Abstract
Human cytosolic sialidase (Neuraminidase 2, NEU2) catalyzes the removal of terminal sialic acid residues from glycoconjugates. The effect of siastatin B, known as a sialidase inhibitor, has not been evaluated toward human NEU2 yet. We studied the regulation of NEU2 activity by siastatin B in vitro and predicted the interaction in silico. Inhibitory and stabilizing effects of siastatin B were analyzed in comparison with DANA (2-deoxy-2,3-dehydro-N-acetylneuraminic acid) toward 4-umbelliferyl N-acetylneuraminic acid (4-MU-NANA)- and α2,3-sialyllactose-degrading activities of recombinant NEU2 produced by E. coli GST-fusion gene expression. Siastatin B exhibited to have higher competitive inhibitory activity toward NEU2 than DANA at pH 4.0. We also revealed the stabilizing effect of siastatin B toward NEU2 activity at acidic pH. Docking model was constructed on the basis of the crystal structure of NEU2/DANA complex (PDB code: 1VCU). Molecular docking predicted that electrostatic neutralization of E111 and E218 residues of the active pocket should not prevent siastatin B from binding at pH 4.0. The imino group (1NH) of siastatin B can also interact with D46, neutralized at pH 4.0. Siastatin B was suggested to have higher affinity to the active pocket of NEU2 than DANA, although it has no C7–9 fragment corresponding to that of DANA. We demonstrated here the pH-dependent affinity of siastatin B toward NEU2 to exhibit potent inhibitory and stabilizing activities. Molecular interaction between siastatin B and NEU2 will be utilized to develop specific inhibitors and stabilizers (chemical chaperones) not only for NEU2 but also the other human sialidases, including NEU1, NEU3 and NEU4, based on homology modeling.
Keywords: Neuraminidase 2, Siastatin B, Docking model, Inhibitor, Chemical chaperone
Highlights
-
•
Siastatin B was found to have potent inhibitory effect on human NEU2 at acidic pH.
-
•
Siastatin B could also effectively stabilize the NEU2 from denaturation at acidic pH.
-
•
We found D46, E111 and E218 regulating the enzymatic functions of NEU2 at acidic pH.
-
•
Findings on siastatin B will be utilized to develop novel modulators for sialidases.
1. Introduction
Sialidases (neuraminidases) are exo-glycosidases, which catalyze the initial step of catabolism of sialylglycoconjugates by hydrolyzing the terminal non-reducing sialic acid residues from the oligosaccharides attached to glycoproteins and glycolipids, and have marked effects on the biological functions of these glycoconjugates [1], [2]. There are four human neuraminidases (NEUs) different in their enzymatic properties including substrate specificities and pH optima [1], [3]. They are also classified based on their major subcellular distributions as intralysosomal NEU1, cytosolic NEU2, plasma membrane NEU3 and mitochondria/lysosomal NEU4. The four human NEUs have the unique amino acid sequences common to the sialidase family, including the arginine triad [1], [2], [4], Asp box [1], [2], [5], and RIP motif [6]. The sequence identity (average) among NEUs2–4 is 34 (±5.2)%, although that of NEU1 with NEUs2–4 [23 (±0.4)%] is relatively low. At present, the crystal structure of NEU2 is the only one elucidated among them [7], which is utilized for homology modeling for other NEUs [8], [9].
Sialidase inhibitors have been applied as a useful tool to characterize the catalytic activities including substrate specificity, and elucidate their biological functions and disease processes [10]. DANA, as a sialic acid transition-state analog, is a well-used inhibitor and exhibits moderate inhibitory activity toward a variety of sialidases, although it is not selective for influenza virus neuraminidases with Ki values in micromolar range [11], [12] compared to those of zanamivir (Relenza) and oseltamivir (Tamiflu) in nanomolar range [13], [14], [15], [16], [17]. The inhibitory effects of DANA toward human NEU2 were also reported by Magesh et al. [9] and Chavas et al. [7] at around neutral pH with Ki value in the mM order. Siastatin B is a sialidase inhibitor, initially isolated from a Streptomyces strain and identified as 6-acetamido-3-piperidine carboxylate structurally similar to sialic acid (N-acetylneuraminic acid) [18]. Siastatin B has broad spectra and inhibits sialidases isolated from various microorganisms, animal tissues and viruses [18], [19], [20]. The charge distribution in siastatin B as a zwitterion resembles that in the N-acetylneuraminate oxocarbenium ion as the putative intermediate form in the enzyme-catalyzed reaction. This feature may account for its binding to the active pockets of sialidases and inhibitory activity [19]. Siastatin B has been also reported to have the inhibitory effects toward mammalian sialidases with IC50 values in the µM range including cytosolic sialidases purified from rat liver and skeletal muscle cells, although the sensitivities of sialidases to the inhibitor differ dependently on species [18]. However, the effect of siastatin B toward human NEU2 has not been evaluated yet. In this study, we first demonstrated the inhibitory effects of siastatin B in vitro toward recombinant human NEU2. The pH dependency of inhibitory activity of siastatin B was also examined in comparison with that of DANA. We also found the stabilizing property of siastatin B toward human NEU2 under acidic pH conditions. Moreover, we also performed molecular docking of siastatin B as to the active pocket of human lysosomal NEU1 model based on the three-dimensional structure of NEU2. Relationship between inhibitory and stabilizing potency of siastatin B, and interaction with amino acid residues located around active pocket of NEU2 and NEU1 model was also discussed.
2. Materials and methods
2.1. Substrates and inhibitors
4-Methylumbelliferyl-N-acetyl-α-d-neuraminic acid (4-MU-NANA) was purchased from Nacalai Tesque, Inc. (Kyoto, Japan). α2,3-Sialyllactose was purchased from Sigma-Aldrich (St. Louis, MO). Siastatin B and DANA were purchased from Peptide Institute, Inc. (Osaka, Japan) and Calbiochem (CA), respectively.
2.2. Expression and purification of recombinant human NEU2
Expression and purification of NEU2 proteins had been described elsewhere [20]. Briefly, E. coli Origami B cells (Takara, Tokyo, Japan) were transformed with plasmid pGEX-2 T-NEU2, and the transformed cells were grown in LB-ampicillin(+) medium at 37 °C until the optical density became 0.6 before the addition of isopropyl-α-d-thiogalactopyranoside (IPTG). After 40 h growth at 27.5 °C in the presence of 0.1 mM IPTG, cells were harvested by centrifugation and suspended in phosphate-buffered saline (PBS). The cells were then lysed in PBS (pH 7.4) by sonication, and a supernatant was obtained by centrifugation during which Triton X-100 was added to a final concentration of 1%. The resultant supernatant containing the GST-NEU2 fusion proteins was applied to a GSH-Sepharose (GE Healthcare, Uppsala, Sweden) affinity chromatography column equilibrated with PBS (pH 7.4) containing 1% Triton X-100. Cleavage of the thrombin-recognition sequence located between the GST and NEU2 in fusion proteins was carried out by adding 100 µL of a thrombin solution comprising 1 IU in PBS, and then incubating the column for 16 h at 22 °C. The eluted NEU2 was further applied to a diethylaminoethyl (DEAE)-Sepharose ion exchange chromatography column equilibrated with 10 mM sodium phosphate buffer (pH 8.0) containing 1 M NaCl, and the NEU2 proteins were eluted as the pass-through fraction using an AKTA apparatus (GE Healthcare). The crude cell lysate and fractions obtained during the purification procedures were assayed for NEU2 activity, and the protein concentration of each fraction was also determined according to the method of Lowry [21]. The purity of the enzyme fraction was determined by SDS-PAGE [22] and Coomassie brilliant blue (CBB R-350) staining.
Western blotting was performed to analyze the molecular weight of purified monomeric NEU2, protein samples were subjected to SDS-PAGE on a 10% acrylamide gel. The protein was transferred to polyvinylidine difluoride membrane (Millipore Billerica, MA). On immunoblotting, membrane was incubated with 50% (v/v) Blocking One (Nacalai Tesque, Japan) in Tris-buffered saline (TBS) (pH 7.4) for 2 h at room temperature. The membrane was treated with anti-NEU2 rabbit polyclonal antibody (Sigma St. Louis, MO, USA) diluted with Blocking One/TBS (1:1,000 dilutions) overnight at 4 °C. After washing with TBS containing 0.1% Tween 20, the membrane was treated with horseradish peroxidase-conjugated anti-rabbit IgG (1:1,000 dilution) (Sigma) as secondary antibodies for 1 h at room temperature. After washing with same buffer, detection of antibody binding was carried out with ECL (PerkinElmer Inc. MA) according to the manufacturer's instructions.
2.3. Enzyme and inhibition assaying of recombinant human NEU2
NEU2 enzyme activity was measured using the synthetic substrate 4-MU-NANA by fluorimetric assay [20]. Routinely, the reaction mixture (40 μL) was composed of 8 μL of 0.2 M sodium acetate buffer (pH 4.0 or pH 6.0), 2 μL of 50 mg/mL bovine serum albumin (BSA), 5 μL of 2 mM 4-MU-NANA, 20 μL of an appropriately diluted inhibitor solution and 5 μL of recombinant human NEU2. In the inhibition assay for NEU2, the enzyme activity in this mixture was maintained at 9.5 nmol/30 min/5 μL. After incubation for 30 min at 37 °C, 770 μL of glycine–NaOH (pH 10.7) was added to stop the reaction. The fluorescence of the released 4-methylumbelliferone (4-MU) was measured using a spectrofluorometer (Fluoroskan ascent microplate fluorometer, Finland) at 360 nm (excitation) and 455 nm (emission). The concentration causing 50% inhibition (IC50) of each inhibitor for NEU2 activity also determined. To evaluate the mode of inhibition by each inhibitor, the concentration of inhibitor versus the reaction rate was plotted, and Ki value was determined by the method of Lineweaver and Burk [23].
The NEU2 activity toward α2,3-sialyllactose was determined by an improved Warren method [24] using sialic acid assay kit (Abnova, Taiwan, Version: 03), in which sialic acid is oxidized to formylpyruvic acid, which reacts with thiobarbituric acid to form a pink colored product. The color intensity at 549 nm is directly proportional to sialic acid concentration in the sample. Reactions were set up for positive control sample by using 5 µg of purified NEU2, in 25 mM of sodium-citrate-phosphate buffer at pH 4.0 and or 6.0, containing 50 µg of α-(2,3) sialyllactose in a final volume of 100 µL (0.75 mM final concentration) [25]. The inhibitory effects of siastatin B and DANA toward NEU2 were measured in the presence of 600 µM inhibitors in a final volume of 100 µL. After incubation at 37 °C for 1 h, total sialic acid (bound sialic acid) and released sialic acid (hydrolyzed sialic acid) were determined according to the procedures recommended by the manufacturer.
2.4. In vitro stabilization assay for NEU2 by siastatin B and DANA
Purified NEU2 proteins (8 µg/mL) were pre-incubated in 0.1 M sodium phosphate buffers at various pH at 37 °C with or without siastatin B or DANA for several time intervals. Pre-incubation was terminated by the addition of two volumes of 0.2 M sodium phosphate buffer (pH 6.0) and kept on ice immediately before enzyme assay. The enzyme activity was determined at pH 6.0 with 4-MU-NANA as the substrate.
2.5. Molecular docking study
Molecular docking of siastatin B to NEU2 was performed on the basis of X-ray crystal structure of human NEU2 in complex with the inhibitor DANA (PDB-ID: 1VCU) at pH 4.0 and 6.0, which involved three steps: calculation of protein ionization of NEU2 at pH 4.0 and 6.0, ligand binding prediction of siastatin B, and binding free energy calculation. Protein ionization of NEU2 at pH 4.0 and 6.0 was predicted using Discovery Studio 3.1 (Accelrys, Inc.) utilizing the theory developed by Bashford and Karplus [26]. Results from assignment of ionization states of titratable amino acids in NEU2, following amino acids were protonated at pH 4.0: E10, E39, E50, D58, E72, E111, D131, D141, E153, E218, E223, D251, D254, E257, D336, D345, E360, E361 and E376, and at pH 6.0: D336. Two pH-dependent models of NEU2/DANA complex were refined for ligand docking and binding energy calculation using the Protein Preparation Wizard Script within Maestro (Schrödinger, LLC). Siastatin B was docked into the reference position of the DANA of DANA/NEU2 complex models at pH 4.0 and pH 6.0 using the Glide ver. 5 SP mode (Schrödinger, LLC) [27]. Finally, the binding energies between each ligand and NEU2 for at pH 4.0 and pH 6.0 were estimated using CHARMm based energies and implicit solvation methods on Discovery Studio 3.1. Moleculer docking of siastatin B was also performed as to the human NEU1 model constructed by homology modeling using two reference proteins, human NEU2 and bacterial sialidase from Micromonospora viridifaciens [28]. Using the model, calculation of protein ionization of NEU1 at pH 4.0, ligand binding prediction of siastatin B and binding free energy calculation were performed according to the above docking procedure for NEU2.
3. Results
3.1. Purification of recombinant human NEU2 produced by Escherichia coli gene expression system
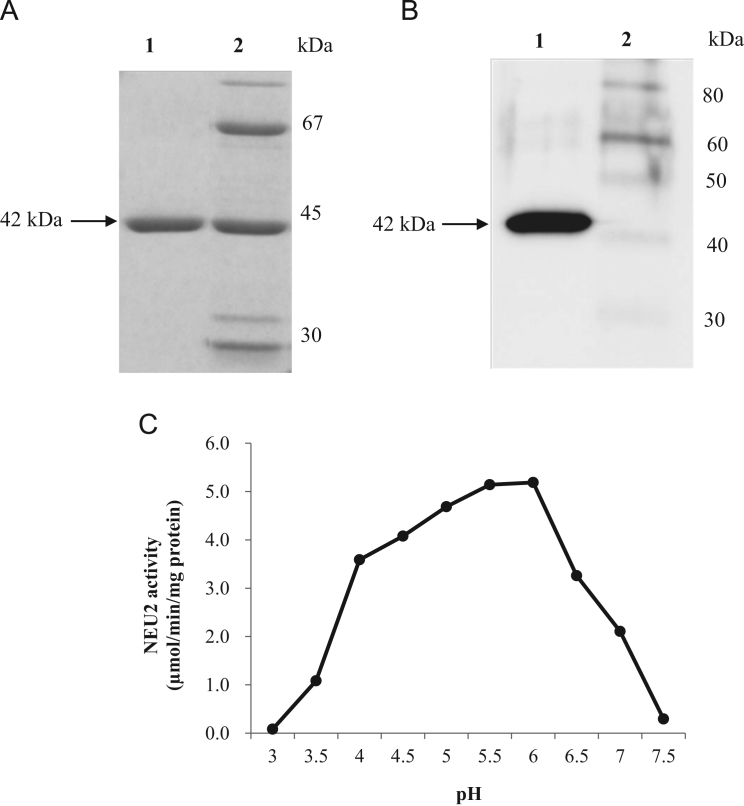
In order to obtain a large amount of recombinant human NEU2 protein, we expressed the GST-NEU2 fusion gene with pGEX-2 T vector in E. coli (Origami B strain) as described under Section 2. As shown in Fig. 1(A), the purified protein appeared as a single band on SDS-PAGE, with an apparent molecular mass of about 42 kDa, indicating a high degree of purity. In addition, the 42-kDa protein was immunostained by using anti-NEU2 antibody on western blotting (Fig. 1(B)). Purified NEU2 exhibited 4-MU-NANA-degrading activity under broad pH conditions (pH 4.0–6.5) with optimal pH range (5.5–6.0) (Fig. 1(C)). The specific activities were 5.19 at pH 6.0, and 3.59 μmol/min/mg protein at pH 4.0. The Km and apparent Vmax values were 0.45 mM and 38.8 μmol/min/mg protein at pH 6.0, and 0.28 mM and 26.5 μmol/min/mg protein at pH 4.0, respectively.
Fig. 1.
Analysis of purified recombinant human NEU2 produced by E. coli (Origami B strain) gene expression system. (A) SDS-PAGE of the fractions obtained during purification, after DEAE column chromatography (lane 1) and eluate treated with thrombin after GSH-Sepharose column chromatography (lane 2). Proteins were stained with CBB R350. (B) Western blot analysis of purified protein samples, the molecular weight of the monomeric NEU2 (42 kDa) detected by specific polyclonal antibody as described under “Materials and methods” (lane 1) and biotinylated protein standards (lane 2). (C) pH dependency of puriified NEU2 activity toward 4-MU-NANA was assayed as described under “Section 2”.
3.2. Inhibitory effects of siastatin B toward recombinant human NEU2
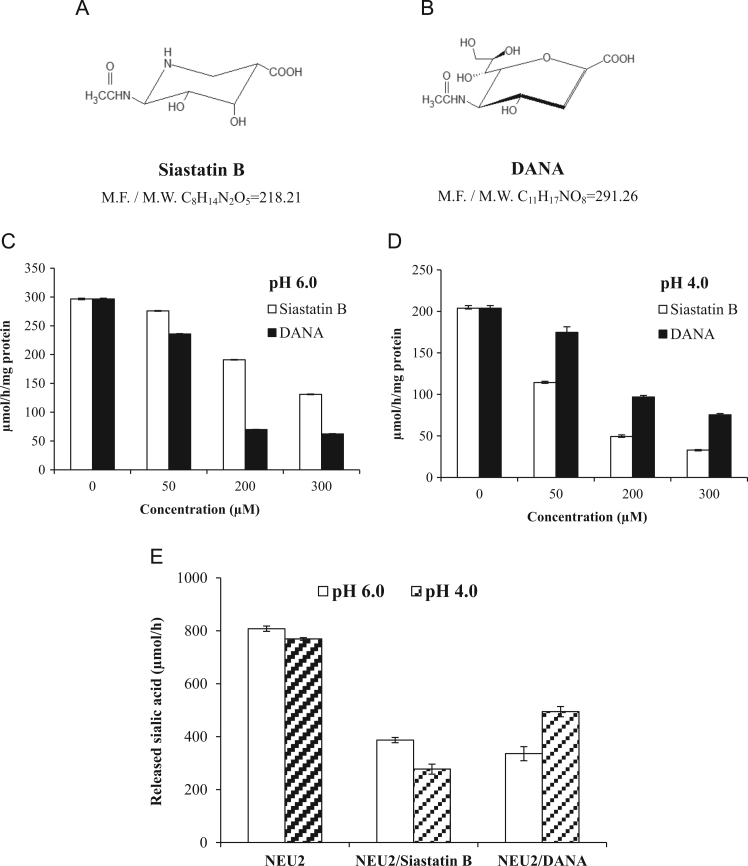
We first examined the inhibitory effects of siastatin B in vitro toward NEU2 activity as to the artificial substrate 4-MU-NANA as well as natural substrate α2,3-sialyllactose. Fig. 2(A and B) shows the structures of siastatin B and DANA, respectively, tested in this study. As shown in Fig. 2(C), DANA exhibited the dose-dependent inhibition of NEU2 activity at pH 6.0 as previously reported [9]. Siastatin B also had the inhibitory effect on NEU2, although the efficacy was less than that of DANA at pH 6.0. In contrast, siastatin B exhibited remarkable dose-dependent inhibition of NEU2 at pH 4.0, while DANA had less effect (Fig. 2(D)). IC50 values for siastatin B and DANA toward 4-MU-NANA-degrading activity, as summarized in Table 1, were 284 µM and 69 µM at pH 6.0, while 71 µM and 146 µM at pH 4.0, respectively. At pH 4.0 siastatin B inhibited 89% of NEU2 activity at 300 µM concentration, whereas DANA did 63% at the same concentration. At pH 6.0, the same concentration of siastatin B inhibited 56%, whereas DANA did 79% under the same experimental conditions. At higher pH range (7.0–7.5), DANA inhibited NEU2 more strongly (data not shown).
Fig. 2.
Structures of siastatin B (6-acetamido-3-piperidine carboxylate) [18] (A) and DANA (2-deoxy-2,3-dehydro-N-acetylneuraminic acid) [11] (B). M.F.: Molecular formula; M.W.: Molcular weight. Inhibitory effects of siastatin B (6-acetamido-3-piperidine carboxylate) and DANA (2,3-didehydro-2-deoxy-N-acetylneuraminic acid) on 4MU-NANA-degrading activity of recombinant human NEU2 at pH 6.0 (C) and pH 4.0 (D), and on α2,3-sialyllactose-degrading activity at pH 6.0 and 4.0 (E).
Table 1.
Inhibitory effects of siastatin B and DANA toward recombinant human NEU2.
| Inhibitor |
IC50(µM) |
Ki(µM) |
Binding free energy (kcal/mol) |
|||
|---|---|---|---|---|---|---|
| pH 4.0 | pH 6.0 | pH 4.0 | pH 6.0 | pH 4.0 | pH 6.0 | |
| Siastatin B | 71.0 | 284 | 150 | 250 | −117 | −108 |
| DANA | 146 | 69.0 | 360 | 100 | −83.9 | −143 |
The modes of inhibition by siastatin B were competitive at both pH 4.0 and 6.0, similar to those by DANA [9]. The Ki values of siastatin B and DANA at different pHs were summarized in Table 1. At pH 6.0 the Ki value for siastatin B was 250 µM, which was 2.5-fold higher than DANA (100 µM). In contrast, the Ki value for siastatin B (150 µM) was 2.4-fold lower than that for DANA (360 µM) at pH 4.0.
NEU2 activity toward a natural substrate α2,3- sialyllactose was also demonstrated to be inhibited by siastatin B (Fig. 2(E)). As shown in Table 2, NEU2 had released sialic acid from α2,3- sialyllactose in an average 808 µmol/h at pH 6.0 and 769 µmol/h at pH 4.0, whereas, the bound sialic acid (sialic acid content) was in an average 884.81 µmol. Siastatin B and DANA both at 600 µM concentration inhibited 52% and 58% of NEU2 activity at pH 6.0 toward α2,3-sialyllactose, respectively. In comparisons, these values at lower acidic pH 4.0 were 64% and 36%, respectively, for siastatin B and DANA both at the same concentration.
Table 2.
Inhibitory effects of siastatin B and DANA toward recombinant human NEU2 as to the natural substrate α2,3-sialyllactosea.
| Sample | pH | Sialic acid |
Relative rate of hydrolysis (%) | |
|---|---|---|---|---|
| Bound | Released | |||
| µmol | ||||
| NEU2 | 6.0 | 885 | 808 | 100 |
| 4.0 | 867 | 769 | 100 | |
| NEU2/siastatin Bb | 6.0 | 866 | 387 | 47.8 |
| 4.0 | 890 | 277 | 36.1 | |
| NEU2/DANAb | 6.0 | 855 | 336 | 41.6 |
| 4.0 | 882 | 494 | 64.3 | |
Substrate was incubated for 1 h under the standard assay conditions.
Siastatin B and DANA: final concentration 600 µM.
3.3. Stabilizing effects of siastatin B toward recombinant Human NEU2
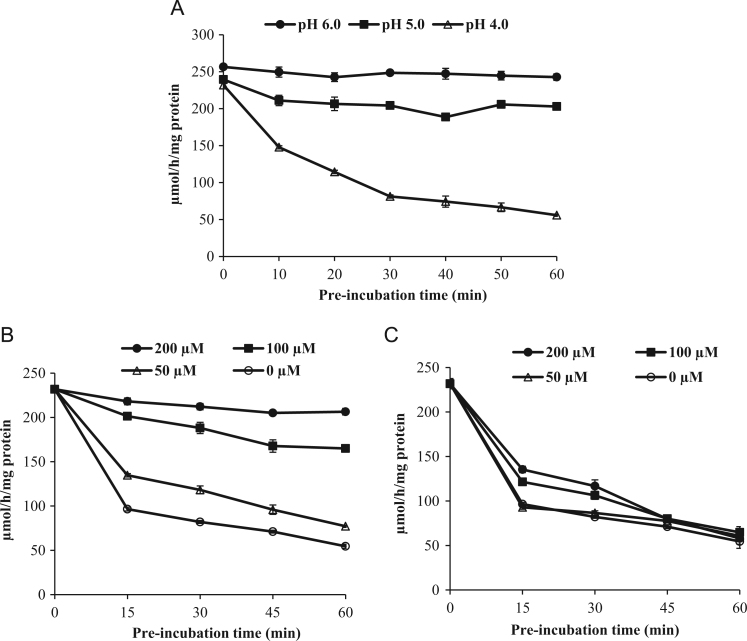
Fig. 3 shows the effect of siastatin B on thermostability of NEU2 activity in vitro. The purified NEU2 was pre-incubated at 37 °C for indicated time at pH 6.0, pH 5.0 and pH 4.0, respectively, and then the remaining enzyme activity was assayed at pH 6.0 with 4-MU-NANA as a substrate. The purified NEU2 activity produced by E. coli (Origami B strain) was found to be thermostable after pre-incubation at pH 6.0 and pH 5.0, but was lost substantially at pH 4.0 (Fig. 3(A)). During pre-incubation at pH 4.0 siastatin B stabilized NEU2 activity dose-dependently. After pre-incubation with 100 µM and 200 µM of siastatin B at pH 4.0 for 1 h at 37 °C, NEU2 retained the 4MU-NANA degrading activity by 60% and 87%, respectively (Fig. 3(B)). In contrast, DANA had no stabilizing effects at pH 4.0 on purified NEU2 at any concentrations tested in this experiment (Fig. 3(C)).
Fig. 3.
In vitro stabilization of NEU2 by siastatin B and DANA. Thermostability of 4-MU-NANA-degrading activity of purified human NEU2 after pre-incubation in 0.1 M sodium acetate buffer at pH 6.0 (●), 5.0 (■) or 4.0 (Δ) at 37 °C for different time intervals (A). Residual NEU2 activity after pre-incubation in 0.1 M sodium acetate buffer (pH 4.0) in the presence of siastatin B (B) and DANA (C) at different concentrations of 50 µM (Δ), 100 µM (■), 200 µM (●) or 0 (○) µM.
3.4. Molecular docking of siastatin B as to the active pockets of human NEU2 and NEU1 model
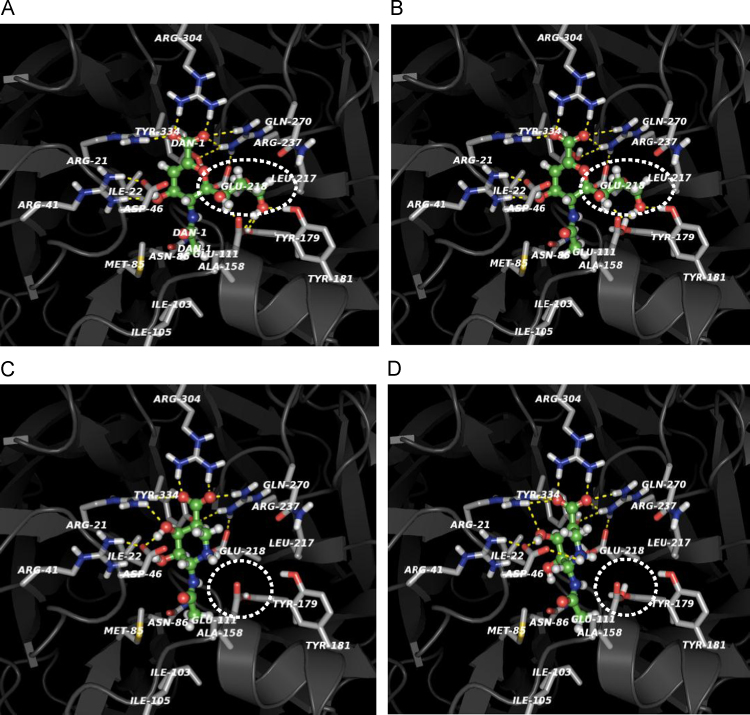
In order to predict the molecular interaction between siastatin B and NEU2, the docking model of siastatin B as to the active pocket of NEU2 was constructed on the basis of the X-ray crystal structure of NEU2/DANA complex (PDB code: 1 VCU). The in silico conditions under which an almost unique configuration with the least steric conflict and proper hydrogen bonds should be formed between siastatin B and amino acid residues located around the active pocket were also screened as described in “Section 2”. The local structure and protonation of amino acid residues around inhibitor binding site of NEU2 were compared at pH 4.0 and pH 6.0, respectively (Fig. 4(A–D)). The carboxyl groups of E111 and E218 residues around the active pocket were predicted to be electrostatically neutralized at pH 4.0. As for the whole molecule of NEU2, only the D336 is neutralized at pH 6.0 in contrast to E10, E39, E50, D58, E72, E111, D131, D141, E153, E218, E223, D251, D254, E257, D336, D345, E360, E361 and E376 at pH 4.0. The binding manners of DANA to the active pocket of NEU2 were compared at pH 4.0 and pH 6.0 (Fig. 4(A and B)). It was predicted that the pH-dependent neutralization at E111 and E218 could reduce the binding affinity of the C7–9 fragment of DANA through influencing the local structure of active pocket necessary for the binding of the DANA fragment (Fig. 4(A and B)). Higher binding affinity of DANA at pH 6.0 was also suggested by the calculated binding energy at pH 6.0 (Table 1). In contrast, siastatin B exhibited relatively higher affinity toward the active pocket of NEU2 at pH 4.0, although it lacks chemical fragment corresponding to the C7–9 fragment of DANA, which should be necessary for interaction with E111 and E218, as shown in Fig. 4(C and D). Additionally, the imino group (1NH) should interact with the side chain of D46, which was also not influenced by neutralization at pH 4.0. The stabilizing effect of siastatin B through binding to the active pocket of NEU2 was supported by the calculated binding energy at pH 4.0 (Table 1).
Fig. 4.
Molecular docking of the DANA and siastatin B to the active pocket of NEU2 based on the crystal structure of the NEU2/DANA complex (PDB code: 1VCU) without steric hindrance. Protonation of amino acid residues is indicated around inhibitor binding site compared at pH 4.0 and pH 6.0. Comparison of binding manners of DANA at pH 4.0 (A) and pH 6.0 (B). pH-dependent neutralization of E111 is predicted to have influence on binding of DANA C7–9 fragment (dashed oval). The E218 interacts with R237 via ionic bond. Neutralization of E218 may possibly have the effect on local structure of NEU2 supporting the binding of DANA C7–9 fragment. Comparison of binding manners of siastatin B at pH 4.0 (C) and pH 6.0 (D). There is no chemical groups in siastatin B corresponding to those of DANA C7–9 fragment, which may not influence the interaction between siastatin B and the neutralized amino acid residues (E111 and E218, dashed circle) at pH 4.0. The imino group (1NH) can interact with D46, which is not influenced by neutralization at pH 4.0.
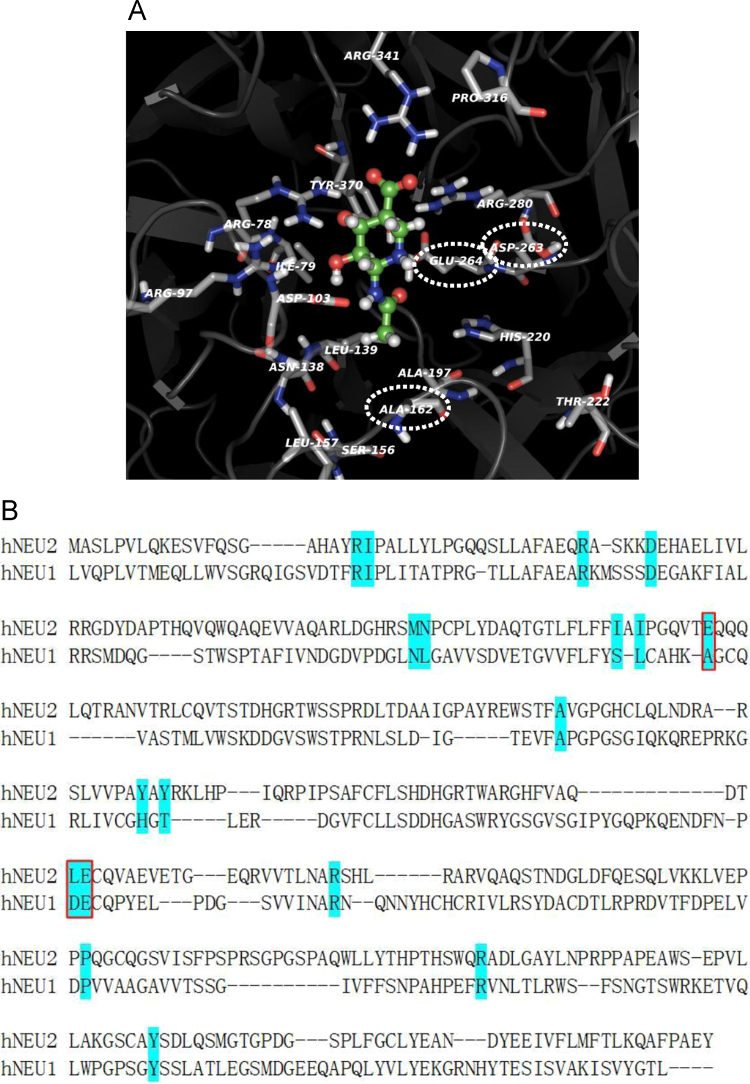
We also predicted the interaction between siastatin B and human lysosomal NEU1 with acidic pH optimum by the molecular docking of siastatin B as to the active pocket of NEU1 model, which was constructed as described under “Section 2”. Fig. 5(A) shows the binding manners of siastatin B as to the active pocket of NEU1 model at acidic pH 4.0. Results from assignment of ionization states of tritratable amino acids in homology model of human NEU1 predicted that the following amino acids were protonated at pH 4.0: E61, D75, E95, E104, D130, D135, D145, E147, D176, D190, E194, D226, D263, E264, E269, D300, D306, E312, D315, E377, D381, E383 and E384. Note the amino acid numbers were defined in case of the N-terminal methionine of human NEU1 precursor protein containing leader sequence as one (M1). Fig. 5(B) shows the putative binding sites with siastatin B at pH4.0. The D103 in NEU1 model corresponding to the D46 in NEU2 was predicted to interact with the imino group (1NH) of siastatin B, which was also not influenced by neutralization at pH 4.0, while the D263 was electrostatically neutralized at pH 4.0 but bound to siastatin B, although the D263 was adjacent to the E264 in NEU1 model corresponding to the L217 of NEU2 adjacent to the E218. The binding affinity of siastatin B as to the active pocket of NEU1 model was also supported by the calculated binding free energy of siastatin B (−113.91 kcal/mol) at pH 4.0.
Fig. 5.
Molecular docking of the siastatin B as to the active pocket of human lysosomal sialidase NEU1. (A) Predicted binding manners of siastatin B as to the active pocket of NEU1 model based on 3D-structure of human NEU2 and bacterial sialidase of M. viridifaciens. (B) Sequence alignment between human NEU2 and NEU1. Amino acid residues colored cyan indicate the putative binding site with siastatin B at acidic pH 4.0. Dashed ovals are shown as in (A) corresponding to those indicated by red squares are shown as in (B).
4. Discussion
In this study, we demonstrated for the first time the inhibitory effects of siastatin B toward recombinant human NEU2 in vitro (IC50 value, 284 µM at pH 6.0). In previous reports, siastatin B was demonstrated to inhibit bacterial, viral and mammalian sialidases at different concentrations ranged from µM to mM [18], [29], [30]. It inhibits bacterial sialidases with the IC50 values, which ranges from 27 to 80 µM [15], [16] and viral sialidases in the 10 µM range [31]. Umezawa et al. demonstrated that the IC50 values toward cytosolic sialidase from rat mammary gland, brain and liver, were 0.99, 3.60 and 1.53 mM, respectively [18]. In contrast, Miyagi et al. reported that siastatin B had less inhibitory activity against partially purified cytosolic sialidase from rat liver and skeletal muscle [10].
Interestingly, the inhibitory activity of siastatin B against human NEU2 was more potent under acidic pH conditions (IC50, 71 µM at pH 4.0) compared to that under higher pH conditions (IC50, 284 µM at pH 6.0) in this study. DANA is a well-known selective inhibitor for human NEU2 and exhibits inhibitory potency at around neutral pH [7], [9]. Lower inhibitory effects of DANA were reported toward acidic sialidases including human lysosomal NEU1 compared to that of cytosolic human NEU2 [9], [32]. In this study we observed the lower efficacy of DANA toward human NEU2 at acidic pH (IC50, 146 µM at pH 4.0) than that at higher pH (IC50, 69 µM at pH 6.0).
The mode of inhibition of siastatin B against human NEU2 was competitive as to the substrate 4-MU-NANA at pH 4.0 (Ki, 150 µM) and pH 6.0 (Ki, 250 µM). Similar inhibition mechanism by siastatin B was reported against bacterial sialidases at around pH 6.0 with a range of Ki value from 17 to 43 µM as to the sialolactose [18]. The lower Ki value (150 µM) of siastatin B at acidic pH indicated higher affinity of siastatin B to the active pocket of human NEU2, whereas lower affinity of DANA toward NEU2 was observed at pH 4.0, as shown by 2.4-fold higher Ki value (360 µM) than siastatin B (Ki, 150 µM) (Table 1). These results suggested higher inhibitory potency of siastatin B toward human NEU2 compared to that of DANA under acidic pH conditions.
Docking model of the NEU2/siastatin B complex was constructed on the basis of NEU2/ DANA crystal structure for prediction of molecular interaction between siastatin B and the active pocket of NEU2 at different pHs. The residues D46, E111 and E218 of NEU2 were reported to be crucial for substrate binding and catalytic activity as an enzymatic acid–base catalyst during hydrolysis thereby facilitating the coordination of the inhibitor [4], [5]. A recent study by comparative analysis of the active sites of sialidases derived from different sources along with NEU2 confirmed the role of the E218 as member of the nucleophile pair (334Y/218E) essential for catalysis [25]. Here we also predicted that the protonation of amino acid residues of NEU2, including E10, E39, E50, D58, E72, E111, D131, D141, E153, E218, E223, D251, D254, E257, D336, D345, E360, E361 and E376, by in silico analysis as the electrostatically neutralized residues at pH 4.0. Among them, E111 and E218 residues located around the active pocket. The E111 residue could be involved in the coordination of the C7–9 chemical fragment of DANA influenced by pH-dependent neutralization. The E218 can also interact with R237 via ionic bond. The neutralization of E218 may possibly have effect on local structure of the active pocket of NEU2 supporting the binding of C7–9 fragment of DANA. The binding energy of DANA (−142.69 kcal/mol) at pH 6.0 and (−83.91 kcal/mol) at pH 4.0 suggested more stable binding of DANA at pH 6.0 compared at pH 4.0. On the other hand, the neutralization at pH 4.0 may not influence the interaction between siastatin B and the active pocket of NEU2 because there are no chemical groups in siastatin B corresponding to those of DANA C7–9 fragment. Additionally, the D46 of NEU2 coordinated the imino group (1NH) of siastatin B, which should also not be influenced by neutralization at pH 4.0, although the binding energy (−107.58 kcal/mol) for siastatin B at pH 6.0 is not so different from −111.66 kcal/mol at pH 4.0. In order to confirm the contribution of the E111 residue to the interaction predicted by docking models, we tried to produce the recombinant mutant NEU2 by site-directed mutagenesis causing single substitution E111A. However, the NEU2 mutant (E111A) had completely lost the catalytic activity toward 4-MU-NANA (data not shown). Mozzi et al. reported NEU2 (E218A) mutant had little catalytic activity [25]. Therefore, we could not validate the prediction of the roles of E111 and E218 at acidic pH.
In this study we revealed that the human NEU2 produced by E. coli (Origami B strain) exhibits striking thermostability at pH 6.0 at least for 60 min but not at pH4.0, suggesting it can retain the proper folding and active conformation at pH 6.0 but lose at pH 4.0. Interestingly, siastatin B had stabilizing activity toward NEU2, which exhibit catalytic function under acidic conditions (pH 4.0–5.5), probably by preventing NEU2 protein from denaturation at acidic pH. The interaction between imino group (1NH) of siastain B and the active pocket of NEU2 including the D46 may also be involved in the stabilizing effect.
Homology modeling studies on not only human NEUs based on amino acid sequence identity and the crystal structure of NEU2 but also viral and bacterial sialidases, suggest structural similarity among NEU2, NEU3 and NEU4 [3], [8], [17] and different undelying mechanisms of inhibitors on these sialidases. Hata et al. reported that the inhibition of oseltamivir (Tamiflu) and zanamivir (Relenza) of influenza viral sialidases N1–3 at the nanomolar levels, while they exhibited the limited inhibitory effects on human sialidases including NEU1-4, although zanamivir had a little inhibitory effect of NEU2 at the micromolar level (16.4 μM at pH 5.5) [16]. We previously predicted using ab initio MO calculations [17] that DANA with the C4–OH group possibly interacts with the E111 of NEU2 due to hydrogen bonding, and the positively charged guanidino group of zanamivir is also involved in electrostatic interactions with the E39 and D46 in NEU2, which may be responsible for the inhibitory effects on NEU2 activity. In contrast, the active form of oseltamivir with NH3+ group at the corresponding position does not exhibit the inhibitory effect on NEU2 because it is not able to form a hydrogen bond to the E111 of NEU2. On the other hand, oseltamivir has remarkable inhibitory activity towards influenza viral sialidase N1-NA [16] because of the possible electrostatic interaction between the NH3+ group and the negatively charged E119 of N1-NA (PDB ID: 2HU4) [17]. As the amino acid residues E113 and E225 of NEU3 as well as E111 and E222 of NEU4 corresponding to E111 and E218 of NEU2, respectively are conserved by homology modeling [3]. Interactions with these glutamic acid residues of NEUs in pH-dependent manner should be considered on molecular design of inhibitor and stabilizer (chemical chaperone) based on the chemical structures of DANA derivatives.
In this study, we observed reverse inhibitory effects between DANA and siastatin B towards NEU2 at pH 6.0 and 4.0. At pH 6.0, the hydrogen bonding interaction between the C4–OH group of DANA and the E111 of NEU2 as well as the C7–9 fragment of DANA and the E218 of NEU2 may be reduced at pH 4.0. In contrast, the local interactions at the active pocket of NEU2, especially between the imino (1NH) group of siastatin B and the D46 of NEU2, may become stronger even at pH 4.0 than those at pH 6.0. Here we also predicted that siastatin B could bind to the active pocket of human lysosomal NEU1 model because the local conformation around the active pocket should be similar to that of NEU2. Siastatin B was suggested to interact with not only the D103 but also the D263 and E264 residues of NEU1 model, corresponding to the D46, L217 and E218 in NEU2, although the E111 in NEU2 was replaced with the A162 in NEU1 model. Therefore, siastatin B may have inhibitory effect on NEU1 at pH 4.0. Siastatin B derivatives retaining the imino group (1NH) will be seed compounds for more specific inhibitors and stabilizers for variety of sialidases, including mammalian, microbial and viral origins, by comparison of the pH-dependent effects of siastatin B.
In conclusion, we found the pH-dependent affinity of siastatin B toward human cytosolic sialidase NEU2 to exhibit potent inhibitory and stabilizing activities. It was supported by molecular docking that the interaction between siastatin B and the D46, E111 and E218 in the active pocket of NEU2 should be the key amino acid residues regulating the enzymatic functions of NEU2 especially at acidic pH.
Acknowledgments
We thank Prof. Soichi Wakatsuki, Drs. Ryu-ichi Kato and Leonard M.G. Chavas for providing the recombinant plasmid pGEX-2T-NEU2. This study was financially supported by the Ministry of Education, Culture, Sports, Science and Technology, Government of Japan (MEXT) (Nos. 23390140 and 40212201), and also by the Agri-health Translational Research Project (No. 5130) of the Ministry of Agriculture, Forestry and Fisheries of Japan.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.09.017.
Appendix A. Supplementary material
Supplementary material
References
- 1.Monti E., Bonten E., Azzo A.D.’, Bresciani R., Venerando B., Borsani G., Schauer R., Tettamanti G. Sialidases in vertebrates: a family of enzymes tailored for several cell functions. Adv. Carbohydr. Chem. Biochem. 2010;64:403–479. doi: 10.1016/S0065-2318(10)64007-3. [DOI] [PubMed] [Google Scholar]
- 2.Miyagi T., Yamaguchi K. Mammalian sialidases: physiological and pathological roles in cellular functions. Glycobiology. 2012;22:880–896. doi: 10.1093/glycob/cws057. [DOI] [PubMed] [Google Scholar]
- 3.Monti E., Bassi M.T., Bresciani R., Civini S., Croci G.L., Papini N., Riboni M., Zanchetti G., Ballabio A., Preti A., Tettamanti G., Venerando B., Borsani G. Molecular cloning and characterization of NEU4, the fourth member of the human sialidase gene family. Genomics. 2004;83:445–453. doi: 10.1016/j.ygeno.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr. Opin. Struct. Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 5.Copley R.R., Russell R.B., Ponting C.P. Sialidase-like Asp-boxes: sequence-similar structures within different protein folds. Prot. Sci. 2001;10:285–292. doi: 10.1110/ps.31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roggentin P., Schauer R., Hoyer L.L., Vimr E.R. The sialidase superfamily and its spread by horizontal gene transfer. Mol. Microbiol. 1993;9:915–921. doi: 10.1111/j.1365-2958.1993.tb01221.x. [DOI] [PubMed] [Google Scholar]
- 7.Chavas L.M.G., Tringali C., Fusi P., Venerando B., Tettamanti G., Kato R., Monti E., Wakatsuki S. Crystal structure of the human cytosolic sialidase Neu2. J. Biol. Chem. 2005;280:469–475. doi: 10.1074/jbc.M411506200. [DOI] [PubMed] [Google Scholar]
- 8.Magesh S., Suzuki T., Miyagi T., Ishida H., Kiso M. Homology modeling of human sialidase enzymes NEU1, NEU3 and NEU4 based on the crystal structure of NEU2: hints for the design of selective NEU3 inhibitors. J. Mol. Graph. 2006;25:196–207. doi: 10.1016/j.jmgm.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Magesh S., Savita V., Moriya S., Suzuki T., Miyagi T., Ishida H., Kiso M. Human sialidase inhibitors: design, synthesis, and biological evaluation of 4-acetamido-5-acylamido-2-fluoro benzoic acids. Bioorg. Med. Chem. 2009;17:4595–4603. doi: 10.1016/j.bmc.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 10.Miyagi T., Hata K., Hasegawa A., Aoyagi T. Differential effect of various inhibitors on four types of rat sialidase. Glycoconj. J. 1993;10:45–49. doi: 10.1007/BF00731186. [DOI] [PubMed] [Google Scholar]
- 11.Meindl P., Bodo G., Palese P., Schulman J., Tuppy H. Inhibition of neuraminidase activity by derivatives of 2-deoxy 2,3-dehydro-N-acetylneuraminic acid. Virology. 1974;58:457–463. doi: 10.1016/0042-6822(74)90080-4. [DOI] [PubMed] [Google Scholar]
- 12.Holzer C.T., Itzstein M.V., Jin B., Pegg M.S., Stewart W.P., Wu W.Y. Inhibition of sialidases from viral, bacterial and mammalian sources by analogues of 2-deoxy-2,3-dihydro-N-acetylneuraminic acid modified at the C-4 position. Glycoconj. J. 1993;10:40–44. doi: 10.1007/BF00731185. [DOI] [PubMed] [Google Scholar]
- 13.Chavas L.M.G., Kato R., Suzuki N., Itzstein M.V., Mann M.C., Thomson R.J., Dyason J.C., Breschkin J.M., Fusi P., Tringali C., Venerando B., Tettamanti G., Monti E., Wakatsuki S. Complexity in influenza virus targeted drug design: interaction with human sialidases. J. Med. Chem. 2010;53:2998–3002. doi: 10.1021/jm100078r. [DOI] [PubMed] [Google Scholar]
- 14.Umezawa H., Aoyagi T., Komiyama H., Morishima H., Hamada M., Takeuchi T. Purification and characterization of a sialidase inhibitor, siastatin, produced by Streptomyces. J. Antibiot. 1974;27:963–969. doi: 10.7164/antibiotics.27.963. [DOI] [PubMed] [Google Scholar]
- 15.Kudo T., Nishimura Y., Kondo S., Takeuchi T. Synthesis and activities of N-substituted derivatives of siastatin B. J. Antibiot. 1992;45:1662–1668. doi: 10.7164/antibiotics.45.1662. [DOI] [PubMed] [Google Scholar]
- 16.Hata K., Koseki K., Yamaguchi K., Moriya S., Suzuki Y., Yingsakmongkon S., Hirai G., Sodeoka M., von Itztein M., Miyagi T. Limited inhibitory effects of oseltamivir and zanamivir on human sialidases, antimicrobial. Agent Chemother. 2008;52:3484–3491. doi: 10.1128/AAC.00344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hitaoka S., Matoba H., Harada M., Yoshida T., Tsuji D., Hirokawa T., Itoh K., Chuman H. Correlation analyses on binding affinity of sialic acid analogues and anti-influenza drugs with human neuraminidase using ab initio MO calculations on their complex structures – LERE-QSAR analysis (IV) J. Chem. Inf. Model. 2011;51:2706–2716. doi: 10.1021/ci2002395. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura Y., Wang W., Kondo S., Aoyagi T., Umezawa H., Siastatin B. a potent neuraminidase inhibitor: the total synthesis and absolute configuration. J. Am. Chem. Soc. 1988;110:7249–7250. [Google Scholar]
- 19.Knapp S., Zhao D. Synthesis of the sialidase inhibitor siastatin B. Org. Lett. 2000;2:4037–4040. doi: 10.1021/ol0066680. [DOI] [PubMed] [Google Scholar]
- 20.Tringali C., Papini N., Fusi P., Croci G., Borsani G., Preti A., Tortora P., Tettamanti G., Venerando B., Monti E. Properties of recombinant human cytosolic sialidase HsNEU2. J. Biol. Chem. 2004;279:3169–3179. doi: 10.1074/jbc.M308381200. [DOI] [PubMed] [Google Scholar]
- 21.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the folin-phenol reagents. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Burk D., Lineweaver H. The influence of fixed nitrogen on Azotobacter. J. Bacteriol. 1930;19:389–414. doi: 10.1128/jb.19.6.389-414.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren L. The thiobarbituric acid assay of sialic acids. J. Biol. Chem. 1959;234:1971–1975. [PubMed] [Google Scholar]
- 25.Mozzi A., Mazzacuva P., Zampella G., Forcella M.E., Fusi P.A., Monti E. Molecular insight into substrate recognition by human cytosolic sialidase NEU2. Proteins. 2012;80:1123–1132. doi: 10.1002/prot.24013. [DOI] [PubMed] [Google Scholar]
- 26.Bashford D., Karplus M. Multiple-site titration curve of proteins: an analysis of exact and approximate methods for their calculation. J. Phys. Chem. 1991;95:9556–9561. [Google Scholar]
- 27.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 28.Hitaoka S., Shibata Y., Matoba H., Kawano A., Harada M., Rahman M.M., Tsuji D., Hirokawa T., Itoh K., Yoshida T., Chuman H. Modeling of human neuraminidase-1 and its validation by LERE-correlation analysis. Chem.-Bio. Inform. J. 2013;13:30–44. [Google Scholar]
- 29.Kudo T., Nishimura Y., Kondo S., Takeuchi T. Synthesis of the potent inhibitors of neuraminidase, N-(1,2-dihydroxypropyl) derivatives of siastatin B and its 4-deoxy analogs. J. Antibiot. 1993;46:300–309. doi: 10.7164/antibiotics.46.300. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura Y., Kudo T., Kondo S., Takeuchi T., Tsuruoka T., Fukuyasu H., Shibahara S. Totally synthetic analogues of siastatin B. III. Trifluoroacetamide analogues having inhibitory activity for tumor matastasis. J. Antibiot. 1994;47:101–107. doi: 10.7164/antibiotics.47.101. [DOI] [PubMed] [Google Scholar]
- 31.Montreuil J., Vliegenthart J.F.G., Schachter H. Vol. 1000. Elsevier Science B; AE Amsterdam, The Netherlands: 1997. Glycoproteins II. [Google Scholar]
- 32.Magesh S., Moriya S., Suzuki T., Miyagi T., Ishida H., Kiso M. Design, synthesis, and biological evaluation of human sialidase inhibitors. Part 1: selective inhibitors of lysosomal sialidase (NEU1) Bioorg. Med. Chem. Lett. 2008;18:532–537. doi: 10.1016/j.bmcl.2007.11.084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material