Abstract
Genetic disruption of the dystrophin complex produces muscular dystrophy characterized by a fragile muscle plasma membrane leading to excessive muscle degeneration. Two genetic modifiers of Duchenne Muscular Dystrophy implicate the transforming growth factor β (TGFβ) pathway, osteopontin encoded by the SPP1 gene and latent TGFβ binding protein 4 (LTBP4). We now evaluated the functional effect of these modifiers in the context of muscle injury and repair to elucidate their mechanisms of action. We found that excess osteopontin exacerbated sarcolemmal injury, and correspondingly, that loss of osteopontin reduced injury extent both in isolated myofibers and in muscle in vivo. We found that ablation of osteopontin was associated with reduced expression of TGFβ and TGFβ-associated pathways. We identified that increased TGFβ resulted in reduced expression of Anxa1 and Anxa6, genes encoding key components of the muscle sarcolemma resealing process. Genetic manipulation of Ltbp4 in dystrophic muscle also directly modulated sarcolemmal resealing, and Ltbp4 alleles acted in concert with Anxa6, a distinct modifier of muscular dystrophy. These data provide a model in which a feed forward loop of TGFβ and osteopontin directly impacts the capacity of muscle to recover from injury, and identifies an intersection of genetic modifiers on muscular dystrophy.
Author summary
Genetic modifiers for muscular dystrophy have been identified through transcriptomic and genomic profiling in humans and mouse models. Two modifiers, Ltbp4 and Spp1, encode extracellular proteins while a third modifier, Anxa6, specifies a membrane-associated protein. Using a model of muscle injury, we assessed the interaction of these modifiers, identifying a feed forward loop between Ltbp4 and Spp1 that promotes TGFβ signaling. This feed forward loop is expected to contribute to the progressive nature of muscular dystrophy. We also evaluated the interaction between Anxa6 and Ltbp4, identifying an additive effect of these two genetic modifiers.
Introduction
Muscular dystrophies are inherited diseases that cause progressive muscle wasting [1]. Many muscular dystrophies are caused by mutations in genes encoding for components of the dystrophin glycoprotein complex, which anchors the actin cytoskeleton of myofibers to their cell membrane, the sarcolemma. Loss of function mutations in the DMD gene cause Duchenne muscular dystrophy (DMD), while mutations in the SGCG gene, which encodes the dystrophin associated protein γ-sarcoglycan, cause limb-girdle muscular dystrophy type 2C (LGMD 2C) [2, 3]. Disruption of the dystrophin complex results in loss of membrane integrity, leading to chronic injury and necrosis of myofibers [4]. Detrimental remodeling, with replacement by fibrofatty tissue, leads to ongoing, progressive impairment of muscle function [1]. This pathological process begins with disruption of the sarcolemma, and mechanisms to enhance sarcolemmal repair may provide insight in possible therapeutic targets for treating muscular dystrophy.
Disease progression in the muscular dystrophies is variable even in the presence of the same primary mutation, suggesting that secondary genetic variants, or genetic modifiers, can considerably impact the outcome of muscle wasting [5]. The effect of modifiers is evident in murine models of muscular dystrophy, where the same genetic mutation results in significantly different outcomes dependent on the genetic background of the mouse strain [6]. Dystrophin deficiency is modeled in mice by the mdx mutation, a premature stop codon in exon 23 of the dystrophin gene, while γ-sarcoglycan deficiency is modeled by mice lacking exon 2 of the Sgcg gene [3, 7]. mdx and Sgcg mutations have been shown to cause muscular dystrophy with strain-dependent variable pathology, which is severe in the DBA/2J genetic background, intermediate in the C57/Bl6-Bl10 strains, and more mild in the 129T2/SvEmsJ (129T2) background [6, 8, 9].
Identification of genetic modifiers and their mechanisms of action is a useful approach to refine prognosis and potentially discover novel therapeutic targets. Several candidate modifiers act as extracellular agonists of signaling cascades, including osteopontin, encoded by the SPP1 gene, and latent TGFβ binding protein 4 (LTBP4). Osteopontin is a secreted glycoprotein that signals through integrin and CD44 receptors [10]. SPP1 expression is highly upregulated in affected muscles of humans and animals with muscular dystrophy [11–19]. Genetic loss of Spp1 in mdx mice correlates with greater strength, less fibrosis and milder pathology, as compared to control mdx littermates [13]. Moreover, Spp1 ablation has been linked to a shift in macrophage polarization towards a regenerative phenotype in mdx muscles [20]. In humans with DMD, a single nucleotide polymorphism (SNP) in the SPP1 promoter (GG/TG) correlates with increased grip strength and later loss of ambulation compared to patients with the more prevalent SNP (TT), especially in DMD individuals who are steroid treated [19, 21, 22]. Some genetic cohort studies have not shown this same effect [23, 24]. The manner in which the SPP1 SNP affects osteopontin expression with disease progression is complex, and it is unclear in which SPP1-expressing cell type(s) this modifier SNP acts.
Ltbp4 was identified as genetic modifier of several pathologic traits in the Sgcg mouse model, including sarcolemmal damage and fibrosis [25]. Latent TGFβ binding protein (LTBP4) is an extracellular protein that binds TGFβ, releasing it upon proteolysis of its hinge region [26]. The LTBP4 modifier also correlates with differential outcomes in humans with muscular dystrophy [21, 23, 24]. In mice, the “risk” Ltbp4 allele encodes a shorter hinge region that is more susceptible to proteolysis, and this risk allele is found in the DBA/2J strain correlating with more severe muscular dystrophy. In contrast, most mouse strains including 129T2 and C57 substrains have the protective Ltbp4 allele encoding a longer hinge region that is more resistant to proteolytic cleavage. Overexpression of the protective Ltbp4 allele in the mdx mouse reduces fibrosis and promotes muscle growth [26]. However, the specific molecular effects of protective and deleterious Ltbp4 alleles on sarcolemmal resealing and repair are still unknown.
Intriguingly, it has been shown that the Spp1 promoter is susceptible to TGFβ-driven transcriptional activation [27, 28]. In mesenchymal cells, osteopontin signaling activates Tgfb1 transcription via the myeloid zinc finger 1 (MZF1) transcriptional factor [29, 30]. These data support a potential interaction between these two modifiers, Spp1 and Ltbp4, in muscular dystrophy. Examining the combinatorial effects of genetic modifiers requires a large population, which is challenging for a rare human disorder like DMD. Here we asked whether genetic manipulation of osteopontin and Ltbp4 impacts sarcolemmal resealing and repair. To specifically address this question, we relied on optimized conditions of sarcolemmal micro-injury, and real-time detection of sarcolemmal damage and repair cap formation in isolated live myofibers [31, 32]. Laser-mediated injury results in larger sarcolemmal damage in dystrophic myofibers than in wildtype myofibers using age- and background-matched conditions [33]. We found that both osteopontin and Ltbp4 modify sarcolemmal repair in wildtype and dystrophic myofibers. Furthermore, we documented how osteopontin and the deleterious Ltbp4 isoform converge in a feed-forward TGFβ signaling loop that correlated with transcriptional repression of annexin genes and impaired sarcolemmal repair. In addition, we dissected the relative impact of the different alleles of Ltbp4 and a third modifier Anxa6 on resealing and repair of injured sarcolemma in background-strain matched conditions. These results indicate that osteopontin and LTBP4 regulate a TGFβ signaling loop to modify sarcolemmal repair in normal and dystrophic muscle.
Results
Recombinant osteopontin worsens sarcolemmal injury
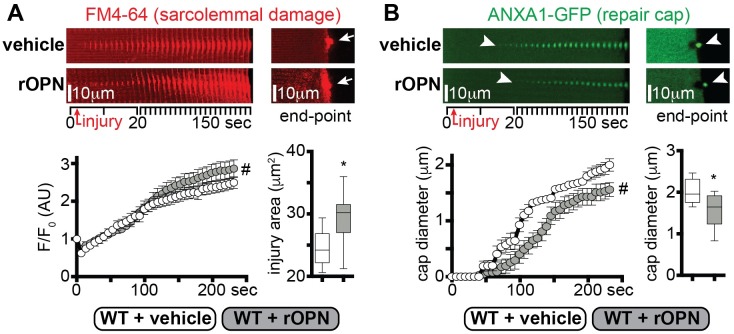
To gain insight in the role of osteopontin (OPN) in myofiber repair, we assessed the effect of recombinant osteopontin (rOPN) on sarcolemmal repair of isolated myofibers [32]. We tested the repair capacity after laser-mediated sarcolemmal injury and quantified two parameters in real time: (i) accumulation of the FM4-64 dye at the injury site, marking the extent of damage; (ii) formation of the repair complex monitoring GFP-tagged annexin A1 (ANXA1-GFP) protein [31]. DBA/2J wildtype (WT) mice were injected intramuscularly with 10μg rOPN at 1, 3 and 5 days after electroporation with the plasmid encoding ANXA1-GFP. The activity of injected rOPN was confirmed by monitoring expression of Mmp2 and Mzf1, factors known to be downstream of OPN [30, 34]. Mmp2 and Mzf1 were increased 48 hours after rOPN injection while vehicle injection did not stimulate this response (S1A Fig). Seven days after electroporation, live myofibers from treated and vehicle injected control muscles were subjected to laser injury. FM4-64 accumulation, which marks the area of injury, was greater in rOPN injected muscle at 240 seconds after laser injury, as compared to vehicle-treated myofibers (Fig 1A). Moreover, the onset of the annexin A1 repair cap formation was significantly slower in rOPN-injected muscle than control myofibers, resulting in a significantly smaller annexin A1 cap size at end-point (Fig 1B). Thus, intramuscular injection of DBA/2J WT muscle with rOPN exacerbated sarcolemmal injury and slowed formation of the annexin-containing repair complex.
Fig 1. Excess osteopontin exacerbates sarcolemmal injury and delays repair cap formation.
The sarcolemmal wounding assay was performed on flexor digitorus brevis muscles from WT muscle in the presence or absence of recombinant osteopontin (rOPN; 10μl at 1μg/μl). A) The extent of sarcolemmal damage, monitored as FM4-64 area, was greater in rOPN-treated myofibers compared to vehicle-treated control, both over time and at end-point (arrows). B) Repair cap formation, monitored by GFP-tagged annexin A1 (GFP-ANXA1) was delayed in rOPN-treated myofibers (arrowheads). Repair cap diameter at end-point was smaller in treated compared to control myofibers (arrowheads). Over time image series represents stacked consecutive images of the injured site at 10° orientation to reveal the extent of dye accumulation or repair cap formation (S1B Fig). FM4-64 and ANXA1-GFP pictures were acquired simultaneously. Marked line plots, avg±sem; box plots, Tukey distribution; n = 50 myofibers (5 mice)/group; #, P<0.05 vs vehicle, 2way ANOVA + Bonferroni; *, P<0.05 vs vehicle, unpaired t-test with Welch’s correction.
Genetic ablation of osteopontin modifies sarcolemmal injury of dystrophic myofibers
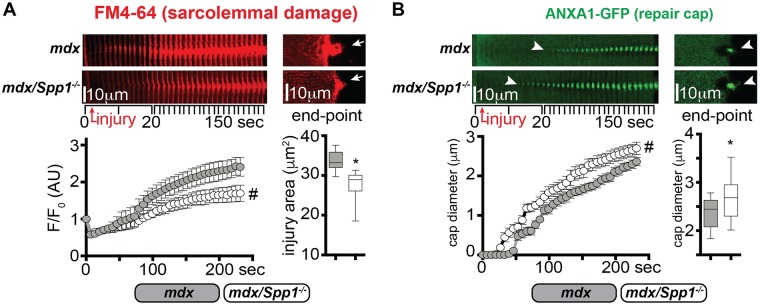
Genetic ablation of Spp1 in mdx mice results in decreased fibrosis and muscle pathophysiology in mdx mice [13]. We asked whether Spp1 loss had a measurable effect on sarcolemmal repair of mdx myofibers. To this end, we compared myofibers from 20 week-old mdx (Spp1+/+; control animals) and age-matched Spp1-deficient mdx (mdx/Spp1-/-) littermates for the extent of sarcolemmal damage after laser injury. Myofibers from both mice groups were electroporated with the ANXA1-GFP-encoding plasmid prior to harvest and laser-injury assay. Spp1-deficient dystrophic myofibers showed significantly reduced FM4-64 accumulation at the injury site over time, and a reduction of injury area at end-point when compared to control myofibers (Fig 2A). We monitored annexin A1 repair cap appearance at the site of injury and observed faster rate of cap appearance in mdx/Spp1-/- compared to mdx control myofibers, resulting in a larger cap size at analysis end-point (Fig 2B). Furthermore, expression levels of endogenous Anxa1 and Anxa6, encoding the repair cap proteins annexins A1 and A6, were significantly upregulated in muscle of mdx/Spp1-/- mice, as compared to littermate control animals (S2A Fig). Moreover, improved sarcolemmal repair in isolated fibers was also reflected in in vivo findings in muscle. Spp1 deficiency correlated with lower levels of fibrosis, quantified as hydroxyproline content, in both quadriceps and diaphragm muscles, as well as with reduced levels of circulating creatine kinase, a marker of striated muscle damage (S2B and S2C Fig), consistent with previous characterization of mdx/Spp1-/- mice [13] [20].
Fig 2. mdx mice lacking osteopontin (mdx/Spp1-/-) recover from sarcolemmal injury more efficiently.
A) The sarcolemmal wounding assay was performed on flexor digitorum brevis myofibers from mdx or mdx/Spp1-/- animals, after electroporation with the ANXA1-GFP-encoding plasmid. The extent of sarcolemmal damage, monitored through accumulation of FM4-64 dye after laser injury, was significantly reduced in mdx/Spp1-/- myofibers compared to control dystrophic myofibers. This was quantified over time as dye accumulation at site of injury (left) and area of injury at end-point (right; arrows). The time series (left images) represent stacked consecutive images of the injured site at 10° orientation to reveal the extent of dye accumulation. B) Repair cap onset, monitored by GFP-tagged ANXA1, was significantly faster in mdx/Spp1-/- myofibers (arrowheads), resulting in a larger cap diameter at end-point (arrowheads). Time series of stacked consecutive images of the injured site at 10° orientation revealed the extent of dye accumulation or repair cap formation. FM4-64 and ANXA1-GFP pictures were acquired simultaneously. Marked line plots, avg±sem; box plots, Tukey distribution; n = 40 myofibers (4 mice)/group; #, P<0.05 vs mdx control, 2way ANOVA + Bonferroni; *, P<0.05 vs mdx control, unpaired t-test with Welch’s correction.
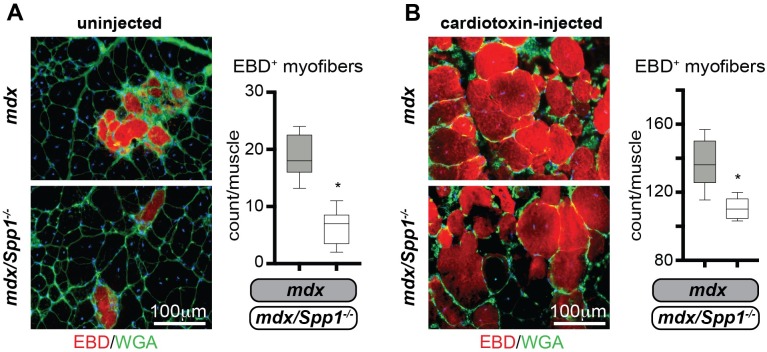
In addition to Spp1’s role in dystrophic muscle remodeling, osteopontin exacerbates injury in toxin-injured wildtype muscles [35]. To assess Spp1’s role in chronic and acute injury of dystrophic muscle, the tibialis anterior (TA) muscles of mdx and mdx/Spp1-/- mice were injected with cardiotoxin along the muscle axis, immediately after systemic delivery of Evans Blue Dye (EBD). Each mouse received cardiotoxin in one muscle, while the contralateral remained uninjected. After three hours, muscles were harvested and injury extent was quantified as EBD-positive myofibers per muscle using serial cross-sections. There were fewer EBD-positive myofibers in mdx/Spp1-/- muscles than in mdx muscles, both under basal conditions and after toxin-induced injury (Fig 3A and 3B). Thus, these data suggest that sarcolemmal repair is enhanced and muscle injury is reduced after genetic ablation of osteopontin.
Fig 3. Spp1 ablation in mdx muscle resulted in fewer disrupted myofibers in vivo.
A) Three hours after systemic EBD delivery, the number of EBD-positive myofibers (red appearance) was significantly less in mdx/Spp1-/- TA muscles than in control mdx muscles (left, representative immunofluorescence panels; right, quantitation). B) Cardiotoxin injection was performed in the contralateral muscles, increasing the number of EBD-positive fibers. After cardiotoxin injury, there were fewer EBD-positive myofibers in mdx/Spp1-/- compared to mdx muscles (left, representative immunofluorescence panels; right, quantitation). Box plots, Tukey distribution; n = 4 mice/group; *, P<0.05 vs mdx control, unpaired t-test with Welch’s correction.
Spp1 deletion correlates with decreased TGFβ signaling in dystrophic muscle
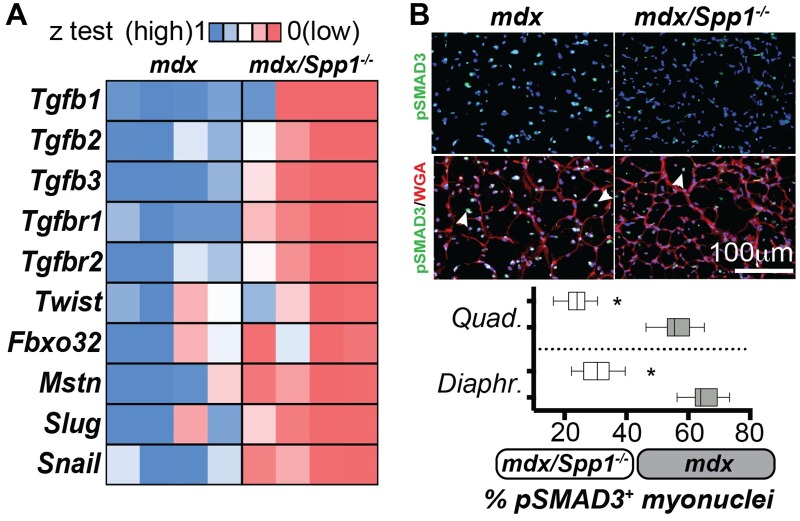
The Spp1 promoter is responsive to TGFβ signaling, and the TGFβ1 ligand increases Spp1 levels [27, 28]. We therefore asked whether TGFβ signaling was altered in mdx/Spp1-/- muscles. To this end, we compared mdx/Spp1-/- and control mdx muscles for expression levels of TGFβ pathway genes and for enrichment of myonuclei positive for phosphorylated SMAD3 (pSMAD3), a known effector of active TGFβ signaling [36]. Quantitative PCR (qPCR) analysis of TA muscles showed that ligands, receptors and downstream factors of the TGFβ pathway, including transcriptional repressors Slug and Snail [37, 38], were significantly downregulated in mdx/Spp1-/- mice as compared to mdx (Fig 4A). Immunofluorescence microscopy (IFM) of quadriceps and diaphragm muscles showed that the relative ratio of pSMAD3+ myonuclei was significantly reduced in both muscles of mdx/Spp1-/- mice compared to mdx control animals (Fig 4B). Thus, genetic loss of Spp1 correlated with decreased TGFβ signaling in multiple mdx muscles.
Fig 4. Spp1 ablation in mdx muscle is associated with decreased TGFβ gene expression and signaling.
A) Heat-map of qPCR analysis of gene expression, normalized to control muscles, showed downregulation of genes associated with the TGFβ pathway and atrophic remodeling in mdx/Spp1-/- dystrophic muscle (tibialis anterior). All genes shown in the heatmap were significantly different (*). B) Percentage of phosphorylated-Smad3-positive nuclei within myofibers (myonuclei; arrowheads) was significantly decreased in quadriceps (representative immunofluorescence panels) and diaphragm muscles of mdx/Spp1-/- mice when compared to mdx mice. Box plots, Tukey distribution; heat-map, z-test average values of fold change to control (ctrl); n = 4 mice/group; *, P<0.05 vs mdx control, 1way ANOVA + Bonferroni.
Osteopontin decreases annexin gene expression via TGFβ pathway stimulation
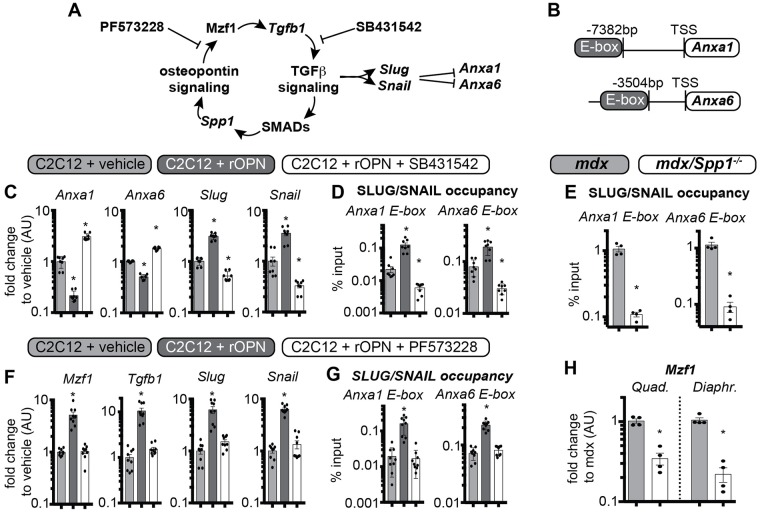
In non-muscle mesenchymal cells, OPN has been shown to stimulate TGFβ1 upregulation via integrin signaling and the transcriptional factor Mzf1 [29, 30]. We hypothesized a feed-forward model, in which OPN sustains TGFβ signaling in muscle and its downstream transcriptional factors Slug and Snail, which bind E-box DNA elements of target genes (Fig 5A) [39]. Through sequence alignment, we identified a predicted E-box element (GTCGAC motif) [39] upstream of the transcriptional start site of Anxa1 (-7382bp) and Anxa6 genes (-3504bp) (Fig 5B). We tested this pathway in C2C12 myoblasts as well as in the myofiber fraction from mdx muscles using qPCR and chromatin immunoprecipitation (ChIP). C2C12 cells were exposed to 1μg/ml rOPN, and after 48 hours of rOPN treatment, both Anxa1 and Anxa6 were downregulated, while Slug and Snail were upregulated, as compared to vehicle-treated cells. These responses were reversed when C2C12 cells were co-treated with rOPN along with 10μM SB431542, a chemical compound specifically inhibiting TGFβ signaling activation (Fig 5C) [40]. ChIP-qPCR analysis showed that occupancy of Anxa1 and Anxa6 E-box elements by the SLUG/SNAIL transcriptional complex was increased after rOPN treatment, but decreased in the presence of TGFβ inhibitor (Fig 5D). In the myofiber fractions from mdx/Spp1-/- muscle where Anxa1 and Anxa6 expression were increased, these promoters had reduced occupancy of the E-box elements when compared to myofibers from mdx littermates (Fig 5E). When C2C12 cells were co-treated with rOPN and 10μM PF573228, a chemical inhibitor of the OPN-driven signaling leading to Mzf1-Tgfb1 axis activation [41], there was blunting of rOPN-associated upregulation of Mzf1, Tgfb1, Slug, and Snail (Fig 5F). Consistent with this, SLUG/SNAIL occupancy of Anxa1/6 E-box elements reverted to vehicle-like levels when cells were co-treated with rOPN and PF573228 (Fig 5G). Finally, expression of Mzf1 was significantly lower in Spp1-deficient hindlimb and respiratory muscles than in control mdx muscles (Fig 5H). Thus, excess OPN is associated with repression of Anxa1 and Anxa6, and increased occupancy of their putative E-box elements by the transcriptional repressor complex SLUG/SNAIL. Repression by SLUG/SNAIL was associated with increased Mzf1 and Tgfb1 levels. Moreover, OPN-associated events were reversed in the presence of a chemical TGFβ inhibitor and were blunted by a chemical inhibitor of OPN signaling, thus supporting the hypothesis of a feed-forward circuitry that involves osteopontin and TGFβ pathways suppressing Anxa1 and Anxa6 and impairing sarcolemmal repair.
Fig 5. Excess osteopontin and TGFβ activation inhibits Anxa1 and Anxa6 gene expression.
A) Model depicting the feed-forward loop in which osteopontin promotes TGFβ, which in turn promotes Spp1/osteopontin expression. Slug/Snail expression, downstream of TGFβ activation, results in Anxa1/Anxa6 gene repression. B) Diagram of putative E-box elements upstream of transcriptional start site (TSS) of Anxa1 and Anxa6 genes in the murine genome. C) qPCR analysis of C2C12 cells after treatment with recombinant osteopontin (rOPN) or rOPN plus the TGFβ inhibitor SB431452. rOPN increased Slug/Snail expression, and this was reversed in the presence of SB431452. D) Correspondingly, ChIP-qPCR showed that the E-box occupancy of the Anxa1 and Anxa6 promoters by the SLUG/SNAIL protein complex was increased by rOPN, and this was reversed in the presence of SB431452. E) Anxa1 and Anxa6 promoter occupancy by SLUG/SNAIL was decreased in the myofiber fraction of mdx/Spp1-/- muscle compared to control mdx muscle (gastrocnemius). F) PF573228, a chemical inhibitor of OPN-driven signaling leading to Mzf1-Tgfb1 axis activation, ablated rOPN-induced upregulation of Slug, Snail, Mzf1, and Tgfb1. G) Accordingly, PF573228 blunted rOPN-dependent increase in SLUG/SNAIL occupancy on Anxa1/6 E-boxes in C2C12 myoblasts. H) Mzf1, transcriptional regulator linking the OPN and TGFβ signaling cascades, was downregulated in both hindlimb (quadriceps) and respiratory (diaphragm) muscles of mdx/Spp1-/- compared to mdx. Histograms, single values & avg±sem; n = 8 independent replicates/group for C2C12 myoblast analyses, n = 4 mice/group for mdx/Spp1-/- mice analyses; (C-D; E-G) *, P<0.05 vs vehicle, 1way ANOVA + Bonferroni; (D) *, P<0.05 vs mdx control, unpaired t-test with Welch’s correction.
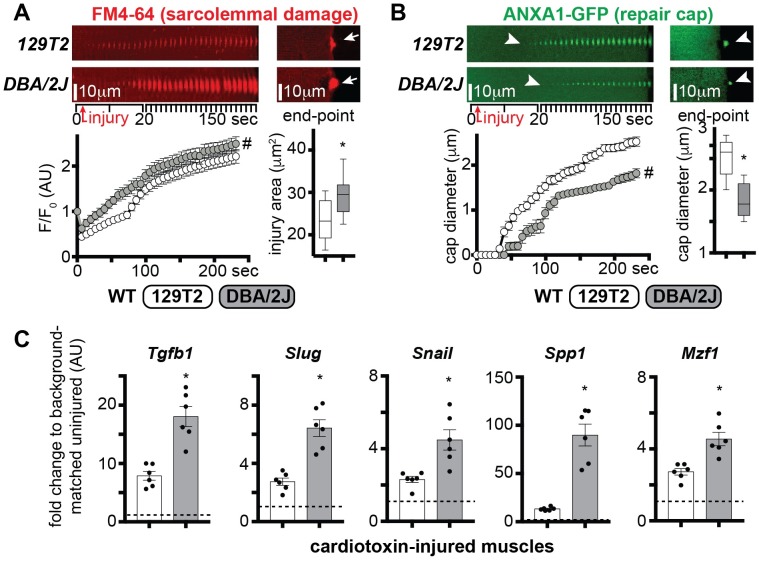
The DBA/2J genetic background is associated with higher levels of osteopontin and TGFβ signaling, and increased sarcolemmal damage
In Sgcg (γ-sarcoglycan) or DMD (mdx) mice, the DBA/2J background exacerbates the dystrophic phenotype [6, 9]. We sought to determine whether the difference in global outcomes associated with those genetic backgrounds was correlated with the efficiency of sarcolemma repair. To exclude effects associated with dystrophic remodeling, we tested WT mice from 129T2 and DBA/2J background using laser-induced sarcolemmal injury. DBA/2J myofibers showed more severe sarcolemmal damage than 129T2 myofibers (Fig 6A). Annexin A1 cap formation, seen as ANXA1-GFP, appeared more slowly in DBA/2J than in 129T2 myofibers and resulted in smaller cap size at end-point (Fig 6B).
Fig 6. The DBA/2J genetic background is associated with increased sarcolemmal damage and higher levels of OPN and TGFβ pathway genes.
The DBA/2J background exacerbates the muscular dystrophy phenotype [6, 9]. A) The extent of sarcolemmal damage was significantly higher in WT DBA/2J myofibers, as compared to age-matched WT 129T2 myofibers, both over time and at end-point (arrows). B) Repair cap formation, monitored by GFP-tagged ANXA1, appeared significantly delayed in DBA/2J myofibers (arrowheads), with a smaller cap diameter at end-point (arrowheads). The time series shows stacked consecutive images of the injured site at 10° orientation to reveal the extent of dye accumulation or repair cap formation. FM4-64 (red) and ANXA1-GFP (green) pictures were acquired simultaneously. C) qPCR analysis of cardiotoxin-injured tibialis anterior muscle tissue at 3 days post-injury from 129T2 and DBA/2J mice versus strain-matched uninjured littermates showed that injury-associated upregulation of Tgfb1, Slug, Snail, Spp1, and Mzf1 was significantly higher in the DBA/2J strain than in the 129T2 strain (dotted line: strain-matched control values, = 1). Marked line plots, avg±sem; box plots, Tukey distribution; histograms, single values & avg±sem; n = 5 mice/group; #, P<0.05 vs vehicle, 2way ANOVA + Bonferroni; *, P<0.05 vs 129T2, unpaired t-test with Welch’s correction.
Although many genetic loci contribute to the DBA/2J background effect, DBA/2J mice feature the Ltbp4 risk allele, which is estimated to contribute to at least 40% of the variance of the muscular dystrophy phenotype in Sgcg mice [42]. RNA sequencing of WT and Sgcg muscle from the severe DBA/2J background was associated with a marked increase in Spp1 expression compared expression in the 129T2 background (S3 Fig). We monitored gene expression changes relevant to the TGFβ pathway three days after cardiotoxin-mediated injury to the TA muscles of age-matched 129T2 or DBA/2J in order to assess the effect of genetic background. After muscle injury, the DBA/2J background was associated with increased expression of Tgfb1, Slug, Snail, Spp1, and Mzf1 compared to injured 129T2 muscle (Fig 6C). Thus, the DBA/2J genetic background associated with increased sarcolemmal damage, and higher levels of osteopontin and TGFβ pathway genes.
LTBP4 modifies sarcolemmal repair of dystrophic myofibers
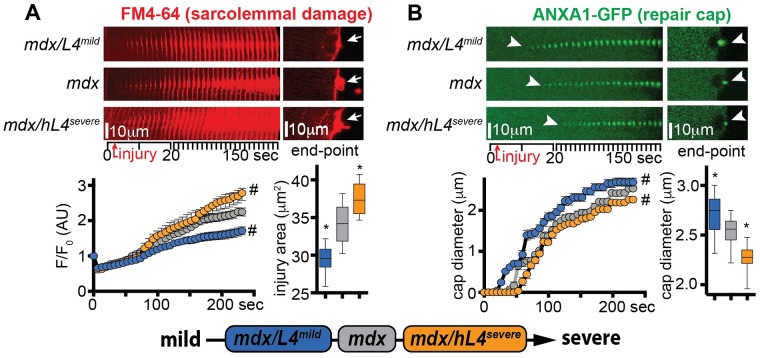
Muscle specific overexpression of the protective Ltbp4 allele (L4mild) in mdx mice reduces fibrosis and increases muscle size [43]. Conversely, expression of the risk human LTBP4 allele (hL4severe) exacerbates dystrophic remodeling in mdx mice [44]. We tested sarcolemmal damage and repair cap formation in myofibers from transgenic age-matched mdx mice overexpressing either the L4mild (mdx/L4mild), or the hL4severe isoform (mdx/hL4severe) at 20 weeks of age. After laser-mediated sarcolemmal injury, the extent of sarcolemmal injury was smaller in myofibers of mdx/L4mild mice than from mdx control animals. Conversely, sarcolemmal damage was increased in mdx/hL4severe myofibers as compared to control muscles (Fig 7A). Repair cap formation, monitored through GFP-labelled annexin A1 (ANXA1-GFP), appeared faster in mdx/L4mild and slower in mdx/hL4severe myofibers than in mdx control myofibers. These trends associated with a bigger cap size in mdx/L4mild and a smaller cap size in mdx/hL4severe, when compared to mdx repair caps at end-point (Fig 7B). Thus, the protective isoform with less TGFβ release associated with reduced sarcolemmal damage and faster cap formation, while the proteolysis-prone, risk allele with higher TGFβ release associated with increased damage and delayed repair cap assembly.
Fig 7. Disease-modifying LTBP4 isoforms differentially modify sarcolemmal repair in dystrophic muscle.
Overexpression of the protective L4mild allele in mdx mice (mdx/L4mild) protects against many features of muscular dystrophy [6]. Expression of the deleterious human LTBP4 allele in mdx mice (mdx/hL4severe) worsens muscular dystrophy [44]. A) Sarcolemmal damage was significantly enhanced in mdx/hL4severe myofibers, as compared to littermate, background-matched mdx myofibers, both over time and at end-point (arrows). Conversely, sarcolemmal damage was reduced in age-matched mdx/L4mild myofibers. B) Repair cap formation, monitored by GFP-tagged ANXA1, was significantly delayed in mdx/hL4severe myofibers (arrowheads), with a smaller cap diameter at end-point (arrowheads). Opposite trends were quantitated in age-matched mdx/L4mild myofibers (arrowheads). Time series of stacked consecutive images of the injured site at 10° orientation revealed the extent of dye accumulation or repair cap formation. FM4-64 and ANXA1-GFP pictures were acquired simultaneously. Marked line plots, avg±sem; box plots, Tukey distribution; n = 50 myofibers (5 mice)/group; #, P<0.05 vs mdx, 2way ANOVA + Bonferroni; *, P<0.05 vs mdx, 1way ANOVA + Bonferroni.
Dissecting the effects of mild and severe alleles of Anxa6 and Ltbp4 on sarcolemmal repair and muscle injury in vivo
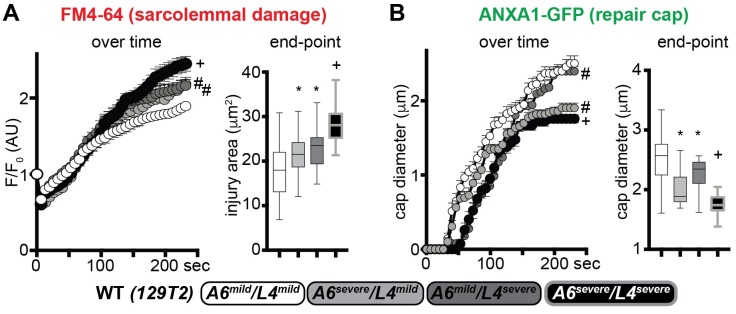
Anxa6, was also identified as a modifier of muscular dystrophy by its action on sarcolemmal repair itself [45], and the DBA/2J background harbors the risk allele for Anxa6. In order to discriminate the effects elicited by the Anxa6 and Ltbp4 genetic modifiers on sarcolemmal repair, we generated WT mice in the 129T2 background strain carrying the four homozygous combinations of mild and severe isoforms of Anxa6 and Ltbp4 (A6mild/L4mild; A6severe/L4mild; A6mild/L4severe; A6severe/L4severe). We assessed sarcolemmal damage and annexin A1 cap formation after laser injury in myofibers from age-matched mice from the four cohorts. Muscles homozygous for either A6severe or L4severe had increased sarcolemmal damage, when compared to A6mild/L4mild myofibers. Moreover, the effect of the two severe isoforms appeared additive on the sarcolemmal damage phenotype, as A6severe/L4severe myofibers presented the highest levels of FM4-64 accumulation across the groups (Fig 8A). Repair cap formation was also regulated by the additive effects of the severe isoforms. The A6severe isoform associated with smaller cap size at end-point, while the L4severe isoform associated with a delay in initial onset of formation, when compared to the A6mild/L4mild genotype. Similarly, the A6severe/L4severe correlated with onset delay and smaller size of the ANXA1 repair cap at end-point (Fig 8B). Thus, the polymorphism affecting LTBP4 function modifies sarcolemmal damage and repair cap formation in WT myofibers, and its effects are additive with respect to the Anxa6 polymorphism.
Fig 8. Relative contribution of Anxa6 and Ltbp4 alleles on sarcolemmal repair in wildtype muscle.
In addition to Ltbp4, Anxa6 has also been shown to modify muscular dystrophy in mice [25, 45]. The deleterious alleles of Ltbp4 (L4) or Anxa6 (A6) (severe) were compared to those from the protective 129 strain (mild) in the sarcolemma injury assay. A) Doubly homozygous A6mild/L4mild myofibers had the least injury while doubly homozygous A6severe/L4severe fibers had the greatest injury, marked by FM4-64. Myofibers with mixed homozygous genotypes were intermediate with respect to FM4-64 marked injury. B) A similar pattern was observed for Annexin A1 (ANXA1) repair caps, where doubly homozygous mild alleles of L4 and A6 assembled caps more rapidly and produced larger repair caps than doubly homozygous severe alleles. However, ANXA1 repair caps were smaller with the A6 homozygous severe allele, despite the presence of the mild L4 allele, suggesting that repair cap formation is dominated by the A6 genotype. FM4-64 and ANXA1-GFP images used for the analyses in A-B were acquired simultaneously. Marked line plots, avg±sem; box plots, Tukey distribution; n = 50 myofibers (5 mice)/group; marked line plots: #, P<0.05 vs control (A6mild/L4mild), +, P<0.05 vs A6severe/L4mild and A6mild/L4severe groups, 2way ANOVA + Bonferroni; boxplots: *, P<0.05 vs control (A6mild/L4mild), +, P<0.05 vs A6severe/L4mild and A6mild/L4severe groups, 1way ANOVA + Bonferroni.
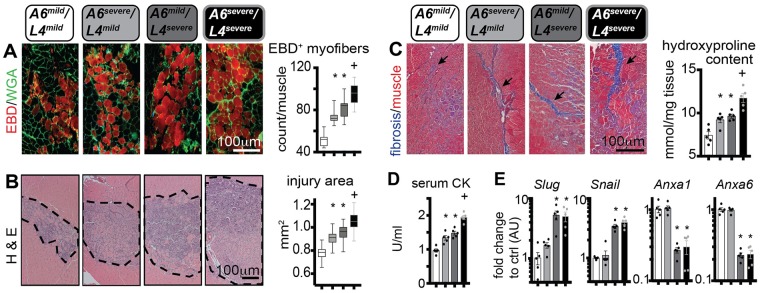
We next subjected mice from all four allele combinations to intra-muscular injury with cardiotoxin, targeting both TA and gastrocnemius muscles. To quantify the number of injured myofibers, mice were intra-peritoneally injected with EBD immediately before intramuscular toxin injection. Three hours after dye delivery, the number of dye-positive myofibers in injured muscles was comparably higher in mice with one severe allele, and the highest in mice with both severe alleles, with respect to A6mild/L4mild mice (Fig 9A). Seven days after injury, the injury area at muscle mid-point and fibrotic scarring followed similar trends, as quantified by histologic analyses (Fig 9B and 9C). Serum creatine kinase (CK) at 24 hours after injury was higher in the presence of either A6severe, or L4severe alleles, and the highest in mice with both severe alleles, as compared to A6mild/L4mild mice (Fig 9D). Seven days after injury, muscle tissue was analyzed for gene expression trends. Transcriptional levels of Slug and Snail were significantly upregulated in the presence of the L4severe allele regardless of the A6 allele status. Expression of Anxa1 and Anxa6 was also downregulated in the presence of L4severe isoforms (Fig 9E). Thus, Anxa6 and Ltbp4 isoforms modify the extent of damage in acute muscle injury in an additive fashion, and the severe isoform of Ltbp4, characteristic of the DBA/2J genetic background, correlated with upregulation of Slug and Snail, and downregulation of Anxa1 and Anxa6.
Fig 9. Relative contribution of Anxa6 and Ltbp4 alleles on muscle injury in vivo.
A) Three days post-injury, the number of dye+ myofibers (red) in injured tibialis anterior muscles was increased in single homozygous mice, either A6severe or L4severe, compared to double homozygous A6mild/L4mild muscle, which displayed the fewest dye+ fibers. Doubly homozygous animals A6severe/L4severe had the greatest number of dye+ fibers. B) The same pattern was seen when measuring injury area from cardiotoxin injected gastrocnemius muscles measured 7 days post-injury. C) Scarring monitored by Masson’s trichrome staining (left panels; arrows) and hydroxyproline content (right chart) was also greatest in doubly mutant (A6severe/L4severe) gastrocnemius muscles 7 days post-injury. D) 24 hours after cardiotoxin-injury of hindlimb muscles, serum CK levels were increased in single homozygous mice A6severe or L4severe, and further increased in double homozygous A6severe/L4severe animals. E) Seven days after cardiotoxin injury, Slug, Snail were upregulated while Anxa1, and Anxa6 expression levels were significantly downregulated in muscle in the presence of homozygous L4severe allele (tibialis anterior) indicating that the L4severe allele impacts Anxa1/Anxa6 expression likely via Slug/Snail regulation. *, P<0.05 vs A6mild/L4mild, +, P<0.05 vs A6severe/L4mild and A6mild/L4severe groups, 1way ANOVA + Bonferroni.
Discussion
A feed forward loop between TGFβ and osteopontin in muscular dystrophy and muscle injury
Genetic modifiers of muscular dystrophy influence outcome through multiple pathways. However, their combinatorial effects, and specifically whether they are additive, synergistic or even opposing in action is challenging to address at a human population level in a rare disorder. Here we utilized a surrogate endpoint, sarcolemmal repair, to begin to assess how osteopontin and LTBP4 modify myofiber repair through a convergent TGFβ pathway circuitry. Furthermore, we showed that upregulated OPN/TGFβ results in transcriptional repression of annexin genes, and this provides one possible means by which the OPN/TGFβ pathway contributes to impaired sarcolemmal resealing. Given the broad gene expression effects of the OPN/TGFβ pathway, we expect that multiple genes mediate the in vivo effect in muscular dystrophy. In vivo, in the absence of osteopontin, mdx muscle had fewer disrupted myofibers, which likely reflected this feed forward loop altering sarcolemmal stability, repair, and even muscle regeneration.
Osteopontin and TGFβ signaling constitute a common marker of dystrophic muscle when compared to healthy controls [11–19, 46–49]. Conversely, genetic manipulation to stifle both cascades reduces dystrophic pathology [13, 36, 43, 50]. To date, these signaling pathways have been mainly associated with activation and tissue remodeling by immune cell infiltrates and resident fibroblasts [10, 51]. These results indicate that osteopontin and TGFβ pathways act synergistically to directly influence the ability of myofibers to repair after sarcolemmal injury. There is an expected crosstalk among myofibers, fibroblasts, and immune cells during both acute and chronic injury, which further modifies dystrophic features.
We found that the presence of either osteopontin or the deleterious form of Ltbp4 resulted in annexin gene repression and increased sarcolemmal damage. A limitation of the sarcolemmal repair assays used for this study is the use of electroporation to express GFP-tagged annexins in the presence of native annexins. To address this, we relied on electroporation of the same construct (ANXA1-GFP) in all different genetic contexts, and the results observed in the sarcolemmal repair assay paralleled the results seen when examining muscle injury in vivo, in both dystrophic and WT settings. Importantly, sarcolemmal repair assays were conducted in the presence of Ca2+, as annexin repair cap formation is known to be Ca2+-dependent [32]. Further analyses in Ca2+-free settings, in combination with finer characterization of membrane composition, may discriminate Spp1- and Ltbp4-dependent effects on repair capacity and membrane mechanical stability, as recently investigated for the repair protein MG53 [52].
In both genetic settings, either Spp1 ablation or the deleterious Ltbp4 alleles, transcriptional regulation of Anxa1 and Anxa6 inversely correlated with expression levels of Slug/Snail and occupancy of the TGFβ-related SLUG/SNAIL repressive complex on the E-box elements upstream of Anxa1 and Anxa6. These findings corroborate the hypothesis that both genetic modifiers contribute to a self-reinforcing, TGFβ-reliant loop in muscle, consistent with traditional observations of a feed-forward regulation of TGFβ signaling [37, 38]. The deleterious VTTT haplotype in the LTBP4 gene correlated with more rapid disease progression in DMD patients, while the rs28357094 polymorphism in the SPP1 promoter has been reported as a disease determinant in some studies, but not in others [19, 21, 22, 24, 53]. However, it is still unclear whether the combination of deleterious polymorphisms at both loci significantly associates with worsened disease outcome. Given the rare nature of DMD, assembling sufficient cohorts for these genetic studies in humans may not be possible.
Insights on sarcolemmal stability and repair
Genome-wide screening for disease traits identified both Ltbp4 and Anxa6 as sarcolemmal damage modifying genes. The DBA/2J mouse strain carries both risk alleles and produces enhanced muscular dystrophy pathology in mice [25, 45]. Anxa6 alleles regulate annexin repair cap size. However, LTBP4 content determines the efficiency of cap formation after injury, with the deleterious Ltbp4 alleles associated with delayed cap formation. These detrimental effects were additive in the presence of both alleles, and translated in additive effects on the in vivo response to muscle injury. In addition to the transcriptional regulation of annexin genes, it is possible that LTBP4-mediated TGFβ overload impacts sarcolemmal repair through post-transcriptional regulation and/or protein interactions. Ltbp4 genotype also appears to be a modifier of the extent of injury, which may reflect enhanced stability of the myofiber prior to injury.
The protective LTBP4 allele in humans has been associated with delayed loss of ambulation in humans with DMD, and this effect was greater in the presence of glucocorticoid steroid regimen in those study cohorts [24]. Interestingly, steroid-associated additive beneficial effects were also reported for the SPP1 polymorphism in DMD patients [21]. We recently reported that glucocorticoid steroids, such as prednisone and deflazacort, improve sarcolemmal repair and annexin cap formation in normal and dystrophic muscle [54]. It has been suggested that glucocorticoid steroids act in muscular dystrophy by synchronizing an asynchronous repair milieu in muscular dystrophy [55]. These data suggest that genetic modifiers beyond SPP1/OPN may contribute to this disorganized repair process and support the development of agents to resynchronize this process.
In summary, we now show that both osteopontin and LTBP4 directly modify sarcolemmal repair in myofibers of normal and dystrophic muscles. In the model, excess osteopontin and deleterious LTBP4 converge to sustain TGFβ-mediated gene expression changes, including the repression of annexins, and likely, many other repair genes. These findings indicate a direct role of those genetic modifiers in myofiber damage regulation and support their consideration for novel therapeutic avenues for treating dystrophic muscle.
Materials and methods
Ethics statement
Mice were housed in a specific pathogen free facility in accordance with Institutional Animal Care and Use Committee (IACUC) regulations. Euthanasia was performed through carbon dioxide or anesthetic gas inhalation followed by cervical dislocation and removal of the heart. All methods using living animals in this study were performed in ethical accordance with the American Veterinary Medical Association (AVMA) and under protocols fully approved by both the Institutional Animal Care and Use Committee (IACUC) at Northwestern University Feinberg School of Medicine (protocol number ISO00000911). Consistent with the approvals stipulated by these protocols, all efforts were made to minimize suffering.
Animals
mdx and mdx/Spp1-/- littermates from a mixed BL/6-BL/10 background were previously described [13]. 129T2/SvEmsJ (129T2) and DBA/2J WT inbred mice were purchased from the Jackson Laboratory (Bar Harbor, ME; Stock # 002065 and 000671, respectively). Anxa6 and Ltbp4 alleles from DBA/2J background were bred on the 129T2 background by means of initial 129T2 x DBA/2J breeding, followed by seven generations of mating the compound heterozygotes with 129T2 mice. All mice used for experiments with DBA/2J Anxa6 and Ltbp4 alleles were conducted on littermates obtained from mating pairs of compound heterozygotes from the 129T2 background. Mdx mice bearing the BAC-hLTBP4 transgene or the HSA::Ltbp4129 transgene were previously described [43, 44]. Both mdx transgenic lines were generated and bred on a mixed BL/6-BL/10 genetic background; the control cohort of mice for the experiments was created by pooling transgene-deficient littermates from both transgenic lines. Both females and males were used in 129T2 and DBA/2J mice experiments, while only males in experiments with mdx mice. Age of mice at the time of experiment was 8 weeks, unless otherwise specified. Mice were maintained on a 12 hour light/dark cycle and fed ad libitum.
Plasmids
The plasmid encoding human annexin A1 with a carboxy-terminal GFP was obtained from Origene (Rockville, MD; Cat# RG201569).
Electroporation, myofiber isolation, and laser injury
Myofibers from the flexor digitorum brevis (FDB) muscles were electroporated in vivo, as previously described in [56] with modifications described in [31]. Briefly, the footpad was injected with 10μl of hyaluronidase (8units) (Cat #H4272, Sigma, St. Louis, MO). After 2 hours, the footpad was injected with 20μl of 2μg/μl endotoxin-free plasmid. Electroporation was conducted with following parameters: 20 pulses, 20 ms in duration/each, at 1Hz, at 100 V/cm. Recovery was allowed for seven days after electroporation to avoid electroporation-induced damage and to allow plasmid expression [57]. Individual myofibers were then explanted and isolated as previously detailed [31].
Live myofibers were ablated with a laser as described [31, 32, 45]. Briefly, myofibers were dissociated in PBS supplemented with 0.2% BSA and 4mg/ml collagenase type II (Cat # 17101, Life Technologies, Grand Island, NY) at 37 degrees in 10% CO2. Muscle was triturated (20–30 pipetting motions through edge-cut 1000μl filter tip) after 60 and 120 minutes. Fibers were then seeded on MatTek dishes (Cat # P35G-1.5-14-C, MatTek, Ashland MA) in Ringers solution and, after 30 minutes, prepared for imaging by adding FM 4–64 dye (T-13320, Molecular Probes, Grand Island, NY) to a final concentration of 2.5μm.
Laser ablation and subsequent real-time imaging were performed at room temperature using a Nikon A1R laser scanning confocal equipped with GaSP detectors through a 60x Apo lambda 1.4 NA objective driven by Nikon Elements AR software. A single pixel set as 120 nm (0.0144 μm2) was ablated using the 405 nm laser at 100% power for up to 5 seconds. Images were acquired as follows: one image prior to damage (0 seconds; reference for relative fluorescence analyses), one image right after laser injury (bleach point), 10 images every 2 seconds after injury, and then one image every 10 seconds for up to 240 seconds after injury. Quality control for myofibers selected for laser ablation relied on following parameters. Only myofibers adherent to the MatTek dish from end to end and not contracting during imaging were used. Imaged fibers were required to have intact sarcomeres and unruptured sarcolemma. The region of the myofiber selected for laser injury was required to be linear without visible deformation or peripheral nuclei. ANXA1-GFP fluorescence within the myofiber body at time 0 was required to be between 200 and 2000 relative light units (RLUs), as per ImageJ analyses.
Relative fluorescence from an 85μm2 circular region encompassing the lesion area and ANXA1 cap diameter (perpendicular to myofiber axis) over time were calculated from images acquired as described above (FM4-64 and ANXA1 images acquired simultaneously), normalizing values to the pre-injury intensity. This method allows inter-group comparisons and reduces variability. Prism Graphpad was used to calculate averages, and values were normalized to the pre-bleach intensity (F/F0). All measurements were from n = 4 or 5 mice per group (depending on experiment), with ≥10 myofibers per mouse (total 40–50 per genotype or condition). Data analysis was conducted blinded to genotype/treatment group. Stack rendering of time course image sequences was conducted by using the “3D project” built-in feature of Image J with default parameters (Projection method: Brightest Point; Slice spacing: 1.00μm). Stacks were then tilted by 10° in order to show extent of sarcolemmal damage or repair cap formation over time.
Recombinant osteopontin (rOPN) and chemical compound treatments
rOPN was purchased as lyophilized powder (Cat #441-OP; R&D Systems, Minneapolis, MN), resuspended as per manufacturer’s instructions, and stored at -80°C. Treatment of FDB myofibers with rOPN was conducted injecting the footpad with 10μg rOPN in 10μl PBS at 1, 3, and 5 days after electroporation. Laser injury was conducted seven days after electroporation. SB431542 and PF573228 (Cat # S4317 and PZ0117, respectively; Sigma-Aldrich; St. Louis, MO) were resuspended in DMSO (Cat #D2650; Sigma-Aldrich; St. Louis, MO) and stored at -20°C. C2C12 myoblasts (ATCC #CRL-1772; Manassas, VA; all experiments performed at passages 5–15 after ATCC batch thawing) were cultured in DMEM supplemented with 10% FBS (Cat #2442 (lot #14E332; Sigma, St. Louis, MO) 1% P/S (Cat #15070; Thermo Fisher Scientific, Waltham, MA). On treatment start day, myoblasts at ~40% confluence were treated with different protein/compound combinations, or with equal amounts of vehicle, as diluted supplement in growth medium. Myoblasts were harvested for qPCR of ChIP-qPCR analyses 48 hours after treatment onset. Chemical compounds were used at a final concentration of 10μM, while rOPN was diluted to a final concentration of 1μg/ml [40, 58]. Effectiveness of dosing in cells and muscles was tested in pilot experiments by means of qPCR analysis of Slug, Snail, and Mzf1 mRNA at 24 hours after treatment onset. Eight independent replicates of C2C12 treatment groups were used for analyses. Analysis was conducted blinded to treatment group.
Muscle injury and Evans-Blue Dye (dye) staining
Cardiotoxin injury was performed by injecting 20μl of a 10μM cardiotoxin (Cat #TXL1376-1; Accurate Chemical & Scientific Corporation, Westbury, NY) solution in PBS in target muscles in sedated animals (3% isoflurane, 0.8 l/min O2). Cardiotoxin was injected bilaterally in both tibialis anterior and gastrocnemius muscles. Cardiotoxin was released in the center of the muscle through the whole major axis, in order to have a homogenous area of injury at the center of the muscle. EBD staining (10mg/ml in PBS, sterile filtered; cat #E2129; Sigma-Aldrich; St. Louis, MO) was injected in a dedicated cohort of toxin-injured mice intra-peritoneally immediately before toxin-injury. Muscles were collected for fluorescence microscopic analysis 3 hours after toxin-injury.
Serum collection and creatine kinase (CK) analysis
Serum was analyzed as previously reported [59] from animals 24 hours after toxin injury. Serum creatine kinase was analyzed in triplicates for each mouse using the EnzyChrom Creatine Kinase Assay (Cat # ECPK-100; BioAssay Systems, Hayward, CA) following manufacturer’s instructions. Synergy HTX multi-mode plate reader (BioTek®, Winooski, VT) was used to collect data, expressed as U/ml. Analysis was conducted blinded to treatment group.
Histology, immunofluorescence microscopy (IF) and antibodies
Explanted muscles were fixed in 10% formaldehyde (Cat #245–684; Fisher Scientific, Waltham, MA) for histologic processing, or frozen in liquid nitrogen, inside pre-cooled Nalgene cryovials, and stored at -80°C for molecular analyses, or embedded in tissue freezing medium (Cat #TFM-5; Triangle Biomedical Sciences, Durham, NC) for IF analyses. Seven μm sections from the center of paraffin-embedded muscles were stained with hematoxylin and eosin (H&E; cat #12013B, 1070C; Newcomer Supply, Middleton, WI) and Masson’s trichrome (Cat #HT-15; Sigma-Aldrich; St. Louis, MO). Injury area was quantitated from >30 non-consecutive sections per muscle. Analyses were conducted blinded to treatment group. Ten μm sections from the center of frozen-embedded muscles were collected on the cryostat (chamber, -20°C; sample, -15°C; cat #CM1950; Leica, Wetzlar, Germany) for immunostaining. At least 30 non-consecutive sections were analyzed per muscle per condition. Analyses were conducted blinded to treatment group.
IF staining was performed using the following conditions: 4% PFA fixation (10 minutes, room temperature); permeabilization with 0.2% Triton (Cat #X-100; Sigma-Aldrich; St. Louis, MO), 1% bovine serum albumin (Cat #A7906; Sigma-Aldrich; St. Louis, MO) PBS (30 minutes, room temperature); blocking in 1% BSA, 10% FBS PBS (30 minutes at room temperature). For pSmad3+ myonuclei detection, rabbit polyclonal primary antibody (Cat #ab51451; Abcam, Cambridge, MA) staining was counterstained with 1μg/ml WGA conjugated to AlexaFluor594 (Cat #W11262; Thermo Fisher Scientific, Waltham, MA) at room temperature for 1 hour, to outline myofibers. For EBD+ myofiber detection, sections were counterstained with 1μg/ml WGA conjugated to AlexaFluor488 (Cat #W11261; Thermo Fisher Scientific, Waltham, MA) at room temperature for 1 hour; nuclei were counterstained with 0.5μg/ml Hoechst PBS (45 minutes, room temperature). EBD is spontaneously fluorescent in the TRITC channel. Imaging was performed using a Zeiss Axio Observer A1 microscope, using 10X and 20X objectives. Gryphax software (version 1.0.6.598; Jenoptik, Jena, Germany) was used for brightfield pictures, while ZEN 2 software (version 2011; Zeiss, Jena, Germany) was used for immunofluorescence images. Quantitation of injury area and myofiber count was based on sections collected throughout the major muscle axis (at least 10 sections per muscle per animal) and was performed using ImageJ (NIH).
Hydroxyproline quantification
Frozen quadriceps muscles (100mg) was used to measure hydroxyproline content, as previously described [25]. Analyses were conducted blinded to treatment group. Results were reported as mmol (HOP)/mg (tissue).
Quantitative RT-PCR
Total RNA was extracted by means of Trizol (Cat #15596018; Life Technologies, Grand Island, NY) from 30mg tissue as per manufacturer’s instructions. Reverse transcription used two μg of RNA with the qScript cDNA kit (Cat #95048; Quanta Biosciences, Beverly, MA) following kit’s instructions. cDNA was diluted 1:7 and 2μl was used per 10μl qPCR reaction. Each qPCR reaction contained 100nM primers and 5μl iTaq SybrGreen Mix (Cat #1725124; Bio-Rad, Hercules, CA). The list of primers and sequences is provided in S1 Table. CFX96 RealTime System (Bio-Rad, Hercules, CA) was used to run the qPCR reaction (95°C, 15sec; 59°C, 60sec; 40 cycles) and quantitate fluorescence. Relative fold change among biological groups was calculated using Pgk as internal normalizer.
Chromatin immunoprecipitation (ChIP)-qPCR and luciferase assays
ChIP-qPCR was performed according to previously reported conditions [60] and adjustments [61]. Forty-eight hours after treatment, 106 myoblasts were collected, washed, fixed in 1% PFA. Fixation was quenched with 0.1375 mmol glycine (Cat #G7126; Sigma-Aldrich; St. Louis, MO). After lysis of cells and nuclei, chromatin was sonicated for 15 cycles (30 sec, high power; 30 sec pause) in a water bath sonicator set at 4°C (Bioruptor 300; Diagenode, Denville, NJ). One μg chromatin was used for pull-down or for input control samples. The primary antibody (anti-SLUG/SNAIL, cat #ab180714, Abcam, Cambridge, MA) were added at a 1:100 dilution in 300μl final volume, while shaking overnight at 4°C. Chromatin complexes were precipitated with proteinA/G beads (cat #20421; Thermo Scientific, Waltham, MA). DNA was de-complexed with 0.07μg/μl proteinase K (cat #19131; Qiagen, Hilden, Germany) at 55°C and purified through QIAQuick PCR purification kit (cat #28106; Qiagen, Hilden, Germany). qPCR amplification was conducted as described for gene expression analysis, with a dedicated thermal profile (95°C, 30 sec; 55°C, 30 sec; 72°C, 30 sec; 50 cycles). Results were expressed as % of raw expression of the respective input. ChIP-qPCR on isolated myofibers was performed as above, with the following adjustments before chromatin sonication. Freshly-isolated gastrocnemius muscle was finely minced and digested in 5ml/muscle of PBS supplemented with 1mM CaCl2 and 100U/ml collagenase II (Cat # 17101, Life Technologies, Grand Island, NY) at 37°C for 1 hour with shaking. After filtration through a 40μm strainer (Cat # 22363547, Fisher Scientific, Waltham, MA), the unfiltered fraction (enriched in myofibers) was kept for further procedures. Separation of mononuclear fraction from myofibers in the filtered suspension was confirmed at a microscope. Myofiber lysis was performed in lysis buffer, using 700μl per muscle, with ~250μl 2.3mm zirconia/silica beads (Cat # 11079125z, BioSpec, Bartlesville, OK). Lysis buffer consisted of 10mM HEPES (pH 7.3; Cat # H3375), 10mM KCl (Cat # P9541), 5mM MgCl2 (Cat # M8266), 0.5mM DTT (Cat # 646563), 3μg/ml cytochalasin B (C6762; all reagents from Sigma, St. Louis, MO); protease inhibitor cocktail (Cat # 11852700, Roche, Mannheim, Germany)). Myofibers were homogenized at the Mini-BeadBeater-16 (Cat # 607, Biospec, Bartlesville, OK) for 30 sec, then by rotating at 4°C for 30 min. Samples were then centrifuged at 3000g for 5 minutes at 4°C; pellet was resuspended in cell lysis buffer, as per described protocol [60], supplemented with 3μg/ml cytochalasin B, and incubated on ice for 10 minutes. Nuclei were pelleted at 300g for 10 min at 4°C, and resuspended in 1ml 1% PFA for 5 min at room temperature. Fixation was quenched with 100μl of 1.375M glycine (Cat # BP381-5, Fisher Scientific, Waltham, MA). Nuclei were re-pelleted as before, and then processed following the described procedure [60], as mentioned for myoblasts. However, all solutions were supplemented with 3μg/ml cytochalasin B during chromatin preparation and sonication, antibody incubation, and wash steps.
Statistical analysis and data presentation
Statistical analyses were performed with Prism (Graphpad, La Jolla, CA). Tests used for statistical comparison depended on group number and normality test (Pearson-D’Agostino). Typically, 2way ANOVA with Bonferroni multi-comparison was used to compare treatment or genotype effect on curves over time. When comparing >2 groups for one variable (typically treatment, or genotype), 1way ANOVA + Bonferroni multi-comparison was used. When comparing two groups for one variable, unpaired t-test with Welch’s correction was used, in order to account for skews in standard deviation between groups. For ANOVA and t-test analyses, a P value less than 0.05 was considered significant. Data were presented as single values (dot plots, histograms) when the number of data points was less than 10. In analyses pooling larger data point sets per group, Tukey distribution bars were used to emphasize data range distribution and histograms with error bars were used to emphasize shifts in average values. Analysis pooling data points over time were presented as marked line plots. Dot plots, histograms and marked line plots depict mean ± SEM. Box plots depict the Tukey distribution of the data pool: interquartile distribution; lower whisker, 25th percentile minus 1.5 times the interquartile range; upper whisker, 75th percentile plus 1.5 times the interquartile range.
Supporting information
(DOCX)
A) Seven days after injection of rOPN to FDB muscles, qPCR analysis shows upregulation of Mmp2 and Mzf1:markers downstream of osteopontin-integrin signaling. Histograms, single values & avg±sem; n = 5 mice/group; *, P<0.05 vs vehicle, unpaired t-test with Welch’s correction. B) Diagram depicting the confocal imaging series used in these studies following laser injury to create sarcolemmal disruption. Image complication was used for both FM4-64 and ANXA1 image stacks over time.
(TIF)
A) Expression levels of Anxa1 and Anxa6 were upregulated in TA muscles of mdx/Spp1-/- mice, as compared to control mdx animals. B) Hydroxyproline content, as an indicator of fibrosis, was reduced in mdx/Spp1-/- quadriceps and diaphragm muscles compared to mdx muscles. C) Serum CK levels were significantly decreased in mdx/Spp1-/- mice compared to control mdx animals. Histograms, single values & avg±sem; n = 4 mice/group; *, P<0.05 vs mdx control, unpaired t-test with Welch’s correction.
(TIF)
RNA-Seq analysis of quadriceps muscle tissue from 129T2-Sgcg-/- and DBA/2J-Sgcg-/- mice versus strain-matched WT littermates. Fold change analysis showed that Spp1 was upregulated in the presence of dystrophic remodeling in both strains. Moreover, Spp1 was consistently upregulated in DBA/2J muscle, as compared to the 129T2 muscle, in both wildtype and dystrophic conditions. Histograms, single values & avg±sem; n = 3 mice/group; *, P<0.05 vs designated group, 1way ANOVA + Bonferroni.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by U54AR052646 (NIH), RO1NS047726 (NIH), R01HL61322 (NIH), P30AR057230 (NIH) and Parent Project for Muscular Dystrophy. MQ is supported by the Muscular Dystrophy Association and the AANEM Foundation (Development Grant #479350). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Emery AE. The muscular dystrophies. Lancet. 2002;359(9307):687–95. doi: 10.1016/S0140-6736(02)07815-7 . [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Knudson CM, Campbell KP, Kunkel LM. Subcellular fractionation of dystrophin to the triads of skeletal muscle. Nature. 1987;330(6150):754–8. doi: 10.1038/330754a0 . [DOI] [PubMed] [Google Scholar]
- 3.Hack AA, Ly CT, Jiang F, Clendenin CJ, Sigrist KS, Wollmann RL, et al. Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin. J Cell Biol. 1998;142(5):1279–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin J, Tajrishi MM, Ogura Y, Kumar A. Wasting mechanisms in muscular dystrophy. The international journal of biochemistry & cell biology. 2013;45(10):2266–79. Epub 2013/05/15. doi: 10.1016/j.biocel.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quattrocelli M, Spencer MJ, McNally EM. Outside in: The matrix as a modifier of muscular dystrophy. Biochim Biophys Acta. 2017;1864(3):572–9. doi: 10.1016/j.bbamcr.2016.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heydemann A, Huber JM, Demonbreun A, Hadhazy M, McNally EM. Genetic background influences muscular dystrophy. Neuromuscul Disord. 2005;15(9–10):601–9. doi: 10.1016/j.nmd.2005.05.004 . [DOI] [PubMed] [Google Scholar]
- 7.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244(4912):1578–80. . [DOI] [PubMed] [Google Scholar]
- 8.Calyjur PC, Almeida Cde F, Ayub-Guerrieri D, Ribeiro AF Jr., Fernandes Sde A, Ishiba R, et al. The mdx Mutation in the 129/Sv Background Results in a Milder Phenotype: Transcriptome Comparative Analysis Searching for the Protective Factors. PLoS One. 2016;11(3):e0150748 Epub 2016/03/10. doi: 10.1371/journal.pone.0150748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coley WD, Bogdanik L, Vila MC, Yu Q, Van Der Meulen JH, Rayavarapu S, et al. Effect of genetic background on the dystrophic phenotype in mdx mice. Hum Mol Genet. 2016;25(1):130–45. doi: 10.1093/hmg/ddv460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giachelli CM, Steitz S. Osteopontin: a versatile regulator of inflammation and biomineralization. Matrix Biol. 2000;19(7):615–22. . [DOI] [PubMed] [Google Scholar]
- 11.Chen YW, Zhao P, Borup R, Hoffman EP. Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology. J Cell Biol. 2000;151(6):1321–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haslett JN, Sanoudou D, Kho AT, Bennett RR, Greenberg SA, Kohane IS, et al. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc Natl Acad Sci U S A. 2002;99(23):15000–5. doi: 10.1073/pnas.192571199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vetrone SA, Montecino-Rodriguez E, Kudryashova E, Kramerova I, Hoffman EP, Liu SD, et al. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J Clin Invest. 2009;119(6):1583–94. doi: 10.1172/JCI37662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tkatchenko AV, Le Cam G, Leger JJ, Dechesne CA. Large-scale analysis of differential gene expression in the hindlimb muscles and diaphragm of mdx mouse. Biochim Biophys Acta. 2000;1500(1):17–30. . [DOI] [PubMed] [Google Scholar]
- 15.Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, et al. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum Mol Genet. 2002;11(3):263–72. . [DOI] [PubMed] [Google Scholar]
- 16.Turk R, Sterrenburg E, van der Wees CG, de Meijer EJ, de Menezes RX, Groh S, et al. Common pathological mechanisms in mouse models for muscular dystrophies. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20(1):127–9. Epub 2005/11/25. doi: 10.1096/fj.05-4678fje . [DOI] [PubMed] [Google Scholar]
- 17.Galindo CL, Soslow JH, Brinkmeyer-Langford CL, Gupte M, Smith HM, Sengsayadeth S, et al. Translating golden retriever muscular dystrophy microarray findings to novel biomarkers for cardiac/skeletal muscle function in Duchenne muscular dystrophy. Pediatric research. 2016;79(4):629–36. Epub 2015/12/18. doi: 10.1038/pr.2015.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zanotti S, Gibertini S, Di Blasi C, Cappelletti C, Bernasconi P, Mantegazza R, et al. Osteopontin is highly expressed in severely dystrophic muscle and seems to play a role in muscle regeneration and fibrosis. Histopathology. 2011;59(6):1215–28. Epub 2011/12/20. doi: 10.1111/j.1365-2559.2011.04051.x . [DOI] [PubMed] [Google Scholar]
- 19.Pegoraro E, Hoffman EP, Piva L, Gavassini BF, Cagnin S, Ermani M, et al. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76(3):219–26. doi: 10.1212/WNL.0b013e318207afeb [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capote J, Kramerova I, Martinez L, Vetrone S, Barton ER, Sweeney HL, et al. Osteopontin ablation ameliorates muscular dystrophy by shifting macrophages to a pro-regenerative phenotype. J Cell Biol. 2016;213(2):275–88. doi: 10.1083/jcb.201510086 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bello L, Kesari A, Gordish-Dressman H, Cnaan A, Morgenroth LP, Punetha J, et al. Genetic modifiers of ambulation in the Cooperative International Neuromuscular Research Group Duchenne Natural History Study. Annals of neurology. 2015;77(4):684–96. Epub 2015/02/03. doi: 10.1002/ana.24370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bello L, Piva L, Barp A, Taglia A, Picillo E, Vasco G, et al. Importance of SPP1 genotype as a covariate in clinical trials in Duchenne muscular dystrophy. Neurology. 2012;79(2):159–62. Epub 2012/06/30. doi: 10.1212/WNL.0b013e31825f04ea [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanigan KM, Ceco E, Lamar KM, Kaminoh Y, Dunn DM, Mendell JR, et al. LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Annals of neurology. 2013;73(4):481–8. Epub 2013/02/27. doi: 10.1002/ana.23819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Bergen JC, Hiller M, Bohringer S, Vijfhuizen L, Ginjaar HB, Chaouch A, et al. Validation of genetic modifiers for Duchenne muscular dystrophy: a multicentre study assessing SPP1 and LTBP4 variants. J Neurol Neurosurg Psychiatry. 2015;86(10):1060–5. doi: 10.1136/jnnp-2014-308409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heydemann A, Ceco E, Lim JE, Hadhazy M, Ryder P, Moran JL, et al. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J Clin Invest. 2009;119(12):3703–12. doi: 10.1172/JCI39845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamar KM, Miller T, Dellefave-Castillo L, McNally EM. Genotype-Specific Interaction of Latent TGFbeta Binding Protein 4 with TGFbeta. PLoS One. 2016;11(2):e0150358 doi: 10.1371/journal.pone.0150358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hullinger TG, Pan Q, Viswanathan HL, Somerman MJ. TGFbeta and BMP-2 activation of the OPN promoter: roles of smad- and hox-binding elements. Exp Cell Res. 2001;262(1):69–74. doi: 10.1006/excr.2000.5074 . [DOI] [PubMed] [Google Scholar]
- 28.Kubota T, Zhang Q, Wrana JL, Ber R, Aubin JE, Butler WT, et al. Multiple forms of SppI (secreted phosphoprotein, osteopontin) synthesized by normal and transformed rat bone cell populations: regulation by TGF-beta. Biochem Biophys Res Commun. 1989;162(3):1453–9. . [DOI] [PubMed] [Google Scholar]
- 29.Driver J, Weber CE, Callaci JJ, Kothari AN, Zapf MA, Roper PM, et al. Alcohol inhibits osteopontin-dependent transforming growth factor-beta1 expression in human mesenchymal stem cells. J Biol Chem. 2015;290(16):9959–73. doi: 10.1074/jbc.M114.616888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber CE, Kothari AN, Wai PY, Li NY, Driver J, Zapf MA, et al. Osteopontin mediates an MZF1-TGF-beta1-dependent transformation of mesenchymal stem cells into cancer-associated fibroblasts in breast cancer. Oncogene. 2015;34(37):4821–33. doi: 10.1038/onc.2014.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demonbreun AR, McNally EM. DNA Electroporation, Isolation and Imaging of Myofibers. J Vis Exp. 2015;(106):e53551 doi: 10.3791/53551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Demonbreun AR, Quattrocelli M, Barefield DY, Allen MV, Swanson KE, McNally EM. An actin-dependent annexin complex mediates plasma membrane repair in muscle. J Cell Biol. 2016. doi: 10.1083/jcb.201512022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vila MC, Rayavarapu S, Hogarth MW, Van der Meulen JH, Horn A, Defour A, et al. Mitochondria mediate cell membrane repair and contribute to Duchenne muscular dystrophy. Cell Death Differ. 2017;24(2):330–42. Epub 2016/11/12. doi: 10.1038/cdd.2016.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi Z, Guo H, Russell MB, Liu Y, Sullenger BA, Kuo PC. RNA aptamer blockade of osteopontin inhibits growth and metastasis of MDA-MB231 breast cancer cells. Mol Ther. 2009;17(1):153–61. doi: 10.1038/mt.2008.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paliwal P, Pishesha N, Wijaya D, Conboy IM. Age dependent increase in the levels of osteopontin inhibits skeletal muscle regeneration. Aging. 2012;4(8):553–66. Epub 2012/08/24. doi: 10.18632/aging.100477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstein JA, Bogdanovich S, Beiriger A, Wren LM, Rossi AE, Gao QQ, et al. Excess SMAD signaling contributes to heart and muscle dysfunction in muscular dystrophy. Hum Mol Genet. 2014;23(25):6722–31. doi: 10.1093/hmg/ddu390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278(23):21113–23. doi: 10.1074/jbc.M211304200 . [DOI] [PubMed] [Google Scholar]
- 38.Naber HP, Drabsch Y, Snaar-Jagalska BE, ten Dijke P, van Laar T. Snail and Slug, key regulators of TGF-beta-induced EMT, are sufficient for the induction of single-cell invasion. Biochem Biophys Res Commun. 2013;435(1):58–63. doi: 10.1016/j.bbrc.2013.04.037 . [DOI] [PubMed] [Google Scholar]
- 39.Prokop JW, Liu Y, Milsted A, Peng H, Rauscher FJ 3rd. A method for in silico identification of SNAIL/SLUG DNA binding potentials to the E-box sequence using molecular dynamics and evolutionary conserved amino acids. J Mol Model. 2013;19(9):3463–9. doi: 10.1007/s00894-013-1876-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62(1):65–74. . [DOI] [PubMed] [Google Scholar]
- 41.Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, et al. Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem. 2007;282(20):14845–52. doi: 10.1074/jbc.M606695200 . [DOI] [PubMed] [Google Scholar]
- 42.Swaggart KA, Heydemann A, Palmer AA, McNally EM. Distinct genetic regions modify specific muscle groups in muscular dystrophy. Physiol Genomics. 2011;43(1):24–31. Epub 2010/10/21. doi: 10.1152/physiolgenomics.00172.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamar KM, Bogdanovich S, Gardner BB, Gao QQ, Miller T, Earley JU, et al. Overexpression of Latent TGFbeta Binding Protein 4 in Muscle Ameliorates Muscular Dystrophy through Myostatin and TGFbeta. PLoS Genet. 2016;12(5):e1006019 doi: 10.1371/journal.pgen.1006019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ceco E, Bogdanovich S, Gardner B, Miller T, DeJesus A, Earley JU, et al. Targeting latent TGFbeta release in muscular dystrophy. Sci Transl Med. 2014;6(259):259ra144 doi: 10.1126/scitranslmed.3010018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swaggart KA, Demonbreun AR, Vo AH, Swanson KE, Kim EY, Fahrenbach JP, et al. Annexin A6 modifies muscular dystrophy by mediating sarcolemmal repair. Proc Natl Acad Sci U S A. 2014;111(16):6004–9. doi: 10.1073/pnas.1324242111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen YW, Nagaraju K, Bakay M, McIntyre O, Rawat R, Shi R, et al. Early onset of inflammation and later involvement of TGFbeta in Duchenne muscular dystrophy. Neurology. 2005;65(6):826–34. Epub 2005/08/12. doi: 10.1212/01.wnl.0000173836.09176.c4 . [DOI] [PubMed] [Google Scholar]
- 47.Vidal B, Serrano AL, Tjwa M, Suelves M, Ardite E, De Mori R, et al. Fibrinogen drives dystrophic muscle fibrosis via a TGFbeta/alternative macrophage activation pathway. Genes & development. 2008;22(13):1747–52. Epub 2008/07/03. doi: 10.1101/gad.465908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akhurst RJ, Hata A. Targeting the TGFbeta signalling pathway in disease. Nature reviews Drug discovery. 2012;11(10):790–811. Epub 2012/09/25. doi: 10.1038/nrd3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacDonald EM, Cohn RD. TGFbeta signaling: its role in fibrosis formation and myopathies. Current opinion in rheumatology. 2012;24(6):628–34. Epub 2012/08/25. doi: 10.1097/BOR.0b013e328358df34 . [DOI] [PubMed] [Google Scholar]
- 50.Accornero F, Kanisicak O, Tjondrokoesoemo A, Attia AC, McNally EM, Molkentin JD. Myofiber-specific inhibition of TGFbeta signaling protects skeletal muscle from injury and dystrophic disease in mice. Hum Mol Genet. 2014;23(25):6903–15. Epub 2014/08/12. doi: 10.1093/hmg/ddu413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wahl SM, Allen JB, Costa GL, Wong HL, Dasch JR. Reversal of acute and chronic synovial inflammation by anti-transforming growth factor beta. The Journal of experimental medicine. 1993;177(1):225–30. Epub 1993/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gushchina LV, Bhattacharya S, McElhanon KE, Choi JH, Manring H, Beck EX, et al. Treatment with Recombinant Human MG53 Protein Increases Membrane Integrity in a Mouse Model of Limb Girdle Muscular Dystrophy 2B. Mol Ther. 2017. Epub 2017/07/29. doi: 10.1016/j.ymthe.2017.06.025 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffman EP, Gordish-Dressman H, McLane VD, Devaney JM, Thompson PD, Visich P, et al. Alterations in osteopontin modify muscle size in females in both humans and mice. Medicine and science in sports and exercise. 2013;45(6):1060–8. Epub 2013/01/01. doi: 10.1249/MSS.0b013e31828093c1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quattrocelli M, Barefield DY, Warner JL, Vo AH, Hadhazy M, Earley JU, et al. Intermittent glucocorticoid steroid dosing enhances muscle repair without eliciting muscle atrophy. J Clin Invest. 2017. Epub 2017/05/10. doi: 10.1172/jci91445 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dadgar S, Wang Z, Johnston H, Kesari A, Nagaraju K, Chen YW, et al. Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. J Cell Biol. 2014;207(1):139–58. Epub 2014/10/15. doi: 10.1083/jcb.201402079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiFranco M, Quinonez M, Capote J, Vergara J. DNA transfection of mammalian skeletal muscles using in vivo electroporation. J Vis Exp. 2009;(32). doi: 10.3791/1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerr JP, Ziman AP, Mueller AL, Muriel JM, Kleinhans-Welte E, Gumerson JD, et al. Dysferlin stabilizes stress-induced Ca2+ signaling in the transverse tubule membrane. Proc Natl Acad Sci U S A. 2013;110(51):20831–6. doi: 10.1073/pnas.1307960110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Many GM, Yokosaki Y, Uaesoontrachoon K, Nghiem PP, Bello L, Dadgar S, et al. OPN-a induces muscle inflammation by increasing recruitment and activation of pro-inflammatory macrophages. Exp Physiol. 2016;101(10):1285–300. doi: 10.1113/EP085768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demonbreun AR, Allen MV, Warner JL, Barefield DY, Krishnan S, Swanson KE, et al. Enhanced Muscular Dystrophy from Loss of Dysferlin Is Accompanied by Impaired Annexin A6 Translocation after Sarcolemmal Disruption. Am J Pathol. 2016;186(6):1610–22. doi: 10.1016/j.ajpath.2016.02.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carey MF, Peterson CL, Smale ST. Chromatin immunoprecipitation (ChIP). Cold Spring Harbor protocols. 2009;2009(9):pdb.prot5279. Epub 2010/02/12. doi: 10.1101/pdb.prot5279 . [DOI] [PubMed] [Google Scholar]
- 61.Quattrocelli M, Swinnen M, Giacomazzi G, Camps J, Barthelemy I, Ceccarelli G, et al. Mesodermal iPSC-derived progenitor cells functionally regenerate cardiac and skeletal muscle. J Clin Invest. 2015;125(12):4463–82. doi: 10.1172/JCI82735 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
A) Seven days after injection of rOPN to FDB muscles, qPCR analysis shows upregulation of Mmp2 and Mzf1:markers downstream of osteopontin-integrin signaling. Histograms, single values & avg±sem; n = 5 mice/group; *, P<0.05 vs vehicle, unpaired t-test with Welch’s correction. B) Diagram depicting the confocal imaging series used in these studies following laser injury to create sarcolemmal disruption. Image complication was used for both FM4-64 and ANXA1 image stacks over time.
(TIF)
A) Expression levels of Anxa1 and Anxa6 were upregulated in TA muscles of mdx/Spp1-/- mice, as compared to control mdx animals. B) Hydroxyproline content, as an indicator of fibrosis, was reduced in mdx/Spp1-/- quadriceps and diaphragm muscles compared to mdx muscles. C) Serum CK levels were significantly decreased in mdx/Spp1-/- mice compared to control mdx animals. Histograms, single values & avg±sem; n = 4 mice/group; *, P<0.05 vs mdx control, unpaired t-test with Welch’s correction.
(TIF)
RNA-Seq analysis of quadriceps muscle tissue from 129T2-Sgcg-/- and DBA/2J-Sgcg-/- mice versus strain-matched WT littermates. Fold change analysis showed that Spp1 was upregulated in the presence of dystrophic remodeling in both strains. Moreover, Spp1 was consistently upregulated in DBA/2J muscle, as compared to the 129T2 muscle, in both wildtype and dystrophic conditions. Histograms, single values & avg±sem; n = 3 mice/group; *, P<0.05 vs designated group, 1way ANOVA + Bonferroni.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.