphylactoid reaction to an unknown component during lentiviral vector infusion. Given that dogs with hemophilia B receive frequent plasma transfusions in response to spontaneous bleedings, they may be pre-sensitized to developing allergic reactions.
Our prior studies in mice with hemophilia B indicated that an optimal lentiviral vector dose to achieve >5% of normal factor IX activity is 2.5x1010 transducing units/kg. According to manufacturing capacity at the beginning of this study and precautions dictated by the testing of lentiviral vectors in large animals, we administered a 45-fold lower dose, 5.7x108 transducing units/kg, to the first dog (M57). Given that canine factor IX activity approached 0.1% of normal, this outcome suggests that predictions based on the dose-response in the mouse are reliable in the canine model. Through improvements in large-scale vector production and transgene codon-usage optimization, we could administer a 4-fold higher dose to a second dog (O21). This dog achieved about 1% of normal canine factor IX activity, which was more than 10-fold higher than that for the first dog. This can be explained by the 2-3 fold gain in potency of the codon-optimized canine factor IX as had previously been observed in mice (14). By exploiting the Padua mutation that confers 7-fold higher activity on activated factor IX (14, 23), we could reconstitute 1% of normal canine factor IX activity in a third dog, even though it was administered a 2-fold lower dose compared to dog O21. Overall, these results show that by delivering a codon-optimized and hyper-functional factor IX transgene one can reduce the effective lentiviral vector dose by more than 1 log. Importantly, all dogs experienced a long-lasting therapeutic benefit from the single treatment, as shown by a marked decrease in spontaneous bleedings recorded throughout the follow-up period of several years. Moreover, the treatment did not induce anti-factor IX inhibitory antibodies consistent with the observed stable long-term factor IX activity in plasma. However, it should be noted that the dogs treated in this study, although totally devoid of circulating factor IX (29), do bear a missense mutation in their F9 gene and thus are not prone to developing anti-factor IX antibodies.
There are a number of limitations to our study. The main limitation is the small number of dogs treated, each treated with a different transgene and vector dose. In addition relatively low doses of lentiviral vector were administered, giving rise to transgene expression at the threshold of therapeutic activity. Thus, further dose-escalating studies in dogs as well as in non-human primates are warranted to investigate the potential occurrence of dose-limiting acute toxicity and the lentiviral vector stability in blood and biodistribution in multiple tissues. These further studies will be crucial to establish the safety of in vivo administration of lentiviral vectors to large animal models and to establish that there is no immune response to the transgene.
If we extrapolate the observed dose-response in dogs to humans, the current lentiviral vector manufacturing capacity (20) could support the treatment of adult patients with hemophilia, provided that a hyper-functional factor IX transgene is used. The manufacturing of lentiviral vectors for clinical use should become less onerous through further improvements in procedures and scale-up and the availability of stable packaging cell lines.
The occurrence of vector-induced oncogenesis in hematopoietic stem cell gene therapy trials based on gamma-retroviral vectors necessitates stringent preclinical assessment of potential vector genotoxicity (30–33). This caution also applies to liver gene therapy as hepatocytes are susceptible to insertional mutagenesis (27, 34–37). Accumulating evidence from non-clinical studies and recent clinical trials supports the view that lentiviral vectors entail a lower genotoxic risk compared to gamma-retroviral vectors (19, 21, 22, 38). Given that most studies of vector-induced genotoxicity have focused on hematopoietic stem and progenitor cells, we undertook a preclinical analysis of lentiviral vector-induced genotoxicity in the mouse liver. We did not find evidence of genotoxicity of our therapeutic lentiviral vectors in mice with hemophilia B and in two ad hoc mouse models with enhanced sensitivity to hepatocellular carcinogenesis (>100 mice and >9,000 integration sites analyzed).
The distribution of lentiviral vector integrations in liver DNA of mice with hemophilia showed that deviations from random were already evident early after gene therapy, thus most likely representing intrinsic integration biases of the vector and not the outcome of in vivo selection. This contention was supported by the substantial overlap between the CIS identified in “early” and “late” liver harvests and the lack of new CIS that passed the Grubb’s test for outliers in the “late” dataset. Note that our sampling and depth of analysis were not designed to attain saturation of CIS, thus we did not expect full overlap between the early and late CIS.
The low genotoxicity of the SIN.ET lentiviral vector design was further demonstrated by administration to tumor-prone mice where this vector did not increase the spontaneous occurrence of HCC. This was at variance with an aptly designed genotoxic lentiviral vector serving as a positive control. Lentiviral vectors were administered to newborn mice, thus increasing the likelihood that proliferating hepatocytes would accumulate additional mutations complementing an eventual oncogenic lentiviral insertion resulting in the induction of hepatocyte transformation. The observation that SIN.ET CIS have lower integration power, integration density and representation in the whole dataset compared to ET.LTR CIS indicates that SIN.ET integration sites are not subject to the same process of in vivo selection as ET.LTR integration sites. We found 3 previously validated liver oncogenes (Braf, Fign and Rtl1) as significant CIS after Grubb’s test in the ET.LTR dataset. Moreover, for some oncogenes such as Fign, we could observe clustering of ET.LTR integration sites in a significant CIS even in non-tumoral tissues (see Fig. 5B), indicating that our experimental design could detect the occurrence of clonal selection due to an oncogenic vector even before overt neoplasia. Therefore, it is unlikely that the lack of evidence for in vivo selection of SIN.ET insertions was due to a limited sensitivity of our assay. Rather, our studies did not uncover any biological or molecular evidence of SIN.ET-induced genotoxicity even in the “worst-case scenario” of transduction of newborn tumor-prone mice.
In conclusion, our study positions liver-directed lentiviral gene therapy for further pre-clinical development. Lentiviral vectors may complement other available vectors to address some of the outstanding challenges posed by liver gene therapy for the treatment of hemophilia and conceivably other diseases.
Materials and methods
Study design
Dog studies were necessarily limited in sample size for feasibility and ethical reasons. Sample size of mouse studies was chosen according to the previously observed tumor incidence in the positive control group (27). No sample or animal administered the intended dose was excluded from the analysis. Mice were randomly attributed to each experimental group. Investigators involved in histopathology analysis and initial integration sites mapping were blinded. Investigators performing mouse handling, sampling, euthanasia and raw data analysis were not blinded.
Large-scale lentiviral vector production
Large-scale lentiviral vector production for dog studies was outsourced to MolMed S.p.A. or Genethon. The vector batches were produced by using a large-scale validated process (19, 20) and following pre-GMP guidelines. Briefly, lentiviral vectors are produced by transient 4 plasmid transfection of 293T cells in 10-tray cell factories by calcium phosphate precipitation. Twenty-four hours after removal of the transfection medium, the cell supernatant is harvested and stored at 4°C. The culture medium is replaced and after further 24 hours a second harvest is performed. The medium collected from the two harvests is pooled and filtered through 5/0.45 μm filters to discard cell debris. The downstream purification process includes a benzonase treatment overnight at 4°C, followed by a DiEthylAmino Anion Exchange (DEAE) chromatography step, concentration and gel filtration in PBS or PBS 5% dimethyl sulfoxide (DMSO). The resulting lentiviral vector preparation undergoes 0.2 or 0.45 μm filtration and aseptic filling. The purified vector preparation is stored at –80°C.
Dog experiments
Hemophilia B dogs (carrying a E379G single amino-acid substitution in the factor IX protein) were maintained at the Francis Owen Blood Research Laboratory, which provides for breeding, whelping, housing, treating and performing the experiments in the dogs on site. Complete blood counts and platelet counts were performed on EDTA-anti coagulated blood with a cell counter (Heska) calibrated for canine cells. Serum liver enzymes were performed by Antech Diagnostics, a commercial veterinarian diagnostic laboratory.
Mouse experiments
Founder C57BL/6 F9 KO mice were kindly obtained from the laboratory of Dr. Inder Verma at the Salk Institute (39). Cdkn2a-/-Ifnar1-/- mice were generated to couple the sensitivity to genotoxic mutations conferred by the Cdkn2a deficiency (21, 22) to the increased permissiveness to liver gene transfer by lentiviral vectors conferred by the Ifnar1 deficiency (28). Additionally, this non-inflammatory tumor-prone mouse model has a clinical relevance, since CDKN2A and its targets – pRB and p53 - are frequently inactivated or silenced in HCC (40). Cdkn2a-/- (C57BL6/J) mice were obtained from NCI-Frederick MMHCC Repository, while Ifnar1-/- (129SVEV) mice were obtained from B&K Universal Limited. Wildtype C57BL/6 mice were purchased by Charles River. Eight-week old wildtype mice transduced or not with ET.LTR at neonatal stage, were administered CCl4 1 mg/kg twice weekly for 6 weeks in a 10% mineral oil solution (Sigma). CCl4 administration results in waves of hepatocytes necrosis and regeneration that cause liver damage (41). Both mouse models were previously described (27).
All the mice were maintained in specific-pathogen free conditions. Vector administration was carried out by tail-vein injections in adult (7-10 weeks old) mice and by temporal vein injection in newborns (1-2 days old). Mice were bled from the retro-orbital plexus using capillary tubes and blood was collected into 0.38% sodium citrate buffer, pH 7.4. Mice were anesthetized with tribromoethanol (Avertin) and euthanized by CO2 inhalation. All animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee. At necropsy we collected masses in the liver parenchyma as well as non-tumoral liver for microscopic and molecular analyses.
Statistical analysis
Statistical analyses were performed upon consulting with professional statisticians. When data were %, Log Odds were calculated to perform tests assuming normal distribution (42). Standard statistical tests were performed using Student’s t test (2 experimental groups) or ANOVA with Bonferroni multiple comparison’s post-test (>2 experimental groups) at α=0.05 level of confidence.
Supplementary Material
Overline: Gene Therapy.
Short One Sentence Summary
Lentiviral vector-mediated gene therapy targeted to hepatocytes provides stable reconstitution of clotting activity in dogs with hemophilia B.
Long One Sentence Summary
Gene therapy with lentiviral vectors targeting transgene expression to hepatocytes provides stable reconstitution of clotting activity in dogs with hemophilia B and does not show genotoxicity in tumor-prone mice.
Accessible Summary.
Advances in gene therapy for hemophilia
Hemophilia is an inherited bleeding disorder caused by deficiency in a blood clotting factor. The current treatment requires lifelong intravenous administration of the missing clotting factor every few days, a costly and demanding regimen for the patients. Gene therapy can provide a single-shot treatment option by replacing a functional gene in the liver cells that naturally produce the factor. Here we report a study of the efficacy and safety of liver-directed in vivo gene therapy in large and small animal models using lentiviral vectors. The strategy described here promises to represent a new gene therapy option, which may complement other available vectors for hemophilia and conceivably other diseases.
Acknowledgements
The authors thank S. Bartolaccini, M. Milani and S. Annunziato for help with some experiments; G. Paolo Rizzardi, Claudia Benati, Giuseppina Marano, Francesca Rossetti, Giuliana Vallanti and Claudio Bordignon (MolMed S.p.A), Sandrine Genries-Ferrand, Laetitia Duranton and Fulvio Mavilio (Genethon) for large-scale lentiviral vector production; Stefania Delai, Ilaria Visigalli and Alessandra Biffi for setting-up lentiviral vector PCR on the canine genome; B.D. Brown, A. Annoni and all members of the Naldini and Montini laboratories for helpful discussions.
Funding: This work was supported by Telethon (TIGET D3 to L.N. and Eu.M.), European Union 7th Framework Programme (grant agreement 222878, PERSIST to L.N., T.V. and M.S.), National Institutes of Health (NIH grant HL063098 to T.N.) and ERC advanced grant (TARGETINGGENETHERAPY to L.N.).
Footnotes
Author contributions: A.C. and M.R. designed and performed experiments, analyzed data and wrote the paper. C.B. performed integration site analysis and analyzed data. M.V. performed linear-amplification mediated-PCR. P.D.V. performed coagulation assays. F.S. performed histopathology analysis. L.S.S., P.G., Fa.B. provided technical assistance. D.B., R.R., El.M. performed dog experiments. C.D. supervised and performed histopathology analysis. Fr.B. and S.M. supervised large-scale lentiviral vector production. A.DA. supervised coagulation assays. T.V. and M.K.C. designed, validated and provided reagents and contributed intellectual input. M.S. supervised integration site analysis and analyzed data. T.N. supervised dog experiments, analyzed data and revised the paper. Eu.M. supervised safety and integration site studies, analyzed data and revised the paper. L.N. coordinated the work, analyzed data and wrote the paper.
Competing interests: LN is an inventor on pending and issued patents on lentiviral vector technology and miR-regulated lentiviral vectors (Gene Vector, WO2007000668).
References
- 1.Bolton-Maggs PH, Pasi KJ. Haemophilias A and B. Lancet. 2003;361:1801–1809. doi: 10.1016/S0140-6736(03)13405-8. [DOI] [PubMed] [Google Scholar]
- 2.High KH, Nathwani A, Spencer T, Lillicrap D. Current status of haemophilia gene therapy. Haemophilia. 2014;20(Suppl 4):43–49. doi: 10.1111/hae.12411. [DOI] [PubMed] [Google Scholar]
- 3.Berntorp E, Shapiro AD. Modern haemophilia care. Lancet. 2012;379:1447–1456. doi: 10.1016/S0140-6736(11)61139-2. [DOI] [PubMed] [Google Scholar]
- 4.Astermark J, Santagostino E, Keith Hoots W. Clinical issues in inhibitors. Haemophilia. 2010;16(Suppl 5):54–60. doi: 10.1111/j.1365-2516.2010.02294.x. [DOI] [PubMed] [Google Scholar]
- 5.Franchini M, Mannucci PM. Past, present and future of hemophilia: a narrative review. Orphanet J Rare Dis. 2012;7:24. doi: 10.1186/1750-1172-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nichols TC, Dillow AM, Franck HW, Merricks EP, Raymer RA, Bellinger DA, Arruda VR, High KA. Protein replacement therapy and gene transfer in canine models of hemophilia A, hemophilia B, von willebrand disease, and factor VII deficiency. ILAR J. 2009;50:144–167. doi: 10.1093/ilar.50.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petrus I, Chuah M, VandenDriessche T. Gene therapy strategies for hemophilia: benefits versus risks. J Gene Med. 2010;12:797–809. doi: 10.1002/jgm.1500. [DOI] [PubMed] [Google Scholar]
- 8.Nathwani AC, Tuddenham EG, Rangarajan S, Rosales C, McIntosh J, Linch DC, Chowdary P, Riddell A, Pie AJ, Harrington C, O'Beirne J, et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N Engl J Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mingozzi F, High KA. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binny C, McIntosh J, Della Peruta M, Kymalainen H, Tuddenham EG, Buckley SM, Waddington SN, McVey JH, Spence Y, Morton CL, Thrasher AJ, et al. AAV-mediated gene transfer in the perinatal period results in expression of FVII at levels that protect against fatal spontaneous hemorrhage. Blood. 2012;119:957–966. doi: 10.1182/blood-2011-09-377630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 12.Brown BD, Cantore A, Annoni A, Sergi LS, Lombardo A, Della Valle P, D'Angelo A, Naldini L. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144–4152. doi: 10.1182/blood-2007-03-078493. [DOI] [PubMed] [Google Scholar]
- 13.Matrai J, Cantore A, Bartholomae CC, Annoni A, Wang W, Acosta-Sanchez A, Samara-Kuko E, De Waele L, Ma L, Genovese P, Damo M, et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantore A, Nair N, Della Valle P, Di Matteo M, Matrai J, Sanvito F, Brombin C, Di Serio C, D'Angelo A, Chuah M, Naldini L, et al. Hyperfunctional coagulation factor IX improves the efficacy of gene therapy in hemophilic mice. Blood. 2012;120:4517–4520. doi: 10.1182/blood-2012-05-432591. [DOI] [PubMed] [Google Scholar]
- 15.Annoni A, Cantore A, Della Valle P, Goudy K, Akbarpour M, Russo F, Bartolaccini S, D'Angelo A, Roncarolo MG, Naldini L. Liver gene therapy by lentiviral vectors reverses anti-factor IX pre-existing immunity in haemophilic mice. EMBO Mol Med. 2013;5:1684–1697. doi: 10.1002/emmm.201302857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt F, Remy S, Dariel A, Flageul M, Pichard V, Boni S, Usal C, Myara A, Laplanche S, Anegon I, Labrune P, et al. Lentiviral vectors that express UGT1A1 in liver and contain miR-142 target sequences normalize hyperbilirubinemia in Gunn rats. Gastroenterology. 2010;139:999–1007. doi: 10.1053/j.gastro.2010.05.008. 1007 e1001-1002. [DOI] [PubMed] [Google Scholar]
- 17.Matsui H, Hegadorn C, Ozelo M, Burnett E, Tuttle A, Labelle A, McCray PB, Jr, Naldini L, Brown B, Hough C, Lillicrap D. A microRNA-regulated and GP64-pseudotyped lentiviral vector mediates stable expression of FVIII in a murine model of Hemophilia A. Mol Ther. 2011;19:723–730. doi: 10.1038/mt.2010.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cartier N, Hacein-Bey-Abina S, Bartholomae CC, Veres G, Schmidt M, Kutschera I, Vidaud M, Abel U, Dal-Cortivo L, Caccavelli L, Mahlaoui N, et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 19.Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, Dionisio F, Calabria A, Giannelli S, Castiello MC, Bosticardo M, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, Baldoli C, Martino S, Calabria A, Canale S, Benedicenti F, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 21.Montini E, Cesana D, Schmidt M, Sanvito F, Ponzoni M, Bartholomae C, Sergi Sergi L, Benedicenti F, Ambrosi A, Di Serio C, Doglioni C, et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- 22.Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, Benedicenti F, Sergi LS, Ambrosi A, Ponzoni M, Doglioni C, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simioni P, Tormene D, Tognin G, Gavasso S, Bulato C, Iacobelli NP, Finn JD, Spiezia L, Radu C, Arruda VR. X-linked thrombophilia with a mutant factor IX (factor IX Padua) N Engl J Med. 2009;361:1671–1675. doi: 10.1056/NEJMoa0904377. [DOI] [PubMed] [Google Scholar]
- 24.DePolo NJ, Reed JD, Sheridan PL, Townsend K, Sauter SL, Jolly DJ, Dubensky TW., Jr VSV-G pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Mol Ther. 2000;2:218–222. doi: 10.1006/mthe.2000.0116. [DOI] [PubMed] [Google Scholar]
- 25.Abel U, Deichmann A, Bartholomae C, Schwarzwaelder K, Glimm H, Howe S, Thrasher A, Garrigue A, Hacein-Bey-Abina S, Cavazzana-Calvo M, Fischer A, et al. Real-time definition of non-randomness in the distribution of genomic events. PLoS One. 2007;2:e570. doi: 10.1371/journal.pone.0000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biffi A, Bartolomae CC, Cesana D, Cartier N, Aubourg P, Ranzani M, Cesani M, Benedicenti F, Plati T, Rubagotti E, Merella S, et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- 27.Ranzani M, Cesana D, Bartholomae CC, Sanvito F, Pala M, Benedicenti F, Gallina P, Sergi LS, Merella S, Bulfone A, Doglioni C, et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat Methods. 2013;10:155–161. doi: 10.1038/nmeth.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown BD, Sitia G, Annoni A, Hauben E, Sergi LS, Zingale A, Roncarolo MG, Guidotti LG, Naldini L. In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood. 2007;109:2797–2805. doi: 10.1182/blood-2006-10-049312. [DOI] [PubMed] [Google Scholar]
- 29.Herzog RW, Arruda VR, Fisher TH, Read MS, Nichols TC, High KA. Absence of circulating factor IX antigen in hemophilia B dogs of the UNC-Chapel Hill colony. Thromb Haemost. 2000;84:352–354. [PMC free article] [PubMed] [Google Scholar]
- 30.Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, Glimm H, Kuhlcke K, Schilz A, Kunkel H, Naundorf S, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 31.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, Asnafi V, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, Gilmour KC, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M, Modlich U, Beier R, Gohring G, Steinemann D, Fronza R, et al. Gene therapy for Wiskott-Aldrich syndrome--long-term efficacy and genotoxicity. Sci Transl Med. 2014;6:227ra233. doi: 10.1126/scitranslmed.3007280. [DOI] [PubMed] [Google Scholar]
- 34.Themis M, Waddington SN, Schmidt M, von Kalle C, Wang Y, Al-Allaf F, Gregory LG, Nivsarkar M, Holder MV, Buckley SM, Dighe N, et al. Oncogenesis following delivery of a nonprimate lentiviral gene therapy vector to fetal and neonatal mice. Mol Ther. 2005;12:763–771. doi: 10.1016/j.ymthe.2005.07.358. [DOI] [PubMed] [Google Scholar]
- 35.Donsante A, Miller DG, Li Y, Vogler C, Brunt EM, Russell DW, Sands MS. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 36.Condiotti R, Goldenberg D, Giladi H, Schnitzer-Perlman T, Waddington SN, Buckley SM, Heim D, Cheung W, Themis M, Coutelle C, Simerzin A, et al. Transduction of Fetal Mice With a Feline Lentiviral Vector Induces Liver Tumors Which Exhibit an E2F Activation Signature. Mol Ther. 2013;22:59–68. doi: 10.1038/mt.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowrouzi A, Cheung WT, Li T, Zhang X, Arens A, Paruzynski A, Waddington SN, Osejindu E, Reja S, von Kalle C, Wang Y, et al. The fetal mouse is a sensitive genotoxicity model that exposes lentiviral-associated mutagenesis resulting in liver oncogenesis. Mol Ther. 2013;21:324–337. doi: 10.1038/mt.2012.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suerth JD, Maetzig T, Brugman MH, Heinz N, Appelt JU, Kaufmann KB, Schmidt M, Grez M, Modlich U, Baum C, Schambach A. Alpharetroviral self-inactivating vectors: long-term transgene expression in murine hematopoietic cells and low genotoxicity. Mol Ther. 2012;20:1022–1032. doi: 10.1038/mt.2011.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Zoppe M, Hackeng TM, Griffin JH, Lee KF, Verma IM. A factor IX-deficient mouse model for hemophilia B gene therapy. Proc Natl Acad Sci U S A. 1997;94:11563–11566. doi: 10.1073/pnas.94.21.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tannapfel A, Busse C, Weinans L, Benicke M, Katalinic A, Geissler F, Hauss J, Wittekind C. INK4a-ARF alterations and p53 mutations in hepatocellular carcinomas. Oncogene. 2001;20:7104–7109. doi: 10.1038/sj.onc.1204902. [DOI] [PubMed] [Google Scholar]
- 41.Weber LW, Boll M, Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: carbon tetrachloride as a toxicological model. Crit Rev Toxicol. 2003;33:105–136. doi: 10.1080/713611034. [DOI] [PubMed] [Google Scholar]
- 42.Santoni de Sio FR, Cascio P, Zingale A, Gasparini M, Naldini L. Proteasome activity restricts lentiviral gene transfer into hematopoietic stem cells and is down-regulated by cytokines that enhance transduction. Blood. 2006;107:4257–4265. doi: 10.1182/blood-2005-10-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanta-Boussif MA, Charrier S, Brice-Ouzet A, Martin S, Opolon P, Thrasher AJ, Hope TJ, Galy A. Validation of a mutated PRE sequence allowing high and sustained transgene expression while abrogating WHV-X protein synthesis: application to the gene therapy of WAS. Gene Ther. 2009;16:605–619. doi: 10.1038/gt.2009.3. [DOI] [PubMed] [Google Scholar]
- 45.Vigna E, Amendola M, Benedicenti F, Simmons AD, Follenzi A, Naldini L. Efficient Tet-dependent expression of human factor IX in vivo by a new self-regulating lentiviral vector. Mol Ther. 2005;11:763–775. doi: 10.1016/j.ymthe.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 46.Russell KE, Olsen EH, Raymer RA, Merricks EP, Bellinger DA, Read MS, Rup BJ, Keith JC, Jr, McCarthy KP, Schaub RG, Nichols TC. Reduced bleeding events with subcutaneous administration of recombinant human factor IX in immune-tolerant hemophilia B dogs. Blood. 2003;102:4393–4398. doi: 10.1182/blood-2003-05-1498. [DOI] [PubMed] [Google Scholar]
- 47.Farson D, Witt R, McGuinness R, Dull T, Kelly M, Song J, Radeke R, Bukovsky A, Consiglio A, Naldini L. A new-generation stable inducible packaging cell line for lentiviral vectors. Hum Gene Ther. 2001;12:981–997. doi: 10.1089/104303401750195935. [DOI] [PubMed] [Google Scholar]
- 48.Escarpe P, Zayek N, Chin P, Borellini F, Zufferey R, Veres G, Kiermer V. Development of a sensitive assay for detection of replication-competent recombinant lentivirus in large-scale HIV-based vector preparations. Mol Ther. 2003;8:332–341. doi: 10.1016/s1525-0016(03)00167-9. [DOI] [PubMed] [Google Scholar]
- 49.Nichols TC, Franck HW, Franck CT, De Friess N, Raymer RA, Merricks EP. Sensitivity of whole blood clotting time and activated partial thromboplastin time for factor IX: relevance to gene therapy and determination of post-transfusion elimination time of canine factor IX in hemophilia B dogs. J Thromb Haemost. 2012;10:474–476. doi: 10.1111/j.1538-7836.2011.04613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 51.Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, Braun S, Glimm H, von Kalle C. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.