Abstract
Asthma is associated with exposure to a wide variety of allergens and adjuvants. The extent to which overlap exists between the cellular and molecular mechanisms triggered by these various agents is poorly understood, but it might explain the differential responsiveness of patients to specific therapies. In particular, it is unclear why some, but not all, patients benefit from blockade of TNF. Here, we characterized signaling pathways triggered by distinct types of adjuvants during allergic sensitization. Mice sensitized to an innocuous protein using TLR ligands or house dust extracts as adjuvants developed mixed eosinophilic and neutrophilic airway inflammation and airway hyperresponsiveness (AHR) following allergen challenge, whereas mice sensitized using proteases as adjuvants developed predominantly eosinophilic inflammation and AHR. TLR ligands, but not proteases, induced TNF during allergic sensitization. TNF signaled through airway epithelial cells to reprogram them and promote Th2, but not Th17, development in lymph nodes. TNF was also required during the allergen challenge phase for neutrophilic and eosinophilic inflammation. In contrast, TNF was dispensable for allergic airway disease in a protease-mediated model of asthma. These findings might help to explain why TNF blockade improves lung function in only some patients with asthma.
Keywords: Immunology, Inflammation
Keywords: Asthma, Mouse models, Th2 response
Introduction
Allergic asthma is a chronic inflammatory disease of the airways characterized by reversible airway obstruction, airway hyperresponsiveness (AHR), and inflammation (1). This disease has been traditionally associated with the type 2 cytokines IL-4, IL-5, and IL-13, which promote immunoglobulin class switching to IgE, eosinophilia, and AHR, respectively (2, 3). However, approximately half of patients with asthma have a noneosinophilic form of this disease, many with predominantly neutrophilic inflammation of the airway (4, 5). Unfortunately, these patients respond very poorly to inhaled corticosteroids, the gold standard asthma therapy (6), and there is a need for novel therapeutic approaches. It is likely that in these individuals, steroid-resistant IL-17–producing Th17 cells (7) drive asthma by promoting recruitment of neutrophils to the airway (8–12). Still other patients display both neutrophilic and eosinophilic inflammation, and this form of asthma is associated with particularly low lung function and high health care costs (13). An improved mechanistic understanding of the induction and regulation of Th2 and Th17 cells should lead to novel therapies that selectively target different types of asthma without compromising other protective immune responses.
Allergens are derived from living organisms, including plants, insects, fungi, and mammals. The wide variety of physical and biochemical properties displayed by different allergens suggests that they can activate multiple innate immune response pathways (14). In this regard, allergens are similar to adjuvants, which also activate innate immunity and for this reason are commonly used in vaccines to strengthen immune responses to a coadministered antigen. Indeed, some allergens can also promote responses to innocuous bystander proteins, such as ovalbumin (OVA) (15). Furthermore, preparations of allergens can also contain associated molecules with adjuvant activity. For example, house dust mite allergens contain LPS that contributes to their allergenicity (16), and the presence of endotoxin in homes is a risk factor for asthma (17). Other well-studied allergens include those that display protease activity. Some examples of protease allergens include the house dust mite allergens Der p1 and Der p9, allergens from the mold Aspergillus, and papain (PAP), a cysteine protease found in papaya. Proteases are not directly recognized by TLRs, but PAP (18) and a protease-containing extract from Aspergillus oryzae (ASP) (19) can also promote allergic responses to inhaled OVA. The extent to which mechanistic overlap exists between TLR ligand– and protease-induced Th2- and Th17-mediated inflammation of the airway is poorly understood, but this knowledge is critical to developing therapies targeting allergen-specific pathways.
It has long been known that TNF mRNA is increased in patients with asthma (20), suggesting a role for TNF in the pathogenesis of this disease. Data from clinical trials suggest that although antagonizing TNF is not helpful to all patients, it is beneficial to a subgroup of asthmatics, including those who have severe disease (21, 22). It will be important to identify individuals whose known environmental exposures and genetic predisposition are predictive of a positive response to TNF pathway blockade. Furthermore, blockade of TNF itself is associated with increased infections (23), and an improved understanding of how other molecules in the TNF pathway contribute to asthma is necessary for design of novel therapies that ameliorate asthma exacerbations without this complication. Here, we compared molecular mechanisms by which 2 different classes of adjuvants, TLR ligands and proteases, promote allergic responses to bystander proteins. We found that TNF signaling through airway epithelial cells (AECs) induced a suite of genes in those cells and was required for TLR ligand–mediated development of antigen-specific Th2 cells, but not antigen-specific Th17 cells. TNF was also required during the challenge phase for eosinophil and neutrophil recruitment to the airway. By contrast, this cytokine was dispensable for protease-mediated Th2 cell development and allergic airway inflammation. These findings might help to explain why TNF blockade ameliorates lung function in only some patients, as well as reveal novel molecules in this pathway that might be targeted by therapeutic intervention.
Results
TLR ligands and proteases promote distinct forms of allergic responses to innocuous inhaled proteins.
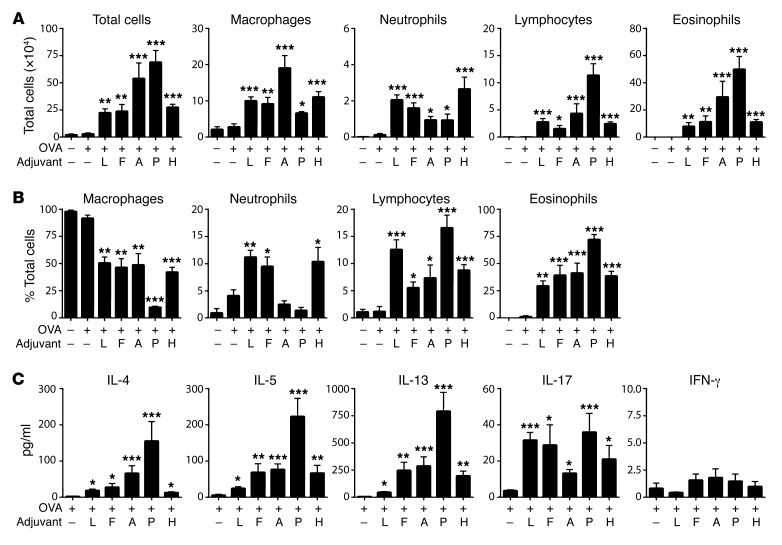
Highly purified OVA is not allergenic in mice when instilled directly into their airways. However, when mice inhale OVA together with certain adjuvants, the animals develop OVA-specific allergic responses and, following subsequent challenge with OVA, develop inflammation of the airway (12, 24). To better understand adjuvant-mediated allergic sensitization, we studied the abilities of 2 TLR ligands, LPS and flagellin (FLA), and 2 proteases, ASP and PAP, to promote allergic sensitization to OVA. We also tested the adjuvant activity of a house dust extract (HDE). HDEs are complex and contain multiple allergens and adjuvants, but they were included in the current study because they are arguably more representative of ambient exposures than are purified allergens or adjuvants (25). As expected, inhalation of OVA alone failed to induce sensitization in mice, since subsequent exposure of these animals to aerosolized OVA did not trigger airway inflammation (Figure 1A). However, mice that inhaled OVA together with any of the adjuvants tested became sensitized, as inferred from the airway inflammation that developed upon subsequent OVA challenge. The number (Figure 1A) and percentages (Figure 1B) of the various leukocyte types following OVA challenge depended on which adjuvant had been used to sensitize the animals. Mice sensitized to OVA using either of the TLR ligands, LPS or FLA, developed both eosinophilic and neutrophilic inflammation, whereas mice sensitized using either of the proteases displayed strong eosinophilic responses with comparatively few neutrophils. Animals sensitized with OVA using HDE as an adjuvant resembled mice sensitized using LPS/OVA or FLA/OVA in that they had relatively high numbers of airway neutrophils. Cytokines in the bronchoalveolar lavage fluid (BALF) of animals were generally consistent with the type of inflammation they displayed, with the PAP/OVA mice having the highest amounts of eosinophils (Figure 1A) and type 2 cytokines, IL-4, IL-5, and IL-13 (Figure 1C). IL-17 levels in the BALF generally correlated with the extent of neutrophilic inflammation, although mice sensitized using PAP/OVA had relatively high amounts of IL-17 but little neutrophilic inflammation. Together, these data show that when inhaled into the airway, TLR ligands elicit immune responses leading to both eosinophilic and neutrophilic inflammation, whereas proteases promote immune responses that primarily drive eosinophilia.
Figure 1. TLR ligands and proteases function as adjuvants to promote distinct forms of allergic inflammation.
Mice were sensitized to OVA using the indicated adjuvants and subsequently challenged with aerosolized OVA. Cell numbers (A) and percentages (B) for the indicated leukocyte subsets, and cytokine levels in BALF after challenge (C). Data shown represent mean ± SEM (n = 6–9 mice per group) from a single experiment, representative of 2. *P < 0.05, **P < 0.01, ***P < 0.001; “OVA only” group vs. other groups; Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test. Adjuvants: L, LPS; F, flagellin; A, Aspergillus protease; P, papain protease; H, house dust extract.
TLR ligands and proteases induce distinct innate immune cytokines during allergic sensitization.
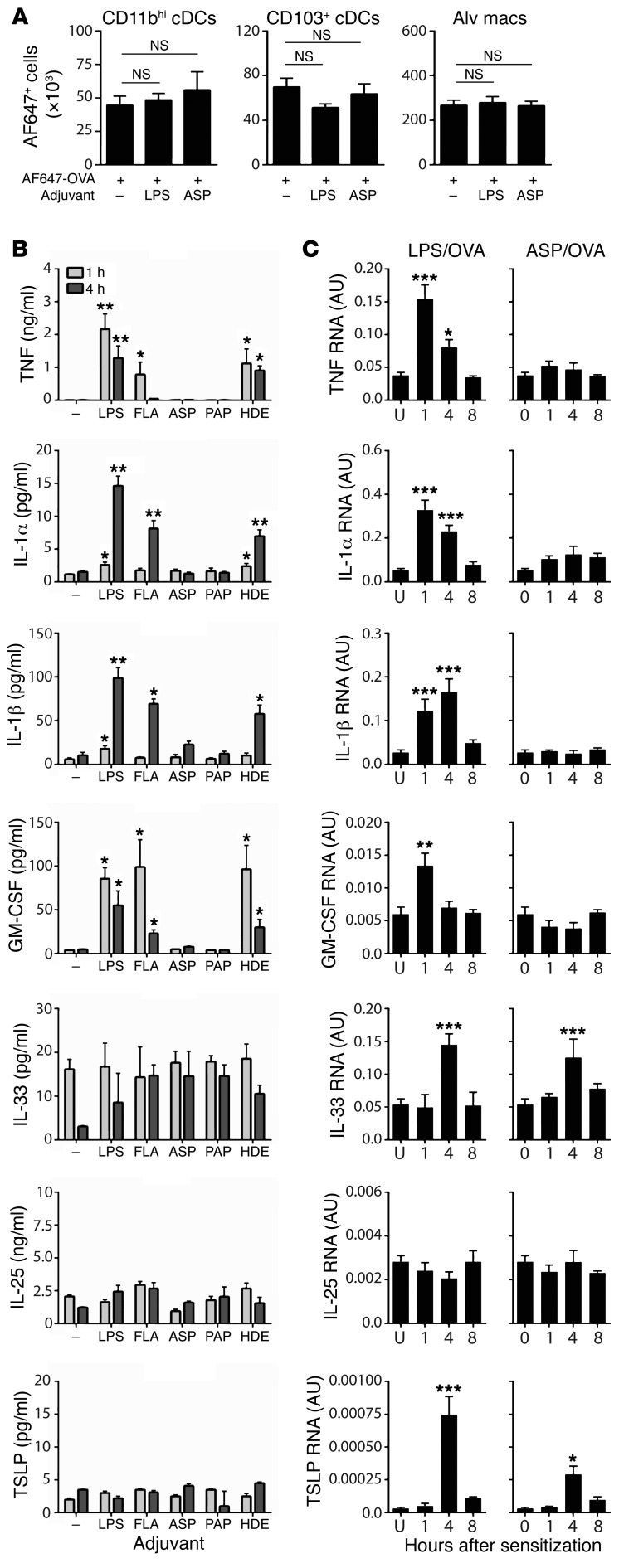
Identifying the mechanisms by which adjuvants promote allergic responses to innocuous proteins is critical to prevent this from occurring. Some adjuvants, including proteases, can disrupt the epithelial cell barrier (26), and it has been proposed that this can increase exposure of conventional DCs (cDCs) on the abluminal side of the airway to inhaled allergens, thereby strengthening the immune response to them (27, 28). However, the number of lung CD103+ cDCs and CD11bhi cDCs taking up Alexa Fluor 647–labeled OVA was similar in the presence or absence of these adjuvants (Figure 2A, and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI90890DS1), excluding increased numbers of antigen-acquiring DCs as a major mechanism by which LPS or ASP promotes type 2 responses in this model. As expected, the adjuvant activity of LPS was dependent on its receptor TLR4 (data not shown), whereas the adjuvant activity of ASP was not (Supplemental Figure 2), ruling out contaminating LPS as being important for the adjuvant activity of this protease. TLR2 and TLR5 were also found to be dispensable for ASP-mediated allergic sensitization, and the adjuvant activity of ASP was heat sensitive, consistent with its protease activity being critical for its adjuvant activity (data not shown).
Figure 2. Innate cytokines in the airways of mice following inhalation of a TLR ligand, a protease, or an HDE.
(A) Numbers of CD11bhi DCs, CD103+ DCs, and alveolar macrophages (Alv macs) that acquired Alexa Fluor 647 (AF647)–OVA following its instillation alone or with the indicated adjuvants. Values represent mean ± SEM (n = 3 mice per group) from a single experiment. NS, not significant (P > 0.05) between indicated groups; Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test. (B) Mean cytokine levels ± SEM in BALF at 1 or 4 hours after treatment with OVA alone (–), or with OVA together with the indicated adjuvants (n = 6 mice per group). *P < 0.05, **P < 0.01; OVA alone vs. other groups; 2-way ANOVA with Dunnett’s multiple comparison test. (C) Relative amounts of cytokine-specific mRNA ± SEM in whole lung from untreated (U) mice and from mice at indicated times following sensitization to LPS/OVA or ASP/OVA. Values shown in arbitrary units (AU) after normalization to Gapdh mRNA (n = 8–12 mice per group). *P < 0.05, **P < 0.01, ***P < 0.001; untreated vs. sensitized; 1-way ANOVA with Dunnett’s multiple comparison test. Data shown are from 1 of 2 experiments yielding similar results, except C, in which the data from 2 experiments were pooled.
To compare pathways used by the different classes of adjuvants, we instilled OVA together with TLR ligands, proteases, or HDE into the airways, and measured cytokines associated with innate immunity. The TLR ligands, LPS and FLA, rapidly induced accumulation in the BALF of TNF, IL-1α, IL-1β, and granulocyte-macrophage CSF (GM-CSF) (Figure 2B). HDE, which contains both LPS and protease activity, also induced those cytokines, whereas neither of the proteases tested, ASP or PAP, did so. Similar results were observed for mRNAs encoding these cytokines following LPS/OVA and ASP/OVA sensitization (Figure 2C). IL-33, IL-25, and thymic stromal lymphopoietin (TSLP) have all been proposed to function during allergic sensitization (reviewed in ref. 29), but none of the adjuvants tested caused measurable amounts of those cytokines to be released into the airway. Il33 mRNA was modestly increased by treatment with LPS or ASP, whereas levels of Tslp mRNA were extremely low, in agreement with a previous report (30). These data show that TLR ligands and proteases induce the secretion of distinct innate immune cytokines into the airway during allergic sensitization.
Requirement of TNF signaling for TLR ligand– but not protease-mediated airway inflammation, mucus production, and AHR.
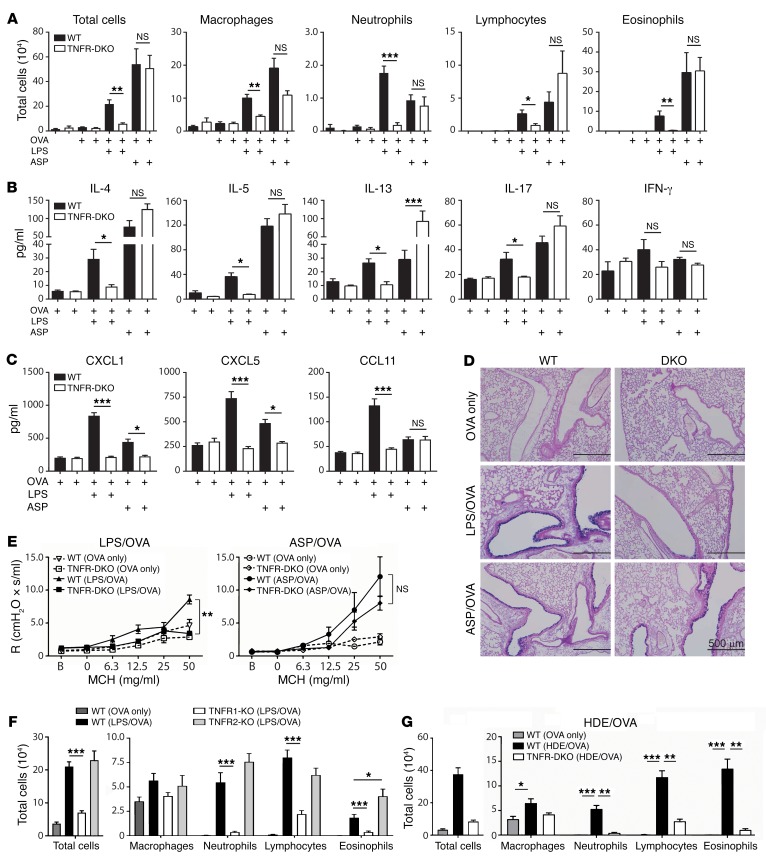
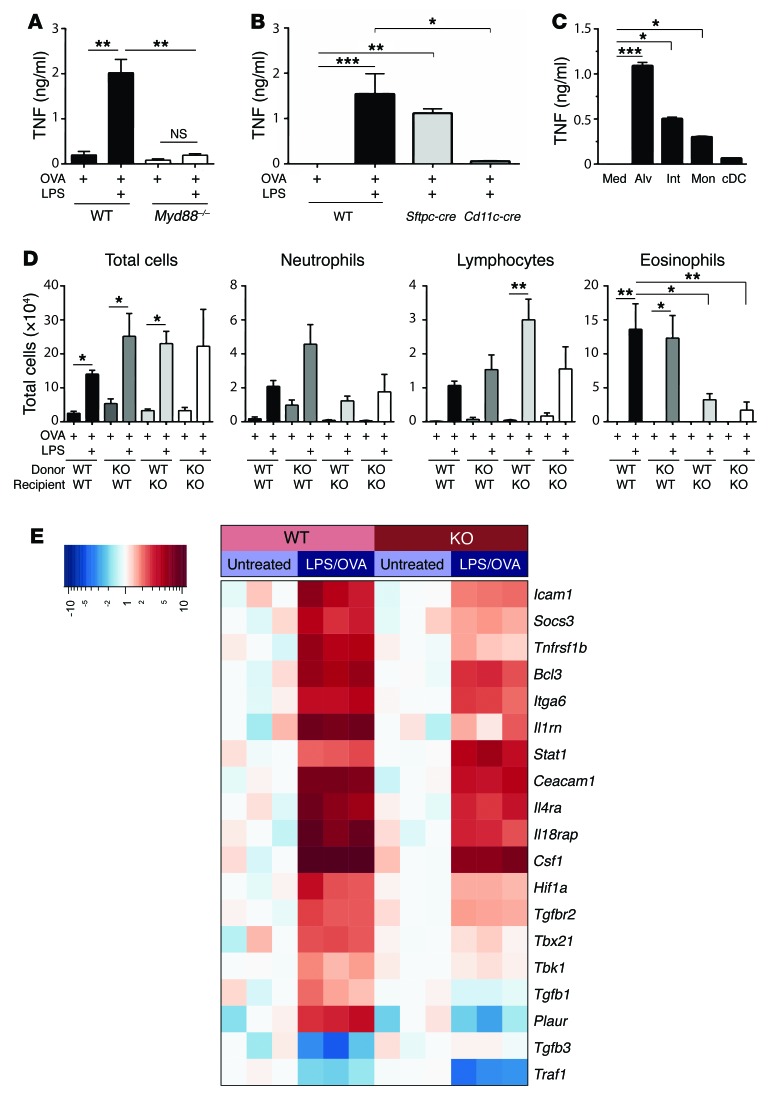
TNF is initially produced as a membrane-bound protein that can be cleaved by TNF-α–converting enzyme (TACE) to yield soluble TNF (31). In mice, soluble TNF is sufficient to promote allergic sensitization to OVA when the 2 proteins are coadministered to the airways, and upon OVA challenge these mice develop allergic airway disease (24). However, it is not known whether TNF is required for allergic airway disease in mice, and whether that depends on which model is used. To investigate this, we initially studied animals lacking both TNF receptors, TNFR1 and TNFR2 (TNFR1/2-DKO mice), and therefore unable to respond to soluble or membrane-bound TNF. Following LPS/OVA sensitization and OVA challenge, TNFR1/2-DKO mice had fewer total cells, including neutrophils, lymphocytes, and eosinophils, than did WT mice (Figure 3A), showing that TNF signaling is indeed required for the recruitment of multiple inflammatory cell types to the airway in this model. Analysis of cytokines in the BALF revealed that the type 2 cytokines IL-4, IL-5, and IL-13, as well as IL-17, were all decreased in TNFR1/2-DKO mice, compared with WT mice, whereas IFN-γ was similar in the 2 strains (Figure 3B). Similar results were obtained using Tnf-deficient mice (Supplemental Figure 3, A and B). The neutrophil-attracting chemokines CXCL1 and CXCL5, as well as the eosinophil-attracting chemokine CCL11 (eotaxin), were also decreased in TNFR1/2-DKO mice compared with WT mice in this model (Figure 3C), consistent with the reduced numbers of these cells. Examination of stained lung sections by a pathologist blinded to their identities revealed that, compared with WT mice, airways of TNFR1/2-DKO had significantly less inflammation and fewer mucus-producing cells (Figure 3D and Supplemental Figure 4). We also observed that whereas WT mice developed AHR, similarly treated TNFR1/2-DKO mice did not (Figure 3E). Together, these data show that in the LPS/OVA model of asthma, TNF is required for multiple features of allergic airway disease, including inflammation, mucus production, and impaired lung function.
Figure 3. TNF is required in a TLR ligand–mediated model of asthma.
(A–E and G) WT and TNFR1/2-DKO mice were sensitized to OVA using the indicated adjuvants, and subsequently challenged with aerosolized OVA. Shown are cell numbers (A) and cytokine (B) and chemokine (C) concentrations in BALF, mucin-producing cells in airways (×100 magnification; scale bars: 500 μm) (D), and airway resistance (R) after allergen challenge (E). Resistance values were obtained at baseline (B) and after administration of the indicated doses of methacholine (MCH). (F) Cell numbers for the indicated leukocytes in BALF of WT and TNFR1- and TNFR2-single-KO mice following LPS/OVA sensitization and subsequent challenge with OVA. (G) Cell numbers for the indicated leukocytes in BALF after challenge. *P < 0.05, **P < 0.01, ***P < 0.001; WT vs. KO mice treated with the same adjuvant; Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test. Values shown represent mean ± SEM (n = 6–12 mice per group) from 1 of 2 experiments yielding similar results, except F and G, which show combined data of 2 experiments yielding similar results (n = 12 mice per group).
The 2 TNF receptors, TNFR1 and TNFR2, are differentially expressed and are associated with distinct activities. TNFR1 is found on most cells, can respond to either soluble or membrane-bound TNF, and can promote either cell death or inflammation. By contrast, TNFR2 is on a smaller number of cell types, including immune cells. It signals primarily in response to membrane-bound TNF, and is associated with cell survival (recently reviewed in ref. 32). To determine which of these 2 receptors has a larger role in the TLR-mediated model of asthma, we sensitized TNFR1- and TNFR2-single-KO mice with LPS/OVA and challenged them with aerosolized OVA. We observed that TNFR1-single-KO mice displayed markedly fewer neutrophils, lymphocytes, and eosinophils than did WT mice (Figure 3F). Lung explants of challenged TNFR1-KO mice also produced significantly lower amounts of IL-4, IL-5, and IL-17 than did their WT counterparts (Supplemental Figure 3C). In contrast to these diminished responses of TNFR1-KO mice, TNFR2-single-KO mice and WT mice had similar numbers of neutrophils and lymphocytes in their airways, and TNFR2-single-KO mice had even more eosinophils than did WT mice (Figure 3F). There were no significant differences between the latter 2 strains in amounts of cytokines in lung explants (Supplemental Figure 3C). These data show that TNF signaling through TNFR1 is critical for allergic airway inflammation when a TLR ligand is used as the adjuvant during allergic sensitization, whereas TNFR2 is dispensable.
LPS can be found in common house dust, and HDEs can promote allergic sensitization to innocuous proteins, such as OVA (25). To investigate whether TNF signaling is also required in this more environmentally relevant model of asthma, we sensitized WT and TNFR1/2-DKO mice to OVA using HDE as an adjuvant. Following subsequent challenge with OVA, TNFR1/2-DKO mice had significantly fewer neutrophils, lymphocytes, and eosinophils in their airways than did WT mice (Figure 3G). A similar reduction in airway inflammation was observed when Tnf-deficient mice were compared with WT animals in the HDE model of asthma (Supplemental Figure 3D). Thus, TNF is required for allergic inflammation of the airway in multiple models of allergic asthma, including one that mimics ambient exposure of humans to house dust.
In contrast to the requirement for TNF signaling in the LPS/OVA and HDE/OVA models of asthma, we found that this cytokine was dispensable for allergic airway disease when ASP was used as an adjuvant. TNFR1/2-DKO mice developed robust eosinophilic and neutrophilic inflammation in this model (Figure 3A), and type 2 cytokines were as high, or higher, in BALF of TNFR1/2-DKO mice than in BALF from WT mice (Figure 3B). Although CXCL1 and CXCL5 were reduced in TNFR1/2-DKO mice compared with WT mice, the 2 strains had similar amounts of the eosinophil-attracting chemokine CCL11 (Figure 3C). WT and TNFR1/2-DKO mice also had similar numbers of mucus-staining cells in the ASP/OVA model of asthma (Figure 3D), and there were no significant differences between these 2 strains in AHR (Figure 3E). Collectively, these data show that TLR ligands and proteases can both act as adjuvants to promote allergic sensitization, but do so by activating distinct signaling pathways, with TNF signaling being required for multiple features of asthma in the TLR ligand–mediated model, but dispensable when a protease is used as the adjuvant.
Requirement of TNF signaling for Th2, but not Th17, development in mediastinal lymph nodes during TLR ligand–mediated allergic sensitization.
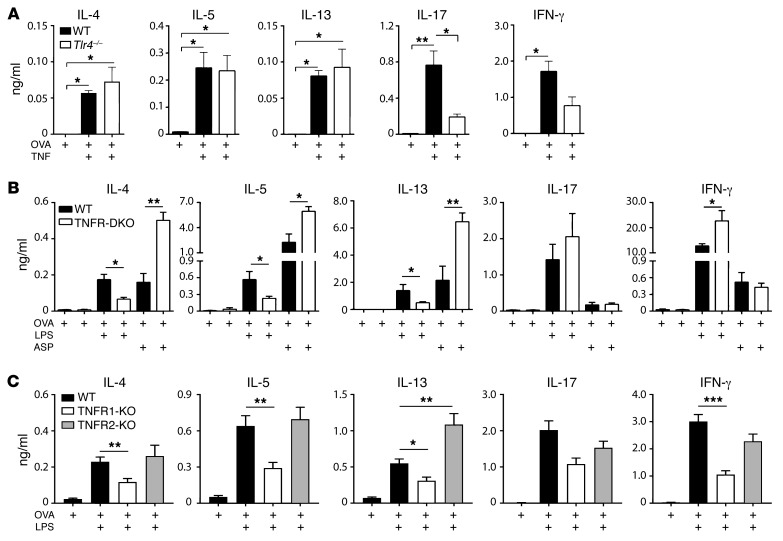
Our experiments thus far indicated that TNF signaling is required in a TLR ligand–mediated model of asthma, but did not reveal whether this cytokine is critical during allergic sensitization, or during the challenge phase of the model, or both. To specifically investigate the role of TNF during the early, sensitization phase of this model, we first performed a dose-response experiment in which mice were sensitized to OVA using different amounts of recombinant mouse (rm)TNF as an adjuvant, and subsequently challenged with OVA. These animals developed eosinophilic inflammation of the airway, although lymphocytes and a smaller number of neutrophils were also seen (Supplemental Figure 5). The dose of the rmTNF used to sensitize the mice affected the degree of allergic inflammation following challenge, with the greatest numbers of eosinophils in the airways of mice that had been sensitized to OVA using 0.67 μg rmTNF. This dose was therefore used in subsequent experiments. To indirectly investigate the effect of TNF on allergen-specific T cell responses, we adoptively transferred naive CD4+ T cells from OT-II mice bearing an OVA-specific T cell receptor (hereafter referred to as OT-II cells) into naive WT mice, sensitized them to OVA using rmTNF as the adjuvant, and cultured cells from excised lung-draining mediastinal lymph nodes (mLNs) in the presence of OVA. Tlr4–/– mice were also included in this experiment to assess the contribution to immune responses of any contaminating endotoxin in the TNF preparation. The type 2 cytokines IL-4, IL-5, and IL-13 were produced in mLNs of both WT and Tlr4–/– mice (Figure 4A). Some IL-17 was also present in these cell cultures, but its production was strongly reduced in Tlr4–/– mice, suggesting that small amounts of LPS in the rmTNF preparation, not the cytokine itself, were primarily responsible for induction of IL-17. Together, these data suggest that inhaled TNF can promote the development of allergen-specific Th2 cells, but not Th17 cells.
Figure 4. TNF signaling through TNFR1 promotes type 2, but not type 17, cell differentiation.
(A–C) Mice receiving adoptive transfer of OVA-specific CD4+ T cells were sensitized to inhaled OVA using the indicated adjuvants. Lung-draining LNs were excised 4 days later, cells from them were restimulated ex vivo with OVA, and the indicated cytokines in the culture supernatants were measured. Shown are cytokines from LN cultures of WT and Tlr4–/– mice sensitized to OVA using rmTNF as an adjuvant (n = 5 mice per group) (A), WT and TNFR1/2-DKO mice sensitized to OVA using either LPS or ASP as an adjuvant (n = 6 mice per group) (B), and WT and TNFR1- and TNFR2-single-KO mice sensitized to OVA using LPS as an adjuvant (n = 12 mice per group) (C). Data represent mean ± SEM from a single experiment, representative of 2, except C, which shows the combined data of 2 experiments yielding similar results. *P < 0.05, **P < 0.01, ***P < 0.001 for indicated comparisons (A) or WT vs. KO mice that were similarly treated (B and C); Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test.
To determine whether TNF signaling is necessary during the sensitization phases of TLR ligand– or protease-mediated models of asthma, OT-II cells were transferred into WT and TNFR1/2-DKO mice before their sensitization to OVA using either LPS or ASP as the adjuvant. Lung-draining mLNs were excised and cells from them cultured in the presence of OVA. When LPS was used as the adjuvant, mLNs of TNFR1/2-DKO mice produced significantly smaller amounts of IL-4, IL-5, and IL-13, but more IFN-γ, than did mLNs of WT mice, whereas IL-17 was produced at similar levels in these 2 strains (Figure 4B). Virtually identical findings were observed in experiments with mice lacking the cytokine TNF (Supplemental Figure 6). However, when ASP was used as the adjuvant, mLNs of TNFR1/2-DKO mice produced even higher amounts of IL-4, IL-5, and IL-13 than did cells from WT mice (Figure 4B), consistent with our observation that large amounts of IL-13 are present in the BALF of TNFR1/2-DKO mice in the ASP/OVA model of asthma (Figure 3B), and confirming that TNF is dispensable for protease-mediated allergic sensitization. To further define the role of TNF in TLR ligand–mediated sensitization, we analyzed cytokines in mLNs of single-TNFR-KO mice following LPS/OVA inhalation. Compared with WT mice, TNFR1 had reduced amounts of type 2 cytokines in mLNs, whereas TNFR2 mice had as high or higher (IL-13) amounts of these cytokines (Figure 4C). No differences between these genotypes were seen for IL-17. Taken together, these data suggest that during LPS/OVA sensitization, TNF signaling through TNFR1 promotes Th2 development in regional LNs, but has little effect on Th17 development.
Interstitial and alveolar macrophages are major sources of TNF in vivo.
A time-course analysis of BALF collected from mice sensitized with LPS/OVA revealed that TNF is released into the airways as early as 30 minutes and reaches its highest concentration between 1 and 4 hours after sensitization (Supplemental Figure 7A). Although neutrophils and lymphocytes can produce TNF in some settings, both cell types arrived after peak production of TNF (Supplemental Figure 7B), suggesting that neither are the primary cellular source of TNF. In support of this, TNF production was not significantly decreased in mice lacking neutrophils, T and B lymphocytes, or mast cells (Supplemental Figure 7, C–E).
Myeloid differentiation primary response protein 88 (MyD88) is an adaptor molecule downstream of all TLRs except TLR3. As expected, TNF production was markedly diminished in Myd88–/– mice following LPS/OVA inhalation (Figure 5A). Mice selectively lacking Myd88 expression in AECs had similar amounts of TNF to those of WT mice (Figure 5B), whereas it was markedly diminished in mice lacking Myd88 in Cd11c-expressing cells (Figure 5B). Because macrophages and cDCs in the lung both express Cd11c, we instilled LPS into the airways of mice, separately purified these cell types from the lungs by FACS, and cultured the cells ex vivo. Alveolar and interstitial macrophages produced relatively large amounts of TNF, monocytes produced less cytokine, and cDCs produced almost no TNF (Figure 5C). Clodronate treatment, which depleted alveolar macrophages, but not interstitial macrophages or monocytes, did not diminish either TNF production or the efficiency of LPS/OVA sensitization (Supplemental Figure 7, F–H), suggesting that although they can produce TNF, alveolar macrophages are not an essential source of this cytokine. Taken together, these data suggest that during TLR ligand–mediated allergic sensitization, interstitial and alveolar macrophages can be major TNF-producing cells.
Figure 5. TNF from CD11c+ macrophages acts on radioresistant AECs to reprogram them and promote allergic sensitization.
(A and B) TNF in BALF after instillation of LPS/OVA into WT and Myd88–/– mice (A) or Myd88fl/fl mice crossed to Sftpc-cre mice or Cd11c-cre mice to delete Myd88 in AECs or Cd11c-expressing cells, respectively (B) (n = 8 mice per group). (C) TNF in medium alone (Med), or culture supernatants of alveolar (Alv) macrophages, interstitial (Int) macrophages, monocytes (Mon), and cDCs purified by FACS from lungs of LPS/OVA-treated mice (n = 6 mice per group). (D) Cell numbers for the indicated leukocytes in airways of WT and TNFR1/2-DKO (KO) reciprocal bone marrow chimeric mice sensitized with OVA with or without LPS and subsequently challenged with aerosolized OVA (n = 6 mice per group). (A–D) Data represent mean values ± SEM and were confirmed by at least 1 repeat experiment. *P < 0.05, **P < 0.01, ***P < 0.001; Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test. (E) Heatmap showing genes whose induction in AECs by inhaled LPS was more than 1.5-fold different in WT and TNFR1/2-DKO mice, P < 0.01 (see Methods for details on statistical analysis).
Radioresistant cells respond to TNF to enhance eosinophilic inflammation in the LPS-mediated model of asthma.
Having identified the major cell sources of TNF, we next asked which cells were responding to it to promote allergic sensitization. We first determined whether TNF responsiveness in structural cells or hematopoietic cells is required in the asthma model by generating reciprocal bone marrow chimeric mice using WT and TNFR1/2-DKO animals. After allowing 12 weeks for recipient mice to be fully reconstituted, they were sensitized with LPS/OVA and challenged with aerosolized OVA. As expected, neutrophils and eosinophils were recruited to the airways of WT animals that received WT bone marrow cells (WT→WT mice) (Figure 5D). Similar numbers of inflammatory cells were seen in WT mice that received TNFR1/2-DKO marrow (DKO→WT), indicating that TNF responsiveness of hematopoietic cells is dispensable for airway inflammation. By contrast, DKO mice receiving either DKO or WT bone marrow (DKO→DKO and WT→DKO, respectively) had significantly reduced eosinophils compared with the 2 WT recipient groups. These findings demonstrate that TNF responsiveness in a radioresistant cell type, such as AECs, is required for eosinophilic inflammation in this TLR ligand–dependent model of asthma. However, neutrophils were not dependent on TNF in the bone marrow chimeric mice, possibly because radiation triggers production of proinflammatory cytokines that remain high for many weeks after irradiation (33).
TNF signaling responses contribute to AEC transcriptional reprogramming during TLR ligand–mediated allergic sensitization.
To investigate the consequences of TNF signaling in AECs during TLR ligand–mediated allergic sensitization, we used flow cytometry–based cell sorting to purify these cells from WT and TNFR1/2-DKO mice 4 hours after sensitization with LPS/OVA. RNA from the AECs was analyzed by a highly quantitative, multiplexed hybridization assay (NanoString Technologies) to evaluate expression of 547 genes known to be associated with immune responses. Of the 200 genes whose expression in AECs was high enough to be reliably measured, 23 displayed at least a 1.5-fold difference in expression between WT and TNFR1/2-DKO mice, with a P value less than or equal to 0.01 (Figure 5E). As expected, Tnf was not among these genes, in agreement with our finding that macrophages, not AECs, are the major source of this cytokine. Several genes with plausible roles in TNF-dependent allergic airway disease were differentially expressed in WT and TNFR1/2-DKO AECs. The most differentially expressed gene between these genotypes was Plaur, which was 6-fold higher in WT AECs than in TNFR1/2-DKO AECs. Plaur encodes the urokinase plasminogen activator receptor (uPAR), and SNPs in this gene have been associated with asthma and lung function (34). Other genes whose expression was also decreased in AECs of TNFR1/2-DKO mice included Il1b, Ccl20, Csf2, s100a9, and Il4ra. Il1 and Csf2 (which encodes GM-CSF) have been previously associated with allergic pulmonary inflammation (reviewed in ref. 35), and CCL20 is a Th17 cell–attracting chemokine (36). IL-4RA is a receptor required for both IL-4 and IL-13 signaling, and S100A9 is an alarmin protein (37). Interestingly, Tgfb1 and Tgfb3 were regulated in a reciprocal manner, with Tgfb1 expression being increased and Tgfb3 decreased in AECs of WT mice following LPS/OVA treatment. Neither of these changes was observed in AECs of TNFR1/2-DKO mice, indicating that TNF signaling controls the reciprocal expression of both Tgfb1 and Tgfb3.
TNF produced during allergen challenge promotes airway inflammation.
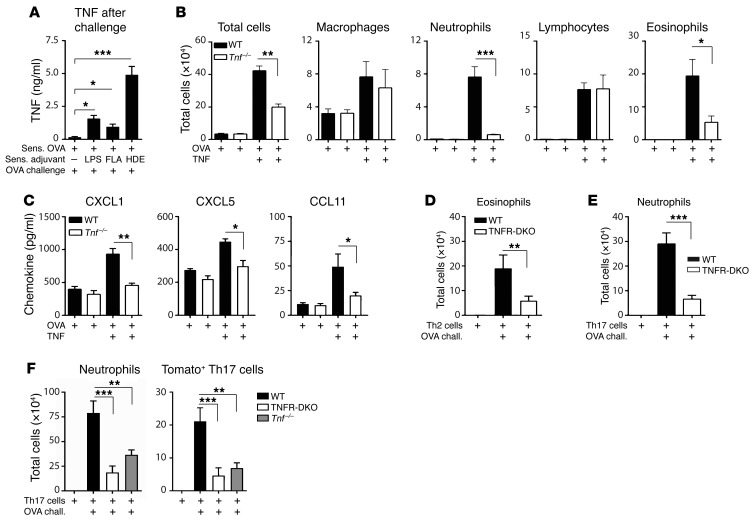
Our studies thus far had shown that TNF is required for type 2 cytokine production in regional LNs (Figure 4B and Supplemental Figure 6) and for both eosinophilic and neutrophilic inflammation in the TLR ligand–mediated mouse model of asthma (Figure 3A and Supplemental Figure 3A). However, this cytokine was dispensable for IL-17 production in regional LNs following allergic sensitization (Figure 4B and Supplemental Figure 6). Together, these data suggest that TNF likely acts after sensitization to promote neutrophilic inflammation of the airway. Analysis of BALF following OVA challenge revealed that TNF was present in the airways of mice that had been previously sensitized to OVA with TLR ligands or HDE, but not in BALF of mice that were sensitized to OVA without an adjuvant (Figure 6A). To specifically test the requirement for endogenous TNF during the challenge phase of an asthma model, we bypassed the requirement for endogenous TNF during sensitization by instilling exogenous rmTNF and OVA into the lungs of WT and Tnf–/– mice to sensitize them. Analysis of mLNs confirmed that rmTNF is sufficient to promote OVA-specific Th2 and Th17 responses in both strains (Supplemental Figure 8). Upon OVA challenge, however, there were striking differences between WT and Tnf–/– animals (Figure 6B). Although both strains displayed robust infiltration of lymphocytes into the lung, Tnf–/– mice had significantly fewer neutrophils and eosinophils than did WT mice. Consistent with this finding, the neutrophil-attracting chemokines CXCL1 and CXCL5, as well as the eosinophil-attracting chemokine CCL11, were also strongly reduced in the airways of Tnf–/– mice following the OVA challenge compared with WT mice (Figure 6C). This suggests that in addition to its requirement for Th2 development during TLR ligand–mediated allergic sensitization, TNF also acts during the challenge phase to promote recruitment of both eosinophils and neutrophils into the airway, in part by eliciting production of chemokines.
Figure 6. TNF is required during the challenge phase of allergic airway disease for eosinophilic and neutrophilic inflammation.
(A) TNF concentrations in BALF of mice previously sensitized (Sens.) with indicated adjuvants and challenged (chall.) with OVA (n = 8 mice per group). (B and C) WT and Tnf–/– mice were sensitized using rmTNF/OVA, and challenged with OVA aerosol. Cell numbers for the indicated leukocytes (B) and chemokine levels in BALF after challenge (C) (n = 6 mice per group). (D–F) Cell numbers for indicated leukocytes in airways of OVA-challenged mice following adoptive transfer of in vitro–polarized, OVA-specific Th2 cells (D), Th17 cells (E), or fluorescent, IL-17 fate-mapping cells (F) (n = 6–12 mice per group). Values shown represent mean ± SEM from 1 of 2 experiments yielding similar results, except A and D, in which data from 2 experiments were pooled. *P < 0.05, **P < 0.01, ***P < 0.001; “OVA only” group vs. other groups (A) or WT vs. Tnf–/– or TNFR1/2-DKO mice that were treated similarly (B–F); Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test.
To confirm that TNF functions during the challenge phase of allergic airway disease to promote eosinophilic and neutrophilic inflammation, we again bypassed the role of TNF during sensitization, this time by generating OVA-specific, Th2 or Th17 T cells in vitro, and separately transferring them into naive WT or TNFR1/2-DKO recipient animals before challenge with OVA. WT mice that received the OVA-specific Th2 cells developed strong eosinophilic inflammation upon OVA challenge, but this response was strongly reduced in similarly treated TNFR1/2-DKO mice (Figure 6D). Likewise, adoptive transfer of in vitro–generated, OVA-specific Th17 cells followed by OVA challenge elicited a strong neutrophilic response in WT recipients, but not in TNFR1/2-DKO recipients (Figure 6E). Similar reductions in eosinophilic and neutrophilic inflammation were seen when either Th2 or Th17 cells were adoptively transferred into mice lacking the cytokine TNF (Supplemental Figure 9, A and B). These data indicate that in addition to its role during allergic sensitization, TNF also functions during allergen challenge to exacerbate allergic inflammation of the lung.
We next tested whether the relatively weak neutrophilic response in TNFR1/2-DKO mice receiving OVA-specific Th17 cells was due to a lower number of these cells in the lung following OVA challenge. To do this, we used triple-transgenic mice bearing an OVA-specific TCR transgene from OT-II mice, a Cre recombinase transgene driven by the Il17 promoter, and a Cre-inducible tdTomato transgene (K.H. Shalaby, unpublished observations). In these mice, tdTomato is irreversibly activated following Il17 expression, and the fluorescent cells can be tracked in vivo. We used naive T cells from these mice to generate OVA-specific Th17 cells in vitro, and adoptively transferred them into WT and TNFR1/2-DKO mice. Following challenge with aerosolized OVA, airways of WT recipient mice contained large numbers of neutrophils and tdTomato+ Th17 cells, whereas numbers of both cell types were decreased in TNFR1/2-DKO and Tnf–/– recipients (Figure 6F and Supplemental Figure 10). Together, these data show that TNF signaling during allergen challenge is required for robust Th17 cell accumulation in the airways and for consequent neutrophilic inflammation.
Discussion
The structural and biochemical diversity displayed by allergens suggests that they likely activate distinct signaling pathways that converge on allergic inflammation of the airway. In the current study, we found that the cytokines TNF, IL-1α, and IL-1β were induced by TLR ligands, but not by proteases. Previous work with animal models has shown that TNF is sufficient to promote allergic sensitization to OVA (24), and that TNFR-KO mice have reduced inflammation in a low-dose-LPS, but not a high-dose-LPS, model of asthma (38). We confirmed that TNF is required for pulmonary inflammation in the LPS/OVA model of asthma, and further showed that this cytokine is dispensable for inflammation in the ASP/OVA model of this disease, thereby identifying the TNF signaling pathway as distinguishing TLR ligand–mediated allergic pulmonary inflammation from that triggered by proteases. The requirement of TNF was not restricted to the LPS/OVA model of asthma, however, because it was also required in a model where HDE was used as the adjuvant.
Dissection of the TLR ligand model into the sensitization and challenge phases revealed that TNF affects eosinophilic and neutrophilic inflammation in different ways (Supplemental Figure 11). The reduced accumulation of eosinophils in the airways of challenged mice can be partly explained by a requirement of TNF during allergic sensitization because lung-draining LNs of TNF-deficient mice had significantly lower amounts of Th2 cytokines than did LNs of WT mice. However, similar decrements of IL-17 were not observed in LNs of the TNF-deficient mice, and using TNF as an adjuvant failed to elicit IL-17 production in mLNs. The selective requirement of TNF for Th2 differentiation in this model suggests that this cytokine does not affect lung DC migration because that would be expected to impact both Th2 and Th17 differentiation. Additional experiments will be required to identify the molecular change(s) downstream of the TNF receptor that is selectively required for Th2 differentiation. We detected only very low levels of the epithelial cell–produced cytokines IL-25, IL-33, and TSLP, in agreement with a previous report (30). The technical difficulties in accurately measuring these cytokines in biological fluids (29, 39) might have contributed to the low amounts detected. However, we recently showed that BALF of HDE-treated mice contains an activity that enhances the ability of DCs to direct Th2 differentiation in vitro, and that this activity is independent of IL-33, TSLP, and IL-25 (40). It is possible, therefore, that binding of TNF to its receptor on AECs releases an unidentified factor that contributes to the Th2-promoting activity of lung DCs.
We found that TNFR1/2 was required on radioresistant cells in bone marrow chimeric mice for eosinophilic inflammation. Furthermore, TNFR1, which is expressed in structural cells, was also found to be required for allergic airway inflammation. This suggests that TNF signaling in AECs might be important in the TLR ligand model of asthma, although other radioresistant cells, such as macrophages or fibroblasts, might also contribute. In multiple experiments using the LPS/OVA model of asthma, we consistently observed that compared with WT mice, intact TNFR1/2-DKO mice and TNFR1-single-KO mice had reduced eosinophils and neutrophils. Reduced eosinophils were also seen in bone marrow chimeric mice lacking TNFR on radioresistant cells. However, reductions in neutrophils and total cells were not seen in the latter animals. Although the explanation for this apparent discrepancy is not known, whole-body irradiation causes profound transcriptional changes in the lung (41). This, in turn, can lead to inflammation, parenchymal remodeling, pulmonary fibrosis (42), and increased amounts of proinflammatory cytokines, including IL-1α and IL-6 (16). Because the latter cytokines contribute to Th17 development, changes caused by radiation in bone marrow chimeric mice might circumvent the requirement of TNF for neutrophilic inflammation observed in intact animals.
RNA profiling of AECs revealed that the gene whose expression in these cells was most affected by the absence of TNFR1/2 was Plaur, which encodes uPAR, a serine protease receptor involved in the generation of plasmin from plasminogen. Previous genetic (34) and biological (43, 44) evidence has strongly implicated uPAR in asthma, particularly in severe, nonatopic asthma. Our current work shows that Plaur expression in AECs is dependent on TNF signaling responses in those cells. The precise mechanistic relationship between asthma and uPAR remains unclear, in part because its enzymatic product, plasmin, has multiple activities, including activation of TGF-β1. The latter observation is of particular interest in light of our finding that TNF also induces the transcription of Tgfb1. Thus, TNF signaling in AECs likely increases the activity of TGF at the transcriptional and posttranslational levels. Although relatively little is known of TGF-β3, its effects on fibrosis are opposite to those of TGF-β1 (45). Whether the TNF-dependent reciprocal regulation we observed for TGF-β1 and TGF-β3 is relevant to allergic sensitization or inflammation awaits additional studies.
TNF was dispensable for IL-17 production in lung-draining LNs, yet neutrophilic inflammation following allergen challenge was much lower in TNFR-deficient mice than in WT animals. Together, these data suggest that TNF acts at some point after allergic sensitization. In support of this, 2 different approaches to bypass the requirement of TNF during allergic sensitization revealed that production of this cytokine during the challenge phase promoted the recruitment of both eosinophils and neutrophils to the airway. This result is consistent with a previous study in which anti-TNF antibodies reduced eosinophilic inflammation following challenge of mice that had been previously sensitized by an extrapulmonary route (46). The precise mechanism by which TNF promotes inflammation during the challenge phase remains unknown and will be an objective of future studies. It is possible that TNF promotes T cell production of cytokines, either directly or indirectly. IL-23 has been proposed to stabilize Th17 cells and promote their production of IL-17, but also to promote Th2 differentiation and allergic inflammation (47). Furthermore, in some situations, TNF can promote both Th2 and Th17 inflammation by inducing IL-23 (48). However, we found that WT and TNFR-deficient mice had similar amounts of IL-23 in their airways after allergen challenge (G.S. Whitehead, unpublished observations), suggesting that in this model, TNF likely promotes airway inflammation through a different pathway.
The increased levels of TNF seen in bronchial biopsies and induced sputum from human subjects with asthma suggest that this cytokine likely has an important role in clinical disease. Furthermore, inhaled TNF promotes airway neutrophilia and AHR in both asthmatics and healthy human subjects (49, 50), and polymorphisms in Tnf are associated with asthma (51). However, results of clinical trials aimed at blockade of the TNF receptor have been mixed, with some studies showing a benefit (21, 22), but others not (23, 52). It seems likely that this is because blockade of TNF is efficacious in only a subgroup of patients, which is consistent with our current finding that TNF is required in some, but not all, models of asthma. Most humans are likely exposed to several adjuvants and allergens, which trigger multiple pathways that converge on allergic inflammation of the airway. Although blockade of TNF might prevent or attenuate asthma where TLR ligand exposures are the primary factor in allergic sensitization and asthma exacerbations, other pathways, triggered by protease allergens, for example, would be expected to remain unaffected, or even enhanced. It is noteworthy that after ASP/OVA-mediated allergic sensitization, Tnf–/– mice had considerably higher amounts of IL-13 in the airway than did WT mice. Although the explanation for this observation is not yet known, TNFR2 is expressed on Tregs and can contribute to their expansion or function (53). Thus, in the absence of TNFR2 signaling, decreased Treg number or function might lead to stronger effector immune responses. In support of this, we found that TNFR2-single-KO mice displayed stronger eosinophilic inflammation than WT mice in the LPS/OVA model, whereas TNFR1-single-KO mice had weaker responses. Thus, TNFR1 and TNFR2 have opposing actions in this model of asthma, suggesting that blockade of TNF might exacerbate allergic inflammation in some individuals, and be protective in others, depending on the relative contribution of those 2 receptors to an individual’s disease. A comprehensive endotyping of individual patients and analysis of their environmental exposures should be helpful in identifying asthma subgroups likely to be responsive to TNF blockade.
In addition to environmental exposures, genetic susceptibility is also likely to impact the effectiveness of anti-TNF therapy. The genes we have shown to be induced in AECs by TNF represent novel candidates that can be tested not only for their potential mechanistic role in asthma, but also in SNP-based studies for predicting responsiveness to anti-TNF therapy. The ability to ameliorate the features of asthma by inhibiting genes downstream of TNF would be preferable to blockade of TNF itself because the latter is associated with increased risk of infection (23). It will therefore be important to investigate these genes as potential targets in novel interventional therapies aimed at reducing the severity of asthma in defined populations while minimizing risk of infection.
Methods
Mice.
Male mice of the following mouse strains were purchased from Jackson Laboratory: C57BL/6J, B6129SF2/J, TNFR1–/– (C57BL/6-Tnfrsf1atm1Imx/J), TNFR2–/– (B6.129S2-Tnfrsf1btm1Mwm/J), TNFR1/2-double-KO (DKO) (B6.129S-Tnfrsf1atm1Imx Tnfrsf1btm1Imx/J), Tnf–/– (B6.129S-Tnftm1Gkl/J), OT-II (B6.Cg-Tg[TcraTcrb]425Cbn/J), Tlr2–/– (B6.129S4-Ccr2tm1Ifc/J), Tlr4–/– (B6.129P2-Tlr4tm1Aki), Tlr5–/– (B6.129S1-Tlr5tm1Flv/J), Myd88fx/fx [B6.129P2(SJL)-Myd88tm1Defr/J], Cd11c-cre [B6.Cg-Tg(Itgax-cre)1-1Reiz/J], KitW-sh (B6.Cg-KitW-sh/HNihrJaeBsmGlliJ), Rag1–/– (B6.129S7-Rag1tm1Mom/J), and B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J. Brigid Hogan (Duke University, Durham, North Carolina, USA) provided B6;D2-Tg (Sftpc-cre)1Blh (Sftpc-cre) mice (54). Mice lacking Myd88 in Cd11c-expressing cells were generated by crossing conditionally mutant Myd88fl/fl mice to Itgax-cre mice. Animals lacking Myd88 in AECs were generated by crossing the Myd88fl/fl mice to Sftpc-Cre mice. Briefly, mice lacking MyD88 in AECs and DCs were generated by crossing of conditionally mutant Myd88fl/fl mice to Sftpc-cre mice and Itgax-cre mice, respectively. OVA-specific, Il17 fate-mapping T cells were obtained from mice that carried three transgenes: an OVA-specific TCR from OT-II mice, a Cre recombinase-YFP fusion protein-encoding gene inserted into the 3′ region of the Il17 gene, and the Cre-inducible tdTomato gene from B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J mice. They were crossed to OT-II mice to generate mice with OVA-specific T cells that permanently acquire tdTomato fluorescence upon Il17 expression. In all experiments, age-matched and genetically matched mice from the same commercial source were used as controls. Mice were housed in pathogen-specific conditions and used between 6 and 12 weeks of age.
Animal models of asthma.
To induce allergic sensitization, mice were lightly anesthetized with isoflurane and given 2 oropharyngeal (o.p.) administrations, 1 week apart, of 50 μg LPS-free OVA (Worthington Biomedical) together with an adjuvant. The adjuvants tested included 100 ng LPS from E. coli 0111:B4 (Sigma-Aldrich), 1,250 ng recombinant FLA (InvivoGen), 20 μg protease from Aspergillus oryzae (Sigma-Aldrich), 33 μg protease from papaya (papain), 20 μl of HDE, and rmTNF (Gemini Bio-Products), which according to the vendor typically contains ≤1 endotoxin unit/μg LPS. All instillations were in a total volume of 50 μl with PBS as the diluent. In some experiments, ASP was heat-inactivated at 95°C for 30 minutes to inactivate protease activity. Seven days after the second sensitization, mice were challenged by exposure to an aerosol of 1% OVA (Sigma-Aldrich) in PBS for 1 hour. Unless otherwise indicated, BALF was collected 4 hours after challenge for analysis of cytokines in the airway, whereas airway inflammation and AHR were assessed at 48 hours after challenge.
OVA uptake by lung DCs.
OVA uptake by lung cDCs and AMs was evaluated following instillation of Alexa Fluor 647–labeled OVA (Life Technologies), as described previously (55).
Analysis of RNA.
Lungs were perfused with PBS through the right ventricle, excised, and flash-frozen in liquid N2, and RNA was isolated from them using a Qiagen RNeasy Mini Kit (Qiagen). Quantitative PCR was performed per the instructions of the manufacturer (Applied Biosystems), and values were normalized to expression of the housekeeping gene Gapdh. To study gene expression in AECs, lungs were perfused via right atrium with Ca++- and Mg++-free PBS, excised, rinsed with PBS, and inflated with 4 U/ml elastase (Roche Diagnostics) for 45 minutes at 37°C. Lungs were then minced, digested with 0.5 mg/ml DNase for 15 minutes at 37°C, and pushed through a 70-μm strainer. The resulting cells were incubated with biotinylated antibodies against the lineage markers CD31, CD34, and CD45, and an AutoMACS column was used to deplete nonepithelial cells. AECs were then subjected to flow cytometry–based sorting as 7-AAD–CD31–CD34–CD45–EpCAM+ cells. Purity of the isolated cells was greater than 99%. RNA prepared from these cells was hybridized to the nCounter Mouse Immunology Gene Expression Codeset per the manufacturer’s instructions and then quantified on an nCounter machine (NanoString). Gene expression data were normalized using 14 NanoString nSolver housekeeper genes: Alas1, Eef1g, G6pdx, Gapdh, Gusb, Hprt, Oaz1, Polr1b, Polr2a, Ppia, Rpl19, Sdha, Tbp, and Tubb5. Genes were excluded from the analysis if their minimum intensity threshold was less than 40 counts for all sample groups.
Analysis of cytokines in BALF, regional LNs, and whole lungs.
Concentrations of cytokines and chemokines in BALF were measured as described previously (12), using a multiplexed fluorescent bead–based immunoassay according to the instructions of the manufacturer (Bio-Rad Laboratories). To indirectly study T cell development in lung-draining mLNs, Histopaque (Sigma-Aldrich) gradient–enriched cells were prepared from spleens and LNs of OT-II transgenic mice as previously described (56) and washed 3 times with sterile PBS, and 107 cells were transferred by retro-orbital injection into recipient mice. These animals were then given o.p. administrations of OVA together with a test adjuvant. Four days after sensitization, the mLNs were excised, minced, and pressed through a 70-μm strainer; 1 × 106 LN cells were cultured for 2 days containing 10% FBS, 55 μM 2-mercaptoethanol (Sigma-Aldrich), 10 mM HEPES (Life Technologies), 100 IU/ml Penicillin/100 μg/ml Streptomycin (Life Technologies), and 10 μg/ml OVA. For measurement of cytokines by lung explants following sensitization and challenge of mice, lungs were first perfused in situ through the right atrium with sterile PBS at 48 hours after challenge, excised, and cultured in 1 ml cRPMI medium containing 10 μg/ml OVA for 24 hours. Supernatants were analyzed for cytokines using multiplexed fluorescent bead–based immunoassays.
Histology.
Lungs were fixed in 10% formalin and embedded in paraffin. Left lobe longitudinal sections 5–7 μm thick were stained with Alcian blue and periodic acid–Schiff and examined under a BX51 microscope (Olympus) using a ×10 objective, and images were captured with an Olympus DP70 digital camera. Inflammation was scored by a pathologist in a blinded fashion, based on both the intensity locally and the number of vessels and bronchi exhibiting surrounding infiltrates. Grades were assigned to mucin staining based on the number of goblet cells and their pattern of distribution in the main bronchus, proximal branches, preterminal bronchioles, and terminal bronchioles.
AHR.
Evaluations of AHR were performed as previously described (12), using the FlexiVent mechanical ventilator system (Scireq). A single-compartment model of the lung was used to assess total respiratory system resistance after delivery of aerosolized methacholine using an ultrasonic nebulizer. Data are reported as peak resistance values.
Cell depletion.
Alveolar macrophages were depleted using a Clodrosome macrophage depletion kit (Encapsula NanoSciences). Briefly, anesthetized mice were given o.p. administrations of 50 μl liposomal clodronate suspension containing 5 mg/ml clodronate, or an equivalent amount of control liposomes lacking clodronate. Flow cytometry was used to confirm depletion of the alveolar macrophages at 7 days after treatment. Neutrophils were depleted by i.p. injections of 150 μg of anti–mouse Ly-6G/Ly-6C (Gr-1) antibody (clone RB6-8C5, eBioscience) or anti–mouse Ly-6G (clone 1A8, BioXCell) 16 and 2 hours before sensitization. Isotype controls were rat IgG2b (clone eB149/10H5) and rat IgG2a (clone 2A3), respectively.
Generation of bone marrow chimeric mice.
Reciprocal bone marrow chimeric mice were created using B6129SF2/J and TNFR1/2-DKO mice using standard procedures. Briefly, 107 bone marrow cells were transferred into lethally irradiated (9.25 Gy over 12 minutes) recipient animals by retro-orbital injections. The mice were given acidified water supplemented with 500 μg/ml neomycin for 2 weeks after irradiation. After allowing 12 weeks for hematopoietic reconstitution, recipient mice were used in experiments.
Purification of cell populations in lung for culture.
Alveolar and interstitial macrophages, monocytes, and cDCs were purified from the lung using flow cytometric sorting, as previously described (57), from mice sensitized to inhaled LPS/OVA 16 hours before tissue procurement. Briefly, alveolar macrophages were identified as CD11chiSiglec-FhiCD88hiCD11blo cells, interstitial macrophages as CD11cintermediateCD88hiSiglec-FloLy-6Glo cells, monocytes as CD11bhiLy6-ChiCD88loMHC-IIlo cells, and cDCs as CD11chiMHC-IIhiCD88lo cells. Sorted cells (1 × 105) were cultured for 24 hours in 200 μl complete cRPMI in a 5% CO2 incubator. The supernatants were analyzed for TNF levels using a multiplexed fluorescent bead–based immunoassay.
In vitro generation of OVA-specific Th2 and Th17 cells.
DCs for in vitro cocultures were obtained from spleens of C57BL/6J mice. Minced spleens were digested with Liberase TM (100 μg/ml; Roche), collagenase XI (250 μg/ml), hyaluronidase 1a (1 mg/ml), and DNase I (200 μg/ml; Sigma-Aldrich) for 30 minutes at 37°C. The reactions were stopped with the addition of EDTA (20 mM final concentration), and the DCs were separated from lymphocytes and red blood cells by discontinuous phase-density centrifugation with 14.5% Nycodenz (Accurate Chemical), washed with PBS containing 2% FCS (Hyclone) and 2 mM EDTA, and further enriched by positive selection using anti-CD11c antibodies on an AutoMACS magnetic bead–based cell sorting system (Miltenyi Biotec). Naive CD4+ T cells were prepared from spleens and LNs of OT-II transgenic mice by negative selection as described previously (56), and cultured at 1 × 106 T cells per well together with 0.5 × 106 splenic DCs in 2 ml cRPMI containing OVA323–339 peptide (10 nM). To generate Th2 cells, recombinant IL-4 (rIL-4) (4 ng/ml), rIL-2 (20 ng/ml), anti–IL-12 (10 μg/ml), and anti–IFN-γ (5 ng/ml) were added to the culture. On day 5, the cells were divided again into 2 wells, and cultured in 2 ml of cRPMI with rIL-2 (20 ng/ml). To generate Th17 cells, rhTGF-β (3 ng/ml), rIL-1α (10 ng/ml), rIL-6 (10 ng/ml), anti–IL-2 mAb (10 μg/ml), anti–IL-4 mAb (10 μg/ml), anti–IL-12 mAb (10 μg/ml), and anti–IFN-γ (5 ng/ml) antibodies were added. After 3 days, the cells were divided into 2 wells and then cultured in the same conditions with the addition of IL-23 (10 ng/ml), but without OVA peptide. On day 7, the OVA-specific T cells were counted and washed 3 times with PBS, and 1 × 106 cells were adoptively transferred into recipient mice by retro-orbital injection.
Statistics.
Most statistical calculations were performed using GraphPad Prism 7 (GraphPad Software Inc.). Data are shown as means ± SEM. Differences between groups were identified by ANOVA using Dunnett’s multiple comparison tests or Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison test. Individual comparisons between groups were confirmed by a 2-tailed Student’s t test or the Mann-Whitney U test, and a P value of less than 0.05 was considered statistically significant. Analysis of NanoString data was performed in R version 3.2.1, using limma version 3.27.5 from Bioconductor version 2.31.0. Statistical tests were performed via a moderated t test using limma-voom, which models the mean-variance to assign observational-level weights prior to downstream linear modeling. Genotype-specific hits were defined as (WTLPS/WTuntreated)/(DKOLPS/DKOuntreated) > 1.5, with P less than 0.01.
Study approval.
All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at the National Institute of Environmental Health Sciences, Research Triangle Park, North Carolina, USA.
Author contributions
GSW, SYT, KN, KHS, TPM, and HN performed experiments with cells and animals. HN, KN, KHS, and GSW prepared cells for adoptive transfer. GSW, SYT, KHS, TPM, and HN analyzed cellular and molecular data, JMW analyzed data from NanoString experiments, and GPF evaluated and scored stained lung sections. DNC, GSW, and HN designed experiments. DNC conceived the project. GSW and DNC wrote the manuscript, and all authors made editorial suggestions and approved the final version.
Supplementary Material
Acknowledgments
We thank Ligon Perrow for support with animal experiments, Rick Fannin and Kevin Gerrish for support with NanoString experiments, Maria Sifre for help sorting AECs, Bill Fitzgerald and John McLamb for irradiation of mice, and Michael Fessler and Stavros Garantziotis for critical reading of the manuscript. This work was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ZIA ES102025-09).
Version 1. 07/31/2017
Electronic publication
Version 2. 09/01/2017
Print issue publication
Funding Statement
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences
Footnotes
TPM’s present address is: Department of Pediatrics, University of North Carolina School of Medicine, Chapel Hill, North Carolina, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(9):3313–3326.https://doi.org/10.1172/JCI90890.
Contributor Information
Keiko Nakano, Email: kn64chhnc@gmail.com.
Hideki Nakano, Email: nakanoh@niehs.nih.gov.
References
- 1.Martinez FD, Vercelli D. Asthma. Lancet. 2013;382(9901):1360–1372. doi: 10.1016/S0140-6736(13)61536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrick CA, Bottomly K. To respond or not to respond: T cells in allergic asthma. Nat Rev Immunol. 2003;3(5):405–412. doi: 10.1038/nri1084. [DOI] [PubMed] [Google Scholar]
- 3.Larché M, Robinson DS, Kay AB. The role of T lymphocytes in the pathogenesis of asthma. J Allergy Clin Immunol. 2003;111(3):450–463. doi: 10.1067/mai.2003.169. [DOI] [PubMed] [Google Scholar]
- 4.McGrath KW, et al. A large subgroup of mild-to-moderate asthma is persistently noneosinophilic. Am J Respir Crit Care Med. 2012;185(6):612–619. doi: 10.1164/rccm.201109-1640OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson NC. Novel approaches to the management of noneosinophilic asthma. Ther Adv Respir Dis. 2016;10(3):211–234. doi: 10.1177/1753465816632638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green RH, Brightling CE, Woltmann G, Parker D, Wardlaw AJ, Pavord ID. Analysis of induced sputum in adults with asthma: identification of subgroup with isolated sputum neutrophilia and poor response to inhaled corticosteroids. Thorax. 2002;57(10):875–879. doi: 10.1136/thorax.57.10.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McKinley L, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181(6):4089–4097. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barczyk A, Pierzchala W, Sozañska E. Interleukin-17 in sputum correlates with airway hyperresponsiveness to methacholine. Respir Med. 2003;97(6):726–733. doi: 10.1053/rmed.2003.1507. [DOI] [PubMed] [Google Scholar]
- 9.Hellings PW, et al. Interleukin-17 orchestrates the granulocyte influx into airways after allergen inhalation in a mouse model of allergic asthma. Am J Respir Cell Mol Biol. 2003;28(1):42–50. doi: 10.1165/rcmb.4832. [DOI] [PubMed] [Google Scholar]
- 10.Kaminska M, et al. Airway remodeling in subjects with severe asthma with or without chronic persistent airflow obstruction. J Allergy Clin Immunol. 2009;124(1):45–51.e1. doi: 10.1016/j.jaci.2009.03.049. [DOI] [PubMed] [Google Scholar]
- 11.Molet S, et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J Allergy Clin Immunol. 2001;108(3):430–438. doi: 10.1067/mai.2001.117929. [DOI] [PubMed] [Google Scholar]
- 12.Wilson RH, Whitehead GS, Nakano H, Free ME, Kolls JK, Cook DN. Allergic sensitization through the airway primes Th17-dependent neutrophilia and airway hyperresponsiveness. Am J Respir Crit Care Med. 2009;180(8):720–730. doi: 10.1164/rccm.200904-0573OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastie AT, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J Allergy Clin Immunol. 2010;125(5):1028–1036.e13. doi: 10.1016/j.jaci.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karp CL. Guilt by intimate association: what makes an allergen an allergen? J Allergy Clin Immunol. 2010;125(5):955–960. doi: 10.1016/j.jaci.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi JP, et al. House dust mite-derived chitin enhances Th2 cell response to inhaled allergens, mainly via a TNF-α-dependent pathway. Allergy Asthma Immunol Res. 2016;8(4):362–374. doi: 10.4168/aair.2016.8.4.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, Lambrecht BN. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat Med. 2009;15(4):410–416. doi: 10.1038/nm.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorne PS, Kulhánková K, Yin M, Cohn R, Arbes SJ, Zeldin DC. Endotoxin exposure is a risk factor for asthma: the national survey of endotoxin in United States housing. Am J Respir Crit Care Med. 2005;172(11):1371–1377. doi: 10.1164/rccm.200505-758OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halim TY, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40(3):425–435. doi: 10.1016/j.immuni.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kheradmand F, Kiss A, Xu J, Lee SH, Kolattukudy PE, Corry DB. A protease-activated pathway underlying Th cell type 2 activation and allergic lung disease. J Immunol. 2002;169(10):5904–5911. doi: 10.4049/jimmunol.169.10.5904. [DOI] [PubMed] [Google Scholar]
- 20.Ying S, et al. TNFα mRNA expression in allergic inflammation. Clin Exp Allergy. 1991;21(6):745–750. doi: 10.1111/j.1365-2222.1991.tb03205.x. [DOI] [PubMed] [Google Scholar]
- 21.Berry MA, et al. Evidence of a role of tumor necrosis factor alpha in refractory asthma. N Engl J Med. 2006;354(7):697–708. doi: 10.1056/NEJMoa050580. [DOI] [PubMed] [Google Scholar]
- 22.Howarth PH, et al. Tumour necrosis factor (TNFalpha) as a novel therapeutic target in symptomatic corticosteroid dependent asthma. Thorax. 2005;60(12):1012–1018. doi: 10.1136/thx.2005.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wenzel SE, et al. A randomized, double-blind, placebo-controlled study of tumor necrosis factor-alpha blockade in severe persistent asthma. Am J Respir Crit Care Med. 2009;179(7):549–558. doi: 10.1164/rccm.200809-1512OC. [DOI] [PubMed] [Google Scholar]
- 24.Eisenbarth SC, Piggott DA, Huleatt JW, Visintin I, Herrick CA, Bottomly K. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med. 2002;196(12):1645–1651. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson RH, et al. The Toll-like receptor 5 ligand flagellin promotes asthma by priming allergic responses to indoor allergens. Nat Med. 2012;18(11):1705–1710. doi: 10.1038/nm.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezaee F, et al. Polyinosinic:polycytidylic acid induces protein kinase D-dependent disassembly of apical junctions and barrier dysfunction in airway epithelial cells. J Allergy Clin Immunol. 2011;128(6):1216–1224.e11. doi: 10.1016/j.jaci.2011.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georas SN, Rezaee F. Epithelial barrier function: at the front line of asthma immunology and allergic airway inflammation. J Allergy Clin Immunol. 2014;134(3):509–520. doi: 10.1016/j.jaci.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbert CA, et al. Augmentation of permeability in the bronchial epithelium by the house dust mite allergen Der p1. Am J Respir Cell Mol Biol. 1995;12(4):369–378. doi: 10.1165/ajrcmb.12.4.7695916. [DOI] [PubMed] [Google Scholar]
- 29.Lloyd CM, Saglani S. Epithelial cytokines and pulmonary allergic inflammation. Curr Opin Immunol. 2015;34:52–58. doi: 10.1016/j.coi.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Willart MA, et al. Interleukin-1α controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. J Exp Med. 2012;209(8):1505–1517. doi: 10.1084/jem.20112691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Black RA, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature. 1997;385(6618):729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 32.Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol. 2016;12(1):49–62. doi: 10.1038/nrrheum.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rübe CE, et al. The bronchiolar epithelium as a prominent source of pro-inflammatory cytokines after lung irradiation. Int J Radiat Oncol Biol Phys. 2005;61(5):1482–1492. doi: 10.1016/j.ijrobp.2004.12.072. [DOI] [PubMed] [Google Scholar]
- 34.Barton SJ, et al. PLAUR polymorphisms are associated with asthma, PLAUR levels, and lung function decline. J Allergy Clin Immunol. 2009;123(6):1391–400.e17. doi: 10.1016/j.jaci.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Lambrecht BN, Hammad H. Allergens and the airway epithelium response: gateway to allergic sensitization. J Allergy Clin Immunol. 2014;134(3):499–507. doi: 10.1016/j.jaci.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 36.Hirota K, et al. Preferential recruitment of CCR6-expressing Th17 cells to inflamed joints via CCL20 in rheumatoid arthritis and its animal model. J Exp Med. 2007;204(12):2803–2812. doi: 10.1084/jem.20071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehrchen JM, Sunderkötter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86(3):557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 38.Kim YK, et al. Airway exposure levels of lipopolysaccharide determine type 1 versus type 2 experimental asthma. J Immunol. 2007;178(8):5375–5382. doi: 10.4049/jimmunol.178.8.5375. [DOI] [PubMed] [Google Scholar]
- 39.Ketelaar ME, Nawijn MC, Shaw DE, Koppelman GH, Sayers I. The challenge of measuring IL-33 in serum using commercial ELISA: lessons from asthma. Clin Exp Allergy. 2016;46(6):884–887. doi: 10.1111/cea.12718. [DOI] [PubMed] [Google Scholar]
- 40.Moran TP, Nakano K, Whitehead GS, Thomas SY, Cook DN, Nakano H. Inhaled house dust programs pulmonary dendritic cells to promote type 2 T-cell responses by an indirect mechanism. Am J Physiol Lung Cell Mol Physiol. 2015;309(10):L1208–L1218. doi: 10.1152/ajplung.00256.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Citrin DE, et al. Role of type II pneumocyte senescence in radiation-induced lung fibrosis. J Natl Cancer Inst. 2013;105(19):1474–1484. doi: 10.1093/jnci/djt212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y, et al. Radiation pneumonitis and early circulatory cytokine markers. Semin Radiat Oncol. 2002;12(1 suppl 1):26–33. doi: 10.1053/srao.2002.31360. [DOI] [PubMed] [Google Scholar]
- 43.Portelli MA, et al. Airway and peripheral urokinase plasminogen activator receptor is elevated in asthma, and identifies a severe, nonatopic subset of patients. Allergy. 2017;72(3):473–482. doi: 10.1111/all.13046. [DOI] [PubMed] [Google Scholar]
- 44.Stewart CE, Nijmeh HS, Brightling CE, Sayers I. uPAR regulates bronchial epithelial repair in vitro and is elevated in asthmatic epithelium. Thorax. 2012;67(6):477–487. doi: 10.1136/thoraxjnl-2011-200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu L, et al. Transforming growth factor β3 attenuates the development of radiation-induced pulmonary fibrosis in mice by decreasing fibrocyte recruitment and regulating IFN-γ/IL-4 balance. Immunol Lett. 2014;162(1 pt A):27–33. doi: 10.1016/j.imlet.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 46.Busse PJ, Zhang TF, Schofield B, Kilaru S, Patil S, Li XM. Decrease in airway mucous gene expression caused by treatment with anti-tumor necrosis factor alpha in a murine model of allergic asthma. Ann Allergy Asthma Immunol. 2009;103(4):295–303. doi: 10.1016/S1081-1206(10)60528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wakashin H, et al. IL-23 and Th17 cells enhance Th2-cell-mediated eosinophilic airway inflammation in mice. Am J Respir Crit Care Med. 2008;178(10):1023–1032. doi: 10.1164/rccm.200801-086OC. [DOI] [PubMed] [Google Scholar]
- 48.Lee HS, et al. TNF-α enhance Th2 and Th17 immune responses regulating by IL23 during sensitization in asthma model. Cytokine. 2016;79:23–30. doi: 10.1016/j.cyto.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 49.Thomas PS, Heywood G. Effects of inhaled tumour necrosis factor alpha in subjects with mild asthma. Thorax. 2002;57(9):774–778. doi: 10.1136/thorax.57.9.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thomas PS, Yates DH, Barnes PJ. Tumor necrosis factor-alpha increases airway responsiveness and sputum neutrophilia in normal human subjects. Am J Respir Crit Care Med. 1995;152(1):76–80. doi: 10.1164/ajrccm.152.1.7599866. [DOI] [PubMed] [Google Scholar]
- 51.Huang H, et al. Effects of TNF-α polymorphisms on asthma risk: a systematic review and meta-analysis. J Investig Allergol Clin Immunol. 2014;24(6):406–417. [PubMed] [Google Scholar]
- 52.Holgate ST, et al. Efficacy and safety of etanercept in moderate-to-severe asthma: a randomised, controlled trial. Eur Respir J. 2011;37(6):1352–1359. doi: 10.1183/09031936.00063510. [DOI] [PubMed] [Google Scholar]
- 53.Chen X, Bäumel M, Männel DN, Howard OM, Oppenheim JJ. Interaction of TNF with TNF receptor type 2 promotes expansion and function of mouse CD4+CD25+ T regulatory cells. J Immunol. 2007;179(1):154–161. doi: 10.4049/jimmunol.179.1.154. [DOI] [PubMed] [Google Scholar]
- 54.Okubo T, Knoepfler PS, Eisenman RN, Hogan BL. Nmyc plays an essential role during lung development as a dosage-sensitive regulator of progenitor cell proliferation and differentiation. Development. 2005;132(6):1363–1374. doi: 10.1242/dev.01678. [DOI] [PubMed] [Google Scholar]
- 55.Nakano H, et al. Migratory properties of pulmonary dendritic cells are determined by their developmental lineage. Mucosal Immunol. 2013;6(4):678–691. doi: 10.1038/mi.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakano H, et al. Pulmonary CD103(+) dendritic cells prime Th2 responses to inhaled allergens. Mucosal Immunol. 2012;5(1):53–65. doi: 10.1038/mi.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakano H, Moran TP, Nakano K, Gerrish KE, Bortner CD, Cook DN. Complement receptor C5aR1/CD88 and dipeptidyl peptidase-4/CD26 define distinct hematopoietic lineages of dendritic cells. J Immunol. 2015;194(8):3808–3819. doi: 10.4049/jimmunol.1402195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.