Abstract
Acute myelogenous leukemia (AML) frequently relapses after complete remission (CR), necessitating improved detection and phenotypic characterization of treatment-resistant residual disease. In this work, we have optimized droplet digital PCR to broadly measure mutated alleles of recurrently mutated genes in CR marrows of AML patients at levels as low as 0.002% variant allele frequency. Most gene mutations persisted in CR, albeit at highly variable and gene-dependent levels. The majority of AML cases demonstrated residual aberrant oligoclonal hematopoiesis. Importantly, we detected very rare cells (as few as 1 in 15,000) that were genomically similar to the dominant blast populations at diagnosis and were fully clonally represented at relapse, identifying these rare cells as one common source of AML relapse. Clinically, the mutant allele burden was associated with overall survival in AML, and our findings narrow the repertoire of gene mutations useful in minimal residual disease–based prognostication in AML. Overall, this work delineates rare cell populations that cause AML relapse, with direct implications for AML research directions and strategies to improve AML therapies and outcome.
Keywords: Genetics, Hematology
Keywords: Bone marrow, Leukemias, Molecular genetics
Introduction
Acute myelogenous leukemia (AML) affects 21,000 new patients yearly in the US and causes 10,000 premature deaths. While most patients with AML achieve complete remission (CR) with induction chemotherapy, many will experience disease relapse. This clinical reality, together with findings of shared genomic characteristics and invariant clonal relatedness (1, 2) of AML at diagnosis and relapse, supports the notion that aberrant cells exist that are able to survive chemotherapy and persist in the bone marrow of patients in clinical CR, a phenomenon commonly referred to as minimal residual disease (MRD). Additional evidence connecting MRD with AML relapse includes the detection of cells with persistent, aberrant leukemia-associated immunophenotypes (LAIPs) in CR using multiparameter flow cytometry (3, 4) and of persistent gene mutations in bone marrow cells in CR that are found in matched relapsed specimens (5, 6). Additionally, quantitation of residual leukemia-associated traits in remission such as fusion transcripts (7–10), gene mutations (11–16), and LAIPs (3, 17–19) has been associated with patient survival, suggesting a prognostic utility of MRD monitoring in specific clinical contexts.
Various biological scenarios have been proposed with regard to the underlying mechanisms of AML relapse. One scenario invokes resistant preleukemic progenitors that, through continued molecular evolution, ultimately acquire properties of relapsing AML blasts; another more parsimonious scenario simply invokes surviving drug-resistant blasts; while yet another scenario invokes selection via chemotherapy of rare mutated cells of whatever cellular characteristic that can form AML blasts. There remain, then, significant limitations to the current understanding of the biology, clonal composition, and genomic landscape of residual leukemia during CR, as well as the very definition and prognostic relevance of residual disease. Most studies of MRD defined by genomic aberrations have focused on analysis of single mutated genes in isolation, notably nucleophosmin 1 (NPM1). Mutant NPM1 mRNA transcripts are frequently detected at less than 0.1% of total NPM1 mRNA in CR bone marrow using allele-specific quantitative PCR (qPCR), and its levels are associated with relapse-free survival (RFS) and overall survival (OS) (11, 14, 16). This suggests that the measurement of other mutated genes at this high sensitivity may be useful for characterizing and prognosticating residual AML, but for most genes, current qPCR approaches are not readily feasible (20).
Furthermore, concurrent measurements of multiple leukemia-associated gene mutations in CR samples may provide a critical genomic context that better delineates the burden and composition of MRD and refines its prognostic value. A few studies have addressed this by performing targeted next-generation DNA sequencing (NGS) of CR samples (12, 13, 21, 22), with the notable finding that most DNMT3A mutations and many TET2 and IDH1/2 mutations persist at high levels (>5% variant allele frequency [VAF]) in early CR. However, error rates inherent to current NGS platforms limited the detection of mutated alleles to a 2.5% or greater VAF, preventing the assessment of lower-level but nevertheless persistent mutations and thereby limiting determination of the clonal composition of residual hematopoiesis in CR.
To more fully characterize residual aberrant clonal hematopoiesis in CR and assess its prognostic impact, we optimized droplet digital PCR (ddPCR) (23–26) to allow high-sensitivity detection of both hotspot and private mutations within most recurrently mutated genes in AML. Mutations identified in pretreatment blasts in 72 patients with AML were measured in CR bone marrow specimens by ddPCR to determine residual post-therapy VAFs with a limit of detection as low as 0.002% VAF. Additionally, in 20 of these AML patients for whom paired relapse specimens were available, we determined the VAF of gene mutations before therapy, during CR, and at relapse. These efforts resulted in the identification of very rare cells with genomic similarity to the dominant pretreatment blast clones as a common cellular source of AML relapse. Finally, we determined the association of ultrasensitive genomic MRD measurements with RFS and OS, thereby proving a justification for in-depth study of ddPCR-based mutation analysis as a clinical tool in AML management.
Results
Characteristics of the AML patients studied.
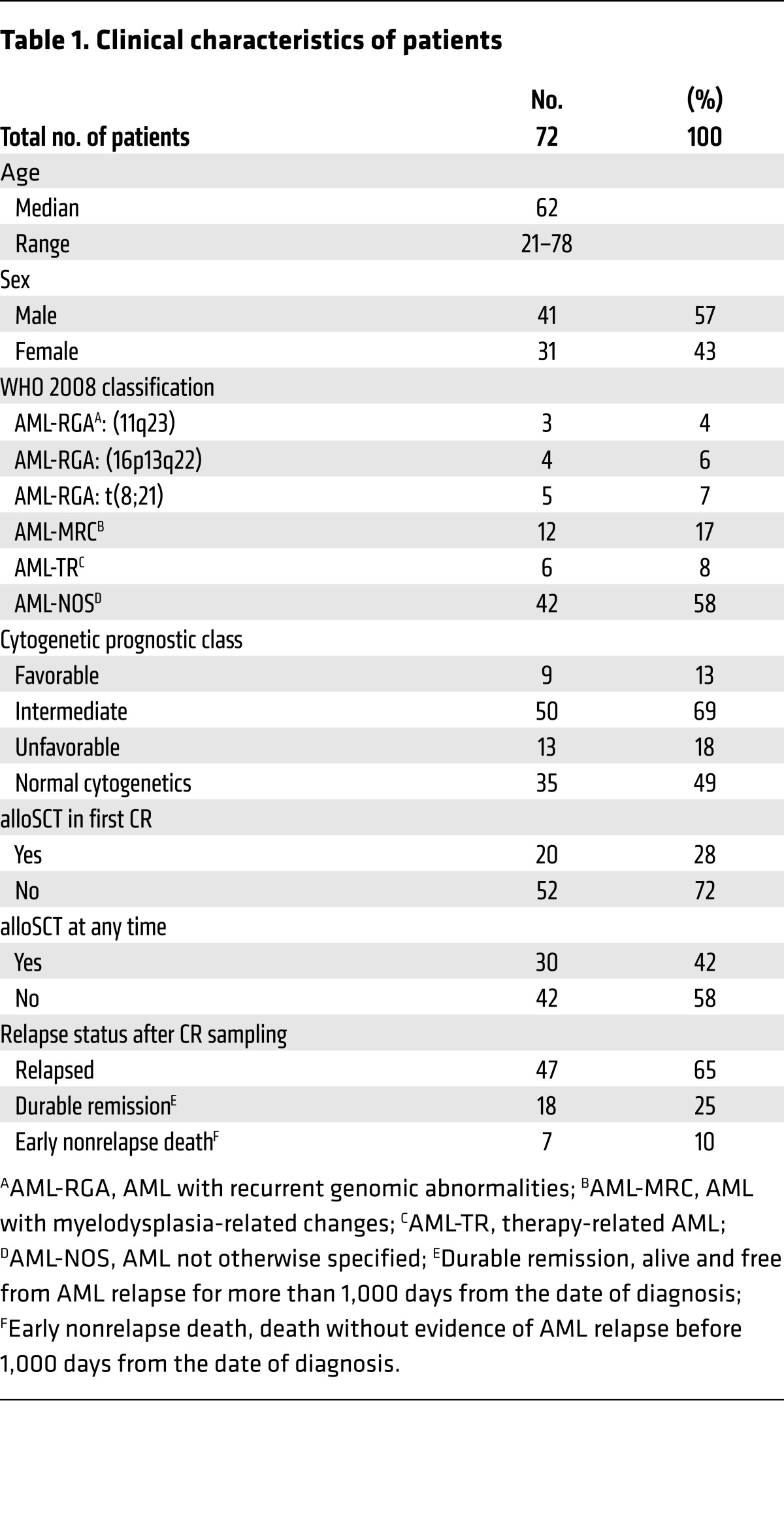
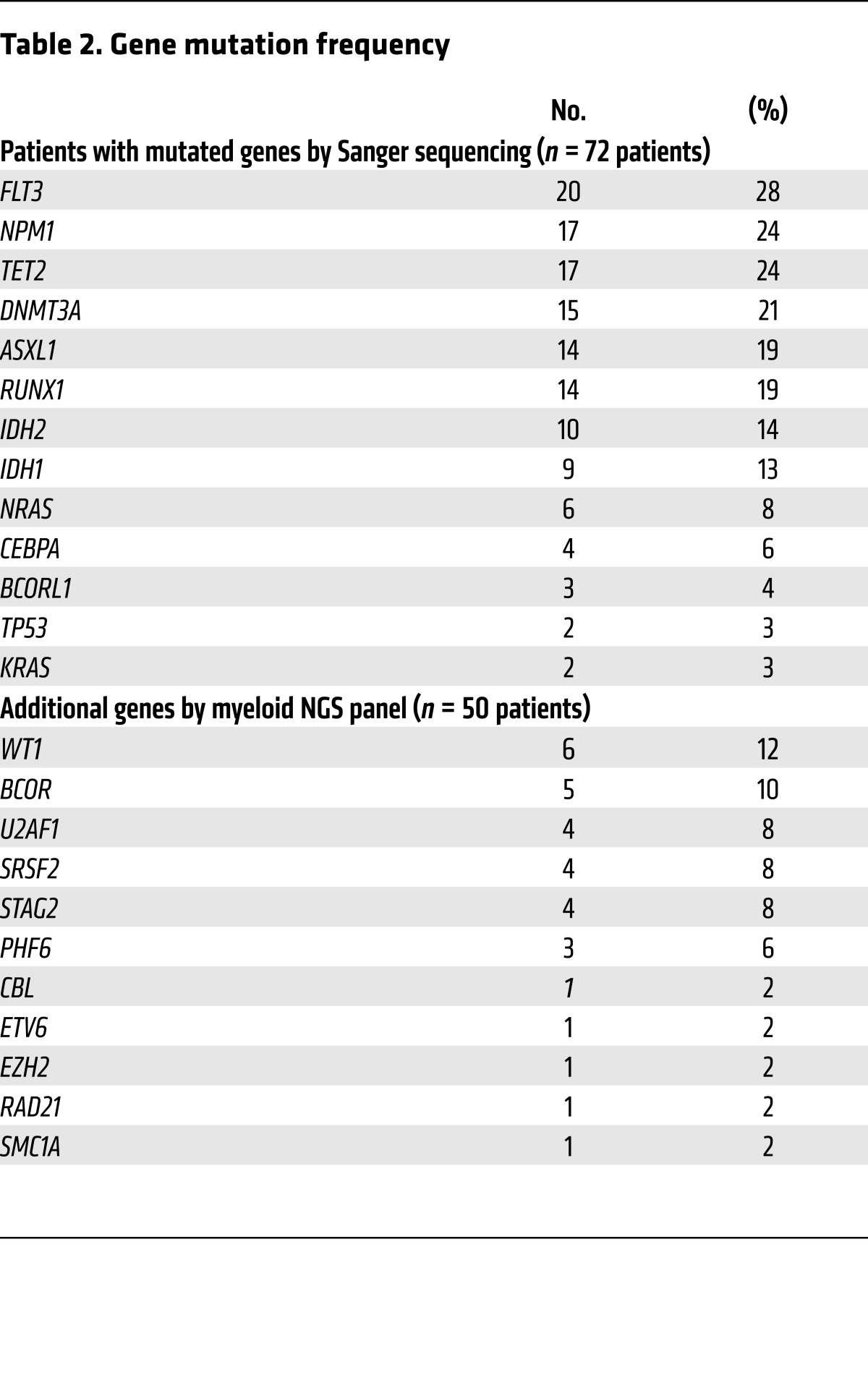
Seventy-two patients were studied on the basis of availability of specimens procured before treatment and after induction of therapy from a single-center AML clinical translational trial (UMCC 2004.072). All but 2 patients were treated with high-intensity induction chemotherapy regimens involving either an anthracycline combined with cytarabine or high-dose cytarabine, and all achieved CR after induction. Sixty-five percent (47 of 72) of patients ultimately relapsed, while twenty-five percent (18 of 72) remained in a durable remission, defined as remaining relapse free for more than one thousand days. Ten percent (7 of 72) of patients died from nonrelapse-related causes before the one thousand–day point. Twenty-eight percent (20 of 72) of patients received an allogeneic stem cell transplantation (alloSCT) during first CR (CR1), while an additional fourteen percent (10 of 72) underwent alloSCT at a time point after the first relapse. The median time to CR bone marrow sampling was 36 days after initiation of induction chemotherapy (range, 26–84 days), referred to hereafter as initial CR samples. Targeted DNA sequencing of 49 recurrently mutated genes in AML in pre-therapy samples identified 168 gene mutations across 72 patients (median 3 mutations/patient; range, 0–6). Whole-exome sequencing (WES) was performed for 19 patients with 1 or fewer mutations found in the 49-gene panel (see Methods), identifying 2 or more gene mutations for all 19 patients. Patients’ characteristics are listed in Table 1 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI91964DS1 Gene mutation frequencies detected in pre-therapy samples with Sanger sequencing and targeted NGS are listed in Table 2, and additional gene mutations detected with WES in pre-therapy samples and subsequently assayed in CR are denoted in Supplemental Table 2.
Table 1. Clinical characteristics of patients.
Table 2. Gene mutation frequency.
Most pretreatment gene mutations are detectable in AML CR.
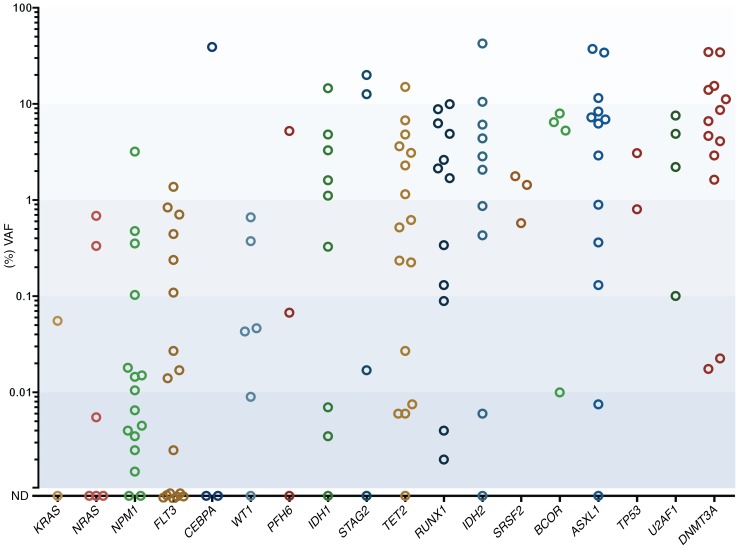
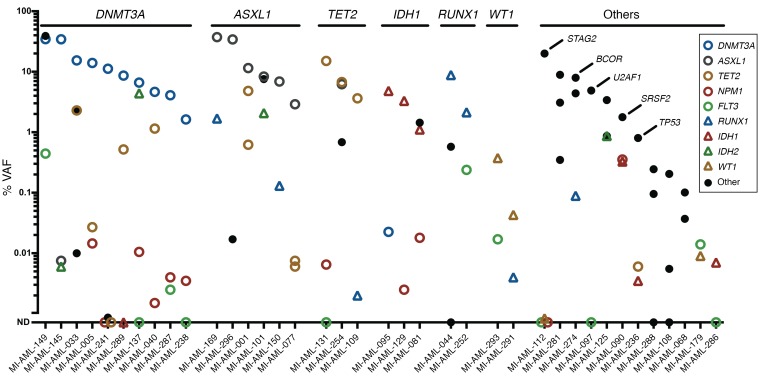
A total of 183 mutations identified in 58 genes detected at diagnosis were measured in paired initial CR samples using ddPCR (median 2.5 measurements/patient; range, 1–5). Eighty-four percent (153 of 183) of these mutations were detectable at VAFs ranging from 0.002% to 42.6%, and at least one persistent gene mutation was detected in 90% of AML remission marrows (65 of 72); the remaining 30 mutations were not detected. The range of VAFs of mutated genes in remission marrows varied widely across most multiply measured genes including NPM1 (not detected [ND], 3.198%); IDH1 (ND, 14.563%); IDH2 (ND, 42.457%); ASXL1 (ND, 37.381%); TET2 (ND, 15.047%); PFH6 (ND, 5.229%); RUNX1 (0.002%–8.814%); DNMT3A (0.017%, 34.607%); and U2AF1 (0.1%–7.570%) (Figure 1, Supplemental Figure 1, and Supplemental Table 2). Notably, a subset of mutated genes frequently demonstrated persistence at high VAF levels (>1%), including 85% (11 of 13) of mutations in DNMT3A (range, 1.631%–34.607%); 62% (8 of 13) in ASXL1 (2.907%–37.381%); 66% (6 of 9) in IDH2 (2.070%–42.457%); 58% (7 of 12) in RUNX1 (1.689%–9.950%); 56% (5 of 9) in IDH1 (1.110%–14.563%); and 44% (7 of 16) in TET2 (1.151%–15.047%). Conversely, another group of genes was typically reduced to low levels (<1%), including 100% (6 of 6) of mutations in NRAS (range ND, 0.685%); 94% (16 of 17) in FLT3 (ND, 0.837%); and 94% (15 of 16) in NPM1 (ND, 0.476%). Overall, VAFs of mutations fell between greater than 1%, greater than 0.1% to 1%, greater than 0.01% to 0.1%, greater than 0.002% to 0.01%, and ND in 36%, 22%, 13%, 13%, and 16% of measurements, respectively.
Figure 1. VAF detected in CR bone marrow samples categorized per gene.
The mean of duplicate measurements is depicted. The VAF percentage is on a log10 scale. VAFs were not corrected for genomic copy number aberrations (aCNAs), as these are rare at sites of mutated genes in AML (39).
Persistence of oligoclonal hematopoiesis is common in AML CR.
Two or more distinct mutations were assayed by ddPCR in 68 AML patients. In 44% (30 of 68) of these patients, all mutations were detected at a similar, concordant VAF, defined as a log10 difference of less than 1 (and often much less) in VAF between mutations (Figure 2). However, in 56% (38 of 68) of cases, the VAFs of various mutations were discordant (>1 log10 difference in VAF), indicating the presence of multiple coexisting clones in CR bone marrow. Within discordant cases, the mutated genes most commonly demonstrating the highest allele frequencies included DNMT3A (10 of 38); ASXL1 (6 of 38); TET2 (3 of 38); IDH1 (3 of 38); RUNX1 (2 of 38); and WT1 (2 of 38), along with single instances of genes including STAG2, BCOR, SRSF2, U2AF2, and TP53 (Figure 3). Seventy-nine percent (30 of 38) of these mutations showed high-level persistence with greater than 1% VAF. The genes most commonly demonstrating the lowest allele frequencies in discordant cases included FLT3 (9 of 38); NPM1 (6 of 38); RUNX1 (5 of 38); TET2 (3 of 38); IDH1 (3 of 38); IDH2 (3 of 38); and NRAS (2 of 38). In most patients (74%, 28 of 38), these mutations were present at a VAF of less than 0.1%.
Figure 2. Concordant mutant gene VAFs in CR bone marrow from 30 AML patients with 2 or more measured genes.
The mean of duplicate measurements is depicted. The VAF percentage is on a log10 scale. Individual patients study IDs are displayed on the x axis.
Figure 3. Discordant mutant gene VAFs in CR bone marrow samples from 38 AML patients with 2 or more measured genes.
Patients with discordant mutant gene VAFs demonstrating the common presence of oligoclonal aberrant hematopoiesis. DNMT3A, ASXL1, TET2, and IDH1 frequently persisted at greater than 1% VAF in CR. The mean of duplicate measurements is depicted. The VAF percentage is on a log10 scale.
AML often relapses from very minor residual cell populations genomically resembling blasts present in AML at diagnosis.
To identify which mutations detected in CR marrows tracked subclones that were ultimately present in the dominant relapse clones, we first determined the mutational status and VAF of 49 genes recurrently mutated in AML in 20 cases using available FACS-sorted, high-purity (>95% tumor) paired pre-therapy and relapsed AML blasts using a targeted NGS panel. We identified 65 gene mutations in the pre-therapy specimens (median of 3 mutations/patient, range 1–7). The VAFs for 77% (50 of 65) of these predominantly heterozygous mutations were greater than 40%, indicating near or full clonal representation before therapy. The VAFs of the remaining 23% (15 of 65) of mutations ranged from 7.4% to 38.5%, representing major subclones in the pre-therapy blasts. Comparison of the mutational spectrum in paired relapsed specimens showed that nearly all mutations (96%, 48 of 50) with more than 40% VAF present at diagnosis were present at relapse. Details of these mutations are listed in Supplemental Table 3 and depicted in Supplemental Figure 2.
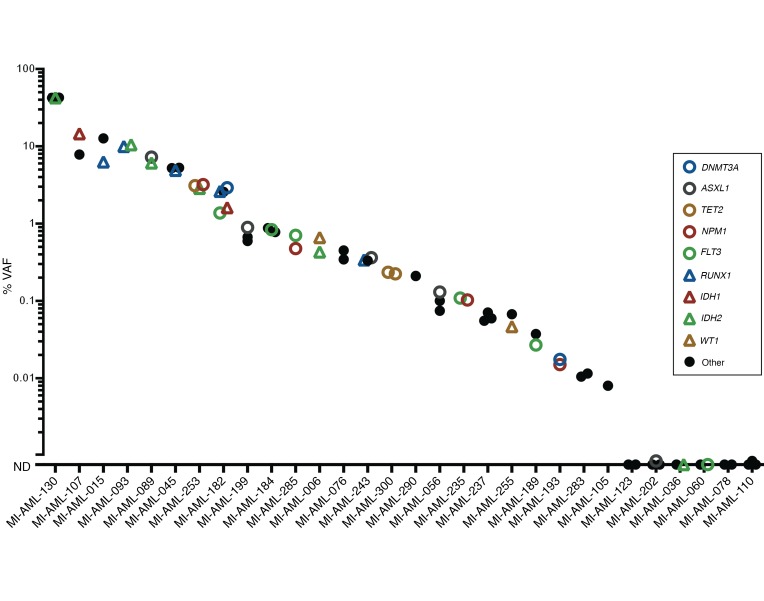
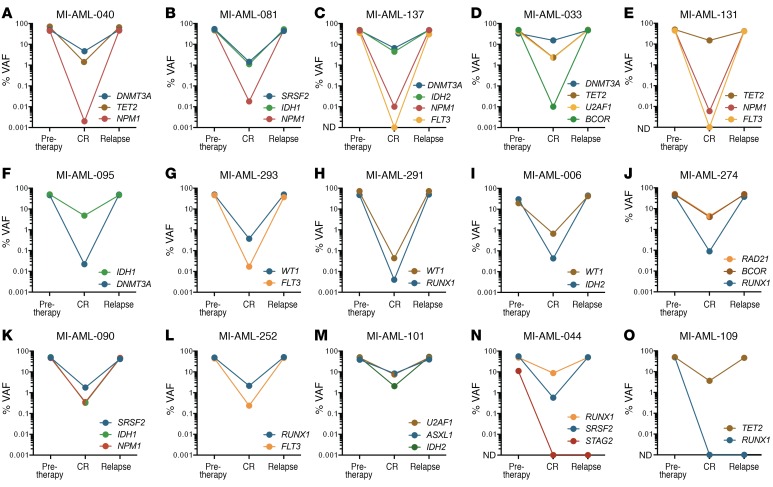
Fifty of these mutations across the twenty patients were measured using ddPCR in intervening initial CR specimens, and all but four were detected (range, 0.003%–42.457%). Two of the undetected mutations in CR involved FLT3 internal tandem duplications (ITDs), which were subsequently detected again at relapse, suggesting their presence in subclones below the detection limit. However, the other 2 undetected mutations — a fully clonal RUNX1 mutation (49.0% VAF) and a subclonal STAG2 mutation (10.1% VAF) in pre-therapy specimens — were not detected at relapse. Importantly, however, all mutations detected in CR marrows, even at levels of less than 0.01% VAF, were present in the dominant clone or major subclones at relapse (VAF 36.6%–99.0%). Furthermore, in 14 cases in which multiple genes were concurrently measured in CR marrow specimens, genes with the lowest VAF in each specimen were fully clonally represented at relapse and had equivalent VAF with other mutations present (Figure 4). This finding suggests that the cell population comprising the small clone demarcated by the low-level residual mutation is responsible for AML disease relapse rather than clones persisting at relatively high levels in CR which lack that mutation. These findings therefore support the persistence of fully transformed leukemic cells (possibly AML blasts) as one common source of relapse. Furthermore, these findings qualify the concept of preleukemic cells defined as those requiring acquisition of additional changes as a common source of clinical AML relapse, since such acquisitions would be characterized by more stochastic mutational spectra and frequent divergence from diagnosis patterns. The relationship of pre-therapy, CR, and relapse VAFs is displayed in Figure 4 and Supplemental Figure 3, with details listed in Supplemental Table 3.
Figure 4. Persistent rare cells that are genomically similar to the presentation disease blasts constitute a common source of relapsed AML.
Pre-therapy and relapse VAFs were measured with deep-coverage NGS (or ddPCR for FLT3-ITD), and CR VAFs were measured with ddPCR. In each panel, the graphs display VAFs of longitudinal measurements of mutationsF. (A–N) Examples of patients with discordant mutations demonstrating fully clonal representation at relapse of cell populations containing mutations with the lowest detected VAF in CR. (N and O) Two patients had mutations not detected in CR that were also not detected at relapse. The mean of duplicate measurements is depicted. The VAF percentage is on a log10scale. The VAFs of genes present on the X chromosome (specifically BCOR and STAG2) were corrected for male patients by a factor of 2 to accurately depict clonal composition of pre-therapy and relapse specimens.
DNMT3A mutations persist at a high VAF, even in cases of durable remission.
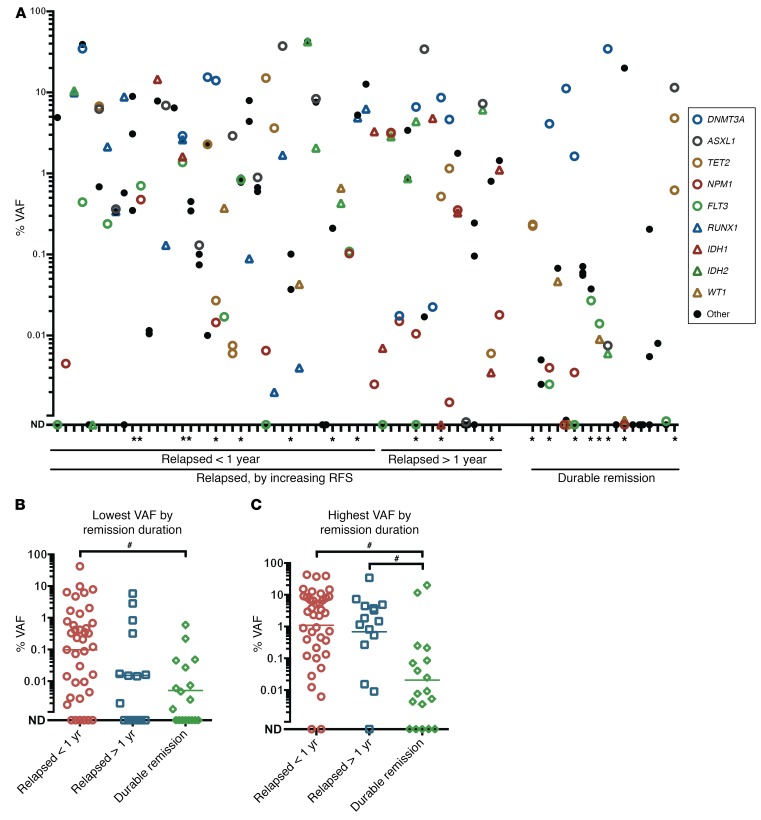
We next evaluated the allelic frequency of mutated genes with respect to categories of relapse status defined as relapse less than 1 year from diagnosis, relapse 1 year or more after diagnosis, or durable remission (relapse free for more than 1,000 days [range, 1,425–3,300 days]). High-level persistence (>1% VAF) of 1 or more gene mutations was noted in 54% (39 of 72) of patients (Figure 5A). Notably, even in patients with durable remissions, recurrent high-level persistence of DNMT3A mutations was observed as well as persistence of ASXL1 and STAG2 mutations, each in a single case (11.50% and 19.98% VAF, respectively) (Figure 5A).
Figure 5. Association of mutant gene VAF with remission duration.
(A) AML patients were grouped by remission duration for those who relapsed within less than 1 year from CR; relapsed after 1 year from CR; or had a durable remission for more than 1,000 days from CR. The VAF of mutated genes measured per patient is arranged by remission duration (left, shortest; right, longest). DNMT3A, ASXL1, TET2, and STAG2 mutations persisted at high VAF in patients who nevertheless experienced a durable remission. The mean of duplicate measurements is depicted. Patients who received alloSCT during the first CR are denoted with an asterisk. (B and C) Comparison of the median VAF for the lowest measured VAF (B) and the highest measured VAF (C) (excluding DNMT3A) in each patient, respectively, for various remission duration groups. The mean of duplicate measurements is depicted. Horizontal lines represent the geometric mean for each group. #P < 0.01; pair-wise comparisons were made using a non-Gaussian, unpaired, 2-tailed Mann-Whitney U test, corrected for multiple hypotheses testing using Bonferroni’s method. The VAF percentage is on a log10 scale.
To elucidate the clinical and biological implications of the persistence of gene mutations with high remission VAFs, we performed parallel clinical outcome analyses. When analyzed using the mutations with the lowest detected VAF for each patient, the geometric VAF means for patient subgroups categorized by relapse within less than 1 year, relapse after more than 1 year, or durable remission were 0.10%, 0.02%, and 0.005%, respectively, with a significant, 10-fold difference between the relapse before 1 year and durable remission groups (P < 0.01) (Figure 5B). This difference in depth-of-mutation persistence in the relapsed groups versus the durable remission group was even more distinct when comparing geometric means of the highest measured VAF for each group, excluding DNMT3A, which, as noted, persisted at high levels across all groups. Specifically, the geometric VAF means for patient subgroups categorized by relapse before 1 year, relapse after 1 year, and durable remission were 1.10%, 0.68%, and 0.02%, respectively, showing a significant, 10-fold difference between both relapse groups compared with the durable remission group (P <0.01 for both) (Figure 5C).
The degree of residual mutation burden correlates with RFS and OS in AML.
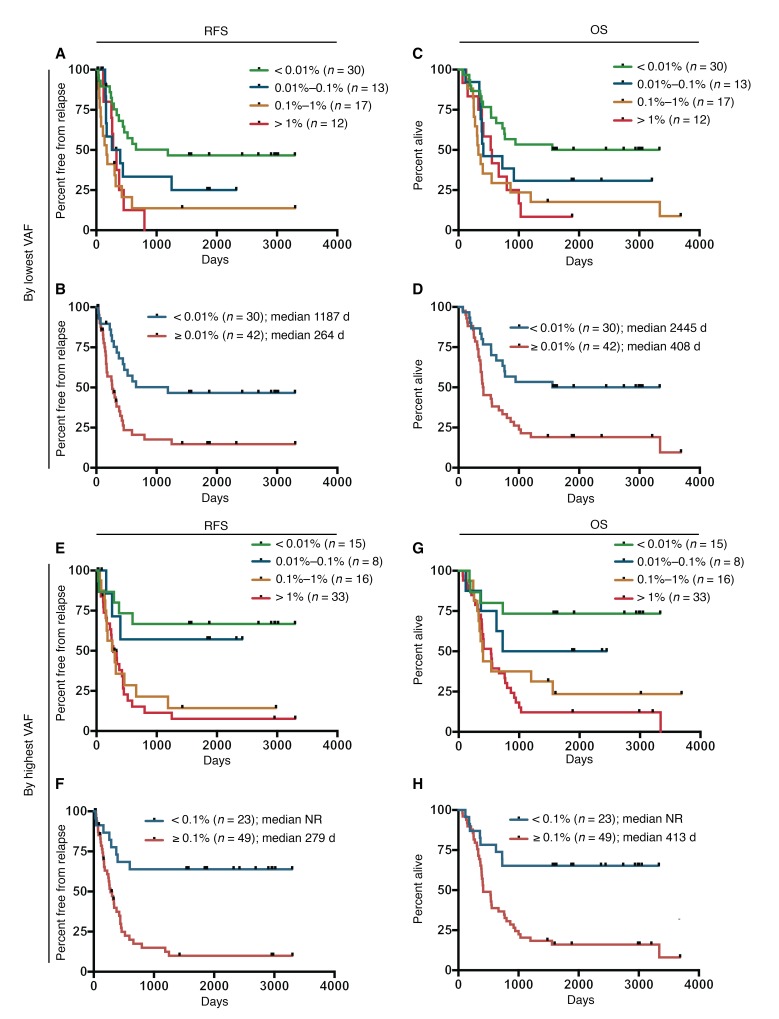
To further determine the extent to which absolute levels of residual mutation burden correlated with patient survival, we first divided patients into groups on the basis of logarithmic partitions (VAF <0.01%, 0.01%–0.1%, 0.1%–1%, and >1%) using the lowest VAF measurement for each. We identified a significant trend associating increasing median RFS with each 10-fold decrease in residual mutation burden for remission bone marrow VAFs of less than 0.01% (1,187 days, n = 30); 0.01%–0.1% (328 days, n = 13); 0.1%–1% (169 days, n = 17); and greater than 1% (306 days, n = 12) (P < 0.01) (Figure 6A). This trend was maintained in the same analysis evaluating OS for VAFs of less than 0.01% (2,445 days, n = 30); 0.01%–0.1% (413 days, n = 13); 0.1%–1% (330 days, n = 17); and greater than 1% (544 days, n = 12) (P < 0.01) (Figure 6C). On the basis of the difference in median RFS and OS between the group with a VAF of less than 0.01% and the other groups, patients were consolidated into groups of VAF of less than 0.01% and VAF of 0.01% or greater. Univariate analysis showed a significant and clinically relevant difference in median RFS for a VAF of less than 0.01% (1,187 days, n = 30) versus a VAF of 0.01% or greater (264 days, n = 42) (P < 0.01) (Figure 6B) and in median OS for a VAF of less than 0.01% (2,445 days, n = 30) versus a VAF of 0.01% or greater (408 days, n = 42) (P < 0.01) (Figure 6D). Five-year OS was 50% for a VAF of less than 0.01% versus 19% for a VAF of 0.01% or greater.
Figure 6. Kaplan-Meier analyses of remission duration and survival based on mutant gene VAF in CR detected with ddPCR.
(A and B) RFS using the lowest measured VAF per patient categorized by 10-fold decreases in VAF or by dichotomous cutoff, respectively. (C and D) OS using the lowest measured VAF per patient categorized by 10-fold decreases in VAF or by dichotomous cutoff, respectively. (E and F) RFS using the highest measured VAF (excluding DNMT3A) per patient categorized by 10-fold decreases in VAF or by dichotomous cutoff, respectively. (G and H) OS using the highest measured VAF (excluding DNMT3A) per patient categorized by 10-fold decreases in VAF or by dichotomous cutoff, respectively. P < 0.01, by log-rank test for trend (A, C, E, and G) and by log-rank Mantel-Cox test (B, D, F, and H).
Next, we performed the same analyses using the highest measured VAF for each patient. Partitioning patients into logarithmic groups revealed no significant differences for RFS or OS (P = 0.25 and 0.15, respectively). However, given the high-level persistence of DNMT3A mutations across the cohort as well as recent studies reporting a lack of prognostic value of DNMT3A mutation persistence in CR (21, 27), we repeated the analysis excluding DNMT3A. Using this refined approach, we identified a significant inverse correlation that associated VAFs of less than 0.01% and 0.01%–0.1% with substantially improved median RFS (not reached [NR], n = 15; and NR, n = 8, respectively) compared with VAFs of 0.1%–1% and greater than 1% (281 days, n = 16; and 279 days, n = 33, respectively) (P < 0.01) (Figure 6E). We observed a similar association with median OS for VAFs of less than 0.01% (NR, n = 15); 0.01%–0.1% (1,588 days, n = 8); 0.1%–1% (398 days, n = 16); and greater than 1% (534 days, n = 33) (P < 0.01) (Figure 6G). On the basis of the separation of median RFS between VAFs of less than 0.01% and 0.01%–0.1% versus VAFs of 0.1%–1% and greater than 1%, patients were consolidated into groups of VAFs of less than 0.1% and 0.1% or greater. Univariate analysis of the highest measured VAF in each sample had a significant and clinically relevant difference in median RFS for a VAF of less than 0.1% (NR, n = 23) versus a VAF of 0.1% or greater (279 days, n = 49) (P < 0.01) (Figure 6F) and in median OS for a VAF of less than 0.1% (NR, n = 23) versus a VAF of 0.1% or greater (413 days, n = 49) (P < 0.01) (Figure 6H). Five-year OS was 65% for a VAF of less than 0.1% versus 16% for a VAF of 0.1% or greater.
Finally, the same survival analyses were performed on the subset of patients with intermediate-risk cytogenetics (n = 50). The association of VAF of gene mutations with RFS and OS remained substantial and highly significant (Supplemental Figure 4). In addition, we evaluated the effect of residual mutation VAF on RFS in the subset of patients who received alloSCT in CR1 and on OS in patients who received alloSCT at any time (Supplemental Figure 5, A–D) as well as RFS in patients who did not receive alloSCT in CR1 and OS in patients who never received alloSCT (Supplemental Figure 5, E–H). The association of gene mutation VAF with RFS and OS remained highly significant when we measured the highest VAF, regardless of transplant status, and was significant or near-significant for RFS and OS when we measured the lowest VAF again, regardless of transplant status.
Discussion
This study represents to our knowledge the first broad quantitative measurement of recurrent gene mutations in any malignancy using high-sensitivity rare variant detection able to detect 1 in 50,000 mutated cells. The data provide several conclusions regarding the biology of residual AML and preleukemia in early CR. First, the vast majority of gene mutations identified in bulk pre-therapy AML blasts persist in initial CR marrows, including the 26% of mutations detected at very low levels of less than 0.1% VAF. Second, 36% of persistent gene mutations were detected at unexpectedly high levels of greater than 1% VAF (indicating mutation presence in at least 2% of marrow cells), particularly mutations in DNMT3A, TET2, ASXL1, RUNX1, and IDH1/2. Third, oligoclonal hematopoiesis was present in CR in most informative cases, suggesting the common scenario of persistence of both low-level residual leukemic cell populations and high-level pre- or nonleukemic clonal cell populations resistant to chemotherapy. Fourth, and importantly, low-level mutations detected in CR (0.003%–0.58% VAF) that were present in the dominant pretreatment blast populations or major subclones were also fully clonally represented at relapse, implying that the cell populations in which they reside, rather than numerically much larger, chemoresistant cell populations, are a common source of AML relapse. Fifth, DNMT3A mutations and frequent ASXL1 and TET2 mutations persist at high levels in initial CR marrows, even in cases in which durable remissions clearly demarcate a non- or preleukemic cellular compartment. These genes are therefore unlikely to be of value in clinical MRD applications. Finally, the allele frequency level of residual mutated genes detected in initial CR marrows is strongly and significantly associated with RFS and OS. Therefore, the much-improved level of detection of variant alleles and subclones harboring them achieved with ddPCR may provide more clinically meaningful risk stratification than that afforded by current genomic MRD platforms.
Previous studies into the clinical relevance of genomic MRD detection have focused either on high-sensitivity detection of single mutated genes, such as qPCR of NPM1 mutations, or less sensitive surveys of large gene panels, such as targeted NGS approaches (11–15, 21, 22). The application of competitive allele-specific ddPCR to MRD detection in this study demonstrates the technical feasibility of detecting single nucleotide variants and insertion-deletion mutations in the vast majority of recurrently mutated genes in AML in clinical samples at levels comparable to or exceeding those detected with qPCR. Furthermore, the use of genomic DNA (gDNA) rather than cDNA favored in established qPCR assays of MRD allows a precise comparison of the relative abundance of mutant alleles of a measured gene and hence the proportion of cells containing those mutations.
The results of this broad, ultrasensitive measurement of residual mutant alleles in AML indicate that “complete remission” is a misnomer, since induction chemotherapy failed to clear the bone marrow of leukemia-associated gene mutations in almost all AML patients studied here. Conversely, not all mutant alleles detected should be considered evidence of residual disease but rather of residual clonal aberrant hematopoiesis of unclear clinical significance. In particular, DNMT3A mutations, which typically persisted at high levels in remission and obscured the prognostic value of VAF levels of other mutated genes when analyzed in aggregate, offer no clear value for MRD monitoring. The individual prognostic value of other genes such as TET2, ASXL1, RUNX1, and IDH1/2 that occasionally persist at high levels in remission will need to be refined in much larger patient cohorts.
The uncertainty regarding the prognostic value of mutations persisting at high levels and the degree to which they mark leukemic or preleukemic cell populations is underscored by the key finding of this study: the cellular reservoir containing mutations with the lowest VAF is fully clonally represented at relapse. This observation, made possible by high-sensitivity detection of multiple mutated genes within each specimen, argues against stochastic reacquisition of mutations in more abundant cell populations marked by high VAF mutations as the source of AML relapse and rather indicates a persistence of cells competent to generate leukemia possibly simply through regrowth.
The nature of chemoresistant, high-level, persistent mutated clones remains unclear. Do these represent preleukemic stem cells or progenitors that have acquired some but not all gene mutations of the dominant pre-therapy AML clone and lack the potential to relapse (28, 29), or are they remnant bystanders of clonal hematopoiesis of indeterminate potential (30–33)? Additional genomic and functional studies of various bone marrow cellular compartments will be necessary to determine the identity of these high-level cell populations and, more importantly, of the low-level cell populations implicated in AML relapse. A critical clinical need to better characterize these residual cells is underscored by this study: patients who achieved a reduction of VAF to less than 0.01% for all measured genes per remission (apart from DNMT3A) were far less likely to relapse by 3 years than those who did not (33% vs. 81%), but nevertheless, a third of these patients relapsed. Similarly, while patients who achieved a reduction of VAF to less than 0.01% in any 1 measured gene had a superior RFS at 3 years compared with those who did not (50% vs. 82%), half of these patients still relapsed. A deeper understanding of the nature of these resistant populations may lead to more effective strategies to maintain remission and eliminate the cell populations responsible for relapse.
Although this is the first study to our knowledge to broadly interrogate CR marrows for low-level residual mutations, there are some limitations. First, most, but not all, mutations identified in pre-therapy samples were measured in CR, raising the possibility that for a given case, there may be additional discordant VAFs and therefore an even higher overall frequency of oligoclonal hematopoiesis than we report. Next, we focused on detecting mutations in CR that were near or fully clonal in pre-therapy disease and so cannot comment on minor subclonal mutations present at diagnosis and their behavior in remission and at relapse. Finally, while the number of cases studied demonstrated a significant association of residual mutation VAF with survival, it was insufficient to perform multivariate survival analyses; additional studies with larger patient cohorts will be necessary to demonstrate an independent effect of ultrasensitive mutation detection on survival and to assess the effect of specific mutated genes on survival.
Methods
Patients.
Between March 2005 and June 2011, 223 patients with AML (all non-M3) were enrolled in a translational study at the University of Michigan Comprehensive Cancer Center. Of the 223 patients, 72 had peripheral blood and bone marrow samples taken shortly before initiating induction chemotherapy and again after achieving morphological CR. Specimens were also collected at the time of relapse for 20 of these 72 patients. Buccal swabs were obtained from all patients upon enrollment in the study to provide a source of normal DNA.
Cell isolation and preparation of sample DNA.
Peripheral blood or bone marrow mononuclear cells from AML patients were isolated using Ficoll gradient separation followed by cryopreservation. To eliminate the influence of nonmalignant cell contamination on VAF measurement in pretreatment and relapse samples, thawed AML cells were highly enriched for blasts using a combination of magnetic column and fluorescence-activated cell sorting, as previously described, to achieve a 90% or greater pure blast population, confirmed by cytospin examination (34). DNA was extracted from this highly purified AML blast preparation and from paired buccal mucosal cell preparations, as previously described (34). For CR specimens, DNA was extracted directly from thawed bone marrow mononuclear cells without additional enrichment.
Genomic MRD target identification in pretreatment samples.
Sanger sequencing was performed on pretreatment gDNA isolated from highly pure blast populations for commonly mutated genes in AML (35, 36), including all coding exons and adjacent intronic sequences of CEBPA, ASXL1, TET2, DNMT3A, RUNX1, and BCORL1, along with exon 12 of NPM1, exons 13–15 and 20 of FLT3, exons 2–10 of TP53, exons 2–3 of KRAS and NRAS, exon 2 of IDH1, and exon 4 of IDH2. For the 50 cases with either fewer than 2 mutations identified in these 13 genes or for which matched relapse samples were available, an additional 36 recurrently mutated genes in AML were sequenced using droplet-based amplicon library preparation (RainDance Technologies) and paired-end NGS on a MiSeq sequencer using MiSeq Reagent Kits, v2 (both from Illumina). The resulting FASTQ files underwent quality filtering, end trimming, reference genome alignment, and variant calling using NextGENe, version 2.4.1 (SoftGenetics). The lower limit of detection using this method was approximately 3% VAF. By including this NGS panel, 2 or more and 1 or more gene mutations were identified in 74% (53 of 72) and 90% (65 of 72) of all patients, respectively. In addition, for the 20 patients for whom relapse samples were available, DNA from pretreatment and relapse specimens underwent NGS using this same method. For the pretreatment samples, 19 patients in whom 1 or fewer gene mutations were identified despite both Sanger and NGS panel screening, we performed WES with an average read depth of 110× on paired tumor and normal samples and identified more than 5 somatic mutations per patient (see Supplemental Methods for full details on library preparation and the bioinformatics variant calling pipeline). Aside from 1 mutation of TET2 and another of SETD2, no recurrent gene mutations were identified, and we therefore selected 2–3 target mutations per patient for ddPCR analysis in CR samples using the following criteria: (a) a presence in major clones defined by a VAF of 35% or greater and (b) a prediction to be damaging to the mutated gene’s protein product either by the presence of a frameshifting indel or by analysis with dbNSFP, version 2.4 (37). All mutation calls from Sanger sequencing, the 49-gene NGS panel, and WES of VAFs of 15% or greater were confirmed by Sanger sequencing using unamplified tumor DNA and paired patient buccal DNA as templates. The mutation detection methods used for pre-therapy, CR, and relapse samples are summarized in Supplemental Figure 6. WES data files have been deposited in the dbGaP public database (https://www.ncbi.nlm.nih.gov/gap; accession number phs001408.v1.p1).
ddPCR of MRD targets in CR samples.
Allele-specific fluorescent oligonucleotide probes using minor groove binding (Life Technologies, Thermo Fisher Scientific) or locked nucleic acid (Integrated DNA Technologies [IDT]) chemistries were developed for each mutation using Primer Express (Life Technologies, Thermo Fisher Scientific) and Oligoanalyzer (IDT), respectively. WT and mutant allele–specific probes (5 μM) were combined with allele-independent forward and reverse PCR primers (20 μM) to generate a probe mix. Each probe mix was optimized for allele discrimination using qPCR to derive an optimal annealing temperature (TA) as follows: 2× TaqMan Genotyping Master Mix (Life Technologies, Thermo Fisher Scientific) was combined with 40× Probe Mix and either 30 ng mutant gDNA derived from a patient’s sample harboring the mutation of interest or homozygous WT gDNA derived from the Ly18 lymphoma cell line and then amplified with a Bio-Rad CFX96 Real-Time System using a TA gradient for 45 cycles and analyzed with Bio-Rad CFX Manager. The temperature at which both WT and mutant probes were amplified in the presence of DNA with a heterozygous mutation but at which the mutant probe was not amplified in the presence of WT-only DNA was selected as the optimal TA to carry forward for ddPCR.
ddPCR was then performed in duplicate on DNA from 72 samples extracted from Ficoll-enriched bone marrow aspirates obtained in CR to determine the VAF for all mutations. Each ddPCR run also included a positive control derived from a pre-therapy patient’s sample harboring the mutation of interest as well as a WT-only control derived from the Ly18 cell line and was performed as follows: a master mix containing 2× TaqMan Genotyping Master Mix, 40× Probe Mix, 25× Droplet Stabilizer (RainDance Technologies), and 50× Restriction Endonuclease (HaeIII or MseI; New England BioLabs) (38) was added to separate wells containing 500 ng gDNA, for a total reaction volume of 25 μl, and then incubated at room temperature for 15 minutes. Samples were then pipetted into a RainDrop Source Chip (RainDance Technologies), and 3.5–4.5 million droplets were generated with the RainDrop Source machine. The resulting emulsion was amplified using the previously derived optimal TA with the following settings: 95°C for 10 minutes, and then 45 cycles at 95°C for 15 seconds and TA for 60 seconds, and then 98°C for 10 minutes. Endpoint fluorescence was then measured with a RainDrop Sense machine and analyzed with RainDrop Analyst software. The WT and mutant signals of each pre-therapy sample were used to draw positive gates to then analyze VAFs of the diagnosis samples and WT-only controls. Of note, for mutations in which both ddPCR and NGS measurements of pretreatment-positive control specimens were available, the resulting VAFs were highly concordant (95% of measurements with less than a 5% difference in VAF percentage), with the exception of FLT3–ITD mutations. NGS variant calling of FLT3-ITD underestimated the VAF by approximately 2-fold compared with ddPCR (data not shown). Because of this discrepancy, NGS was used to quantify the VAF of all mutations in paired pre-therapy and relapse samples, except for FLT3-ITD, for which the values represent ddPCR measurements.
The general level of blank (LOB) across all probes was 0.0082% VAF and 0.0009% VAF for NPM1 probes in particular. Because the LOB could not be determined for probe sets used in a single case, the presence of a mutant signal was conservatively defined as the presence of 5 or more droplets in the mutant gate and a 5-fold higher number of mutant droplets in patients’ samples compared with the concurrently assayed WT control. Additional details on the assay parameters and LOB for all probe sets used in 2 or more cases are listed in Supplemental Table 4.
Statistics.
Pair-wise assessment of differences in median VAF levels between various relapse groups was performed using the Mann-Whitney U test, corrected for multiple hypotheses testing with Bonferroni’s method. OS was defined as the time between AML diagnosis and the patient’s death. RFS was defined as the time between achievement of CR (documented by bone marrow examination and review of complete blood counts) and subsequent relapse. For patients still alive or relapse free, respectively, at the time of data analysis, the date of censoring was June 1, 2015. The log-rank test for trend was used to evaluate the significance of the trend of Kaplan-Meier estimates of improved RFS and OS with sequentially decreasing orders of magnitude of measured VAF. Univariate analyses were based on Kaplan-Meier estimates of survivor functions. Median survival times were estimated directly from the survivor function estimates. Significance levels for group-wise comparisons in the univariate analyses assessed whether the hazard ratio between groups differed from 1 in a Cox proportional hazards model. All statistical tests were 2 tailed, and a P value of less than 0.05 was considered significant.
Study approval.
This study was approved by the IRB of the University of Michigan (IRBMED 2004–1022), and written informed consent was obtained from all patients prior to their enrollment in the study.
Author contributions
BP, ALJ, and QK performed the laboratory research. MT and ADR provided reagents and reviewed the manuscript. BP and SNM conceived the study, analyzed the data, and wrote the manuscript. SNM supervised the work.
Supplementary Material
Acknowledgments
SNM is a Scholar of the Leukemia and Lymphoma Society of America and is supported by the Hartman MDS fund and NIH grant R01-CA171972. BP is a Scholar of the American Society of Hematology and is supported by the Elsa U. Pardee Foundation.
Version 1. 08/21/2017
Electronic publication
Version 2. 09/01/2017
Print issue publication
Footnotes
ALJ’s present address is: Department of Radiation Oncology, University of Alabama Birmingham, Birmingham, Alabama, USA.
ADR’s present address is: Department of Gastroenterology, Hepatology and Nutrition, University of Texas M.D. Anderson Cancer Center, Houston, Texas, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(9):3484–3495.https://doi.org/10.1172/JCI91964.
Contributor Information
Brian Parkin, Email: bparkin@med.umich.edu.
Angelina Londoño-Joshi, Email: angelinalondono@gmail.com.
Qing Kang, Email: qkang@med.umich.edu.
Muneesh Tewari, Email: mtewari@med.umich.edu.
References
- 1.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkin B, et al. Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia. Blood. 2013;121(2):369–377. doi: 10.1182/blood-2012-04-427039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kern W, Voskova D, Schoch C, Hiddemann W, Schnittger S, Haferlach T. Determination of relapse risk based on assessment of minimal residual disease during complete remission by multiparameter flow cytometry in unselected patients with acute myeloid leukemia. Blood. 2004;104(10):3078–3085. doi: 10.1182/blood-2004-03-1036. [DOI] [PubMed] [Google Scholar]
- 4.Freeman SD, et al. Prognostic relevance of treatment response measured by flow cytometric residual disease detection in older patients with acute myeloid leukemia. J Clin Oncol. 2013;31(32):4123–4131. doi: 10.1200/JCO.2013.49.1753. [DOI] [PubMed] [Google Scholar]
- 5.Shlush LI, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111(7):2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jourdan E, et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood. 2013;121(12):2213–2223. doi: 10.1182/blood-2012-10-462879. [DOI] [PubMed] [Google Scholar]
- 8.Yin JA, O’Brien MA, Hills RK, Daly SB, Wheatley K, Burnett AK. Minimal residual disease monitoring by quantitative RT-PCR in core binding factor AML allows risk stratification and predicts relapse: results of the United Kingdom MRC AML-15 trial. Blood. 2012;120(14):2826–2835. doi: 10.1182/blood-2012-06-435669. [DOI] [PubMed] [Google Scholar]
- 9.Corbacioglu A, et al. Prognostic impact of minimal residual disease in CBFB-MYH11-positive acute myeloid leukemia. J Clin Oncol. 2010;28(23):3724–3729. doi: 10.1200/JCO.2010.28.6468. [DOI] [PubMed] [Google Scholar]
- 10.Grimwade D, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol. 2009;27(22):3650–3658. doi: 10.1200/JCO.2008.20.1533. [DOI] [PubMed] [Google Scholar]
- 11.Ivey A, et al. Assessment of Minimal Residual Disease in Standard-Risk AML. N Engl J Med. 2016;374(5):422–433. doi: 10.1056/NEJMoa1507471. [DOI] [PubMed] [Google Scholar]
- 12.Klco JM, et al. Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia. JAMA. 2015;314(8):811–822. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohlmann A, et al. Monitoring of residual disease by next-generation deep-sequencing of RUNX1 mutations can identify acute myeloid leukemia patients with resistant disease. Leukemia. 2014;28(1):129–137. doi: 10.1038/leu.2013.239. [DOI] [PubMed] [Google Scholar]
- 14.Shayegi N, et al. The level of residual disease based on mutant NPM1 is an independent prognostic factor for relapse and survival in AML. Blood. 2013;122(1):83–92. doi: 10.1182/blood-2012-10-461749. [DOI] [PubMed] [Google Scholar]
- 15.Krönke J, et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol. 2011;29(19):2709–2716. doi: 10.1200/JCO.2011.35.0371. [DOI] [PubMed] [Google Scholar]
- 16.Schnittger S, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114(11):2220–2231. doi: 10.1182/blood-2009-03-213389. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, et al. Pre- and post-transplant quantification of measurable (‘minimal’) residual disease via multiparameter flow cytometry in adult acute myeloid leukemia. Leukemia. 2016;30(7):1456–1464. doi: 10.1038/leu.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terwijn M, et al. High prognostic impact of flow cytometric minimal residual disease detection in acute myeloid leukemia: data from the HOVON/SAKK AML 42A study. J Clin Oncol. 2013;31(31):3889–3897. doi: 10.1200/JCO.2012.45.9628. [DOI] [PubMed] [Google Scholar]
- 19.Walter RB, et al. Impact of pretransplantation minimal residual disease, as detected by multiparametric flow cytometry, on outcome of myeloablative hematopoietic cell transplantation for acute myeloid leukemia. J Clin Oncol. 2011;29(9):1190–1197. doi: 10.1200/JCO.2010.31.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimwade D, Freeman SD. Defining minimal residual disease in acute myeloid leukemia: which platforms are ready for “prime time”? Blood. 2014;124(23):3345–3355. doi: 10.1182/blood-2014-05-577593. [DOI] [PubMed] [Google Scholar]
- 21.Debarri H, et al. IDH1/2 but not DNMT3A mutations are suitable targets for minimal residual disease monitoring in acute myeloid leukemia patients: a study by the Acute Leukemia French Association. Oncotarget. 2015;6(39):42345–42353. doi: 10.18632/oncotarget.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnittger S, et al. DNMT3A is a Powerful Follow-up Marker in NPM1 mutated AML. Blood. 2014;124(21):122 [Google Scholar]
- 23.Drandi D, et al. Minimal Residual Disease Detection by Droplet Digital PCR in Multiple Myeloma, Mantle Cell Lymphoma, and Follicular Lymphoma: A Comparison with Real-Time PCR. J Mol Diagn. 2015;17(6):652–660. doi: 10.1016/j.jmoldx.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Laurent-Puig P, et al. Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin Cancer Res. 2015;21(5):1087–1097. doi: 10.1158/1078-0432.CCR-14-0983. [DOI] [PubMed] [Google Scholar]
- 25.Hindson CM, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10(10):1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WW, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatnagar B, et al. Persistence of DNMT3A R882 mutations during remission does not adversely affect outcomes of patients with acute myeloid leukaemia. Br J Haematol. 2016;175(2):226–236. doi: 10.1111/bjh.14254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jan M, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med. 2012;4(149):149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong TN, et al. Rapid expansion of preexisting nonleukemic hematopoietic clones frequently follows induction therapy for de novo AML. Blood. 2016;127(7):893–897. doi: 10.1182/blood-2015-10-677021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkin B, et al. Acquired genomic copy number aberrations and survival in adult acute myelogenous leukemia. Blood. 2010;116(23):4958–4967. doi: 10.1182/blood-2010-01-266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research Network, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papaemmanuil E, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34(9):E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang Q, Parkin B, Giraldez MD, Tewari M. Mutant DNA quantification by digital PCR can be confounded by heating during DNA fragmentation. BioTechniques. 2016;60(4):175–185. doi: 10.2144/000114401. [DOI] [PubMed] [Google Scholar]
- 39.Parkin B, Ouillette P, Yildiz M, Saiya-Cork K, Shedden K, Malek SN. Integrated genomic profiling, therapy response, and survival in adult acute myelogenous leukemia. Clin Cancer Res. 2015;21(9):2045–2056. doi: 10.1158/1078-0432.CCR-14-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.