Abstract
The EGFR belongs to the well-studied ErbB family of receptor tyrosine kinases. EGFR is activated by numerous endogenous ligands that promote cellular growth, proliferation, and tissue regeneration. In the present study, we have demonstrated a role for EGFR and its natural ligand, epiregulin (EREG), in pain processing. We show that inhibition of EGFR with clinically available compounds strongly reduced nocifensive behavior in mouse models of inflammatory and chronic pain. EREG-mediated activation of EGFR enhanced nociception through a mechanism involving the PI3K/AKT/mTOR pathway and matrix metalloproteinase-9. Moreover, EREG application potentiated capsaicin-induced calcium influx in a subset of sensory neurons. Both the EGFR and EREG genes displayed a genetic association with the development of chronic pain in several clinical cohorts of temporomandibular disorder. Thus, EGFR and EREG may be suitable therapeutic targets for persistent pain conditions.
Keywords: Neuroscience
Keywords: Pain
Introduction
Chronic pain is a major human health problem affecting almost one-quarter of the population at any one time (1–3). Chronic pain is difficult to manage, treatment options are limited and associated with unwanted side effects, and the identification of novel pharmacotherapeutic targets remains challenging. Recently, the EGFR has received attention for its therapeutic potential against pain. EGFR inhibition is the first-line treatment for non–small cell lung cancer, and there have been a number of case reports suggesting that EGFR inhibition provides rapid relief of cancer pain (4–8). Cancer patients administered EGFR inhibitors report a significant reduction in pain scores and an overall improvement in quality of life independent of the effect on tumor progression and size (6).
The EGFR is a member of the ErbB family of tyrosine kinase receptors (9) that regulate cellular growth, survival, proliferation, and differentiation of fibroblasts and hepatocytes (10, 11). Several ligands can bind to and activate EGFR, including EGF, TGF-α, heparin-binding EGF-like growth factor (HB‑EGF), amphiregulin, betacellulin, and epiregulin (EREG) (9). The downstream effects of EGFR are mediated by a number of important signaling pathways, including MAPK (8) and PI3K/AKT/mTOR — which are known to regulate pain (12–14). EGFR has been shown to affect receptors important for pain processing, including opioid receptors (15), β‑adrenergic receptors (16), cannabinoid type 1 (CB1), and transient receptor potential cation channel, subfamily V, member 1 (TRPV1) receptors (17). However, none of these previous reports examined these interactions using behavioral or cellular models specifically relevant to pain processing. Here, our primary goal was to understand the EGFR pathway’s role and mechanism of action in pain processing.
We investigated the role of the EGFR and its ligands in nociception using murine behavioral and ex vivo studies to identify mechanistic targets for EGFR signaling in pain that may be generalizable across pain conditions. Specifically, we demonstrate herein that inhibition of the tyrosine kinase site of the EGFR, using both experimental and clinically available compounds, is analgesic against a variety of tonic and chronic pain modalities in the mouse. The genes coding for both EGFR and EREG demonstrate genetic association with a human chronic pain syndrome, temporomandibular disorder (TMD), and genetic inhibition of EGFR modulates pain behavior in both mice and Drosophila. Finally, we show that stimulation of EGFR, by its ligand EREG, specifically, activates dorsal root ganglion (DRG) neurons, producing pain behaviors through a mechanism that involves TRPV1, the PI3K/AKT/mTOR/4E-BP1 signaling pathway, and MMP-9, a molecule known to be involved in inflammation and the early stages of chronic pain (18).
Results
Modulation of EGFR affects pain sensitivity in mice.
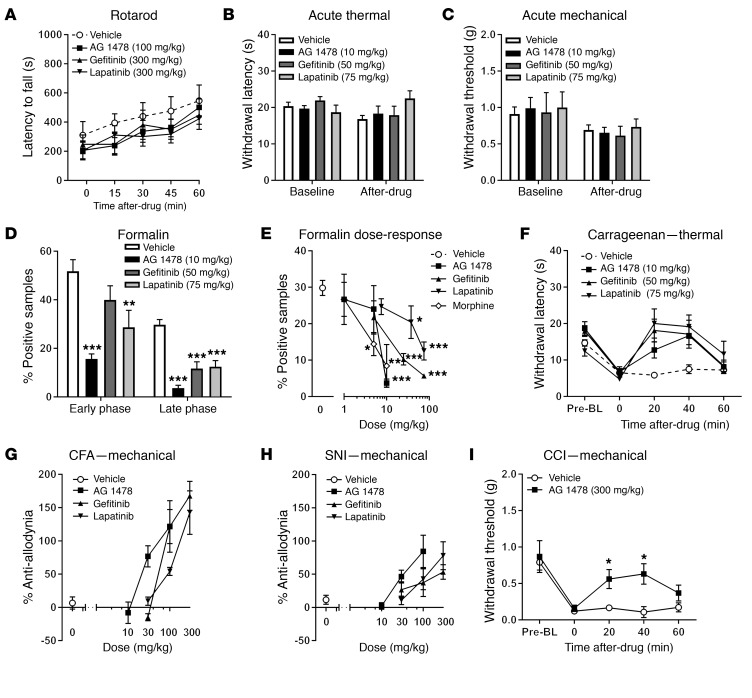
To assess the effect of EGFR inhibition on pain in mice, we tested 1 preclinical (tyrphostin AG 1478; hereinafter, AG 1478; 10 mg/kg) and 2 clinically available (50 mg/kg gefitinib and 75 mg/kg lapatinib) EGFR tyrosine kinase inhibitors in a battery of algesiometric assays. Doses were chosen based on prior in vivo efficacy against stress‑induced necrotic lesions in the heart (AG 1478) and in chemoprevention of lung cancer (gefitinib) or breast cancer (lapatinib) in mice (19–21). We first confirmed that none of the drugs, delivered systemically and at very high doses (AG 1478, 100 mg/kg; gefitinib, 300 mg/kg; lapatinib, 300 mg/kg), produced significant ataxia over a 1-hour testing period on the rotarod (Figure 1A). These drugs did not affect acute noxious thermal (Figure 1B) or mechanical (Figure 1C) sensitivity. In contrast, in the formalin test, EGFR inhibitors produced robust inhibition of tonic inflammatory pain (Figure 1D) without affecting edema (not shown). Compilation of full dose-response curves revealed dose‑dependent analgesia only in the late or tonic phase (Figure 1E), with efficacy and potencies comparable to those of morphine (see Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI87406DS1). Further, EGFR inhibition completely reversed the thermal hypersensitivity produced by an inflammatory mediator, λ‑carrageenan, at 20 to 40 minutes after injection (Figure 1F). Finally, we examined mechanical hypersensitivity (allodynia) after longer-lasting inflammatory and neuropathic injuries. Higher doses were required, but all drugs produced complete and dose‑dependent reversal of allodynia in a CFA model of inflammatory pain (Figure 1G). Similarly, all drugs produced complete and dose‑dependent reversal of allodynia in the spared nerve injury (SNI) model of neuropathic pain (Figure 1H). Half-maximal analgesic doses and confidence intervals for the EGFR inhibitors for CFA and SNI are presented in Supplemental Table 2. In a separate experiment, we used the chronic constriction injury (CCI) model of chronic pain to provide generalizability across neuropathic assays. Similar to results using SNI, EGFR inhibition produced robust antiallodynia using CCI (Figure 1I).
Figure 1. EGFR antagonists produce analgesia and EREG produces hyperalgesia in the mouse.
(A) No sedation or ataxia (2-way ANOVA, drug × repeated measures: F12,80 = 0.5, P = 0.88) produced by high doses of EGFR antagonists. Symbols represent mean ± SEM for latency to fall off rotarod at each time point; n = 6–8/drug. (B) No effect of EGFR antagonists on acute thermal pain measured using the radiant heat paw-withdrawal test (2-way ANOVA, drug × repeated measures: F3,19 = 2.3, P = 0.10). Bars represent mean ± SEM for latency to withdraw from a noxious thermal stimulus before (baseline) and 30 minutes after injection; n = 5–6/drug. (C) No effect of EGFR antagonists on acute mechanical sensation using the von Frey test (2-way ANOVA, drug × repeated measures: F3,19 = 0.3, P = 0.80). Bars represent mean ± SEM for hind paw withdrawal threshold (g) before (baseline) and 30 minutes after injection; n = 5–6/drug. (D) EGFR antagonists produce analgesia during the formalin test in both the early (0–10 minutes; 1-way ANOVA, F3,29 = 7.2, P = 0.001) and late (10–60 minutes; 1-way ANOVA, F3,29 = 15.9, P < 0.001) phases. Bars represent mean ± SEM for percentage of samples featuring licking/biting behavior; n = 7–9/drug. (E) Dose‑dependent analgesia from EGFR antagonists and morphine on the late‑phase formalin test; symbols represent mean ± SEM for percentage of samples featuring licking/biting behavior; n = 6–8/drug/dose. See Supplemental Table 1 for half-maximal analgesic doses and 95% confidence intervals. (F) EGFR antagonists reverse thermal hypersensitivity induced by carrageenan (2-way ANOVA, drug × repeated measures: F9,57 = 2.8, P = 0.01). Symbols represent mean ± SEM for latency to withdraw from a noxious thermal stimulus before carrageenan (Pre‑BL), 3 hours after carrageenan (0), and 20–60 minutes after drug administration; n = 5–6/drug. (G) EGFR antagonists dose‑dependently reverse mechanical allodynia induced by CFA (3 days after injection). Symbols represent mean ± SEM for percentage of maximum possible antiallodynia (i.e., reversal back to baseline withdrawal thresholds at all post-drug time points; see Methods); n = 5–6/drug/dose. (H) EGFR antagonists dose‑dependently reverse mechanical allodynia induced by SNI (7 days after surgery). Symbols as in G; n = 5–6/drug/dose. See Supplemental Table 2 for half‑maximal analgesic doses and 95% confidence intervals relevant to graphs in G and H. (I) AG 1478 reverses mechanical allodynia induced by CCI (14 days after surgery); n = 6/drug (2-way ANOVA, drug × repeated measures: F4,40 = 2.6, P = 0.02). *P < 0.05; **P < 0.01; ***P < 0.001 compared with vehicle (0) group by Dunnett’s case-comparison post hoc test.
Activation of EGFR by EREG, but not other EGFR ligands, promotes nociception.
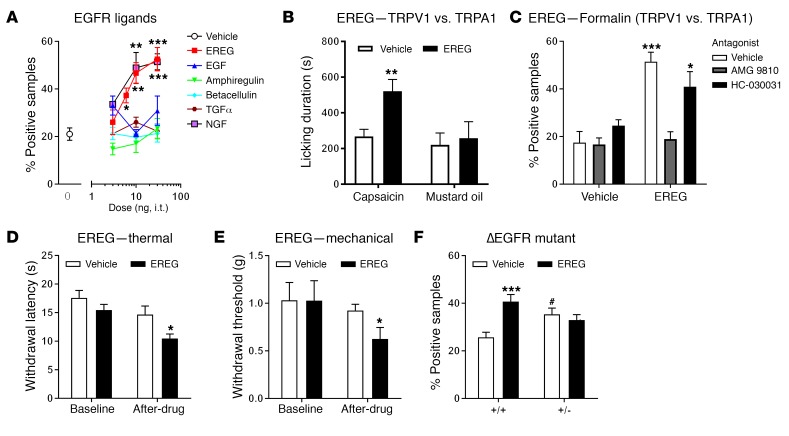
In order to determine whether EGFR activation is sufficient to increase nociception, we screened a number of EGFR ligands for their ability to promote nocifensive behaviors in the formalin test. We found that late-phase formalin-induced nocifensive behaviors were enhanced in a dose-dependent manner with intrathecal (i.t.) injections of EREG, but none of the other tested EGFR ligands, including betacellulin, TGF-α, amphiregulin, or EGF (Figure 2A). EREG produced a robust and dose‑dependent increase in licking behavior that was indistinguishable from that produced by nerve growth factor (NGF), known for its prominent role in pain processing (Figure 2A). Next, we assessed whether EREG enhanced pain behaviors induced by capsaicin or mustard oil, 2 potent algogens known to activate TRPV1 and TRPA1, respectively. We found that i.t. delivery of EREG potentiated nocifensive behavior from intraplantar capsaicin, but not mustard oil (Figure 2B). In addition, the TRPV1 antagonist AMG 9810 (30 mg/kg; i.p.), but not the TRPA1 antagonist HC-030031 (30 mg/kg; i.p.), blocked the hyperalgesic effect of EREG in the late phase of the formalin test (Figure 2C). Spinal delivery of EREG produced both thermal (Figure 2D) and mechanical (Figure 2E) pain hypersensitivity in the absence of injury. EREG’s hyperalgesic effects during the formalin test were independent of other tyrosine receptor kinases, as K252a (an inhibitor of TrkA, TrkB, and TrkC) failed to block EREG‑induced hypersensitivity (Supplemental Figure 1). Furthermore, administration of AG 1478 blocked the hyperalgesic effects of EREG during the formalin test, but did not reverse the hyperalgesia produced by i.t. injections of NGF, a potent activator of TrkA, confirming that EREG-mediated hypersensitivity is dependent on the EGFR and not TrkA (Supplemental Figure 1).
Figure 2. Spinally administered EREG, but not other EGFR ligands, produces hypersensitivity.
(A) Significant and dose‑dependent hypersensitivity from EREG (2-way ANOVA, F3,26 = 6.8, P = 0.002), but not betacellulin, amphiregulin, EGF, or TGF-α (all P values are greater than 0.50) in the late phase (10–60 minutes) of the formalin test. Symbols represent mean ± SEM for percentage of samples featuring licking/biting behavior; n = 6–8/drug/dose. EREG enhancement of formalin-induced licking was equivalent to that of NGF; 2-way ANOVA, F3,22 = 10.9, P < 0.001. (B) EREG (10 ng, i.t.) increases nocifensive behavior when coadministered with intraplantar injections of the TRPV1 agonist capsaicin (2-tailed t test, t10 = 3.4, P = 0.01), but not the TRPA1 agonist mustard oil (2-tailed t test, t13 = 0.34, P = 0.70). Bars represent mean ± SEM for duration of licking behavior over 10 minutes after injection; n = 6‑8/algogen/drug. (C) The TRPV1 antagonist AMG 9810 (30 mg/kg, i.p.), but not the TRPA1 antagonist HC-030031 (30 mg/kg, i.p.), blocks EREG-induced hyperalgesia during the formalin test (2-way ANOVA, drug × antagonist interaction: F2,38 = 7.2, P = 0.002). Bars as in graph A; n = 7-8/group. (D) EREG (10 ng) increases thermal sensitivity by itself (2-tailed paired t test, t10 = 2.6, P = 0.03). Bars represent mean ± SEM for latency to withdraw from a noxious thermal stimulus before (baseline) and 30 minutes after injection; n = 6/drug. (E) EREG (10 ng) increases mechanical sensitivity by itself (2-tailed paired t test, t10 = 2.2, P = 0.05). Bars represent mean ± SEM for hind paw–withdrawal threshold (g) before (baseline) and 30 minutes after injection; n = 6/drug. (F) ΔEGFR mutants have higher baseline pain sensitivity to formalin than WT (vehicle groups), but no longer respond to EREG (10 ng, i.t.). Bars as in graph E; n = 7–8/genotype/drug. For all panels, *P < 0.05; **P < 0.01; ***P < 0.001 compared with vehicle (0) by Dunnett’s case-comparison post hoc test or t test as indicated. #P < 0.01 compared with other genotype (in F) by Dunnett’s case-comparison post hoc test.
EGFR gene mutant effects in mice and Drosophila.
In order to confirm the efficacy of EREG in promoting pain hypersensitivity, we tested a partial loss-of-function EGFR mutant mouse for EREG-induced hypersensitivity. The EGFRvIII/ΔEGFR mutant mouse has a large deletion of the extracellular domain that renders EGFR constitutively active, but with no ability to bind extracellular ligands (22). The homozygous mutation is embryonically lethal, and thus we tested ΔEGFR heterozygotes for EREG-induced pain sensitivity during the formalin test. Basal formalin-induced licking behavior of ΔEGFR heterozygotes was increased, but EREG no longer produced hypersensitivity during the formalin test (Figure 2F). The increased formalin sensitivity of ΔEGFR heterozygotes was likely due to constitutive activity of the EGFR. Since further activation by ligand binding is reduced in this mutant, EREG would not be expected to increase pain behavior any further. In addition, we used genetic knockdown and somatic Drosophila mutants to confirm that EGFR acts via sensory neurons to mediate nociception in response to a 46°C probe (Supplemental Figure 2). Larvae with Egfr knocked down in peripheral ppk+ nociceptor neurons exhibited impaired thermal nociception (Supplemental Figure 2, A–C), and conversely, reintroduction of intact Egfr specifically in ppk+ sensory neurons was sufficient to rescue nociception in whole body Egfr somatic mutant animals (Supplemental Figure 2, D–F). Together, these data establish that the EGFR system is a conserved component of the nociception apparatus, regulating peripheral nociceptor function in vivo.
EREG and EGFR genetic loci are associated with the risk of development of a chronic pain condition.
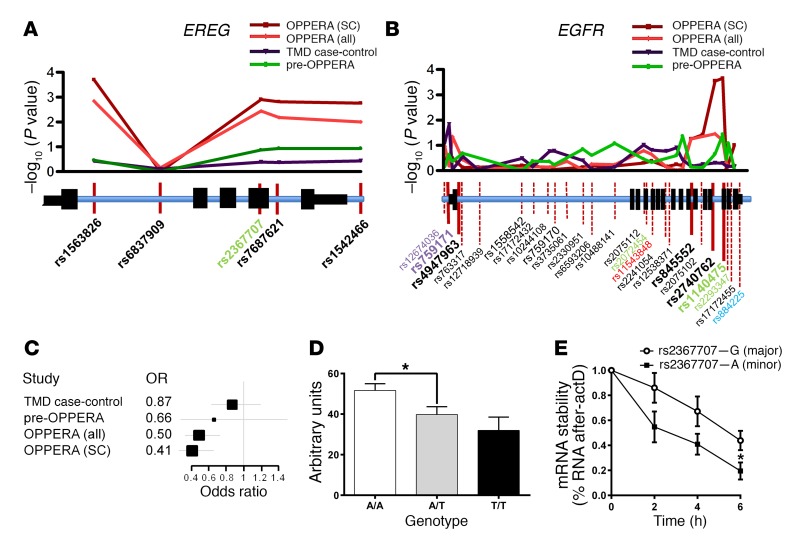
Since our studies in mice indicate a robust role for EREG and EGFR in mediating pain, we next searched for evidence that EGFR contributes to pain in a human clinical population. Three human cohorts of a common chronic pain condition, TMD, were assessed in 4 case-control association analyses (Supplemental Table 3) on a panel of 358 pain-relevant candidate genes. In the first analysis, designed to minimize experimental variance, we contrasted 127 female TMD cases of mixed European descent from the Orofacial Pain: Prospective Evaluation and Risk Assessment (OPPERA) (23) cohort against a subset of 231 demographically matched “super‑controls,” who reported absolutely no procedural pain at exam. Of the genes screened, EREG (rs1563826, odds ratio = 0.4, P = 2.0 × 10-4) (Figure 3A) and EGFR (rs1140475, odds ratio = 2.6, P = 2.2 × 10-4) (Figure 3B) demonstrated the highest association with the development of TMD. In addition, the majority of SNPs deviating from the QQ plot were located in either EREG or EGFR loci (Supplemental Figure 3A). Formal pathway analysis identified the EGFR signaling pathway as significantly associated with TMD (P = 0.0013; Supplemental Table 4). Nominally significant (P < 0.05) associations were also observed for EREG and EGFR in the full OPPERA cohort, in which TMD cases were contrasted with all 731 enrolled TMD-free controls (Figure 3, A and B). We replicated these association results in 2 independent cohorts of females of mixed European descent, including 1 case-control study of 200 TMD cases and 198 controls (TMD case-control cohort) (24) and another prospective study of 186 initially pain-free subjects in which 15 developed TMD over a 3‑year follow-up period (pre-OPPERA cohort) (25). These analyses identified several additional SNPs with either significant or borderline associations (Figure 3, A and B). Whereas the pattern of association within the EREG gene locus was identical in all 4 cohorts (Figure 3A), with SNP rs1563826 showing the strongest association with TMD, the pattern of association for EGFR differed between cohorts (Figure 3B). To explain the discrepancies in single SNP results between cohorts, the EGFR gene locus was subjected to haplotype analysis that produced convergent results (Supplemental Figure 3B and Supplemental Table 5).
Figure 3. Genetic association of EREG and EGFR with TMD pain.
Genetic association of EREG (A) and EGFR (B) SNPs with chronic TMD pain in OPPERA cases vs. super controls (SC), OPPERA cases vs. controls (all), TMD case-control cohort, and pre-OPPERA cohorts (see Methods). Manhattan plots and corresponding gene loci are shown where position of tested SNPs is given relative to gene structure. Nonsynonymous (red), synonymous (green), promoter region (gray), and 3′ intragenic region (blue) SNPs are indicated. The pattern of association within the EREG gene locus was identical in all cohorts. The pattern of association for the EGFR gene locus revealed some differences between cohorts; however, the 5′ and 3′ ends of the gene consistently showed association with elevated TMD risk. The EGFR SNPs showing association in independent SNP tests (marked in bold) were used for haplotype analysis (see Supplemental Table 5). (C) Forest plot depicting odds ratios (OR; with 95% confidence intervals) for minor allele T of rs1563826 in 4 human chronic pain cohorts. (D) Association of EREG mRNA level with EREG rs1563826 in the TMD case-control cohort measured by quantitative RT‑PCR. Bars represent mean ± SEM for EREG expression in leukocytes expressed in arbitrary units relative to GAPDH (see Methods). One-way ANOVA revealed a significant difference among genotypes; F2,247 = 3.7, P = 0.03. *P < 0.05; the A/A versus T/T comparison was P = 0.053, likely due to the small number of T/T homozygotes. (E) Following actD treatment, the rate of mRNA degradation was significantly lower for cells expressing WT (G allele) EREG mRNA compared with those the minor A allele of rs2367707 (2-tailed paired t test, t4 = 2.8, P = 0.05). Symbols represent mean ± SEM for percentage of mRNA expression compared with time 0; n = 3/genotype.
Total RNA isolated from blood leukocytes was collected from participants in the TMD case‑control study. Relatively high expression levels of EREG (but not EGFR) in leukocytes allowed us to measure EREG mRNA using quantitative PCR (qPCR) in all subjects and evaluate its association with EREG genotypes. The minor allele T of SNP rs1563826, which showed the strongest association with decreased odds of TMD (Figure 3C), was also associated with lower EREG mRNA levels (Figure 3D), suggesting that higher levels of EREG contribute to hyperalgesic states in patients.
We then assessed the EREG gene locus for functional SNPs potentially responsible for transcript regulation. No SNPs with minor allele frequency of more than 5% are reported within 5 kB upstream of the EREG promoter. However, the synonymous rs2367707 SNP within the EREG coding region was found to be in close-to-perfect linkage disequilibrium with rs1563826 and with an almost identical P value for TMD risk (Figure 3A). To examine the effect of rs2367707 allelic variants on transcript levels, we transfected HEK293 cells with expression constructs that carried allelic variants of rs2367707. In line with the genetic effect of the EREG haplotype on endogenous mRNA levels, the minor A allelic variant of EREG showed significantly lower transcript stability than the major G allelic variant (Figure 3E). The association of EGFR and EREG with chronic pain in a clinical cohort is an important translational complement to our mouse data and supports the potential value of targeting EGFR for chronic noncancer pain management in humans.
EREG levels and EGFR phosphorylation are upregulated in mouse models of chronic pain.
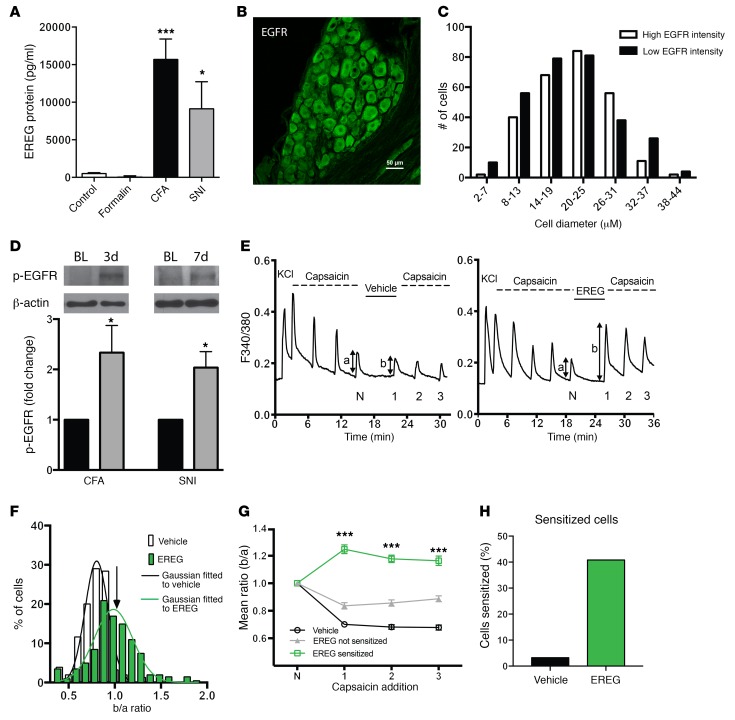
The finding of increased EREG mRNA levels in leukocytes of TMD patients (Figure 3D) prompted us to investigate whether chronic pain states in mice are associated with increased levels of EREG in the blood. CFA and SNI (but not formalin) produced a massive upregulation of EREG, as assessed by ELISA (Figure 4A). To study the site of EREG action, we investigated the distribution of EGFR in DRGs and spinal cord. EGFR was nonhomogeneously expressed by all DRG neurons within the cytoplasm of individual DRG cells, and neuronal size did not affect EGFR expression levels (Figure 4, B and C). In the spinal cord, EGFR expression was very low and was not found on neurons (Supplemental Figure 4). Immunostaining of EREG was not possible due to lack of appropriate antibodies. Based on our findings of increased EREG levels in CFA and SNI, we hypothesized that the activity of the EGFR should be increased in these conditions. To this end, we measured the phosphorylation of EGFR (on Tyr1068), which is reflective of EGFR activation, in lysates prepared from mouse DRG. EGFR phosphorylation was significantly increased following CFA and SNI (Figure 4D), consistent with the hypothesis that chronic inflammatory and neuropathic injury–induced increases in EREG levels enhances EGFR phosphorylation. Since EREG potentiated capsaicin-dependent nocifensive behavior, we next measured whether EREG (200 ng/ml) potentiates TRPV1-dependent calcium transients when capsaicin is applied onto cultured DRG neurons (Figure 4E). EREG caused a sensitization of the response to capsaicin, which was large in some neurons (>2 SD above the mean of vehicle-treated cells) and absent in others (Figure 4, F and G). The variability in the degree of sensitization is likely to reflect the variable expression of the EGFR in the population of DRG neurons (see Figure 4C). In those neurons where sensitization occurred, the calcium peak ratio before and after (b/a) drug administration was significantly enhanced by application of EREG (vehicle, 0.76 ± 0.01; EREG, 1.25 ± 0.03, mean ± SEM, P < 0.001, Figure 4G). Accordingly, based on the 2 SD criterion, sensitization occurred in 40.79% of neurons (Figure 4H), but we estimate that approximately 3.23% of these neurons represent false positive because this is the percentage of vehicle-treated neurons with b/a ratios greater than 2 SD above the mean. Thus, we estimate that approximately 37% of DRG neurons in culture were sensitized by EREG — a proportion similar to that exhibited by NGF (26).
Figure 4. EREG and EGFR are upregulated in chronic pain states, and EREG increases activation of medium-small DRG sensory neurons.
(A) EREG in the blood is upregulated by CFA and SNI, but not formalin (F3,38 = 10.0, P < 0.001), as measured by ELISA. Bars represent mean ± SEM for protein levels (pg/ml); n = 9–10 biological replicates/group. One-way ANOVA followed by Dunnett’s case-comparison post hoc test. *P < 0.05; ***P < 0.001 compared with control group. (B) EGFR (green) is abundantly found in all DRG sensory neurons. Scale bar: 50 μm. (C) The cellular distribution of EGFR is equal among different cell sizes that exhibit either high or low EGFR staining. n = 3 mice. (D) Top: representative Western blots showing p-EGFR and β-actin in the DRG before (BL) (left band) and 3 days or 7 days after CFA or SNI, respectively (right band). Bottom: quantification of Western blot data (n = 5 biological replicates/condition) after normalization to β-actin and compared with baseline values. *P < 0.05 compared with 1.0 by 1-tailed t test (CFA: t4 = 2.5, P = 0.03; SNI: t4 = 3.3, P = 0.02). (E) Representative calcium traces of neurons responsive to multiple capsaicin (500 nM, 15 seconds for every 4 minutes) pulses and treated either with vehicle (HBSS, left panel) or EREG (200 ng/ml, right panel) for 6 minutes before 3 challenging pulses of capsaicin were applied. The ratio of Ca2+ peak heights (b/a) before and after exposure to EREG or vehicle was calculated as a measure of signal enhancement. (F) Collected b/a ratio values obtained from experiments in E. White bars show ratios obtained without exposure to EREG (ncell = 60, nexp = 9). The distribution was well fitted by a Gaussian function with mean of 0.76, SD of 0.12, and upper 95% 2-tailed confidence limit of 1.05 (arrow). Green bars show ratios following 6 minutes exposure to EREG (200 ng/ml; ncell = 101, nexp = 13). Following EREG exposure, 40.79% of ratios exceeded the 95% confidence limit, and the mean ± SEM of these ratio values was 1.25 ± 0.03. (G) Neurons treated with EREG were separated into 2 groups: those in which sensitization was (ncell = 40) or was not (ncell = 61) observed in the first exposure following EREG addition. Data presented as mean ± SEM, n = 40–61/group, from a total of 9/13 experiments for vehicle/EREG. ***P < 0.001, unpaired Student’s t test. (H) The proportion of neurons sensitized in the presence of EREG based on b/a ratio values greater than 2 SD above the mean of the vehicle group.
EREG pain hypersensitivity is mediated by a signal transduction pathway involving mTOR, translational regulation, and MMP-9.
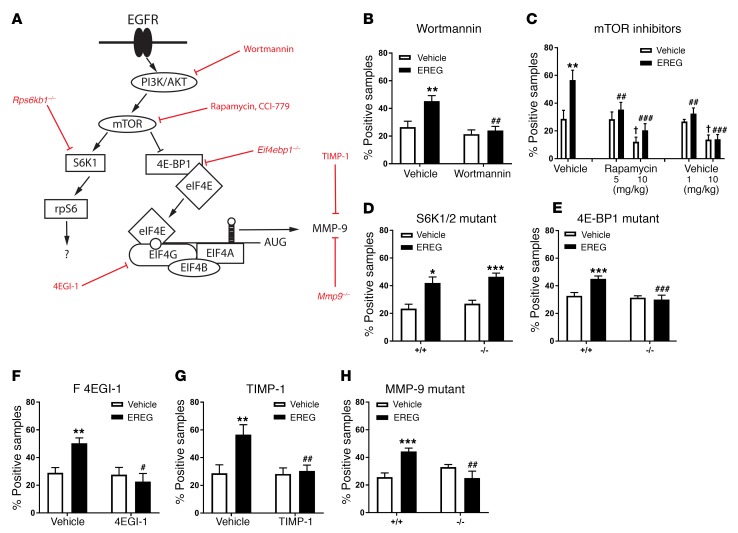
Since our preclinical and clinical data indicated a critical role of EREG (but not other EGFR ligands) and EGFR in pain processing, we sought to determine the intracellular machinery by which EREG-mediated EGFR activation increases pain. EGFR signaling has previously been shown to increase the activity of mTOR (27), a master regulator of mRNA translation (28, 29). Therefore, we hypothesized that mTOR is a downstream effector of EGFR with respect to pain. We investigated the contribution of mTOR signaling to EREG‑induced hypersensitivity by screening nocifensive behaviors during the formalin test and disrupting elements of the mTOR signal transduction pathway, either with pharmacological inhibitors or, where available, using null mutants (See Figure 5A for an overview of the pathway). Pretreatment with wortmannin, a covalent inhibitor of PI3K, completely abolished EREG‑induced hypersensitivity without affecting formalin-induced pain behavior per se (Figure 5B). Next, we tested 2 inhibitors of mTOR, rapamycin and CCI 779. At higher doses, these drugs produced analgesia during the formalin test, as has been reported previously (28, 29). However, at lower doses, both drugs prevented EREG-induced hypersensitivity without affecting formalin‑induced pain behavior per se (Figure 5C). mTOR regulates mRNA translation via 2 downstream effectors: ribosomal protein S6 kinases (S6Ks) and eukaryotic translation initiation factor 4E–binding (eIF4E‑binding) protein 1 (4E-BP1). Mutant mice lacking expression of both S6K1 and S6K2 (S6K1/2 double knockout) showed completely intact EREG-induced hypersensitivity (Figure 5D), whereas 4E‑BP1–deficient mice displayed no EREG‑induced hypersensitivity (Figure 5E). 4E-BP1 represses the formation of the eukaryotic translation initiation factor 4F (eIF4F) complex, which is a critical regulator of cap-dependent translation. An inhibitor of eIF4F complex, 4EGI-1, blocked EREG-induced hypersensitivity (Figure 5F), further supporting the role of mTOR/4E‑BP1/eIF4F in pain. To determine whether EREG hypersensitivity was mediated in part by ERK signaling, we used the MEK1/MEK2 inhibitor PD98059 (30). A low concentration of PD98059 did not block EREG-induced hypersensitivity on either the formalin test or the von Frey test (Supplemental Figure 5), suggesting that EREG does not potentiate pain behavior through enhanced ERK signaling. Since enhanced eIF4F complex formation has been shown to increase endogenous MMP‑9 in blood (31), we used tissue inhibitor of metalloproteinases-1 (TIMP-1), an endogenous blocker of MMP‑9, and Mmp9–/– mice to study whether MMP-9 signaling is important for EREG-induced pain hypersensitivity. TIMP‑1 prevented (Figure 5G, also see Supplemental Figure 6A) and Mmp9–/– mice did not display EREG‑induced hypersensitivity (Figure 5H). We further tested the importance of MMP-9 for EGFR pain signaling by confirming that the analgesic efficacy of gefitinib in the formalin test was abolished in Mmp9–/– mice (Supplemental Figure 6B).
Figure 5. EREG/EGFR increases pain through a PI3K/AKT→mTOR→4E-BP1→eIF4F complex→MMP-9 signaling pathway.
(A) The signaling pathway investigated, with major proteins indicated in black and blocking drugs or mutants shown in red. (B) Treatment with wortmannin (5 μg, i.t.) blocks EREG‑induced increases in late-phase formalin-induced pain behavior (drug × drug: F1,23 = 4.7, P = 0.04). (C) Low doses of rapamycin (5 mg/kg) and CCI 779 (1 mg/kg) block EREG effects without affecting formalin-induced pain per se (rapamycin, drug × drug: F1,27 = 3.6, P = 0.04; CCI 779 drug × drug: F1,28 = 4.2, P = 0.03); higher doses (10 mg/kg) are analgesic (main effects: rapamycin, F1,28 = 22.9, P < 0.001; CCI 779, F1,28 = 30.2, P < 0.001). (D) No effect on EREG increases in formalin-induced pain behavior in SGK1/2 (Rps6kb1/Rps6kb2) double-null mutant mice (Rps6kb1/2–/–; main effect of drug: F1,18 = 25.8, P < 0.001). (E) Lack of EREG effects in 4E-BP1 (Eif4ebp1–/–) null mutant mice (genotype × drug: F1,33 = 7.1, P = 0.01). (F) Treatment with 4EGI-1 (25 μg, i.t.) blocks EREG effects (drug × drug: F1,20 = 7.6, P = 0.01). (G) Treatment with TIMP-1 (4 pmol, i.t.) blocks EREG effects (drug × drug: F1,30 = 5.6, P = 0.02). (H) Lack of EREG effects in MMP-9 null mutants (Mmp9–/–; genotype × drug: F1,20 = 16.1, P = 0.001). In all experiments, EREG was injected at 10 ng i.t. Bars in all graphs represent mean ± SEM for percentage of samples featuring licking/biting behavior; n = 6–8/drug/dose and n = 6–12/drug/genotype (dependent on breeding success). Two-way ANOVA for all panels followed by t test compared with EREG vehicle. *P < 0.05; **P < 0.01; ***P < 0.001, compared with wortmannin, rapamycin, CCI 779, 4EGI-1, or TIMP-1 vehicle, or +/+ genotype. #P < 0.05; ##P < 0.01; ###P < 0.001, compared with rapamycin/CCI 779 vehicle. †P < 0.05 compared with vehicle/vehicle group.
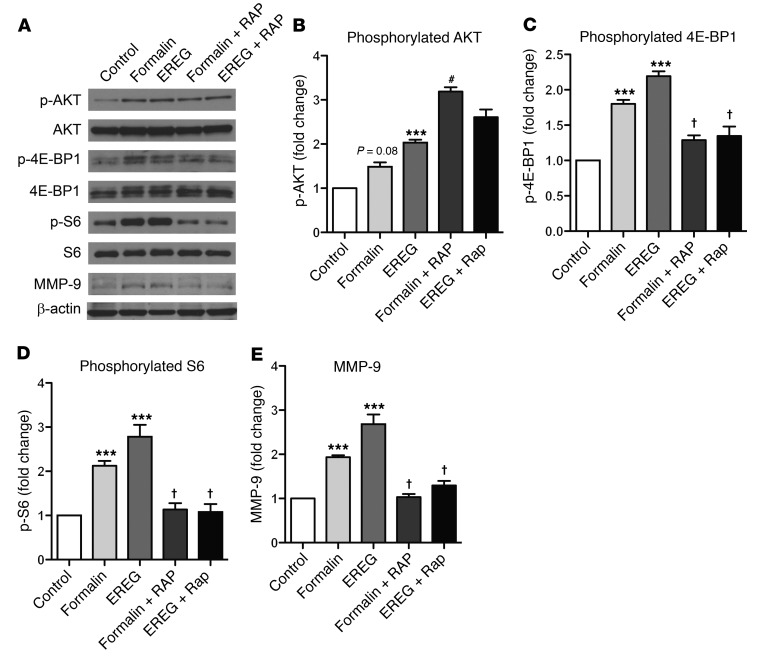
To further study the role of mTOR and MMP-9 in EGFR-mediated pain, we examined the phosphorylation of mTOR pathway components in DRG lysates. Formalin treatment alone did not significantly increase the phosphorylation of AKT (p-AKT), but EREG increased p-AKT in DRG tissue relative to control (Figure 6, A and B). Congruent with our behavioral data, both formalin and EREG increased the phosphorylation of 4E-BP1 (p‑4E-BP1) in DRG tissue, and cotreatment with rapamycin prevented this increase (Figure 6, A and C). Further, p-S6 was significantly increased with EREG treatment, and again, cotreatment with rapamycin prevented the increases (Figure 6, A and D). Finally, MMP-9 protein levels were increased in DRG tissue after formalin or EREG injection, and this increase was blocked with rapamycin inhibition of mTOR (Figure 6, A and E).
Figure 6. EREG and formalin induce phosphorylation of AKT and 4EBP-1 and increase the expression of MMP-9 in lumbar DRG tissue.
EREG (10 ng, i.t.) or 5% formalin (20 μl, intraplantar) was injected and lumbar DRG tissue harvested 40 minutes later. Rapamycin was injected 20 minutes before EREG or formalin to mimic behavioral experiment parameters. (A) Representative Western blots showing the phosphorylated and total protein abundance for AKT, 4E-BP1, and S6. The total amount of MMP-9 is also presented. Quantification (phosphorylated/total) for the percentage of fold increase (compared with the control condition) in phosphorylated AKT, 4E‑BP1, and S6 is presented in panels B–D along with the quantification for total MMP-9 (E). Bars represent mean ± SEM for relative change in protein expression. (B) EREG significantly increases the phosphorylation of AKT in DRG tissue. (C) Both formalin and EREG increase the phosphorylation of 4E-BP-1, and the increases are blocked by rapamycin. (D) The phosphorylation of S6 is significantly elevated relative to control tissue by EREG treatment, an increase blocked by rapamycin. (E) Formalin and EREG significantly increase MMP-9 expression, and these increases are blocked by rapamycin. Sample sizes in all groups are n = 4–6. One-way ANOVA for all panels followed by Dunnett’s case-comparison post hoc test. ***P < 0.001 compared with control tissue. †P < 0.05 decrease compared with EREG or formalin alone group. #P < 0.05 increase compared with EREG or formalin alone group.
To determine whether the increase in MMP-9 protein levels following EREG treatment was the result of increased Mmp9 mRNA translation, DRG extracts were fractionated on sucrose density gradients and the distribution of Mmp9 mRNA across these gradients was determined by qPCR analysis. In DRG extracts, Mmp9 mRNA shifted to the heavy polysome fractions after EREG injection, indicative of enhanced translation (Supplemental Figure 7). Rapamycin blocked this shift, indicating that in the DRG, EREG stimulates Mmp9 mRNA translation in an mTOR-dependent manner. Taken together, our results support a key role for mTOR, the eIF4F translational initiation complex, and MMP-9 signaling in EGFR-mediated pain behavior.
Discussion
Although there are indications from the clinical literature that EGFR inhibition may have analgesic properties (5, 6, 8), this possibility has not heretofore been examined systematically. Here, we used a broad range of techniques and analyses to demonstrate that (a) EGFR inhibition is analgesic and activation of the EGFR by EREG enhances pain; (b) EREG and EGFR display a genetic association with the development of chronic pain in clinical cohorts of TMD; and (c) EREG increases pain behavior and signaling through a mechanism that involves TRPV1 and mTOR/eIF4F/MMP-9–dependent signaling in the DRG.
We find that the EGFR is expressed by all DRG sensory neurons (Figure 4, B and C) and show that EREG potentiates capsaicin-induced calcium influx, suggesting that functional EREG receptors are present in DRG neurons (Figure 4, E–H). In addition, we provide compelling evidence that EREG increases pain sensitivity through EGFRs, whereas other EGFR ligands do not appear to play a role. This is consistent with prior observations that EGF or the heparin-bound EGF do not increase sensitivity to painful stimuli (32). EGF-like growth factors, including EREG, stimulate a variety of biological responses, and it is thought that ligand-induced homo- and heterodimerization can account for the majority of this diversity (33, 34). For instance, EREG is known to bind to and phosphorylate heterodimers of EGFR and ERBB4, but other EGF ligands (i.e., EGF and amphiregulin) only activate EGFR homodimers (35, 36). We suspect that an associated membrane protein, such as ERBB4, is involved in EREG-induced hypersensitivity, but this currently remains unclear.
Our human data further support a role of EREG in pain, as we found that EREG was associated with TMD development and upregulated in the blood of the patients. Moreover, our genetic association results revealed a strong effect of genetic polymorphism in the EREG-EGFR system, distinguishing those with chronic pain from controls. The observed association among TMD risk, EREG mRNA level, and transcript stability suggests that rs2367707 may be the functional SNP in humans. The fact that the TMD protective allele is associated with lower mRNA expression is congruent with our rodent findings. The EGFR SNPs tested in this study are probably only markers of the true effect-producing variants, but both 5′- and 3′-located SNPs within the EGFR locus showed association signals, suggesting both expression differences and transcript stability, respectively, may alter TMD risk. Thus, our results suggest that targeting EGFR and EREG for pain therapies may be an effective strategy. Importantly, since EGFR inhibition is associated with adverse side effects — folliculitis, hair loss and skin rash — inhibition of EREG may constitute an improved therapeutic option for pain management.
At the cellular level, the effects of EREG on EGFR involve TRPV1, as EREG potentiates capsaicin-evoked calcium responses of sensory neurons, and blockade of TRPV1 attenuated EREG hyperalgesia in the formalin assay. TRPV1 has been shown to induce EGFR transactivation in a model of epithelial wound healing, leading to PI3K/AKT stimulation (17). This is in line with our current findings showing disruption of PI3K/AKT/mTOR signaling with specific inhibitors of the AKT/mTOR pathway blocking, and 4E‑BP1 null mice lacking, EREG-induced hypersensitivity. In the DRG, EREG or formalin increased the phosphorylation of both S6 and 4E‑BP1, the 2 main downstream targets of mTOR. However, since S6K1/2 null mutant mice had intact EREG-induced hypersensitivity, we conclude that S6K is not necessary for EREG‑stimulated pain behavior. In addition, our results indicate that EREG-induced pain behavior is not mediated by ERK signaling, an observation that is consistent with data showing that EREG does not activate ERK in DRG neurons (32). Further, our data indicate that the phosphorylation of 4E-BP1 increases the translation of MMP-9, and pharmacological or genetic reduction of MMP-9 activity renders EREG and EGFR antagonists ineffective against inflammatory pain. Together, our results indicate that EREG upregulation in the blood may activate EGFRs on DRG neurons to induce hypersensitivity through transactivation of TRPV1 and the mTOR signaling pathway, which increases MMP-9 translation. It is likely that EREG originates from a cellular source within the blood, as we found elevated levels of EREG following CFA and SNI, which parallel EREG mRNA expression levels in TMD patients.
In summary, we find that clinically available small molecule EGFR inhibitors targeting the tyrosine kinase site of the EGFR receptor, including gefitinib and lapatinib, are effective analgesics in mice for inflammatory and neuropathic pain. These drugs are routinely given to non–small cell lung cancer patients (37, 38) to inhibit tumor growth, but have not been systematically studied for their role in pain management. Since we find that EREG is the primary endogenous activator of EGFR-related pain hypersensitivity, our data suggest that an effective treatment strategy may be the selective inhibition of EREG over other endogenous EGFR ligands. Directly targeting EREG may result in a reduced side-effect profile when compared with currently available EGFR-inhibition strategies.
Methods
Study design
Animals were randomized to drug condition using within-cage randomization, and all behavioral experiments were performed by an experimenter blinded to drug conditions. As we had no a priori expectation of effect sizes, power analyses were not used to calculate sample sizes. Instead, we adhered to standard practices in the field (39). In many cases, sample sizes were dictated by breeding success. Statistical outliers were defined via Studentized residuals of more than 3 and excluded before analyses were run.
Rodent subjects
Most experiments were performed on naive, adult (7–12 weeks of age) outbred CD-1 (ICR:Crl) mice of both sexes, bred in-house (J.S. Mogil and L.J. Martin laboratories) from breeders obtained from Charles River Laboratories. Heterozygote breeding pairs for mutant mice containing a large deletion of the extracellular domain of the EGFR receptor (EGFRvIII/ΔEGFR) were obtained from the laboratory of David Threadgill (North Carolina State University, Raleigh, North Carolina, USA) on a C57BL/6 background. We only tested heterozygote EGFRvIII/ΔEGFR mice because the homozygotes are neonatally lethal. Null mutant mice for 4E‑BP1 (Eif4ebp–/–; C57BL/6 background) and p70 S6K 1/2 double-knockout mice (Rps6kb1/Rps6kb2–/–; mixed 129Sv × C57BL/6 background) and their associated WT were generated and bred in one of our laboratories (N. Sonenberg). The latter mutants were provided by G. Thomas (University of Cincinnati, Cincinnati, Ohio, USA). Mice lacking the MMP-9 gene (Mmp9–/–) were purchased from The Jackson Laboratory on an FVB/NJ background and compared with WT mice of that strain. No overt behavioral abnormalities were noted in any of the mutant mouse strains. All mice were housed with their same-sex littermates (2 to 4 animals per cage) in standard shoebox cages, maintained in a temperature-controlled (20 ± 1°C) environment (14-hour light/10-hour dark cycle), and fed (Harlan Teklad 8604) and watered ad libitum. Mice were assigned to experimental conditions in a randomized fashion within-cage.
Behavioral assays
Subjects were habituated to the testing environment for at least 15 minutes in every assay before testing commenced.
Rotarod test.
Drug effects on motor coordination were tested using an accelerating rotarod treadmill (Acceler Rota-Rod 7650, Ugo Basile) for mice. Mice were placed on the rotarod, which accelerated from 4 to 40 rpm over a period of 5 minutes, and the time spent on the rotating drum was recorded for each mouse. On the test day, 1 preinjection baseline trial (drug-free) was performed before the animals were treated with either saline, AG 1478 (100 mg/kg), gefitinib (300 mg/kg), or lapatinib (300 mg/kg). Performance was indicated by the latency to fall from the rotarod at 15–60 minutes after injection.
Radiant heat paw-withdrawal test.
Mice were placed on a 3/16-inch–thick glass floor within small Plexiglas cubicles (9 × 5 × 5 cm high), and a focused high-intensity projector lamp beam was shone from below onto the midplantar surface of the hind paw (40). The commercial device (IITC model 336) was set to 20% active intensity. Latency to withdraw from the stimulus was measured to the nearest 0.1 second. Baseline measurements consisted of testing both hind paws twice on 3 separate occasions separated by at least 30 minutes. Following drug injection, both hind paws were only tested once at the indicated time.
von Frey test.
The up-down method of Dixon (41) was used. Mice were placed on a perforated metal floor (with 5-mm diameter holes placed 7 mm apart) within small Plexiglas cubicles as described above, and a set of 8 calibrated von Frey fibers (Stoelting Touch Test Sensory Evaluator Kit no. 2 to no. 9; ranging from ~0.015 g to ~1.3 g of force) were applied to the plantar surface of the hind paw until the fibers bowed and then held for 3 seconds. The threshold force required to elicit withdrawal of the paw (median 50% withdrawal) was determined twice on each hind paw (and averaged) for all baseline measurements, with sequential measurements separated by at least 20 minutes. For experiments in which a drug was injected, 1 measurement per hind paw was taken at the indicated time point.
Formalin test.
Mice were placed on a tabletop within Plexiglas cylinders (30 cm high; 30 cm diameter) and allowed to habituate. Then, 20 μl of 5% formalin was injected s.c. into the plantar surface of the left hind paw using a 100-μl microsyringe with a 30‑gauge needle. Mice were then returned to the cylinders and left undisturbed for 60 minutes, with behaviors recorded using digital video. Videos were later coded offline, where the first 10 seconds of every minute were monitored for the presence of licking/biting (positive sample) of the left hind paw for a total of 60 observations. The early phase was defined as the percentage of positive samples during the first 0–10 minutes after injection of formalin, the late phase as the percentage of positive samples during the period 10–60 minutes after injection. For the drug studies, EGFR, TRPV1, TRPA1, and mTOR inhibitors were injected 20 minutes before formalin and TIMP-1 was injected 1 hour before formalin. EGFR ligands and NGF were injected immediately before formalin.
Carrageenan.
Carrageenan (2%, 20 mg/ml; Sigma-Aldrich) was suspended by sonication in saline and injected s.c. in a volume of 20 μl into the left plantar hind paw using a 100-μl microsyringe with a 30-gauge needle. Mice were tested for thermal sensitivity of both hind paws using the radiant heat paw-withdrawal test as described above, before and 3 hours after carrageenan injection. All drugs were injected immediately following the test for carrageenan hypersensitivity at the 3-hour time point, and post‑drug measurements were taken at 20, 40, and 60 minutes.
CFA.
CFA (50%; Sigma-Aldrich) was injected s.c. in a volume of 20 μl into the left plantar hind paw using a 100-μl microsyringe with a 30-gauge needle. Mice were tested for mechanical sensitivity of both hind paws using the von Frey test as described above, before and 3 days after CFA injection. All drugs were injected immediately following the 3-day post-CFA test, and post‑drug measurements were taken at 20, 40, 60, and 90 minutes. Percentage of maximal possible anti-allodynia (i.e., reversal of allodynia back to preinjection baseline values at all post-drug time points) was calculated using the trapezoidal method.
Spared nerve and chronic constriction injury.
SNI and CCI, 2 experimental nerve injury procedures designed to produce neuropathic pain, were performed under isoflurane/oxygen anesthesia as described previously (42, 43). Mice were tested for mechanical sensitivity before and after surgery using the von Frey test as described above, except that the “spared” sural region was targeted for SNI and the mid‑plantar surface was targeted for CCI by applying the fibers to the hind paw. All drugs were injected immediately following the test for SNI- or CCI-induced mechanical allodynia 7 or 14 days following surgery, respectively, and post-drug measurements were taken at 20, 40, 60, and 90 minutes. Percentage of maximal possible anti-allodynia (i.e., reversal of allodynia back to presurgery baseline values at all post-drug time points) was calculated using the trapezoidal method.
Capsaicin and mustard oil.
Mice were allowed to habituate to an observation chamber (see formalin test above) for 15 minutes. Mice then received a s.c. injection of capsaicin (2.5 μg; Sigma-Aldrich) or mustard oil (5%; Sigma-Aldrich) into the plantar left hind paw (20 μl) and were digitally videotaped for 10 minutes. Video files were later scored for the total duration of licking/biting of the injected paw.
Drugs
AG 1478, gefitinib, lapatinib, rapamycin, CCI 779, and wortmannin were purchased from LC Laboratories and dissolved in 30% polyethylene glycol, except for wortmannin, which was dissolved in 10% DMSO. EREG, EGF, betacellulin, amphiregulin, TGF-α, NGF, AMG 9810, K252a, PD 98059, and TIMP-1 were purchased from R&D Systems and were dissolved in sterile saline, except K252a and PD 98059, which were dissolved in 20% DMSO. Capsaicin, mustard oil and HC 030031 were purchased from Sigma-Aldrich. 4EGI-1 was purchased from Santa Cruz Biotechnology Inc. and dissolved in 20% DMSO. Morphine sulfate was obtained from Health Canada and dissolved in saline. Drugs were administered either i.p. (10 ml/kg volume) or i.t. (5 μl volume) (44).
Enzyme-linked immunosorbant assay
Mice received no treatment, formalin injection, CFA injection, or SNI surgery and were euthanized 60 minutes (formalin), 3 days (CFA), or 7 days (SNI) later. Trunk blood was collected into EDTA-coated Vacutainer tubes/heparinized syringes. Blood was centrifuged at 15,000 g for 15 minutes at 4°C to isolate plasma from other blood components. Plasma was aliquoted into tubes, frozen with liquid nitrogen, and stored at –80 °C. Plasma samples were then thawed on ice and EREG measured in duplicate using an ELISA kit from Abcam according to the manufacturer’s instructions.
Immunohistochemistry
Naive mice were deeply anesthetized with sodium pentobarbital (60 mg/kg) and perfused transcardially through the left cardiac ventricle with 100 ml of perfusion buffer, followed by 250 ml of 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB), pH 7.4, at room temperature for 15 minutes. Subsequently, the spinal column was removed and post-fixed in the same fixative for 24 hours at 4°C. Spinal cord lumbar segments L3 and L4 and DRGs at the same levels were extracted and cryoprotected with 30% sucrose in 0.1 M PB. Tissue was embedded in an optimum cutting temperature medium (Tissue Tek OCT; Sakura), and 16-μm and 50-μm transverse DRG and spinal cord sections, respectively, were cut at –20°C on a Leica CM3050 S cryostat. DRG sections were placed directly on gelatin-subbed slides. and spinal cord sections were collected as free-floating sections in PBS. The staining protocols for slides and free-floating tissue were similar. Sections were rinsed 3 times with PBS, with 0.2% Triton X-100 (PBS-T) for 10 minutes, and preincubated with 10% normal goat serum (NGS) for 1 hour. To assess the colocalization between EGFR and markers of primary afferent neuronal populations, sections were incubated overnight at 4°C in 5% NGS with either (a) anti-EGFR raised in rabbit (1:50, Santa Cruz Biotechnology Inc., catalog SC-03, lot F1512) or (b) anti-EGFR and anti-NeuN raised in mouse (1:5000, Millipore, catalog MAB377, lot 2062313). To assess the specificity of the EGFR antibody, the diluted antibody (1:50) was preincubated with the EGFR-blocking peptide (1:5, Santa Cruz Biotechnology Inc., catalog SC-03p, lot E2109) overnight at 4°C in PBS before adding it to the tissue. The next day, tissue was washed 3 times with PBS-T for 10 minutes, incubated in Alexa Fluor 488 anti-rabbit (1:800, Invitrogen, catalog A11034, lot 870976), Alexa Fluor 594 anti–guinea pig (1:800, Invitrogen, catalog A11076, lot 714263), or Alexa Fluor 568 anti-mouse goat secondary antibodies (1:800, Invitrogen, catalog A11031, lot 822389) in the dark for 2 hours and washed 2 times with PBS-T and 1 time with PBS. Free-floating sections were mounted on slides. All slides were coverslipped with Aqua-Poly/Mount (Polysciences). Sections were examined using a Zeiss LSM 510 confocal scanning laser microscope, equipped with argon and helium-neon lasers using a multi-track approach.
Western blots
Tissue extracts for Western blotting were prepared in ice-cold homogenization buffer containing the following: 50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton X-100; 5 mM NaF; 1.5 mM Na3VO4; and protease inhibitor cocktail (complete, EDTA-free, Roche Applied Science). For measuring MMP-9, DRGs were removed from animals that were perfused transcardially with PBS. Following centrifugation at 12,000 g for 10 minutes at 4°C, the supernatant protein concentration was measured and equal protein quantities were boiled for 5 minutes in sample buffer and separated by SDS-PAGE. Following electrophoresis, proteins were transferred to 0.2-mm nitrocellulose membranes. Membranes were blocked in 5% dry milk powder in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) for 1 hour prior to overnight incubation with primary antibody. The membranes were then washed, incubated for 1 hour with HRP-conjugated secondary antibody, washed again, treated with Enhanced Chemiluminescence reagent (PerkinElmer), and exposed to autoradiography films (Denville Scientific Inc.). All signals were obtained in the linear range for each antibody, and densitometric analyses were performed with ImageJ (NIH). Each phosphoprotein was normalized to the expression of the corresponding total protein. The antibodies and dilutions for the Western blots used in these studies were as follows: 4E-BP1 (1:1000, catalog 9644, Cell Signaling Technology), P-4E-BP1 (Thr37/46) (1:1000, catalog 2855, Cell Signaling Technology), AKT (1:1000, catalog 4685, Cell Signaling Technology), p‑AKT (Ser473) (1:1000, catalog 9271, Cell Signaling Technology), EGFR (1:1000, catalog sc-03, Santa Cruz Biotechnology Inc.), p-EGFR (1:1000, catalog 3777, Cell Signaling Technology), MMP-9 (1:1000, catalog AB19016, Chemicon), S6 (1:1000, catalog sc-74459, Santa Cruz Biotechnology Inc.), p-S6 (Ser240/244) (1:1000, catalog 2215, Cell Signaling Technology), and β‑actin (1:5000, catalog A5441, Sigma-Aldrich).
Calcium imaging
Mouse lumbar DRG neurons (L3–L5; at least n = 4 mice per condition) were harvested and cultured as previously described (45). Briefly, DRGs were isolated, transferred into HBSS, and enzyme-digested by incubation with papain and collagenase type II (Worthington Biochemical Corp.). Dissociated neurons were plated on glass coverslips coated with poly-d-lysine and laminin and maintained at 37°C at 5% CO2/95% air in F12 media (Life Technologies) with 10% FBS. After 2–6 hours, dissociated neurons on coverslips were loaded with 1 μM of the cell-permeable calcium-sensitive dye Fura-2, AM (Life Technologies) for 30 minutes and washed with HBSS before use. The protocol for the experiment is shown in Figure 4E and was selected to avoid spurious differences in calcium dye loading and indicator measurements (26). Coverslips were placed in a chamber containing HBSS at room temperature (20–22°C). Following baseline recording, 25 mM KCl was applied for 15 seconds, followed by capsaicin (500 nM in HBSS) application for 15 seconds every 4 minutes, as previously described (26). HBSS (vehicle, 60 cells, 9 different experiments) or EREG (200 ng/ml, 101 cells, 13 different experiments) was applied for 6 minutes after the fourth, fifth, or sixth application of capsaicin, when the response had largely stabilized. After the incubation, 3 more pulses of capsaicin were applied and the ratio of the Ca2+ increases before and after exposure to EREG or HBSS was calculated as an index of enhancement (b/a ratio). In order to determine the degree of sensitization and the proportion of neurons sensitized by EREG, the variability of the calcium signal (b/a ratio) in vehicle-treated neurons was examined and an index of sensitization was calculated as the percentage of neurons where the ratio exceeded 2 SD above the mean for vehicle-treated neurons (Figure 4F, values to the right of the arrow). The b/a ratio was normally distributed with a mean of 0.76, SD 0.12 (Figure 4F). The mean percentage of false positives with ratios above 1.05 (2 SD above the mean) was 3.22%. Fluorescence was detected by a Zeiss Observer Ratio Z1 microscope at 340 nm and 380 nm excitation wavelengths and analyzed with ZEN Black software (Zeiss).
Quantitative real-time PCR
DRGs (L3–L4) were isolated and subjected to RNA extraction using TRIzol (Invitrogen). Reverse transcription was performed using a SuperScript III Reverse-Transcriptase Kit and Random Hexamers (Invitrogen) according to the manufacturer’s instructions. qPCRs were carried out in a CFX96-PCR system using iQ Sybr Green Supermix RT (Bio-Rad) according to the manufacturer’s instructions. The following primers were used: Mmp9 (forward) GATCCCCAGAGCGTCATTC, Mmp9 (reverse) CCACCTTGTTCACCTCATTTTG; Gapdh (forward) TCCATGACAACTTTGGCATTG, and Gapdh (reverse) CAGTCTTCTGGGTGGCAGTGA. Analyses were carried out in triplicate, and the Mmp9 signal was normalized to Gapdh.
Polysomal profile analysis
Lumbar DRGs (L3–L5; pooled from at least n = 10 mice per condition) were isolated and placed in ice‑cold HEPES-KOH HBSS (pH 7.4) containing 100 μg/ml cyclohexamide. HBSS was replaced with the ice-cold hypotonic lysis buffer (46) containing protease (cOmplete EDTA-free, Roche Products) and RNase inhibitors (Rnasin, Promega), and the tissue was subjected to brief homogenization using a glass homogenizer. The homogenated material was spun at 18,000 g for 2 minutes at 4°C, and the supernatant was loaded on a 10%–50% w/w sucrose gradient in 20 mM HEPES‑KOH, pH 7.6, 100 mM KCl, and 5 mM MgCl2, and centrifuged at 35,000 g for 2.5 hours at 4°C in an Optima L-80 XP Ultracentrifuge (Beckman Coulter) using an SW40Ti rotor. Polysome analysis was performed by measuring the OD at 254 nm using ISCO fractionators (Teledyne ISCO), as described previously (46). qPCR analysis was performed as previously described (47). Sucrose gradient fractions were subjected to RNA extraction using TRIzol (Invitrogen). Reverse transcription was performed using a SuperScript III Reverse-Transcriptase Kit (Invitrogen) and Random Hexamers (Invitrogen) according to the manufacturers’ instructions. qPCRs were carried out in a CFX96 (Bio-Rad) RT-PCR system using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions using the following primers: Mmp9 (forward) GATCCCCAGAGCGTCATTC and Mmp9 (reverse) CCACCTTGTTCACCTCATTTTG). For all experiments, n = 4 (technical replicates); results are presented in arbitrary units as relative amounts using serial dilutions of DRG or spinal RNA as qPCR concentration standards.
Human subjects and phenotyping
Genotype and phenotype data from the OPPERA case-control study are available at the Database of Genotypes and Phenotypes (dbGaP phs000762.v1.p1). The OPPERA cohort was recruited and phenotyped as detailed previously (48, 49) and is described briefly here. Volunteers were recruited at 4 US study sites. Cases (n = 127) had examiner-verified TMD at enrollment; controls (n = 731) were individuals who reported no significant history of TMD symptoms. Classification of TMD was based on the Research Diagnostic Criteria (RDC) for Temporomandibular Disorder (50). To increase genetic homogeneity of the cohorts, only subjects of mixed European descent were analyzed in this study; results from the full cohort were extremely similar. An additional subgroup of super controls (n = 231 mixed European descent) was classified post hoc as TMD‑free controls who experienced no tenderness during palpation of 8 masticatory muscles and 2 temporomandibular joints. Their genotypes were contrasted with 129 TMD patients of mixed European descent.
The TMD case-control cohort (24) included 200 TMD cases and 198 controls, using recruitment protocols and diagnostic criteria similar to those used in OPPERA, with the exception that enrollment was open to non-Hispanic females of mixed European descent ages 18 to 45 and cases were recruited through a tertiary care pain clinic rather than from the general population.
The pre-OPPERA cohort (25) included n = 186 initially pain-free females of mixed European descent, ages 18–34, of which 15 developed RDC-verified TMD over the course of the 3 year follow-up. Measures of sensitivity to a multitude of pain-evoking stimuli were also collected in all 3 cohorts (24, 25, 48, 49).
Genetic association
Genetic analysis of the OPPERA cohort was described previously (51). Genotyping was performed on DNA extracted from whole blood, using the Pain Research Panel (Algynomics Inc.). The Pain Research Panel is a microarray platform that assesses 3,295 SNP markers representing 358 genes of potential relevance to pain, inflammation, and/or mood and affect as well as 160 ancestry informative markers used to adjust for population stratification. Duplicate study samples and HapMap reference DNA were genotyped to confirm accuracy and reliability of genotyping, and quality filters were imposed for call rates of more than 95%, reliability of more than 99%, minor allele frequency of more than 1%, and adherence to Hardy-Weinberg equilibrium. The overall call rate was 99.7%, with 2,924 SNPs passing quality filters. Genotyping of the pre-OPPERA cohort was performed separately on the Pain Research Panel using DNA from whole blood after amplification.
PLINK v.1.07 (Broad Institute) software (52) was used to perform case-control association tests by logistic regression, assuming a codominant inheritance model. All tests on the OPPERA cohort controlled for recruitment site, and tests that included subjects not of mixed European descent were also adjusted for race using the first 2 eigenvectors of a principal components analysis (PCA) on the genotype matrix (53).
After initial association tests performed in OPPERA identified EREG and EGFR, 5 SNPs from EREG and 25 SNPs from EGFR were extracted from the full SNP panel. Haplotype blocks were identified in each gene using Haploview v.4.2 (54), and tag SNPs were selected to cover haplotypic variation in EREG (rs2367707, rs7687621, rs1542466) and EGFR (5′ region: rs759171, rs4947963; 3′ region: rs1140475, rs2740762, rs845552). Haplotype testing was performed in the R statistical environment using logistic regression. Omnibus tests were used to detect differences in TMD odds between any major haplotype groups, and post hoc tests were performed contrasting individual haplotypes against all others in order to characterize their effects. Combined P values for haplotype analysis were calculated using the optimally weighted Z test (55).
To explore cellular mechanisms underlying the associations, we applied bioinformatic pathway analysis (based on Pathway Studio, Elsevier) to identify signaling networks implicated by the association results in the discovery cohort. The multiple testing–adjusted significance threshold was determined to be 0.002 for the 5% level, as determined through permutation (56).
Human mRNA studies
Leukocytes were obtained from 6 ml heparinized venous blood from human subjects. Briefly, whole blood was diluted in endotoxin-free RPMI 1640 medium and centrifuged through Ficoll/Histopaque 1077 (Sigma-Aldrich), and the buffy coat cells were washed 5 times with sterile isotonic saline. Total RNA was isolated with TRIzol Reagent (Life Technologies), and RT-PCR was performed with the SABioscience Custom PCR Array System. The cDNAs from all participants were normalized to GAPDH through dilution to the concentration at which GAPDH gave an equal signal in RT-PCR reactions.
Human cDNA amplified from study participants with the major EREG haplotype was cloned into pCDNA3 vectors under a CMV promoter to generate a WT expression EREG plasmid (gift from Oskar Laur, Emory University, Atlanta, Georgia, USA). Briefly, an expression plasmid with the minor allele at rs2367707 was generated by site-directed mutagenesis. Expression plasmids were transiently transfected into HEK293 cells using Lipofectamine 2000 (Life Technologies) in accordance with the manufacturer’s recommendations. The time course of mRNA degradation was measured after actinomycin D (actD) (Sigma-Aldrich) treatment. Thirty-six hours after transfection of EREG plasmids, cells were treated with actD (10 μg/ml) and collected at 0, 2, 4, or 6 hours after treatment. Total RNA was isolated after each time point using TRIzol reagent (Life Technologies). The isolated RNA was treated with RNase Free–DNase I (Promega) and reverse transcribed using a SuperScript III Reverse-Transcriptase Kit (Invitrogen) and Random Hexamers (Invitrogen) according to the manufacturer’s instructions. qRT-PCRs were carried out in a CFX96 (Bio-Rad) RT-PCR system using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s instructions. cDNAs of EREG and the housekeeping gene GAPDH were amplified using forward and reverse PCR primers (GGCTATTGTTTGCATGGACAG and CACGGTCAAAGCCACATATTC, for EREG; and CTTTGGTATCGTGGAAGGACTC and GTAGAGGCAGGGATGATGTTC, for GAPDH). Two independent experiments were conducted in triplicate. Data were normalized to GAPDH.
Drosophila studies
For further details, see Supplemental Methods.
Statistics
Data were analyzed by 2-tailed Student’s t test (unless otherwise indicated) or 1-way or 2‑way ANOVA (or Kruskal-Wallis ANOVA by ranks), followed where appropriate by Tukey’s honest significant difference (HSD) post hoc test or Dunnett’s case‑comparison post hoc test. A P value of less than 0.05 was considered significant. Four data points were excluded based on their identification as statistical outliers (Studentized residuals > 3). Analgesic doses needed to reduce pain by 50% (AD50s) and associated 95% confidence intervals were calculated using the FlashCalc 40.1 macro (M.H. Ossipov, University of Arizona, Tucson, Arizona, USA).
Study approval
Mice were maintained in the animal facilities of McGill University and the University of Toronto, Mississauga. All mouse experiments were approved and performed in accordance with the guidelines of relevant local animal care and use committees according to the Canadian Council on Animal Care (CCAC) guidelines. The OPPERA study was reviewed and approved by Institutional Review Boards at each of the 4 study sites and at the data coordinating center: University at Buffalo, University of Maryland, University of North Carolina at Chapel Hill, University of Florida, and the Battelle Memorial Institute. All participants verbally agreed to a screening interview done by telephone and provided informed, signed consent for all other study procedures.
Author contributions
JSM and LD conceived of the initial project with input from LJM and SBS. LJM and JSM designed the majority of the animal experiments. LJM, SBS, AK, CAM, AS, RES, CC, NY, SS, ST, TC, TMK, EM, DGG, JSW, SGS, and JSA performed research. SBS and LD performed the bioinformatics analysis on the OPPERA data set. LJM, SBS, AK, CAM, CBM, AS, JHG, AGN, ARDS, GGN, DVZ, JSM, and LD analyzed the data. JDG, RBF, RO, GDS, CK, RD, WM, and LD provided clinical data. CG, NS, AGN, ARDS, and GGN oversaw portions of this project. LJM, SBS, JSM, and LD wrote the paper with input from all authors.
Supplementary Material
Acknowledgments
The authors would like thank Oskar Laur for creating EREG-expressing constructs at the Custom Cloning Core Facility, Emory University. This research was supported by the Canadian Institutes for Health Research (CIHR) (NS, ARDS, and JSM), the NIH (JDG, RBF, RO, GDS, CK, RD, WM, and LD), the Australian National Health and Medical Research Council (TC, TMK, GGN), and the Louise and Alan Edwards Foundation (JSM). LJM was supported by postdoctoral fellowships from CIHR and the Canadian Pain Society.
Version 1. 08/07/2017
Electronic publication
Version 2. 09/01/2017
Print issue publication
Footnotes
AGN and WM’s present address is: Department of Anesthesiology, Center for Translational Pain Medicine, Duke University, Durham, North Carolina, USA.
Conflict of interest: S.B. Smith, R.B. Fillingim, G.D. Slade, and J.S. Mogil are consultants and equity stock holders, and William Maixner and Luda Diatchenko are cofounders and equity stock holders in Algynomics, a company providing research services in personalized pain medication and diagnostics.
Reference information: J Clin Invest. 2017;127(9):3353–3366.https://doi.org/10.1172/JCI87406.
Contributor Information
Arkady Khoutorsky, Email: arkady.khoutorsky@mcgill.ca.
Alexander Samoshkin, Email: alex.samoshkin@mcgill.ca.
Robert E. Sorge, Email: rsorge@uab.edu.
Chulmin Cho, Email: chulmin.cho@utoronto.ca.
Sivaani Sivaselvachandran, Email: sivaani.sivaselvachandran@mail.utoronto.ca.
Sarasa Tohyama, Email: sarasa.tohyama@utoronto.ca.
Tiffany Cole, Email: tiffany.cole@sydney.edu.au.
Ellen Mir, Email: ellen.mir@sas.com.
Christos Gkogkas, Email: Christos.Gkogkas@ed.ac.uk.
Nahum Sonenberg, Email: nahum.sonenberg@mcgill.ca.
Richard Ohrbach, Email: ohrbach@buffalo.edu.
Charles Knott, Email: cknott@rti.org.
Ronald Dubner, Email: rdubner@umaryland.edu.
Alfredo Ribeiro-da-Silva, Email: alfredo.ribeiro-da-silva@mcgill.ca.
G. Gregory Neely, Email: greg.neely@sydney.edu.au.
William Maixner, Email: william.maixner@duke.edu.
Luda Diatchenko, Email: luda.diatchenko@mcgill.ca.
References
- 1.Elliott AM, Smith BH, Penny KI, Smith WC, Chambers WA. The epidemiology of chronic pain in the community. Lancet. 1999;354(9186):1248–1252. doi: 10.1016/S0140-6736(99)03057-3. [DOI] [PubMed] [Google Scholar]
- 2.Gereau RW, et al. A pain research agenda for the 21st century. J Pain. 2014;15(12):1203–1214. doi: 10.1016/j.jpain.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Institute of Medicine (US) Committee on Advancing Pain Research, Care, Education. Relieving Pain in America: A Blueprint for Transforming Prevention, Care, Education, and Research. Washington, D.C.: National Academies Press; 2011. [PubMed] [Google Scholar]
- 4.Erschbamer M, Pernold K, Olson L. Inhibiting epidermal growth factor receptor improves structural, locomotor, sensory, and bladder recovery from experimental spinal cord injury. J Neurosci. 2007;27(24):6428–6435. doi: 10.1523/JNEUROSCI.1037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kersten C, Cameron MG, Laird B, Mjåland S. Epidermal growth factor receptor-inhibition (EGFR-I) in the treatment of neuropathic pain. Br J Anaesth. 2015;115(5):761–767. doi: 10.1093/bja/aev326. [DOI] [PubMed] [Google Scholar]
- 6.Kersten C, Cameron MG. Cetuximab alleviates neuropathic pain despite tumour progression. BMJ Case Rep. 2012;2012:bcr1220115374. doi: 10.1136/bcr.12.2011.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moryl N, Obbens EA, Ozigbo OH, Kris MG. Analgesic effect of gefitinib in the treatment of non-small cell lung cancer. J Support Oncol. 2006;4(3):111. [PubMed] [Google Scholar]
- 8.Kersten C, Cameron MG, Laird B, Mjåland S. Epidermal growth factor receptor-inhibition (EGFR-I) in the treatment of neuropathic pain. Br J Anaesth. 2015;115(5):761–767. doi: 10.1093/bja/aev326. [DOI] [PubMed] [Google Scholar]
- 9.Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol. 2006;7(7):505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 10.Pastore S, Mascia F, Mariani V, Girolomoni G. The epidermal growth factor receptor system in skin repair and inflammation. J Invest Dermatol. 2008;128(6):1365–1374. doi: 10.1038/sj.jid.5701184. [DOI] [PubMed] [Google Scholar]
- 11.Shirakata Y, et al. Epiregulin, a novel member of the epidermal growth factor family, is an autocrine growth factor in normal human keratinocytes. J Biol Chem. 2000;275(8):5748–5753. doi: 10.1074/jbc.275.8.5748. [DOI] [PubMed] [Google Scholar]
- 12.Kaganoi J, et al. STAT1 activation-induced apoptosis of esophageal squamous cell carcinoma cells in vivo. Ann Surg Oncol. 2007;14(4):1405–1415. doi: 10.1245/s10434-006-9274-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Antitumor activity of taspine by modulating the EGFR signaling pathway of Erk1/2 and Akt in vitro in vivo. Planta Med. 2011;77(16):1774–1781. doi: 10.1055/s-0030-1271132. [DOI] [PubMed] [Google Scholar]
- 14.Wells A, Gupta K, Chang P, Swindle S, Glading A, Shiraha H. Epidermal growth factor receptor-mediated motility in fibroblasts. Microsc Res Tech. 1998;43(5):395–411. doi: 10.1002/(SICI)1097-0029(19981201)43:5<395::AID-JEMT6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Long H, Wu Z, Jiang X, Ma L. EGF transregulates opioid receptors through EGFR-mediated GRK2 phosphorylation activation. Mol Biol Cell. 2008;19(7):2973–2983. doi: 10.1091/mbc.E07-10-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, et al. Epidermal growth factor-induced esophageal cancer cell proliferation requires transactivation of beta-adrenoceptors. J Pharmacol Exp Ther. 2008;326(1):69–75. doi: 10.1124/jpet.107.134528. [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Wang Z, Capó-Aponte JE, Zhang F, Pan Z, Reinach PS. Epidermal growth factor receptor transactivation by the cannabinoid receptor (CB1) and transient receptor potential vanilloid 1 (TRPV1) induces differential responses in corneal epithelial cells. Exp Eye Res. 2010;91(3):462–471. doi: 10.1016/j.exer.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawasaki Y, et al. Distinct roles of matrix metalloproteases in the early- and late-phase development of neuropathic pain. Nat Med. 2008;14(3):331–336. doi: 10.1038/nm1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pareja M, Sánchez O, Lorita J, Soley M, Ramírez I. Activated epidermal growth factor receptor (ErbB1) protects the heart against stress-induced injury in mice. Am J Physiol Regul Integr Comp Physiol. 2003;285(2):R455–R462. doi: 10.1152/ajpregu.00588.2002. [DOI] [PubMed] [Google Scholar]
- 20.Keith RL, Karoor V, Mozer AB, Hudish TM, Le M, Miller YE. Chemoprevention of murine lung cancer by gefitinib in combination with prostacyclin synthase overexpression. Lung Cancer. 2010;70(1):37–42. doi: 10.1016/j.lungcan.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konecny GE, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing trastuzumab-treated breast cancer cells. Cancer Res. 2006;66(3):1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 22.Lee TC, Threadgill DW. Generation and validation of mice carrying a conditional allele of the epidermal growth factor receptor. Genesis. 2009;47(2):85–92. doi: 10.1002/dvg.20464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maixner W, et al. Orofacial pain prospective evaluation and risk assessment study--the OPPERA study. J Pain. 2011;12(11 Suppl):T4–11.e1. doi: 10.1016/j.jpain.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slade GD, et al. Cytokine biomarkers and chronic pain: association of genes, transcription, and circulating proteins with temporomandibular disorders and widespread palpation tenderness. Pain. 2011;152(12):2802–2812. doi: 10.1016/j.pain.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diatchenko L, Nackley AG, Tchivileva IE, Shabalina SA, Maixner W. Genetic architecture of human pain perception. Trends Genet. 2007;23(12):605–613. doi: 10.1016/j.tig.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Bonnington JK, McNaughton PA. Signalling pathways involved in the sensitisation of mouse nociceptive neurones by nerve growth factor. J Physiol (Lond) 2003;551(Pt 2):433–446. doi: 10.1113/jphysiol.2003.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao C, et al. Galpha(i1) and Galpha(i3) are required for epidermal growth factor-mediated activation of the Akt-mTORC1 pathway. Sci Signal. 2009;2(68):ra17. doi: 10.1126/scisignal.2000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obara I, et al. Systemic inhibition of the mammalian target of rapamycin (mTOR) pathway reduces neuropathic pain in mice. Pain. 2011;152(11):2582–2595. doi: 10.1016/j.pain.2011.07.025. [DOI] [PubMed] [Google Scholar]
- 29.Xu Q, et al. Spinal phosphinositide 3-kinase-Akt-mammalian target of rapamycin signaling cascades in inflammation-induced hyperalgesia. J Neurosci. 2011;31(6):2113–2124. doi: 10.1523/JNEUROSCI.2139-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obata K, et al. Role of mitogen-activated protein kinase activation in injured and intact primary afferent neurons for mechanical and heat hypersensitivity after spinal nerve ligation. J Neurosci. 2004;24(45):10211–10222. doi: 10.1523/JNEUROSCI.3388-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murooka TT, Rahbar R, Platanias LC, Fish EN. CCL5-mediated T-cell chemotaxis involves the initiation of mRNA translation through mTOR/4E-BP1. Blood. 2008;111(10):4892–4901. doi: 10.1182/blood-2007-11-125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andres C, Meyer S, Dina OA, Levine JD, Hucho T. Quantitative automated microscopy (QuAM) elucidates growth factor specific signalling in pain sensitization. Mol Pain. 2010;6:98. doi: 10.1186/1744-8069-6-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heldin CH. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80(2):213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 34.Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94(3):277–280. doi: 10.1016/S0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 35.Riese DJ, Komurasaki T, Plowman GD, Stern DF. Activation of ErbB4 by the bifunctional epidermal growth factor family hormone epiregulin is regulated by ErbB2. J Biol Chem. 1998;273(18):11288–11294. doi: 10.1074/jbc.273.18.11288. [DOI] [PubMed] [Google Scholar]
- 36.Shelly M, et al. Epiregulin is a potent pan-ErbB ligand that preferentially activates heterodimeric receptor complexes. J Biol Chem. 1998;273(17):10496–10505. doi: 10.1074/jbc.273.17.10496. [DOI] [PubMed] [Google Scholar]
- 37.Kurtze I, Sonnemann J, Beck JF. KRAS-mutated non-small cell lung cancer cells are responsive to either co-treatment with erlotinib or gefitinib and histone deacetylase inhibitors or single treatment with lapatinib. Oncol Rep. 2011;25(4):1021–1029. doi: 10.3892/or.2011.1160. [DOI] [PubMed] [Google Scholar]
- 38.Lu PH, Kuo TC, Chang KC, Chang CH, Chu CY. Gefitinib-induced epidermal growth factor receptor-independent keratinocyte apoptosis is mediated by the JNK activation pathway. Br J Dermatol. 2011;164(1):38–46. doi: 10.1111/j.1365-2133.2010.10038.x. [DOI] [PubMed] [Google Scholar]
- 39.Mogil JS, et al. Screening for pain phenotypes: analysis of three congenic mouse strains on a battery of nine nociceptive assays. Pain. 2006;126(1-3):24–34. doi: 10.1016/j.pain.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Hargreaves K, Dubner R, Brown F, Flores C, Joris J. A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain. 1988;32(1):77–88. doi: 10.1016/0304-3959(88)90026-7. [DOI] [PubMed] [Google Scholar]
- 41.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 42.Decosterd I, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 2000;87(2):149–158. doi: 10.1016/S0304-3959(00)00276-1. [DOI] [PubMed] [Google Scholar]
- 43.Bennett GJ, Xie YK. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33(1):87–107. doi: 10.1016/0304-3959(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 44.Hylden JL, Wilcox GL. Intrathecal morphine in mice: a new technique. Eur J Pharmacol. 1980;67(2-3):313–316. doi: 10.1016/0014-2999(80)90515-4. [DOI] [PubMed] [Google Scholar]
- 45.Malin SA, Davis BM, Molliver DC. Production of dissociated sensory neuron cultures and considerations for their use in studying neuronal function and plasticity. Nat Protoc. 2007;2(1):152–160. doi: 10.1038/nprot.2006.461. [DOI] [PubMed] [Google Scholar]
- 46.Mamane Y, et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS One. 2007;2(2):e242. doi: 10.1371/journal.pone.0000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parsyan A, et al. The helicase protein DHX29 promotes translation initiation, cell proliferation, tumorigenesis. Proc Natl Acad Sci USA. 2009;106(52):22217–22222. doi: 10.1073/pnas.0909773106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slade GD, et al. Study methods, recruitment, sociodemographic findings, and demographic representativeness in the OPPERA study. J Pain. 2011;12(11 Suppl):T12–T26. doi: 10.1016/j.jpain.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohrbach R, et al. Clinical findings and pain symptoms as potential risk factors for chronic TMD: descriptive data and empirically identified domains from the OPPERA case-control study. J Pain. 2011;12(11 Suppl):T27–T45. doi: 10.1016/j.jpain.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dworkin SF, LeResche L. Research diagnostic criteria for temporomandibular disorders: review, criteria, examinations and specifications, critique. J Craniomandib Disord. 1992;6(4):301–355. [PubMed] [Google Scholar]
- 51.Smith SB, et al. Potential genetic risk factors for chronic TMD: genetic associations from the OPPERA case control study. J Pain. 2011;12(11 Suppl):T92–101. doi: 10.1016/j.jpain.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 54.Barrett JC, F ry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 55.Zaykin DV. Optimally weighted Z test is a powerful method for combining probabilities in meta-analysis. J Evol Biol. 2011;24(8):1836–1841. doi: 10.1111/j.1420-9101.2011.02297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slade GD, et al. Facial pain with localized and widespread manifestations: separate pathways of vulnerability. Pain. 2013;154(11):2335–2343. doi: 10.1016/j.pain.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.