Abstract
Varicella zoster virus (VZV) typically causes chickenpox upon primary infection. In rare cases, VZV can give rise to life-threatening disease in otherwise healthy people, but the immunological basis for this remains unexplained. We report 4 cases of acute severe VZV infection affecting the central nervous system or the lungs in unrelated, otherwise healthy children who are heterozygous for rare missense mutations in POLR3A (one patient), POLR3C (one patient), or both (two patients). POLR3A and POLR3C encode subunits of RNA polymerase III. Leukocytes from all 4 patients tested exhibited poor IFN induction in response to synthetic or VZV-derived DNA. Moreover, leukocytes from 3 of the patients displayed defective IFN production upon VZV infection and reduced control of VZV replication. These phenotypes were rescued by transduction with relevant WT alleles. This work demonstrates that monogenic or digenic POLR3A and POLR3C deficiencies confer increased susceptibility to severe VZV disease in otherwise healthy children, providing evidence for an essential role of a DNA sensor in human immunity.
Keywords: Genetics, Infectious disease
Keywords: Innate immunity
Introduction
Varicella zoster virus (VZV) is a human pathogenic alphaherpesvirus, and the causative agent of varicella (chickenpox) during primary infection and zoster (shingles) during reactivation from latency in sensory ganglia (1, 2). In rare cases, VZV primary infection or reactivation can cause more severe diseases, including pneumonitis and CNS disease, manifesting as cerebellitis, encephalitis, or stroke (1). The frequencies of VZV CNS infection and pneumonia during primary infection in children are about 0.02% and 0.25%, respectively (3), and certain countries have introduced a VZV vaccine with the Oka strain, resulting in reduced frequency of VZV infection cases (4). Antiviral therapy with acyclovir is used for treatment of VZV encephalitis, pneumonitis, and certain cases of varicella in pregnant women and patients with a known immunodeficiency. However, despite treatment with acyclovir, the incidence and severity of sequelae following severe VZV infection are considerable (3).
The VZV isolates exhibit little sequence variability, which does not correlate with virulence (5), suggesting that defective host immunity is a major determinant of disease severity, rather than differences between virus strains. Several different primary immunodeficiencies (PIDs) within both the innate and adaptive immune system have previously been recognized to predispose to infection with VZV as well as a range of other pathogens. These include disseminated VZV infection in children with SCID or NK cell defect (6–8). More recently, additional conditions conferring increased susceptibility to VZV infection have been described, including loss-of-function mutations in GATA2, DOCK2, DOCK8, IFNGR1 and TYK2 (9–13). Inherited GATA2 haploinsufficiency causes NK cell deficiency, and an NK cell–deficient patient reported with severe VZV infection was subsequently found to have GATA2 deficiency (8, 14). Patients with any of the above underlying conditions are vulnerable to a wide range of severe infections. Otherwise, the cellular basis of severe VZV infection in patients with those conditions remains largely unknown. Moreover, severe VZV disease is often striking in otherwise healthy children and adolescents, in whom a specific impairment of anti-VZV immunity might be involved.
In recent years, it has emerged that single-gene inborn errors of innate or cell-intrinsic immunity can underlie enhanced susceptibility to specific (viral) infections in otherwise healthy individuals (15, 16). A striking example is that mutations in genes controlling Toll-like receptor (TLR) 3-dependent type I and III IFN–mediated immunity confer susceptibility to herpes simplex encephalitis (HSE) (17–22). Notably, these genetic defects often appear to display incomplete penetrance, in which case they can be designated as monogenic but not Mendelian (18, 20, 23). Common to the identified genetic defects in the TLR3 pathway is that they lead to reduced type I and III IFN induction in patients cells after HSV-1 infection or stimulation through the TLR3 pathway ex vivo. In contrast, no PIDs leading to selectively enhanced susceptibility to severe VZV infection have yet been identified. In some patients with PIDs conferring various infectious phenotypes, including severe VZV infection, impaired type I IFN production (autosomal recessive [AR] DOCK2 deficiency) or responses (AR TYK2 deficiency) have been observed among other immunological phenotypes. We hypothesize that severe VZV disease in some otherwise healthy individuals may be caused by monogenic inborn errors of IFN immunity not necessarily displaying complete clinical penetrance.
The innate immune system utilizes pattern recognition receptors (PRRs) to detect pathogen-associated molecular patterns (PAMPs) to mount protective immune responses, including production of cytokines and IFNs (24). Different classes of PRRs are involved in recognition of virus infections, including membrane-associated TLRs; cytosolic retinoic acid–inducible gene 1–like (RIG-I–like) receptors, which sense RNA; and DNA sensors (24). Each of these classes of PRRs stimulates production of IFNs, which exhibit antiviral activity through their ability to induce IFN-stimulated genes (ISGs). With respect to DNA sensors, TLR9 detects unmethylated DNA, RNA polymerase III (POL III) recognizes AT-rich DNA, while gamma-interferon-inducible protein 16 (IFI16) and cyclic GMP-AMP synthase (cGAS) sense double-stranded DNA in a sequence-independent manner (25–29). Based on the current knowledge on the IFN induction pathways, we searched for gene mutations that may underlie susceptibility to severe VZV infection in whole exome sequencing (WES) data of 21 unrelated children with severe acute VZV infection. Here we describe 4 children with severe clinical manifestations of VZV infection who carried heterozygous missense mutations in POLR3A and/or POLR3C encoding two subunits of the intracellular DNA sensor POL III.
Results
Identification of heterozygous mutations in POLR3A and POLR3C in 4 children with acute severe VZV infection.
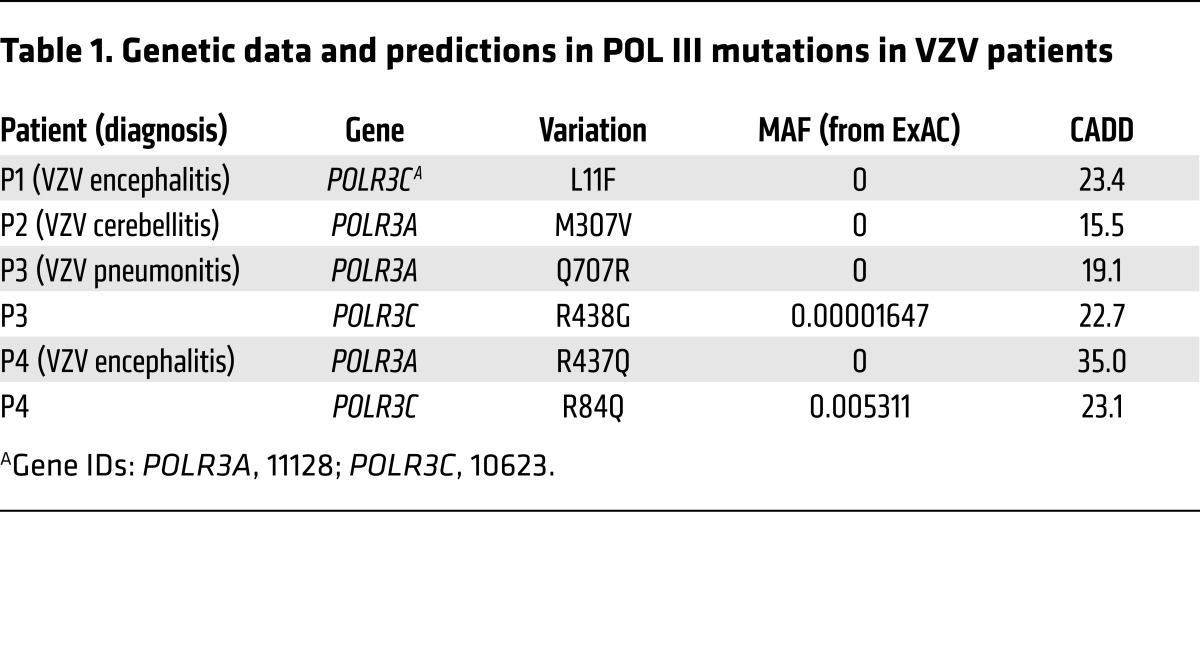
Genomic DNA from 21 children with severe acute VZV infection was subjected to WES. Due to the low incidence of severe acute VZV infection, we decided to first focus on novel or very rare mutations (minor allele frequency [MAF] <0.0001 in the ExAC database (http://exac.broadinstitute.org/), which contains WES data of 60,706 individuals; as well as the in-house WES database of the Human Genetics of Infectious Diseases [HGID] laboratory, which contains WES data of 3,055 individuals with diverse infectious diseases). We searched for rare nonsynonymous (including missense, stop loss or stop gain, start loss, in-frame or frameshift insertions or deletions) and splice site mutations, in a comprehensive list of genes known to be involved in IFN immunity (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI92280DS1). This search led to the identification of 3 heterozygous missense mutations in POLR3A (M307V, Q707R, R437Q) in 3 children (P2, P3, P4), respectively; and 2 heterozygous missense mutations in POLR3C (L11F, R438G) in 2 children (P1, P3), respectively. Interestingly, P3 is heterozygous for both POLR3A and POLR3C missense mutations. We therefore further looked for POLR3A or POLR3C variations with a MAF <0.01 in the ExAC database and <0.005 in the 1000 Genomes database (http://www.internationalgenome.org/), and found that P4 is also heterozygous for an R84Q variation in POLR3C. Therefore, a total of 4 of 21 patients carry heterozygous missense mutations in POLR3A and/or POLR3C encoding two subunits of the intracellular DNA sensor POL III (Table 1). Using the same criteria, no other immunity genes, including genes encoding other subunits of POL III, were found to be mutated in more than two patients in the cohort; there was a significant enrichment of mutations in the exomes of the severe VZV infection cohort compared with (i) exomes of patients in the HGID database with infectious diseases other than severe VZV infection or (ii) exomes in the ExAC database. Importantly, mutations in POLR3A or POLR3C were significantly enriched in the severe VZV infection cohort (VZV cohort vs. control cohort consisting of 1,220 individuals of the HGID cohorts with non-viral infections and 2,504 individuals from the 1000 Genomes database, either under a monogenic inheritance model [ethnic origin–adjusted P = 0.002 and 0.0046 for POLR3A and POLR3C, respectively] or under a digenic inheritance model [ethnic origin–adjusted P = 0.0012]; Supplemental Table 2).
Table 1. Genetic data and predictions in POL III mutations in VZV patients.
Monogenic and digenic POLR3A, POLR3C mutations may underlie severe VZV infection with incomplete clinical penetrance.
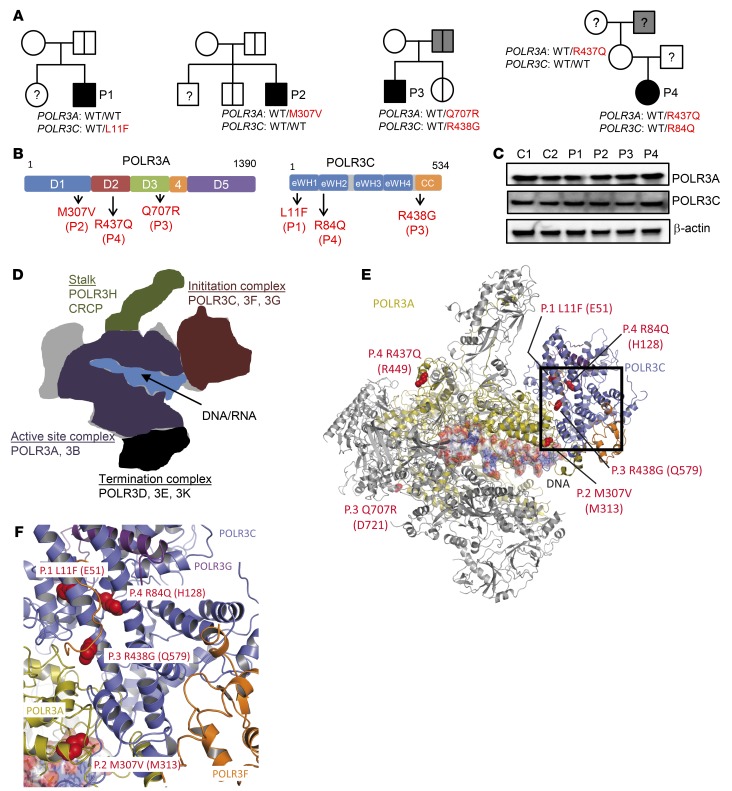
The 4 patients, who did not have a history of unusually severe infections, were of European ancestry, and no consanguinity was reported. P1 and P2, two Belgian boys, were aged 3 and 5 years, respectively when they were seen in hospital due to VZV encephalitis (P1) and cerebellitis (P2). P3 and P4, one boy and one girl, born to French and German parents, respectively, were aged 5 and 11 years when they developed VZV pneumonitis (P3) and encephalitis (P4). A summary of the clinical infection history of the patients and available whole blood immunophenotyping is given in Supplemental Tables 3 and 4. In the patients exomes, no mutations were found in the genes encoding proteins in the TLR3 pathway; other DNA sensing pathways, including cGAS, IFI16, and STING; or other proteins in the signaling pathways leading to IFN production. No mutations were found in other known PID genes. Sanger sequencing confirmed the POLR3A and POLR3C mutations (Supplemental Figure 1), and familial segregation suggested incomplete penetrance in the case of monogenic POLR3A or POLR3C mutations (P1, P2) (Figure 1A). One of the two siblings of P2, a non-identical twin brother, carried the same POLR3A M307V mutation as P2 and experienced chickenpox twice. No other mutation carriers from the two families have experienced severe infection due to VZV or other viruses. Higher but still incomplete clinical penetrance may apply in the case of digenic POLR3A and POLR3C mutations (P3 and P4 families), as the father of P3, who carried the same heterozygous mutations in POLR3A and POLR3C as P3, had cytomegalovirus encephalitis at the age of 20 years. No clinical varicella has been reported for the father of P3. No biological material was available from the father of P4, while her mother carried the same heterozygous POLR3A R437Q mutation as P4. The father of P4’s mother, from whom no biological material was available, experienced suspected viral meningitis at the age of 16 years. Figure 1B depicts the localization of the mutations in the POLR3A and POLR3C proteins, which are part of the active site and initiation subcomplex of POL III, respectively (30, 31). All mutations lie within highly conserved regions of the proteins (Supplemental Figure 2). All the identified mutations have Combined Annotation-Dependent Depletion (CADD) scores above the predicted Mutation Significance Cutoffs (MSC) of 95% confidence (Supplemental Figure 3) (32, 33). The genes themselves are highly conserved and subject to purifying selection, with low gene damage index and purification coefficient (34). Immunoblotting of peripheral blood mononuclear cell (PBMC) lysates from P1–P4 for levels of POLR3A and POLR3C revealed that these proteins were expressed at levels comparable to those in healthy controls (Figure 1C). Collectively, these findings suggest that heterozygous mutations in the POLR3A and POLR3C genes may underlie life-threatening VZV disease, and the inheritance mode may be monogenic (P1, P2) or digenic (P3, P4).
Figure 1. Heterozygous mutations in POLR3A and POLR3C in patients with acute severe VZV infection.
(A) Pedigrees for the families with allele segregation. Family members heterozygous for the same mutations as that of the probands are indicated by a vertical line. Gray shading: individuals affected by a relevant clinical phenotype, although not precisely the same as what affected the probands. (B) Illustration of the localization of the identified mutations in POLR3A and POLR3C. eWH, extended winged helix; CC, coiled coil. (C) PBMC lysates from P1–P4 and two healthy controls were analyzed for protein levels of POLR3A, POLR3C, and β-actin by immunoblotting. (D) Schematic representation of POL III highlighting the subunits in the different subcomplexes. (E) Cryo-EM structure of POL III with the identified mutations highlighted in red. (F) Close-up image of the POLR3A-POLR3C interface harboring 4 of the 6 identified mutations.
Structural modeling of POLR3A and POLR3C mutations.
POL III is a multi-subunit protein complex consisting of 17 subunits, which are organized into different subcomplexes, involved in DNA/RNA binding, initiation of transcription, enzymatic activity, and termination of transcription (Figure 1D) (30). When we modeled the mutations from each of the 4 patients onto the cryo-electron microscopy structure of Saccharomyces cerevisiae POL III, we found that two of the three POLR3A mutations were localized in regions exposed to the surface (P3, Q707R; P4, R437Q) (Figure 1E), while the third POLR3A mutation (P2, M307V) and all of the 3 POLR3C mutations localized to buried parts of the proteins at or close to the POLR3A-POLR3C interface (P2, M307V) (Figure 1F and Supplemental Table 5). To test the impact of the mutations in POLR3C on the biophysical properties of the proteins, we expressed the WT and mutant proteins as His-tagged proteins in bacteria. This was not feasible with POLR3A, since this protein is poorly expressed from bacteria. All POLR3C forms could be expressed, and WT and R438G (P3) were precipitated with cobalt affinity resin (Supplemental Figure 4). By contrast, the L11F (P1) and R84Q (P4) mutants failed to bind the resin. This may suggest that the L11F and R84Q mutants have altered biophysical properties, the nature of which requires further investigation.
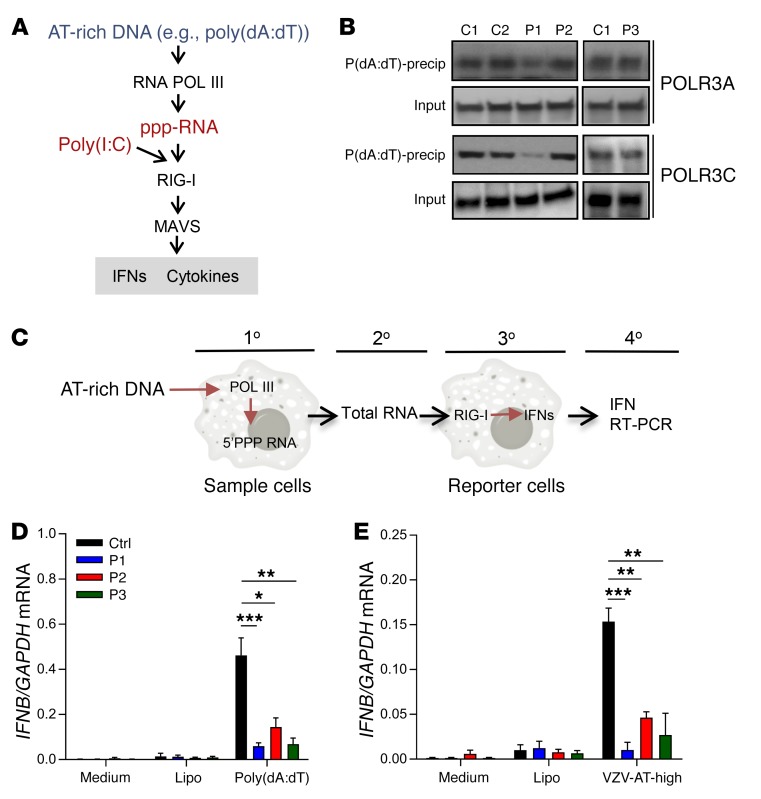
Induction of IFNs by AT-rich DNA in human PBMCs is POL III–dependent.
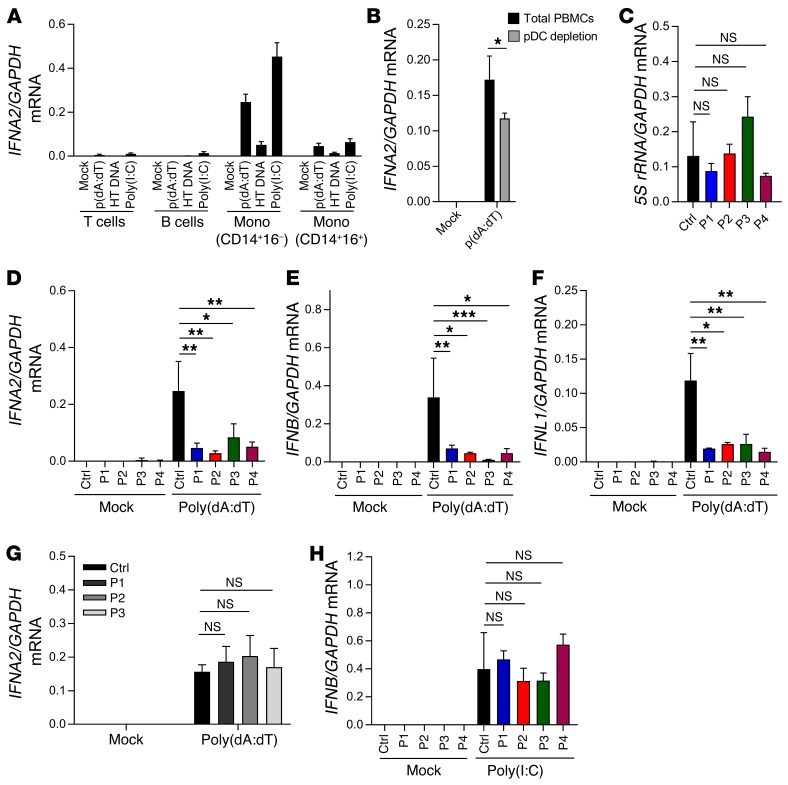
In a first set of experiments, we aimed to evaluate whether stimulation by AT-rich DNA in PBMCs induces IFN responses in a POL III–dependent manner. When PBMCs from healthy controls were incubated in the presence or absence of the POL III inhibitor ML60218, we observed that the inhibitor was able to block IFNA2 and IFNB expression induced by poly(dA:dT) but not by the TLR7/8 agonist R848 (Supplemental Figure 5). Stimulation of sorted PBMC subpopulations revealed that CD14+CD16– monocytes induced the highest levels of IFNA2 in response to poly(dA:dT) (Figure 2A). The frequency of plasmacytoid dendritic cells (pDCs) in the thawed PBMC populations was low (0.2 %), so we evaluated the impact of pDCs on poly(dA:dT)-induced IFN production by depleting this cell type prior to stimulation. The pDC-depleted PBMCs still induced robust, but significantly reduced, expression of IFNA2 (Figure 2B). These data demonstrate that poly(dA:dT) induces IFN expression through the POL III pathway in PBMCs and stimulates specific cell populations, most notably classical monocytes.
Figure 2. Impaired DNA-stimulated induction of IFNs in PBMCs from patients with mutations in POL III genes.
(A and B) PBMCs from 3 healthy controls were sorted into the indicated cell types and transfected with poly(dA:dT) (2 μg/ml), herring testes (HT) DNA (2 μg/ml), or poly(I:C) (2 μg/ml). Total RNA was harvested 6 hours later, and levels of IFNA2 mRNA were determined by RT-qPCR. Mono, monocytes. (C) RNA from untreated PBMCs from P1–P4 and controls was analyzed for levels of 5S rRNA. (D–H) Levels of IFNA2, IFNB, and IFNL1 mRNA were determined by RT-qPCR, in total PBMC RNA harvested 6 hours after stimulation, in PBMCs from P1–P4 and a panel of 4 healthy controls were transfected with (E and F) poly(dA:dT) (2 μg/ml) or (H) poly(I:C) (4 μg/ml); (G) PBMCs from 3 VZV patients not carrying POLR3 mutations and 4 healthy controls were transfected with poly(dA:dT) (2 μg/ml). For all PCR data, the levels of the mRNA of interest were normalized to GAPDH, and data are presented as mean ± SD of 3 replicates from 3 independent experiments. Ctrl, controls. Average of 4 sex- and age-matched controls. NS, P > 0.05; *0.01 < P < 0.05; **0.01 < P < 0.001; ***0.001 < P < 0.0001. Groups were compared pairwise using post hoc t tests based on data from all groups and 1-way ANOVA.
Impaired AT-rich DNA–stimulated IFN production by PBMCs from POLR3A-, POLR3C-mutated patients.
To start investigating the patient cells, we first examined the levels of 5S rRNA, which is transcribed in a POL III–dependent manner. Interestingly, PBMCs from the 4 patients were indistinguishable from control cells with respect to the levels of 5S rRNA (Figure 2C). By contrast, PBMCs from all patients exhibited a strongly impaired ability to induce expression of IFNA2, IFNB, and IFNL1 upon stimulation with poly(dA:dT) (Figure 2, D–F). Importantly, this was not observed when we examined PBMCs from children with VZV infection not carrying mutations in the POL III genes (Figure 2G) or from donors who harbored other heterozygous mutations in POLR3A and POLR3C but had not experienced severe VZV infection (Supplemental Figure 6). Moreover, cells from the patients responded normally to stimulation with transfected polyinosinic-polycytidylic acid [poly(I:C)] (Figure 2H), which stimulates the MAVS pathway at the level of RIG-like receptors downstream of POL III (35). To determine whether the identified phenotype in PBMCs was also observable in other cell types, we compared induction of IFNB expression in fibroblasts from a healthy donor and P3 after stimulation with RNA or DNA or infection with VZV. In contrast to the observation in PBMCs, fibroblasts from P3 responded normally to poly(dA:dT) (Supplemental Figure 7A and data not shown). Interestingly, immunoblotting revealed that the potent IFN-inducing DNA sensor cGAS was expressed at very low levels in PBMCs compared with fibroblasts, monocyte-derived macrophages, and keratinocytes (Supplemental Figure 7, B and C). Collectively, PBMCs from patients with mutations in genes encoding the POLR3A and POLR3C subunits of the POL III complex exhibited a reduced capacity to mount IFN expression in response to AT-rich DNA.
AT-rich regions from the VZV genome stimulate IFN expression in controls but not patient PBMCs.
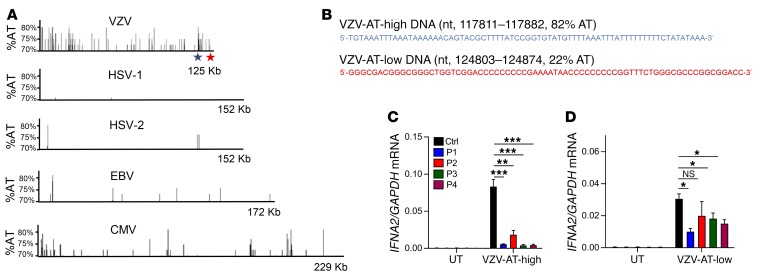
Since POL III initiates promoter-independent transcription from DNA regions with high AT content (26), we analyzed the VZV genome for AT content. The overall AT content in the VZV genome is 54%. Interestingly, the VZV genome included several islands with high AT content, between 70% and 80% (Figure 3A). A comparison with other human pathogenic herpesviruses revealed that the abundance of AT-rich regions was very low in the genomes of HSV-1, HSV-2, and Epstein-Barr virus, whereas several AT-rich regions were found in the CMV genome, although not to the same extent as in the VZV genome (Figure 3A). From the VZV genome we selected two 72mers located in AT-high and AT-low clusters, respectively (Figure 3B). PBMCs from controls or patients were stimulated with VZV-AT-high and VZV-AT-low dsDNA, and induction of IFNA2 was examined. Similar to what was observed after stimulation with poly(dA:dT), the cells from all 4 patients displayed impaired ability to induce IFNA2 expression after stimulation with VZV-AT-high DNA (Figure 3C). In contrast to this, the response to VZV-AT-low DNA was less affected in patient cells (Figure 3D).
Figure 3. AT content in herpesvirus genomes.
(A) The content and distribution of AT-rich regions in the genomes of VZV were compared with those in the genomes of HSV-1, HSV-2, EBV, and CMV using the EMBOSS Isochore algorithm. Data are presented to show regions in the genome with AT content greater than 70% AT. The regions harboring the sequences termed “VZV-AT-high DNA” and “VZV-AT-low DNA” are indicated by blue and red stars, respectively. (B) Sequences and genome localization of “VZV-AT-high” and “VZV-AT-low” DNA. (C and D) PBMCs from P1–P4 and 3 controls were transfected with VZV-AT-high or VZV-AT-low DNA (2 μg/ml). Total RNA was harvested 6 hours later, and levels of IFNA2 mRNA were determined by RT-qPCR. Data are presented as mean ± SD of 3 replicates from 3 independent experiments. UT, untreated. NS, P > 0.05; *0.01 < P < 0.05; **0.01 < P < 0.001; ***0.001 < P < 0.0001. Groups were compared pairwise using post hoc t tests based on data from all groups and 1-way ANOVA.
Impaired ability of patient cells to produce the immunostimulatory RNA PAMP.
POL III transcribes AT-rich DNA to produce an RNA product carrying a tri-phosphate signature in the 5′ end (ppp-RNA) (26, 27). This RNA can serve as agonist for the RIG-I/MAVS pathway (26, 27) (Figure 4A). To functionally characterize the ability of POL III from patient cells to bind DNA, we used an assay where we immunoblotted for POL III subunits in poly(dA:dT) precipitates (Supplemental Figure 8, A and B). This assay showed that interaction between AT-rich DNA and POL III was impaired in lysates from P1, but not P2 and P3, compared with controls (Figure 4B). P4 was not examined due to the limited availability of material from this patient. To examine for production of the immunostimulatory RNA PAMP, we adapted a protocol described previously (26, 27) (Figure 4C and Supplemental Figure 8C). Interestingly, RNA isolated from P1–P3 cells transfected with poly(dA:dT) or VZV-AT-high DNA stimulated significantly less IFNB mRNA expression as compared with RNA isolated from transfected cells from healthy donors (Figure 4, D and E). These data suggested that cells from patients with heterozygous mutations in the POLR3A and POLR3C genes may lead to functional defects in the POL III DNA sensing system and impaired ability to convert AT-rich DNA into the immunostimulatory RNA PAMP, thus providing a molecular mechanism for the impaired IFN responses to AT-rich DNA of the patients’ PBMCs.
Figure 4. Impaired production of immunostimulatory RNA by patient cells after transfection with AT-rich DNA.
(A) Schematic illustration of the pathways through which poly(dA:dT) and poly(I:C) induce IFN expression. (B) Poly(dA:dT) beads incubated with cytoplasmic extracts of PBMCs from healthy controls, P1, P2, and P3 were precipitated (precip) and immunoblotted with anti-POLR3A and anti-POLR3C. (C) Overview of the assay used to evaluate for POL III activity. (D and E) PBMCs from P1, P2, P3 and 4 healthy controls were transfected with poly(dA:dT) or VZV-AT-high DNA (2 μg/ml). Total RNA was isolated 14 hours later and transfected into HEK293 cells. Total RNA was isolated 6 hours later, and levels of IFNB mRNA were determined by RT-qPCR. The levels of IFNB mRNA were normalized to GAPDH, and data are presented as mean ± SD from 3 replicates from 2 independent experiments. Lipo, Lipofectamine. *0.01 < P < 0.05; **0.01 < P < 0.001; ***0.001 < P < 0.0001. Groups were compared pairwise using post hoc t tests based on data from all groups and 1-way ANOVA.
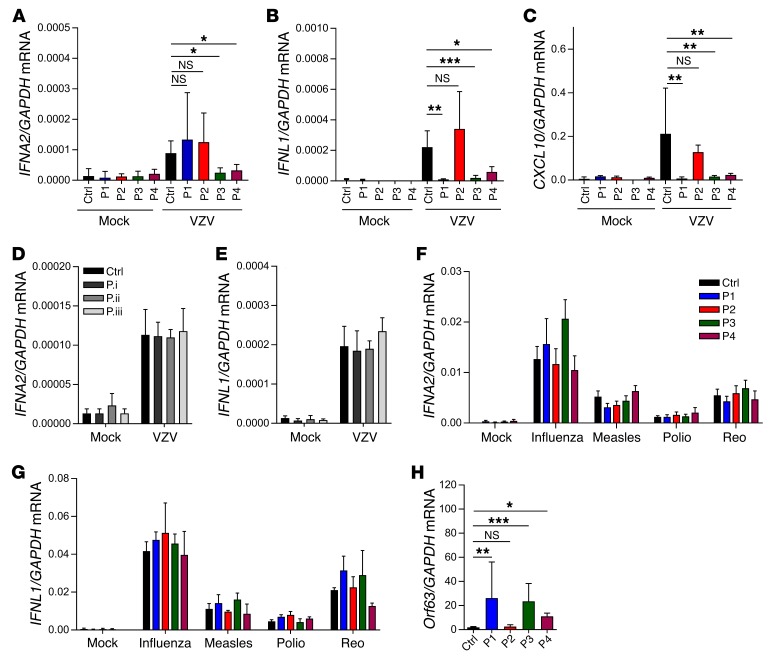
Reduced IFN induction in response to VZV infection in PBMCs from POLR3A-, POLR3C-mutated patients.
To evaluate whether the POLR3A, POLR3C mutations also had an impact on the innate immune response to VZV infection, we treated PBMCs from controls and the 4 patients with VZV-infected MeWo cells and evaluated expression of type I and III IFNs, as well as ISGs. An initial characterization of the MeWo-PBMC coculture system revealed that viral replication occurred in both MeWo cells and PBMCs, whereas IFN expression was observed only in the PBMCs and only upon coculture with infected MeWo cells (Supplemental Figure 9, A and B). We observed that both cell-associated and cell-free virus induced IFN expression, and that full induction of IFN responses was dependent on viral gene expression, as indicated by its inhibition by UV treatment (Supplemental Figure 9, C and D). VZV-infected MeWo cells did not produce detectable levels of IFNL1 mRNA (Supplemental Figure 9B). For IFNA2 expression, PBMCs from P3 and P4, but not P1 and P2, showed reduced ability to induce this response during VZV infection (Figure 5A). For type III IFNs, P1, P3, and P4, but not P2, failed to induce IFNL1 expression in the VZV-infected cultures (Figure 5B). The impaired VZV-induced IFN expression in P1, P3, and P4 was translated into lower induction of the ISG CXCL10 (Figure 5C). The finding demonstrates that although infection of MeWo cells with VZV stimulated only low IFN gene expression in control PBMCs, it lead to ISG expression, which, importantly, was abolished in cells from P1, P3, and P4. For P1, we examined for a wider range of ISGs, since we observed impaired induction of IFNL1 but not IFNA2. Importantly, P1 PBMCs failed to induce a large number of ISGs during VZV infection (Supplemental Figure 10). By comparison, PBMCs from children with VZV infection not carrying mutations in the POL III genes induced expression of IFNA2 and IFNL1 to the same extent as PBMCs from healthy children after VZV infection in vitro (Figure 5, D and E). Interestingly, when PBMCs from the family of P3 were treated with VZV-infected MeWo cells, we observed that both children carrying the mutations failed to mount IFNL1 expression after VZV infection (Supplemental Figure 11A). The impaired IFN response to VZV infection was also observed in PBMCs from the father of P3, who did carry the same POL III mutations as P3, but the impairment was not as pronounced as observed in cells from the children with the mutations (Supplemental Figure 11A). PBMCs from the mother of P4, who is heterozygous for the same R84Q POLR3C mutation as P4 but does not carry P4’s other POLR3A mutation, displayed a normal response to VZV-infected MeWo cells, and hence the role of this R84Q POLR3C mutation in P4’s severe VZV infection remains unclear (major gene vs. modifier). We also noted that PBMCs from adults generally stimulated significantly higher IFN expression after VZV infection than PBMCs from children (Supplemental Figure 11A). Immunoblotting revealed that the levels of cGAS, POLR3A, and POLR3C were expressed at comparable levels among the P3 family members (Supplemental Figure 11B), thus suggesting a more complex explanation for the age-dependent innate response to VZV infection than merely differential expression of DNA sensors. In summary, these data demonstrated that digenic mutations in POLR3A and POLR3C (P3 and P4) were related to severe cellular phenotypes of IFN induction upon VZV infection in PBMCs, while milder cellular phenotypes were related to monogenic POLR3C mutation (P1), and monogenic POLR3A mutation was not related to impaired responses to VZV (P2).
Figure 5. PBMCs from P1, P3, and P4 exhibit defective ability to produce IFNs during VZV infection and to control the virus.
PBMCs from P1–P4 (A–C and F–H), 3 VZV patients not carrying POL III gene mutations (D and E), and age-matched healthy controls (A–H) were treated with VZV-infected MeWo cells. Total RNA was harvested 48 hours after initiation of coculture with VZV-MeWo cells. (F and G) PBMCs from P1–P4 and age-matched healthy controls were treated with IAV, measles virus, poliovirus, or reovirus (Reo) (all MOI 5). Total RNA was harvested 6 hours later. Levels of IFNA2, IFNL1, CXCL10, and Orf63 mRNA were determined by RT-qPCR as indicated. For all PCR data, the levels of the mRNA of interest were normalized to GAPDH, and data are presented as mean ± SD of 3 replicates from 3 independent experiments. Average of 4 age-matched controls. NS, P > 0.05; *0.01 < P < 0.05; **0.01 < P < 0.001; ***0.001 < P < 0.0001. Groups were compared pairwise using post hoc t tests based on data from all groups and 1-way ANOVA.
Responses to other viruses in patients’ PBMCs.
To examine whether the impaired IFN response to virus infection was specific to VZV, we also infected POLR3A-, POLR3C-mutated patient PBMCs with HSV-1 and a panel of RNA viruses and measured accumulation of IFN transcript. While PBMCs from P1 exhibited reduced IFN expression after HSV-1 infection, the response in P2 and P3 was only marginally or not different from control cells (Supplemental Figure 12). Cells from P4 were not examined. With respect to RNA virus infections, all 4 patients’ cells responded with normal expression of type I and III IFNs to all viruses tested (Figure 5, F and G). Examination of the induction of IFNA2 and IFNL1 in cells from patients carrying mutations previously shown to confer susceptibility to HSE revealed that only IRF3 deficiency, but not TLR3 or TRIF deficiency, conferred reduced VZV-stimulated IFN expression (Supplemental Figure 13, A and B) (17, 18, 22). This IRF3 defect also ablated induction of IFNL1 by HSV-1 but did not affect the response to influenza A (IAV) infection (Supplemental Figure 13, C and D).
Impaired control of VZV replication by PBMCs from POLR3A-, POLR3C-mutated patients.
Finally, we evaluated whether patient cells exhibited impaired ability to control VZV replication. VZV-infected MeWo cells were cocultured with PBMCs from controls or each of the 4 patients, and we measured expression of viral Orf63, which is among the most abundant viral transcripts (36). Consistent with the impaired IFN responses to VZV by PBMCs from P1, P3, and P4, we observed that VZV infection in MeWo cells gave rise to significantly higher viral gene expression when cocultured with PBMCs from P1, P3, or P4. By contrast, viral gene expression in cultures with P2 PBMCs was similar to that of the controls (Figure 5H), which is consistent with the normal IFN responses to VZV in P2 cells.
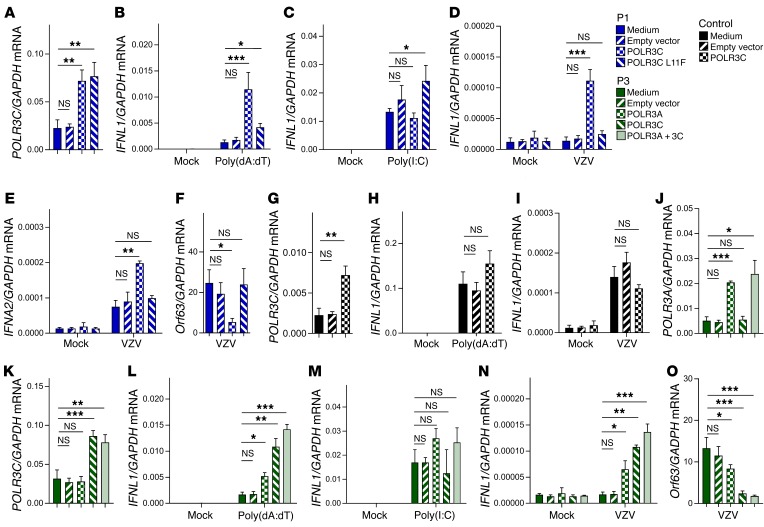
Reconstitution of WT POLR3C expression in P1 PBMCs rescues the control of VZV replication.
To examine whether the impaired responses observed in patient cells were due to the mutations on POL III subunits, we wanted to reconstitute WT gene expression. Transduction of P1 PBMCs with lentivirus encoding WT or L11F POLR3C led to significantly higher levels of POLR3C mRNA compared with those in cells transduced with a control lentivirus (Figure 6A). Importantly, cells from P1 transduced with WT, but not L11F, POLR3C gained the capacity to produce IFNL1 in response to poly(dA:dT), while the response to poly(I:C) was unaffected (Figure 6, B and C). Next, the transduced cells from P1 were infected with VZV, and levels of IFNL1, IFNA2, and Orf63 mRNAs were evaluated. Interestingly, in patient cells transduced with WT POLR3C but not in cells transduced with empty vector or L11F POLR3C, VZV infection induced IFNL1 expression (Figure 6D), and increased levels of IFNA2 (Figure 6E). The elevated induction of IFN in patient cells transduced with WT POLR3C was mirrored in reduced viral expression of Orf63 (Figure 6F). Transduction of WT POLR3C into cells from a healthy donor did elevate POLR3C mRNA levels, but did not affect IFNL1 expression after stimulation with poly(dA:dT) or infection with VZV (Figure 6, G–I). Thus, reconstitution of WT POLR3C expression in P1 PBMCs restored the ability to induce IFN expression upon VZV infection and to control VZV replication.
Figure 6. Rescue of WT POLR3 expression restores IFN response and control of VZV infection.
(A) RNA was isolated from PBMCs from P1, which were either left untreated, or transduced with a lentivirus encoding WT or L11F POLR3C or carrying empty vector. Levels of POLR3C mRNA were determined by RT-qPCR. (B and C) The transduced P1 PBMCs were treated with poly(dA:dT) (2 μg/ml) or poly(I:C) (4 μg/ml). Total RNA was harvested 6 hours later, and levels of IFNL1 mRNA were determined by RT-qPCR. (D–F) The transduced P1 PBMCs were treated with VZV-infected MeWo cells. Total RNA was harvested 48 hours later, and levels of IFNL1, IFNA2, and Orf63 mRNA were determined by RT-qPCR as indicated. (G) PBMCs from a healthy donor was transduced with a lentivirus encoding WT POLR3C or carrying empty vector. Levels of POLR3C mRNA were determined by RT-qPCR. (H and I) The transduced control PBMCs were treated with poly(dA:dT) (2 μg/ml) or with VZV-infected MeWo cells. Total RNA was harvested 6 and 48 hours later, respectively, and levels of IFNL1 mRNA were determined by RT-qPCR as indicated. (J and K) RNA was isolated from PBMCs from P3, which were either left untreated, or transduced with a lentivirus encoding WT POLR3A, WT POLR3C, or carrying empty vector. Levels of POLR3A and POLR3C mRNA were determined by RT-qPCR. (L and M) The transduced P3 PBMCs were treated with poly(dA:dT) (2 μg/ml) or poly(I:C) (4 μg/ml). Total RNA was harvested 6 hours later, and levels of IFNL1 mRNA were determined by RT-qPCR. (N and O) The transduced P3 PBMCs were treated with VZV-infected MeWo cells. Total RNA was harvested 48 hours later, and levels of IFNL1 and Orf63 mRNA were determined by RT-qPCR as indicated. NS, P > 0.05; *0.01 < P < 0.05; **0.01 < P < 0.001; ***0.001 < P < 0.0001. Groups were compared pairwise using post hoc t tests based on data from all groups and 1-way ANOVA.
Reconstitution of WT POLR3A and POLR3C expression in P3 PBMCs rescues the control of VZV replication.
P3 and P4 carry mutations in both POLR3A and POLR3C, and we were interested in examining whether there was a requirement for correction of both mutations to mount innate immune responses. Therefore, cells from P3 were transduced with WT POLR3A and POLR3C (Figure 6, J and K). Transduction of either WT subunit enabled the cells to induce IFNL1 in response to poly(dA:dT) and VZV without significantly affecting the responsiveness to poly(I:C) (Figure 6, L–N). Transduction with both POLR3A and POLR3C led to a marginal, but not statistically significant, further elevation of the response to poly(dA:dT) and VZV. Finally, VZV replication was modestly impaired in cells incubated with patient PBMCs transduced with WT POLR3A, and was largely abrogated if incubated with patient PBMCs transduced with WT POLR3C or both POLR3A and POLR3C (Figure 6O). Collectively, the results indicate that the impaired ability of P1 and P3 PBMCs to produce IFNs in response to VZV was rescued by expression of the WT genes, which correlated with regained control of virus replication. A summary of the cellular phenotypes observed is shown in Supplemental Table 6. Collectively, these reconstitution experiments established causal relationships between monogenic POLR3C mutation (P1) or digenic POLR3A, POLR3C mutations (P3), and impaired antiviral IFN responses to VZV.
Discussion
One of the outstanding questions in human medicine and immunology is why different individuals display differential susceptibility to infections with a given microbe. In recent years, it has emerged that inborn errors in genes encoding proteins of innate or cell-intrinsic immunity can underlie specific infections in otherwise healthy individuals (15, 16). In this work we describe 4 patients with severe VZV infections who carried missense mutations in POLR3A and/or POLR3C, which encode subunits of the innate DNA sensor POL III. Since this study was based on WES, it is possible that some of the remaining 17 patients with severe VZV have genetic variations influencing RNA POL III function in noncoding regions of their genomes. PBMCs from the identified patients had impaired ability to induce expression of IFN in response to synthetic AT-rich DNA only (P2, heterozygous for POLR3A M307V) or to both AT-rich DNA and VZV infection (P1, heterozygous for POLR3C L11F; P3, heterozygous for POLR3A Q707R and POLR3C R438G; P4 heterozygous for POLR3A R437Q and POLR3C R84Q). Reconstitution of expression of WT alleles enabled the P1 and P3 cells to induce IFN expression in response to AT-rich DNA and VZV infection, and to control VZV infection. These immunological deficits probably account for the failure of P1, P3, and P4 to control infection by VZV. In P3 and P4, the POLR3A, POLR3C digenic defect of POL III is the probable genetic cause of severe VZV infection. In P1, and perhaps in P2, POLR3C or POLR3A monogenic defect of POL III conferred the cellular and clinical phenotypes. The finding of milder in vitro defects for P2 is consistent with his milder clinical phenotype, when compared with the other 3 patients. In these 4 families, there is incomplete clinical penetrance, with the possible exception of the P3 family, suggesting that environmental factors, such as virus clades and infectious virus load, or host modifying factors, such as age of infection or modifying genes, are also involved in the pathogenesis.
The POL III complex consists of 17 subunits, 10 of which are specific for POL III (37). One question that arises based on the mutations identified in this study is why we identified mutations only in POLR3A and POLR3C. It is possible that future studies will lead to identification of mutations in other POL III subunits in patients with severe VZV infection and that the apparent bias observed in this study does not reflect the underlying biology of POL III in immunological DNA sensing. It may simply reflect the small size of our sample (21 patients). Second, it is possible that POL III can act in a modular fashion, with the requirement for different subunits being dependent on the DNA to be transcribed, e.g., promoter-dependent versus- independent transcription. Third, since all patients carry heterozygous mutations, and these mutations do not lead to total loss of function of POL III, it is possible that the different functions of POL III have differential tolerance for mutations in the different subunits. POLR3C, POLR3F, and POLR3G form a subcomplex that is required for transcriptional initiation but not elongation by POL III, whereas the core POL III complex is required for elongation (37). We observed that the POLR3C mutants found in P1 and P4 had reduced solubility compared with WT POLR3C. This may imply that a significant fraction of the POL III complexes in P1 and P4 lack POLR3C, hence impairing the ability to transcribe RNA from AT-rich DNA. Lysates from all patients examined were strongly impaired in their ability to convert AT-rich DNA to immune-stimulatory RNA, whereas transcription of 5S rRNA was largely unaffected. Previous studies have identified mutations in POLR3A and POLR3B in patients with autosomal recessive 4H (hypomyelination, hypodontia, hypogonadotropic hypogonadism) leukodystrophy, which is a disease characterized by degeneration of the white matter in the brain (38–40). However, the functional impact of the POLR3 gene mutations in the 4H leukodystrophy patients on POL III activity remains to be established. Since defects in POL III are associated with both degenerative processes in the CNS and susceptibility to VZV in humans, there is a requirement for more knowledge on the biochemistry of POL III in transcription of endogenous as well as exogenous types of DNA.
Our results demonstrated that stimulation of IFN expression by AT-rich DNA was reduced in PBMCs from patients, therefore confirming previous results on POL III as an innate sensor of AT-rich DNA (26, 27). The VZV genome was found to be rich in regions with high AT content, and we found that synthetic DNA derived from an AT-rich region of the genome stimulated significantly less IFN expression in patient compared with control cells. DNA derived from a region of the VZV genome with low AT content stimulated IFN expression in a manner less dependent on POL III, although the response in PBMCs from P1, P3, and P4 was still impaired. This suggests that high AT content renders DNA more potent as a PAMP for POL III, but also that DNA with low AT content can stimulate this pathway to some extent. In this context, it was interesting to note that IFN induction by infection with HSV-1, which has a very low genomic AT content, was also reduced in cells from P1. Since its discovery, cGAS has been thoroughly characterized and is emerging as a dominant DNA sensor (29, 41). Mice deficient in cGAS are highly susceptible to several viral infections, and most cell types fail to respond to DNA in the absence of cGAS (41, 42). However, it was shown that cGAS-deficient murine lung fibroblasts responded normally to poly(dA:dT), but did have an impaired response to other DNA species (41). Together with the results of the present study, these data suggest that cGAS is not the key sensor of AT-rich DNA in certain cell types. Interestingly, we observed that human PBMCs have low expression of cGAS compared with skin fibroblasts and monocyte-derived macrophages. This may explain the in vitro phenotype documented in PBMCs but not skin fibroblasts of patients, and may also provide clues as to why POL III mutations confer selective susceptibility to VZV infection.
A central part of the natural history of VZV infection is a viremic stage in T cells, from which the virus is delivered to the skin, sensory nerve cell bodies, and other locations (2, 43). By contrast, viremia is not believed to be important for the pathogenesis of HSE, and it is worth noting that we did not find significant enrichment of POLR3A or POLR3C mutations in our large data set of 222 patients with HSE. Therefore, we speculate that the elevated susceptibility to severe VZV infections in individuals with POL III mutations might be explained, at least partly, by the combined effect of the high AT content of the VZV genome and the low cGAS expression by blood cells. Impaired POL III–mediated VZV viral DNA sensing and the consequent low levels of IFN responses of POLR3A-, POLR3C-mutated PBMCs might be the key to VZV dissemination in the blood and severe complication in some organs such as the CNS and the lung. Therefore, the present work contributes insight into how individual mutations affecting central antiviral pathways sometimes appear to present with a very narrow and selective infectious phenotype in patients. It will be interesting to explore further the role of POL III in innate immune responses to infection. Finally, the identification of POL III defects in patients with severe disseminated VZV infections, particularly those affecting the CNS and the lung, may have clinical implications, including decisions on prophylactic VZV vaccination or acyclovir treatment, genetic counseling of family members, and possibly management of disseminated VZV infection with type I IFN as previously suggested for HSE (44).
Collectively, the results of this work demonstrate that functional defects in two genes of the POL III pathway confer increased susceptibility to severe VZV infection, which is associated with impaired ability to detect foreign DNA with high AT content and mount antiviral IFN responses. This provides the first direct evidence to our knowledge of a role for an innate DNA sensor in control of infections in humans.
Methods
Clinical information.
Patients were retrospectively recruited using databases from hospitals and pediatric practices in Belgium, or recruited in the United States and in France, in accordance with local regulations and with the approval of the IRB of the Antwerp University Hospital, the Rockefeller University, and INSERM. Informed consent was obtained in the home country of each patient, in accordance with local regulations and with IRB approval. Patients were identified as having a severe VZV-related CNS or respiratory complication if (i) severe clinical course led to hospitalization, (ii) the complication occurred within 1 month after onset of chickenpox and no other reasons for the complication were found, or (iii) CSF or tracheal suction material collected after the complication tested positive for VZV DNA using PCR. Time points for VZV infection and sampling were as follows: P1, episode occurred in 2010, sampling took place in 2014 and 2016; P2, episode occurred in 2011, sampling took place in 2014 and 2016; P3, episode occurred in 2010, sampling took place in 2010 and 2016; P4, episode occurred in 2007, sampling took place in 2007 and 2017.
Whole exome sequencing and bioinformatics.
Genomic DNA was isolated from peripheral blood lymphocytes using standard procedures. For P1 and P2, fragmentation of DNA was carried out on the Covaris M220. The TruSeq DNA Sample Preparation Kit LT (Illumina) was used for adapter ligation and library amplification in order to prepare sample libraries for enrichment. Exome enrichment and capture were carried out using the SeqCap EZ Human Exome Library v3.0 kit (Nimblegen). Whole exome sequencing was performed on an Illumina HiSeq 1500 (Illumina) according to standard protocols for a paired end 2 × 100 bp high-throughput run. Reads were processed using an in-house pipeline as follows. Cutadapt (https://github.com/marcelm/cutadapt) was used to trim adapter sequences, after which quality trimming, with a Phred score of 30 as threshold, was performed. Reads were mapped to the human reference genome (UCSC hg19) using the Burrows-Wheeler Aligner (BWA-MEM with default settings; http://bio-bwa.sourceforge.net/), and duplicates were removed using Picard (http://broadinstitute.github.io/picard/). According to the Genome Analysis Toolkit best practice guidelines local indel realignment and base quality recalibration were performed with recommended settings (GATK, https://software.broadinstitute.org/gatk/). Variant Calling was performed using the Unified Genotyper both on the single-sample and multi-sample level, followed by merging of the resulting variant lists and variant quality recalibration. For P3 and P4, captured libraries were sequenced with the Illumina HiSeq2500 and Illumina HiSeq2000 sequencing platforms, respectively, and reads were mapped to the human reference genome assembly GRCh37 (UCSC hg19) using the Burrows-Wheeler Aligner. Variants were identified with the GATK, Sequence Alignment/Map (SAM) tools (http://samtools.sourceforge.net/), and Picard tools. Variant calls with a read coverage ≤2× and a Phred-scaled SNP quality ≤20 were eliminated.
Variants were annotated and filtered using minor allele frequency <0.005 in the 1000 Genomes database; <0.01 in the ExAC, which contains WES data of 60,706 individuals; as well as the in-house WES database of the HGID laboratory, which contains WES data of 3,055 individuals with diverse infectious diseases. We searched for rare nonsynonymous (including missense, stop loss or stop gain, start loss, in-frame or frame-shift insertions or deletions) and splicing cite mutations, in a comprehensive list of genes known to be involved in IFN immunity (Supplemental Table 1), as well as in known PID genes.
Mutation validation by Sanger sequencing.
For all patients, candidate mutations were verified by capillary sequencing using BigDye chemistry (Life Technologies). More precisely, primers to validate each of the variants were designed using Primer3 and ordered at Integrated DNA Technologies. The desired fragments were amplified by PCR using the GoTaq G2 PCR Master Mix (Promega). All reactions were carried out on a Thermal Cycler 2720 (Applied Biosystems). Primer sequences were POLR3A (P2): forward, 5′-AGCCTGTGACCTCCAATCAG-3′, reverse, 5′-GACCCTCCGTTGTGAGTGAT-3′; POLR3A (P3): forward, 5′-TACAAACTATATCTAGCAGGC-3′, reverse 5′-ACCTCCAGGGTCTCCTCAG-3′; POLR3A (P4): forward, 5′-ATGGAGGCATCTTAGATTTG-3′, reverse, 5′-CAGTGCTACTCACCAGATG-3′; POLR3C (P1): forward, 5′-GCAGGCTTTCTTCATTTTTCCTT-3′, reverse, 5′-GCTTTTTCTTTCTTTGGACACAATAT-3′; POLR3C (P3): forward, 5′-GGCTAAGTAACTCCAGGTAG-3′, reverse, 5′-GTTCTATTTCAGCAGCTCAG-3′; POLR3C (P4): forward, 5′-CCATTTAGGTGAAGAAAGCC-3′, reverse, 5′-CTCCATGGTCTCTGTGAGC-3′. The samples were purified using calf intestinal alkaline phosphatase (Roche Life Science, 20 U/μl) and Exonuclease I (New England BioLabs, 20,000 U/μl). Purified PCR products were bidirectionally sequenced using the BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) and separated on an ABI 3130XL Genetic Analyzer in accordance with the manufacturer’s instructions (Applied Biosystems).
Viral genome analysis.
The content and distribution of AT-rich regions in herpesvirus genomes were analyzed using the EMBOSS Isochore algorithm (http://www.ebi.ac.uk/Tools/seqstats/emboss_isochore/) and publically available viral genome sequences. For the data presented, the parameters for the analysis were: window size, 50 bp; shift, 50 bp.
Cell sorting.
Frozen PBMCs were thawed and stained, and the following populations (monoclonal antibodies used, all from BD Biosciences) were sorted into cell culture plates: T cells (mouse anti–human CD3, clone UCHT1), B cells (mouse anti–human CD19, clone HIB19), classical and inflammatory monocytes (mouse anti–human CD14, clone M5E2; mouse anti–human CD16, clone 3G8). The purity of the cell populations was (donors 1, 2, 3): T cells: 100.0%, 98.2%, 99.2%; B cells: 99.2%, 99.5%, 99.8%; classical monocytes (CD14hiCD16–): 99.8%, 100.0%, 99.8%; non-classical monocytes (CD14+CD16+): 95.4%, 92.2% (not analyzed for donor 3). pDCs were depleted using the Plasmacytoid Dendritic Cell Isolation Kit II, human kit (Miltenyi Biotech). For sorting of PBMCs and MeWo cocultures (ratio 1:1) into the respective populations, the cell culture supernatants were collected, with trypsinization of adherent cells, as well as mechanical collection using a cell scraper (Sigma-Aldrich). Cells and supernatants were centrifuged separately at 250 g for 7 minutes, after which the two pellets were pooled in 10 ml buffer (PBS, 1% BSA, 2.5 mM EDTA, and 25 mM HEPES). Cells were incubated with 5 μl anti-CD45 antibody per 10 × 1010 cells/ml, with 1 × 106 cells/ml kept as unstained control. All cells were incubated at 4°C for 30 minutes. After this, cells were washed in 10 ml buffer, followed by centrifugation at 250 g for 7 minutes. Propidium iodide was used to distinguish between live and dead cell populations. Acquisition and cell sorting were performed on a MoFlo Astrios (Beckman Coulter). A 4-way sort was performed; live PBMCs, dead PBMCs, live VZV-MeWo cells, and dead VZV-MeWo cells were sorted. Data analysis was performed using FlowJo v10. RNA was harvested from each population, and virus presence and IFN production were analyzed by real-time PCR.
Cell culture and stimulation.
Frozen PBMCs were thawed in 50-ml tubes with 20 ml preheated media (RPMI-1640 w/l-glutamine [Biowest], 10% heat-inactivated FBS [Life Technologies], 1% penicillin/streptomycin) before centrifugation at 350 g in 10 minutes. Normal adult human keratinocytes were obtained by trypsinization of skin samples from patients undergoing plastic surgery as previously described (45). Whole cell lysates were made from second-passage keratinocytes grown in serum-free keratinocyte medium supplemented with human recombinant epidermal growth factor, bovine pituitary extract, and gentamicin (all from Life Technologies/Invitrogen). For stimulation experiments, cells were distributed in 24-well tissue culture plates with 0.5 × 106 cells per 300 μl media and incubated overnight at 37°C and 5% CO2. PBMCs were transfected with 2 μg/ml poly(dA:dT) (CAYLA InvivoGen) or 4 μg/ml poly(I:C) using Lipofectamine (Life Technologies). The synthetic analogs were incubated with Lipofectamine for 20 minutes before dropwise addition to the PBMCs. Furthermore, PBMCs were stimulated with different viruses: HSV-1 (F strain), IAV (PR8 strain, H1N1), measles virus (Edmonston strain), polio virus 1 (LSa strain), reovirus type 1 (Lang strain), VZV (ROka strain) (PBMC/MeWo-VZV ratio, 1:1). One vial of VZV-infected MeWo cells was thawed, spun down at 1,000 rpm for 10 minutes, and resuspended in 400 μl preheated media. From this solution, 30 μl was added to the wells to be stimulated with VZV. Cells were incubated for 6 hours, except for MeWo-PBMC cocultures, which were incubated for 48 hours before cell lysis and RNA harvest. The stimulations were performed in duplicate, except IAV stimuli, which represent 5 biological replicates from two experiments. To isolate cell-free VZV, infected MeWo cells were freeze-thawed overnight. Following thawing, cells were scraped from the flask by mechanical collection using a cell scraper (Sigma-Aldrich). Cells were disrupted using a bath sonicator, 3 times for 1.5 minutes at 15-second intervals. The lysate was collected and used as cell-free virus. For UV inactivation, the sonicated solution was treated for 5 minutes on ice under a UV lamp.
Isolation of RNA to use for stimulation of cells.
For isolation of total RNA from PBMCs (2.4 × 106 cells per 12 wells), cells were lysed in TRIzol (Invitrogen) 16 hours after stimulation with DNA (2 mg/ml). Nucleic acids were extracted with phenol and chloroform, and precipitated with isopropanol containing linear polyacrylamide (5 mg/ml). For some experiments, extracted RNA was treated as follows before transfection into recipient cells: DNase I (200 U/ml/mg RNA), RNase A (0.1 mg/ml/mg RNA) (Ambion), or alkaline phosphatase (AP; 100 U/ml; Roche Applied Science) at 37°C for 1 hour. AP-treated RNA was incubated at 65°C for 15 minutes for heat inactivation before further treatment with PNK. Enzyme-treated RNAs were precipitated with isopropanol before transfection.
In vitro DNA binding assay.
Cells (2 × 106) were resuspended in lysis buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 0.1% Nonidet-P40; 5 mM EDTA; and 10% [vol/vol] glycerol), and the lysate was cleared by centrifugation for 15 minutes at 5,000 g. For precipitation with NeutrAvidin beads (Pierce), cell lysates were incubated for 2 hours with biotin-labeled poly(dA:dT). After incubation with NeutrAvidin beads, bound complexes were pelleted by centrifugation and analyzed by immunoblotting. For confirmation of binding specificity, assays were also performed in the presence or absence of increasing amounts (0.5, 5, or 50 μg/ml) of unlabeled poly(dA:dT).
RNA isolation and reverse transcription PCR.
RNA was harvested from patient and control PBMCs using a High Pure RNA Isolation Kit (Roche) and synthesized to cDNA using the QIAGEN QuantiTect Reverse Transcription Kit (catalog 205313) according to the manufacturer’s instructions. Real-time qPCR with TaqMan probes was used for measurement of mRNA levels of IFNA2, IFNB, IFNL1, CXCL10, IL6, 5S rRNA, and GAPDH (PerfeCTa qPCR FastMix II, Quantabio, catalog 95119). The Taqman probes were IFNA2 (Hs00265051_s1), IFNB1 (Hs01077958_s1), IFNL1 (Hs00601677_g1), CXCL10 (Hs01124251_g1), IL6 (Hs00985639_m1), 5S rRNA (Hs03682751_gH), and GAPDH (Hs02758991_g1) (Thermo Fisher Scientific). The samples were analyzed in duplicate using the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) at 95°C for 10 minutes, followed by 45 cycles of 95°C for 30 seconds and 60°C for 1 minute. The levels of Orf63 were determined by RT-qPCR with a QuantiFast SYBR Green Kit (QIAGEN) using the following primer sequences: ORF63 forward: GCGCCGGCATGATATACC and ORF63 reverse: GACACGAGCCAAACCATTGTA; GAPDH forward: TCTTTTGCGTCGCCAGCCGAG and GAPDH reverse: ACCAGGCGCCCAATACGACCA. The reactions were carried out on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) at 95°C for 5 minutes, 45 cycles at 95°C for 10 seconds, and 62°C for 22 seconds.
Fluidigm.
cDNA was prepared using a QuantiTect Reverse Transcription kit (QIAGEN). For pre-amplication, cDNA was mixed with SuperScript III Taq RT Platinum mix (Invitrogen) and a 0.2× mix of all TaqMan gene expression assays (LifeTechnologies) used in the Fluidigm assay (see below) and amplified using the following thermal cycle protocol: one incubation of 15 minutes at 50°C; one incubation of 2 minutes at 95°C; and 18 cycles of 15 seconds at 95°C and 4 minutes at 60°C. The amount of cDNA present in each samples was assessed by regular qPCR using a TaqMan gene expression assay for ACTB (see below) and the TaqMan RT-PCR mix (Applied Biosystems). Fluidigm-based qPCR was then performed on the BioMarker HD instrument using a Flex Six Integrated Fluidic Circuit (IFC) (Fluidigm) according to the manufacturer’s instructions. The Flex Six IFC was loaded using the HX IFC controller (Fluidigm). The Flex Six IFC was then placed in the BioMarker HD instrument. qPCR was performed using the “GE FlexSix Standard v1” thermal cycle protocol. The data were analyzed using Real-Time PCR Analysis software 4.1.3 (Fluidigm). As a Ct threshold method, the “auto (global)” method was used; for baseline correction, the “linear (derivative)” method was used. For all TaqMan gene expression assays, except ISG15, 0.65 was set as a quality threshold; for ISG15, 0.31 was set as a quality threshold. Data failing the quality call were removed from the analysis. Data were normalized to average of ACTB and GAPDH expression. Fold induction of genes due to treatment was calculated in reference to the untreated samples of each test subject. TaqMan gene expression assays used for Fluidigm analysis were: ACTB: Hs00357333_g1; GAPDH: Hs02758991_g1; IFNB: Hs01077958_s1; CXCL10: Hs01124252_g1; IFIT1: Hs03027069_s1; IFIT2: Hs00533665_m1; ISG15: Hs00192713_m1; ISG20: Hs00158122_m1; MX1: Hs00895608_m1; OAS1: Hs00973637_m1.
Western blotting.
Total cell lysates were subjected to SDS-PAGE and immunoblotting. The blots were blocked with 5% milk and probed with the following primary antibodies: POLR3A (Abcam, ab96328, 1:500), POLR3C (Sigma-Aldrich, HPA027516, 1:250), cGAS (Cell Signaling Technology, 15102, 1:500), and β-actin (Abcam, ab49900, 1:10,000). Peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories Inc., HRP–anti-rabbit, 711-035-152, 1:10,000) was used for development.
Generation of lentiviral constructs and reconstitution of POLR3A and POLR3C expression.
For plasmid construction, POLR3C (PlasmID HsCD00321588) and POLR3A (GenScript OHu27038C) cDNAs were acquired commercially. The lentiviral vector constructs pCCL/PGK-POLR3C and pCCL/PGK-POLR3A were produced by PCR amplification of POLR3C and POLR3A cDNA, respectively, and subsequent insertion into XhoI-/BamHI-digested pCCL/PGK-eGFP (46) by NEBuilder HiFi DNA Assembly according to the manufacturer’s instructions (New England BioLabs). pCCL/PGK-POLR3C(L11F) was created by introducing a 36G>T point mutation into pCCL/PGK-POLR3C by NEBuilder HiFi DNA assembly. Production of lentiviral vectors has previously been described in detail (46). Briefly, packaging plasmids pMDlg/p-RRE, pMD.2G, and pRSV-Rev were cotransfected into HEK 293T cells with pCCL/PGK-POLR3C, pCCL/PGK-POLR3A, pCCL/PGK-POLR3C(L11F), or pCCL/PGK-eGFP using calcium phosphate transfection. Vector-containing supernatants were passed through 0.45-μM filters (Sarstedt) and ultracentrifuged through a 20% sucrose cushion for 2 hours at 100,000 g, 4°C. Pellets were resuspended in PBS and stored at –80°C. HIV-1 p24 concentrations were measured by ELISA (XpressBio) according to the manufacturer’s instructions. For transductions, 500,000 PBMCs in 24-well plates were transduced with lentivirus at a concentration of 500 ng p24 per well. Two days later the cells were stimulated as shown.
In silico structural modeling of patient mutations.
In silico structural modeling of patient mutations was performed using the published crystal structure of yeast RNA POL III (30). The mutated residues found in P1 to P4 were located and displayed at the equivalent position in the yeast counterpart subunit using sequence alignment. The figures were prepared using PyMOL (http://www.pymol.org).
Bacterial coprecipitation experiments.
Rosetta2 cells were transformed with a pET-15b–derived plasmid harboring POLR3C with either a WT or mutated open reading frame. Each construct was fully sequenced in order to ensure the integrity of the DNA sequence. Cells were grown at 37°C, and 1/10 of the cell culture was saved as non-induced control (NI). Cells were induced with 1 mM ITPG for an overnight incubation at 15°C. Cells were harvested by centrifugation and resuspended in a buffer containing 1.5× PBS, 1 mM Mg acetate, 0.1% NP-40, 20 mM imidazole, 10% glycerol (lysis buffer). The cell pellet was sonicated for 5 seconds at 7 W and centrifuged. About 10% of the supernatant was kept as the total lysate, and levels of POLR3C in the lysate were determined by Western blotting. The supernatant was incubated with cobalt-affinity resin for 30 minutes at 4°C. Beads were washed 3 times with 1 ml of the lysis buffer. About 5% of the crude extract and 15% of the bound fraction were analyzed on a 12.5% SDS-PAGE and visualized by Coomassie blue staining.
Study approval.
This study was approved by the ethics board of Antwerp University Hospital, and approved by the Institutional review board of the Rockefeller University. Written informed consent was obtained from the parents of all study participants younger than 18 years old and all participants older than 12 years old.
Statistics.
The gene expression data are presented as mean of 4–5 observations ± SD. For statistical analysis, first the hypothesis of a common mean in the groups was tested using an F-test based on a 1-way ANOVA. When there was evidence against the hypothesis of a common mean, groups were compared pairwise using post hoc t tests (i.e., based on data from all groups and 1-way ANOVA). A P value less than 0.05 was considered significant.
Author contributions
BO, SYZ, JLC, SRP, and THM conceived the project. CD, SH, GE, AK, AVA, LV, LL, HV, AJ, BC, GPW, JK, CP, HVB, LL, SB, JF, and SC cared for and isolated material from the patients. KBS, KAS, MCT, SL, TP, YC, JM, EB, GV, CC, and YS performed the experiments. JR, AS, ES, LVL, GM, JGM, JLC, SRP, and THM supervised the work. SF performed the modeling of the POLR3A/C mutations on the POL III structure. BO, NAB, SYZ, GK, RAF, LA, JLC, SRP, and THM analyzed the results. SRP and THM drafted the manuscript with input from BO, JM, GV, LVL, SYZ, and JLC. All authors edited the manuscript.
Supplementary Material
Acknowledgments
The technical assistance of Kirsten S. Petersen and the Aarhus University FACS core Facility is greatly appreciated. We are grateful for the participation of all patients and their families. We thank Benedetta Bigio and Bertrand Boisson for bioinformatics assistance. This work was supported by the Lundbeck Foundation (R198-2015-171 [SRP], R151-2013-14668 [THM]), the Danish Research Council (12-124330 [SRP], 11-107588 [THM]), Aarhus University Research Foundation (SRP and THM), the Research Foundation Flanders (FWO) (BO, JM, and GV; project grant G098911N), the University of Antwerp Concerted Research Action Program (BOF-GOA), the Hercules Foundation Belgium (AUHA/11/008 NGS Antwerp application for medium-scale research infrastructure), the European Society for Paediatric Infectious Diseases (ESPID Grant Award [BO]), the National Center for Advancing Translational Sciences (NCATS), NIH Clinical and Translational Science Award (CTSA) program grant UL1TR000043, and French ANR grant HSEIEIER (SYZ). The PhD scholarships to MCT and SL were funded by the European Commission (Horizon2020 program H2020 MSCA-ITN GA 675278 EDGE) and the Boehringer Ingelheim Fonds, respectively. JLC is an investigator at the Howard Hughes Medical Institute.
Version 1. 08/07/2017
Electronic publication
Version 2. 08/14/2017
Description of supplemental table 2 updated.
Version 3. 08/17/2017
Formatting error corrected from previous version.
Version 4. 09/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(9):3543–3556.https://doi.org/10.1172/JCI92280.
Contributor Information
Benson Ogunjimi, Email: benson.ogunjimi@uantwerpen.be.
Shen-Ying Zhang, Email: shzh289@mail.rockefeller.edu.
Madalina Carter-Timofte, Email: m.carter-timofte@biomed.au.dk.
Gaspard Kerner, Email: gaspard.kerner@inserm.fr.
Stefanie Luecke, Email: s.luecke@biomed.au.dk.
Thaneas Prabakaran, Email: tpra@biomed.au.dk.
Yujia Cai, Email: y.cai@biomed.au.dk.
Josephina Meester, Email: Josephina.Meester@uantwerp.be.
Esther Bartholomeus, Email: Esther.Bartholomeus@uantwerpen.be.
Nikhita Ajit Bolar, Email: nikhita.ajitbolar@uantwerp.be.
Geert Vandeweyer, Email: geert.vandeweyer@uantwerp.be.
Charlotte Claes, Email: charlotte.claes@uantwerp.be.
Yasmine Sillis, Email: yasmine_sillis@hotmail.com.
Soraya Boucherit, Email: soraya.boucherit@inserm.fr.
Charlotte Dielman, Email: charlotte.dielman@zna.be.
Steven Heynderickx, Email: heynderickx.steven@gmail.com.
George Elias, Email: george.elias@uantwerp.be.
Andrea Kurotova, Email: andrea.kurotova@gza.be.
Ann Vander Auwera, Email: Ann.VanderAuwera@gza.be.
Lieve Verstraete, Email: verstraete.lieve@telenet.be.
Lieven Lagae, Email: lieven.lagae@uzleuven.be.
Helene Verhelst, Email: helene.verhelst@ugent.be.
Anna Jansen, Email: anna.jansen@uzbrussel.be.
Jose Ramet, Email: jose.ramet@uza.be.
Arvid Suls, Email: arvid.suls@uantwerp.be.
Evelien Smits, Email: evelien.smits@uza.be.
Berten Ceulemans, Email: berten.ceulemans@uza.be.
Lut Van Laer, Email: lut.vanlaer@uantwerp.be.
Genevieve Plat Wilson, Email: plat.g@chu-toulouse.fr.
Capucine Picard, Email: capucine.picard@inserm.fr.
Horst Von Bernuth, Email: Horst.Von-Bernuth@charite.de.
Stephane Chabrier, Email: stephane.chabrier@chu-st-etienne.fr.
Laurent Abel, Email: laurent.abel@inserm.fr.
Geert Mortier, Email: geert.mortier@uantwerp.be.
Sebastien Fribourg, Email: sebastien.fribourg@inserm.fr.
Jacob Giehm Mikkelsen, Email: giehm@humgen.au.dk.
Jean-Laurent Casanova, Email: Jean-Laurent.Casanova@mail.rockefeller.edu.
References
- 1. Arvin AM, Gilden D. Varicella-zoster virus. In: Knipe DM, Howley P, eds. Fields Virology. Philadelphia, Pennsylvania, USA: Lippincott Williams & Wilkins;2015: 2015–2057. [Google Scholar]
- 2.Zerboni L, Sen N, Oliver SL, Arvin AM. Molecular mechanisms of varicella zoster virus pathogenesis. Nat Rev Microbiol. 2014;12(3):197–210. doi: 10.1038/nrmicro3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Whitley RJ. Chickenpox and herpes zoster (varicella-zoster virus). In: Bennet J, Dolin R, Blaser MJ, eds. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Amsterdam, Netherlands: Elsevier; 2014: 1731–1737. [Google Scholar]
- 4.Gershon AA, Gershon MD. Perspectives on vaccines against varicella-zoster virus infections. Curr Top Microbiol Immunol. 2010;342:359–372. doi: 10.1007/82_2010_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breuer J. VZV molecular epidemiology. Curr Top Microbiol Immunol. 2010;342:15–42. doi: 10.1007/82_2010_9. [DOI] [PubMed] [Google Scholar]
- 6.Wiegering V, et al. Varicella-zoster virus infections in immunocompromised patients — a single centre 6-years analysis. BMC Pediatr. 2011;11:31. doi: 10.1186/1471-2431-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks EG, et al. A novel X-linked combined immunodeficiency disease. J Clin Invest. 1990;86(5):1623–1631. doi: 10.1172/JCI114884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320(26):1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 9.Hsu AP, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118(10):2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobbs K, et al. Inherited DOCK2 deficiency in patients with early-onset invasive infections. N Engl J Med. 2015;372(25):2409–2422. doi: 10.1056/NEJMoa1413462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, et al. Combined immunodeficiency associated with DOCK8 mutations. N Engl J Med. 2009;361(21):2046–2055. doi: 10.1056/NEJMoa0905506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roesler J, et al. Listeria monocytogenes and recurrent mycobacterial infections in a child with complete interferon-gamma-receptor (IFNgammaR1) deficiency: mutational analysis and evaluation of therapeutic options. Exp Hematol. 1999;27(9):1368–1374. doi: 10.1016/S0301-472X(99)00077-6. [DOI] [PubMed] [Google Scholar]
- 13.Kreins AY, et al. Human TYK2 deficiency: mycobacterial and viral infections without hyper-IgE syndrome. J Exp Med. 2015;212(10):1641–1662. doi: 10.1084/jem.20140280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mace EM, et al. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood. 2013;121(14):2669–2677. doi: 10.1182/blood-2012-09-453969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casanova JL. Human genetic basis of interindividual variability in the course of infection. Proc Natl Acad Sci U S A. 2015;112(51):E7118–E7127. doi: 10.1073/pnas.1521644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci U S A. 2015;112(51):E7128–E7137. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317(5844):1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 18.Sancho-Shimizu V, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121(12):4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez de Diego R, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33(3):400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman M, et al. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J Exp Med. 2012;209(9):1567–1582. doi: 10.1084/jem.20111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Audry M, et al. NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J Allergy Clin Immunol. 2011;128(3):610–617.e1. doi: 10.1016/j.jaci.2011.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen LL, et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med. 2015;212(9):1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim HK, et al. TLR3 deficiency in herpes simplex encephalitis: high allelic heterogeneity and recurrence risk. Neurology. 2014;83(21):1888–1897. doi: 10.1212/WNL.0000000000000999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22(2):240–273. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 26.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10(10):1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unterholzner L, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoffmann NA, et al. Molecular structures of unbound and transcribing RNA polymerase III. Nature. 2015;528(7581):231–236. doi: 10.1038/nature16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefèvre S, et al. Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat Struct Mol Biol. 2011;18(3):352–358. doi: 10.1038/nsmb.1996. [DOI] [PubMed] [Google Scholar]
- 32.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itan Y, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13(2):109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itan Y, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A. 2015;112(44):13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 36.Nagel MA, Choe A, Traktinskiy I, Cordery-Cotter R, Gilden D, Cohrs RJ. Varicella-zoster virus transcriptome in latently infected human ganglia. J Virol. 2011;85(5):2276–2287. doi: 10.1128/JVI.01862-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11(10):1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 38.Bernard G, et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89(3):415–423. doi: 10.1016/j.ajhg.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tétreault M, et al. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89(5):652–655. doi: 10.1016/j.ajhg.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitsu H, et al. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am J Hum Genet. 2011;89(5):644–651. doi: 10.1016/j.ajhg.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341(6152):1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoggins JW, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505(7485):691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ku CC, Zerboni L, Ito H, Graham BS, Wallace M, Arvin AM. Varicella-zoster virus transfer to skin by T cells and modulation of viral replication by epidermal cell interferon-alpha. J Exp Med. 2004;200(7):917–925. doi: 10.1084/jem.20040634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.James SH, Kimberlin DW, Whitley RJ. Antiviral therapy for herpesvirus central nervous system infections: neonatal herpes simplex virus infection, herpes simplex encephalitis, and congenital cytomegalovirus infection. Antiviral Res. 2009;83(3):207–213. doi: 10.1016/j.antiviral.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kragballe K, Desjarlais L, Marcelo CL. Increased DNA synthesis of uninvolved psoriatic epidermis is maintained in vitro. Br J Dermatol. 1985;112(3):263–270. doi: 10.1111/j.1365-2133.1985.tb04852.x. [DOI] [PubMed] [Google Scholar]
- 46.Jakobsen M, et al. Amelioration of psoriasis by anti-TNF-alpha RNAi in the xenograft transplantation model. Mol Ther. 2009;17(10):1743–1753. doi: 10.1038/mt.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.