Abstract
The most frequent chromosomal structural loss in hepatocellular carcinoma (HCC) is of the short arm of chromosome 8 (8p). Genes on the remaining homologous chromosome, however, are not recurrently mutated, and the identity of key 8p tumor-suppressor genes (TSG) is unknown. In this work, analysis of minimal commonly deleted 8p segments to identify candidate TSG implicated GATA4, a master transcription factor driver of hepatocyte epithelial lineage fate. In a murine model, liver-conditional deletion of 1 Gata4 allele to model the haploinsufficiency seen in HCC produced enlarged livers with a gene expression profile of persistent precursor proliferation and failed hepatocyte epithelial differentiation. HCC mimicked this gene expression profile, even in cases that were morphologically classified as well differentiated. HCC with intact chromosome 8p also featured GATA4 loss of function via GATA4 germline mutations that abrogated GATA4 interactions with a coactivator, MED12, or by inactivating mutations directly in GATA4 coactivators, including ARID1A. GATA4 reintroduction into GATA4-haploinsufficient HCC cells or ARID1A reintroduction into ARID1A-mutant/GATA4-intact HCC cells activated hundreds of hepatocyte genes and quenched the proliferative precursor program. Thus, disruption of GATA4-mediated transactivation in HCC suppresses hepatocyte epithelial differentiation to sustain replicative precursor phenotype.
Keywords: Genetics, Oncology
Keywords: Epigenetics, Liver cancer
Introduction
Hepatocellular carcinoma (HCC) is the world’s second leading cause of cancer death. In the effort to understand the molecular mechanisms of HCC genesis, a relevant lead is that the most recurrent chromosomal structural loss in HCC is of chromosome 8 (8p), occurring in more than 60% of cases (1, 2). The frequency of this event suggests loss of one or more 8p genes is central to transformation. Of the approximately 500 genes on chromosome 8p, however, none are mutated at a high rate — presumably, complete loss of function by mutation of the remaining alleles might be deleterious, and haploinsufficiency of specific 8p genes is enough to contribute to transformation. The lack of mutations, however, complicates the task of identifying the key tumor-suppressor genes (TSGs) and pathways disrupted by 8p deletion, and the identity of this gene or genes is unknown. One clue is that occasionally, instead of the usual deletion of the entire chromosome 8p, there are smaller deletions targeting the segment from 8p22 to 8p23 (minimal commonly deleted regions), removing tens instead of hundreds of genes (3).
There have been attempts at functional interrogation of this more limited catalog of genes. In one study, FGL1 (also known as LFIRE or HFREP-1), which encodes for a fibrinogen (blood coagulation) family protein, was identified as a candidate 8p TSG because of its location at 8p22 and its lower expression in HCC versus adjacent noncancerous liver and because its further knockdown in HCC cells by antisense oligonucleotides increased proliferation, while its exogenous introduction had the opposite effect (4). Fgl1-knockout mice, however, have no liver phenotype (5). In other studies, DLC1 on chromosome 8p22, which encodes for a GTPase-activating protein, was implicated as a TSG based on a range of observations similar to those generated for FGL1 (6, 7). Again though, Dlc1-haploinsufficient mice have no overt phenotype, liver or otherwise, and the complete knockout is embryonic lethal, with neural tube and cardiac but not liver defects (8). Thus, neither Fgl1 nor Dlc1 have been found to have a usual central role in liver cell fate determination to warrant the frequency of haploinsufficiency observed in HCC.
The epidemiologic link between HCC and inflammatory insults such as viruses suggests participation in transformation of pathways regulating liver regeneration. In several organ systems, a hierarchy of quiescent stem cells, then highly proliferative committed precursors (transit amplifying cells), then nonproliferating specialized mature cells, is well chronicled as mediating tissue homeostasis. At one point, the liver was thought to contrast with these other organs in that mature epithelial cells (hepatocytes and cholangiocytes) might respond to mitotic triggers to mediate regeneration/homeostasis (9). Recent detailed molecular and cellular examination, however, has shown that a cellular hierarchy is present in the liver. Despite difficulties in separating liver cells by morphologic appearance, a cellular subset displays high expression of early hepatocyte differentiation-driving transcription factors Tbx3, Foxa2, and Sox9, low expression of hepatocyte epithelial-differentiation genes, a diploid genome, and replicative potential; it also mediates liver homeostasis by differentiating into cells with a typical hepatocyte epithelial-differentiation gene expression profile and polyploid genome (9–12). Several groups have found that HCC phenocopies this subset of liver precursor cells (10, 13–19). How this happens, however, is unknown. Here, we show that haploinsufficiency of a master transcription factor driver of hepatocyte differentiation, GATA4, located on chromosome 8p, and genetic alterations that compromise the transactivating function of GATA4 are a cause of HCC precursor phenotype. Interestingly, another highly recurrent structural deletion in HCC, that of chromosome 17p, also centers on a gene for a master transcription factor, TP53, the master regulator of apoptosis. Thus, the master regulatory circuits for 2 major MYC-antagonizing, proliferation-terminating metazoan programs, epithelial differentiation and apoptosis, are genetically disrupted in HCC.
Results
Candidate 8p TSGs.
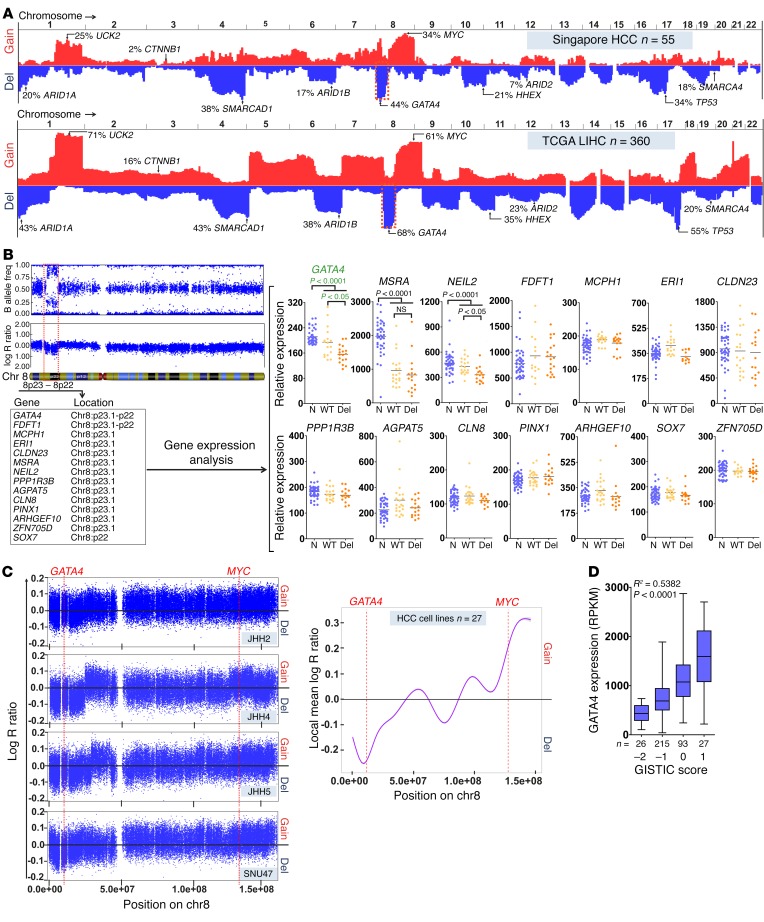
Chromosome 8p was frequently deleted in our HCC series from Singapore (24 of 55, 44% of cases). Deletion of 8p is also the most frequent structural deletion identified by SNP array karyotyping in the large TCGA series of patients with HCC in the USA (245 of 360, 68%) (Figure 1A). One patient in the Singapore series had a smaller deleted 8p segment that contained only 14 genes (Figure 1B). Of these 14 genes, only GATA4 (master transcription factor driver of hepatocyte differentiation) and NEIL2 (1 of 4 DNA glycosylases) had gene expression characteristics expected of candidate TSGs — lower expression in HCC with 8p deletion than without and in HCC versus normal liver (Figure 1B). Karyotyping of 26 HCC cell lines identified several minimal commonly deleted chromosome 8p segments that again centered close to GATA4 (Figure 1C) (Cancer Cell Line Encyclopedia [CCLE], https://portals.broadinstitute.org/ccle/home). In multiple publicly available gene expression databases, GATA4 was consistently less expressed in HCC versus normal liver and in HCC with 8p deletion than without, and a strong correlation (Spearman’s correlation, R = 0.5382, P < 0.0001, n = 361) was observed between GATA4 copy number and GATA4 expression (Figure 1D and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI93488DS1).
Figure 1. GATA4 is a candidate TSG on chromosome 8p.
(A) Chromosome gains and losses in HCC. HCC (n = 55 Singapore series, n = 330 TCGA series) karyotyped by SNP array. Blue plot, frequency of loss by chromosome location; red plot, frequency of gain. (B) Of the 14 genes in the smallest minimal deleted region observed in a patient with HCC, only 2 were less expressed in HCC with 8p deletion (del) than without (WT) and less expressed in HCC than in normal liver (N). Gene expression by microarray. Wilcoxon rank sum test, 2-tailed. (C) Chromosome 8 (chr8) copy number loss peaked in proximity to GATA4,while copy number gain peaked in proximity to MYC in multiple HCC cell lines (n = 27). Representative images for JHH2, JHH4, JHH5, and SNU475 are shown. (D) Correlation between GATA4 mRNA expression and GATA4 GISTIC score (https://software.broadinstitute.org/software/cprg/?q=node/31). Spearman’s rank correlation coefficient, 2-tailed. TCGA, n = 361.
Liver-conditional Gata4 haploinsufficiency produced a phenotype of persistent precursor proliferation and failed hepatocyte differentiation.
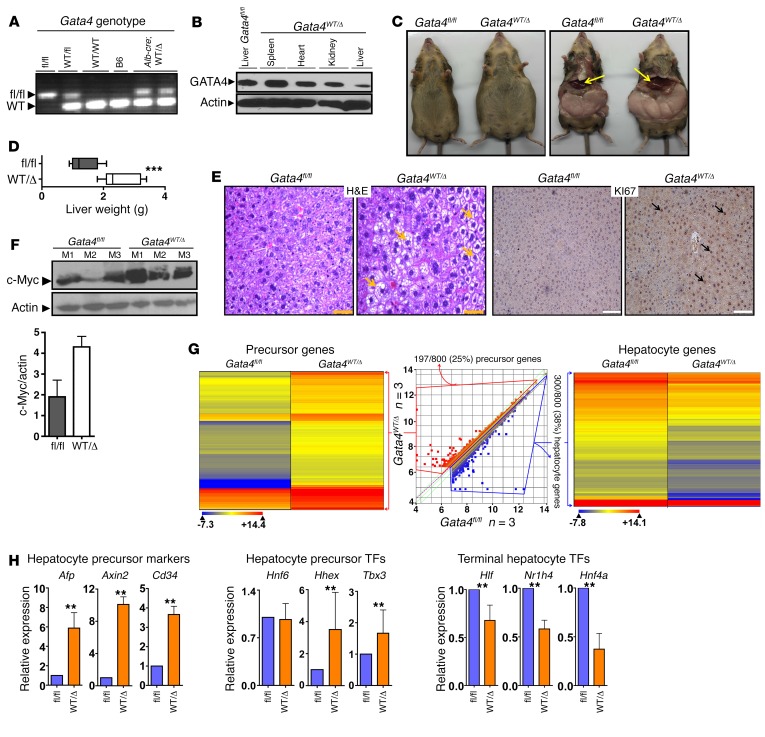
Thus, integrated copy number and gene expression analyses implicated NEIL2 and GATA4 as candidate key 8p TSGs. Neil2-knockout mice, however, have no overt phenotype (20). In contrast, Gata4-knockout mice have liver agenesis, demonstrating a critical role for GATA4 in liver development (21, 22). To model the somatically acquired liver GATA4 haploinsufficiency of HCC, we crossed Gata4fl/fl mice to albumin promoter creatinine recombinase (Alb-cre) mice to produce progeny with liver-conditional deletion of a single Gata4 allele (Gata4WT/Δ) (Figure 2A). Compared with Gata4fl/fl mice, Gata4WT/Δ mice demonstrated substantially decreased GATA4 protein levels only in the liver (Figure 2B). At formal phenotyping at 3 and 8 months, the Gata4WT/Δ mice (n = 11) had strikingly enlarged livers on a normal diet (Figure 2, C and D, and Supplemental Figure 2). Markers of proliferation Ki67 and MYC, measured by immunohistochemistry and Western blot, were substantially increased in Gata4WT/Δ (Gata4 haploinsufficient) versus Gata4fl/fl (Gata4 WT) livers (Figure 2, E and F, and Supplemental Figure 2).
Figure 2. Liver-conditional Gata4 haploinsufficiency (Gata4WT/Δ) produced enlarged livers with a proliferative precursor phenotype.
(A) Genotyping for floxed Gata4 allele in Gata4fl/fl, Alb-cre, progeny of the cross (Gata4WT/Δ), and controls. DNA isolated from tails. PCR analysis (primers in Supplemental Table 6). (B) GATA4 protein reduction in liver, but not other tissues, in Gata4WT/Δ compared with Gata4fl/fl mice. Western blot. (C) Markedly enlarged livers in Gata4WT/Δ versus Gata4fl/fl mice. Arrows indicate liver (n = 11 Gata4WT/Δ mice generated with similar phenotype). (D) Liver weights in Gata4WT/Δ versus Gata4fl/fl mice. ***P < 0.001, Wilcoxon rank sum test, 2-tailed. n = 11 each group. Sacrifice at 8 months. (E) Liver histopathology in Gata4WT/Δ versus Gata4fl/fl mice. Proliferation evaluated by immunohistochemistry for KI67. Arrows indicate fatty changes in H&E stain and positive KI67. Magnification: ×200 (H&E), ×100 (KI67). (F) MYC protein levels were increased in Gata4WT/Δ versus Gata4fl/fl livers. Western blot. Bar graph shows densitometric analysis. (G) Gene ontology analysis of genes 1.5-fold more or less expressed in Gata4WT/Δ versus Gata4fl/fl liver showed increased expression of liver precursor genes, but decreased expression of hepatocyte genes with Gata4 haploinsufficiency. RNA-sequencing at 8 months. n = 3 mice per group. Precursor and hepatocyte-specific genes were identified by their significant differential expression at intermediate versus late stages of liver development (Supplemental Table 2). (H) QRT-PCR confirmation of increase in hepatocyte precursor markers and transcription factors and decrease in terminal hepatocyte differentiation transcription factors. **P < 0.05, Wilcoxon rank sum test, 2-tailed. n = 3 mice per group.
RNA-sequencing was used to compare the gene expression profiles of Gata4-haploinsufficient versus WT livers. Of genes more highly expressed in Gata4-haploinsufficient livers, 25% (197 of 800) were associated with a precursor stage of liver development (Figure 2G and Supplemental Table 1) (23). Of genes less expressed in Gata4-haploinsufficient livers, more than a third (300 of 800, 38%) were late liver development specialized hepatocyte epithelial function genes, such as lipid metabolism genes, coagulation pathway genes, and genes that antagonize MYC to terminate proliferation (24–26). Key hepatocyte precursor marker genes Afp, Axin2, and Cd34 were also measured by quantitative real-time PCR (QRT-PCR), affirming their enrichment with Gata4 haploinsufficiency (Figure 2H).
To better understand the level of the differentiation continuum at which hepatocyte maturation was impeded by Gata4 haploinsufficiency, we also used QRT-PCR to measure expression levels of key transcription factor drivers that operate at early versus late stages of hepatocyte epithelial differentiation, identified by others during hepatocyte generation in vitro and in liver development; expression levels of commitment and early hepatocyte differentiation–driving transcription factors Hnf6, Hhex, and Tbx3 were preserved while those of late-differentiation driving transcription factors Hnf4a, Hlf, and Nr1h4 were significantly suppressed with Gata4 haploinsufficiency (Figure 2H) (17–19, 27–29). GATA4 is a “pioneer” transcription factor, known to initiate remodeling of compacted chromatin for subsequent gene activation (30). We therefore asked whether baseline differences in the chromatin state could explain preserved/high expression of transcription factor genes Hnf6, Hhex, and Tbx3, but repression of Hnf4a, Hlf, and Nr1h4. Epigenetic marks at these genes were therefore examined at the ultimate baseline, embryonic stem cells (ESCs). Hnf6, Hhex, and Tbx3 had open chromatin in ESCs, that is, low DNA CpG methylation and high histone 3 lysine 4 trimethylation (H3K4me3) levels (Supplemental Figure 3). In contrast, suppressed late-differentiation transcription factor genes Hnf4a, Hlf, and Nr1h4 had closed or compacted chromatin at this same baseline, with high CpG methylation and low H3K4me3 levels (Supplemental Figure 3).
Hundreds of hepatocyte epithelial-differentiation genes are also suppressed and precursor genes increased in HCC compared with normal liver.
Gene expression of adjacent noncancerous liver tissue and HCC was then compared by microarray (n = 46 pairs). Approximately 2,486 genes were less expressed (average expression at least 1.5-fold higher in nonmalignant liver) in HCC (n = 46) than in nonmalignant liver (n = 46) (Figure 3A). The top tissue expression association of these suppressed genes was in liver genes, that is, approximately 25% of these genes (600 of 2,486) were specialized hepatocyte genes, such as members of the cytochrome p450 family and lipid metabolism genes (Figure 3A and Supplemental Table 2) (P < 1 × 10–16, Benjamini corrected, DAVID gene ontology analysis; https://david.ncifcrf.gov/). This pattern of suppression of hundreds of specialized hepatocyte genes was seen even in HCCs that appeared to be well differentiated by light microscopy (American Joint Committee on Cancer histologic grade 1; ref. 31.) (Supplemental Figure 4). As in murine conditional liver Gata4 haploinsufficiency, the problem seemed to be with late-stage maturation, since expression of transcription factors that drive hepatocyte commitment and early differentiation (HNF6, HHEX, HNF1A) was preserved, but that of late-differentiation driving transcription factors (CEBPD, HLF, NR1H4, NR2F1) was suppressed (Supplemental Figure 5). More than 400 genes suppressed in HCC versus noncancerous livers were also suppressed in Gata4-haploinsufficient versus WT livers (Figure 3B). Specialized hepatocyte epithelial genes constituted a large fraction of these genes commonly suppressed in both HCC and Gata4-haploinsufficient livers (Figure 3C).
Figure 3. Similar precursor gene enrichment and hepatocyte gene suppression in HCC and Gata4-haploinsufficient livers.
(A) Gene ontology analysis of genes 1.5-fold more or less expressed in HCC (n = 46) versus adjacent noncancerous liver (n = 46). DAVID gene ontology analysis complemented by hepatocyte differentiation-stage–specific expression as per Supplemental Table 1. (B) Overlap in genes more or less expressed in HCC versus noncancerous and in Gata4WT/Δ (haploinsufficient) versus Gata4fl/fl (WT) livers. (C) Examples of specialized hepatocyte epithelial-differentiation genes that are similarly suppressed in Gata4-haploinsufficient murine livers (WT/Δ) and human HCC versus normal livers (N liver).
To complement the above analyses, genes differentially expressed between nonmalignant livers and HCC were identified also by another method, unsupervised hierarchical clustering. Of genes with significantly higher gene expression in HCC versus normal liver (false discovery rate [FDR] < 0.01), 90 of 700 (13%) were liver precursor genes (significantly linked with early stages of liver development) while only 10 of 700 (~1%) were terminal-differentiation genes. In contrast, of genes significantly less expressed in HCC versus normal liver, only 8 of 700 (~1%) were precursor genes, while 175 of 700 (25%) were terminal-differentiation genes (Figure 4, A and B, and Supplemental Table 3). Also, as observed earlier for Gata4-haploinsufficient livers (Supplemental Figure 3), genes highly expressed in HCC, including proliferation genes (MYC target genes), have open or “poised” chromatin at baseline in ESCs, with low CpG methylation and high H3K4me3 levels, while in contrast, genes less expressed in HCC, including hundreds of known hepatocyte epithelial-differentiation genes, begin at this same baseline with closed chromatin characterized by high CpG methylation and low H3K4me3 levels (Figure 4, C–F).
Figure 4. A baseline difference in chromatin at precursor/proliferation genes with high versus hepatocyte epithelial-differentiation genes with low expression in HCC.
(A) Precursor genes were identified by their differential expression at early/intermediate (precursor) versus terminal stages of liver development. Hierarchical clustering by GenePattern analysis (FDR < 0.01) (GEO GSE13149) (23). Gene names are listed in Supplemental Table 1. (B) High expression of precursor and low expression of epithelial-differentiation genes in HCC versus nonmalignant liver (FDR < 0.01). Gene names are listed in Supplemental Table 3. (C) Genes highly expressed in HCC, including MYC target genes, have chromatin that is open/poised for gene activation in the ultimate baseline, ESCs; in contrast, less expressed genes, including known hepatocyte epithelial-differentiation genes, have closed chromatin with low H3K4me3 levels. Aligned H1 ESC H3K4me3 ChIP-Seq data (ENCFF775QSF) were imported, analyzed, and visualized to show number of reads with or without 2,000 bp from the transcription start site using Easeq (80). (D) Close-up of H3K4me3 at cytochrome genes with low (CYP1A2) versus high (CYP2R1) expression in HCC. (E) Quantification of data shown in C. Plotted are medians and interquartile range (box) and minimum/maximum (whiskers). (F) This difference in baseline chromatin state was also evident by CpG methylation analysis. Methylation values (β values) by Illumina 450-k CpG array for CpG linked with these genes in ESC (n = 19) and normal liver (n = 4) (β values from GEO GSE31848) (81). MYC target genes, 356 genes/5716 CpG (82); hepatocyte epithelial-differentiation genes, 600 genes/9238 CpG; genes significantly highly expressed in HCC, 700 genes/10528 CpG; genes significantly less expressed in HCC 700, genes/11616 CpG.
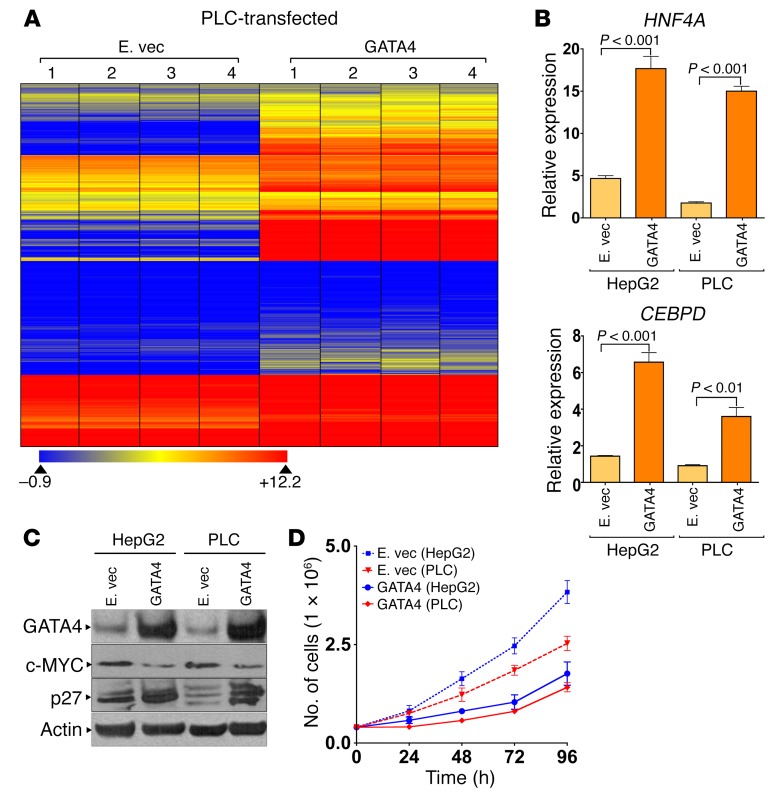
A cause-effect role for GATA4 in the regulation of these suppressed hepatocyte genes was demonstrated by their upregulation upon exogenous correction of GATA4 levels in the HCC cell line PLC, which contains an 8p deletion (Figure 5A and Supplemental Figure 6); GATA4 markedly increased expression of 329 of 600 (55%) of the hepatocyte genes suppressed in HCC (Figure 5A). Of particular interest are hepatocyte genes that antagonize MYC function to terminate proliferation; the hepatocyte factors HNF4A and CEBPD have been shown to have such actions (17–19, 27–29). GATA4 introduction activated both HNF4A and CEBPD quantified by QRT-PCR (Figure 5B). As expected, this was accompanied by downregulation of the MYC protein, upregulation of the p27/CDKN1B protein (p27/CDKN1B is the cyclin-dependent kinase inhibitor that mediates cell-cycle exits by differentiation — Cdkn1b knockout mice have >20% increase in liver mass; refs. 32–34) (Figure 5C), and decreased cell growth without early apoptosis (Figure 5D).
Figure 5. GATA4 activated hundreds of hepatocyte differentiation genes, antagonized MYC, and decreased proliferation.
(A) Introduction of GATA4 into GATA4-haploinsufficient HCC cells activated hundreds of hepatocyte genes. Expression vectors for GATA4 or empty vector control were transfected into GATA4-haploinsufficient (8p deleted) HCC cells (PLC). RNA harvested for gene expression by microarray 96 hours after transfection. Heat map: probe set intensity values for 329 liver differentiation genes suppressed in primary HCC compared with noncancerous liver and upregulated by GATA4 in PLC. Four independent transfections and experiments. (B) GATA4 activated the MYC antagonists HNF4A and CEBPD. QRT-PCR (relative to nontransfected cells). (C) GATA4 decreased MYC and increased p27/CDKN1B protein. Western blot. (D) GATA4 reduced proliferation of HCC cells (PLC and HepG2). Cell counts by automated cell counter.
Atypical HCC containing a rare germline GATA4 loss-of-function mutation.
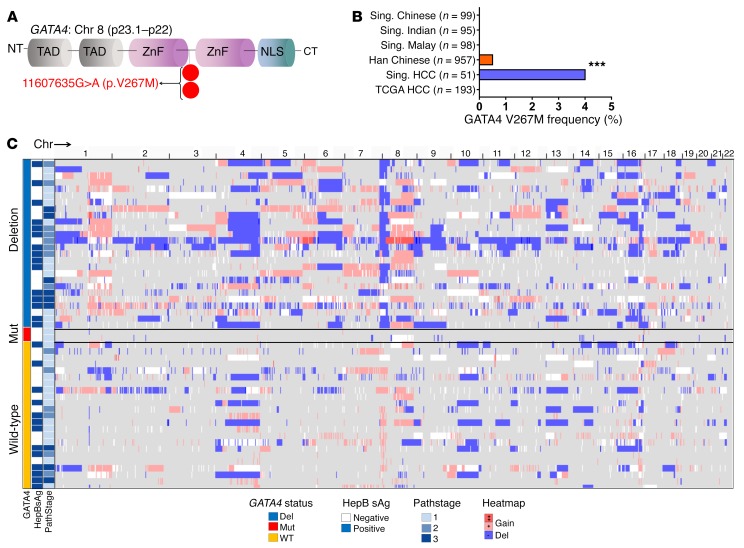
To evaluate whether GATA4 is also inactivated by mutation in HCC, we sequenced all GATA4-coding regions from HCC and paired noncancerous liver. Remarkably, we found an identical germline GATA4 missense mutation (GATA4 V267M) in 2 cases of HCC without 8p deletion (frequency 2 of 51, 4%; DNA from peripheral blood mononuclear cells was sequenced to evaluate the germline) (Figure 6A and Supplemental Figure 7). The amino acid altered by this recurrent mutation was highly conserved across species (Supplemental Figure 7G). Although this mutation has not been noted in the sequencing of the general population of Singapore, it has been noted as a rare, exclusively heterozygous germline variant in ethnic Han Chinese in China (4 of 957, 0.4%, rs116781972), where it has been linked with congenital heart defects (Figure 6B) (35–37). Both HCC cases with the mutation were atypical compared with other HCC patients; both were female, even though HCC rates are 4 times higher in males than females (38), neither patient had hepatitis B or C or cirrhosis or other HCC risk factors, and neither HCC contained frequent chromosomal structural gains or deletions by SNP array karyotyping, contrasting with most other HCCs in the series (Figure 6C). The mutation affected only 1 GATA4 allele (variant allele frequency, ~50%) (Supplemental Figure 7 and Supplemental Table 4).
Figure 6. Atypical HCC containing a rare germline GATA4 mutation.
(A) GATA4 germline missense mutation (V267M) found in 2 of 51 patients (4%). Sanger and targeted deep sequencing of HCC, adjacent nonmalignant liver, and peripheral blood mononuclear cells (Supplemental Figure 7). (B) Frequency of this mutation in healthy individual populations from Singapore (Sing.) and China (34–36) ***P < 0.001, Fisher’s exact test. (C) HCC cases with GATA4 mutation had near-normal karyotype. Karyotype analyzed by SNP array and imaged by IGV. Pathstage, American Joint Committee on Cancer staging system. HepBsAg, hepatitis B surface antigen; Del, GATA4 locus deletion.
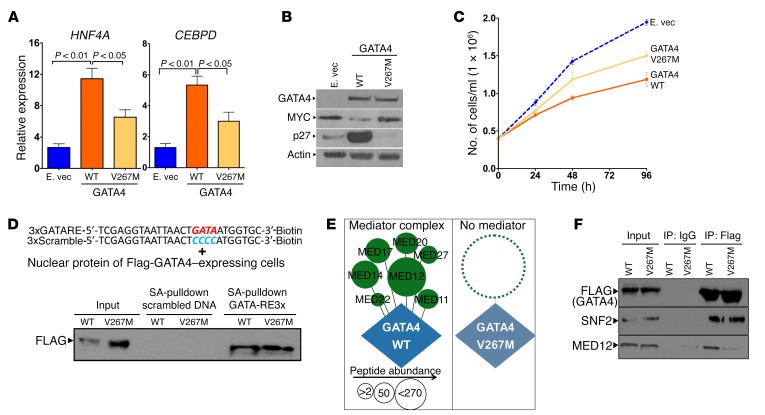
Unlike gene deletion, missense alteration is not unmistakably a loss-of-function event. When we used expression vectors to express similar amounts of WT GATA4 or GATA4 V267M protein in GATA4-haploinsufficient HCC cells (PLC), however, GATA4 V267M was less able than GATA4 to activate HNF4A and CEBPD, downregulate MYC, upregulate p27/CDKN1B, or slow HCC cell growth (Figure 7, A–C). We therefore looked for biochemical reasons for decreased transactivating function of GATA4 V267M. Decreased DNA binding was not the explanation, since GATA4 V267M bound GATA-response elements with affinity equal to that of WT GATA4 in DNA-binding assays (Figure 7D). Nor was GATA4 V267M mislocalized to the cytoplasm instead of the nucleus by Western blot of cell fractions (Supplemental Figure 8). Amino acid 267 is located between the zinc finger DNA-binding domains of GATA4, a region thought to mediate protein-protein interactions. Thus, we comprehensively analyzed the GATA4 V267M and WT GATA4 protein interactomes by immunoprecipitation followed by liquid chromatography tandem mass spectrometry (LC-MS/MS). The unbiased protein interactome analysis revealed a striking absence of the mediator complex from the GATA4 V267M versus the WT GATA4 protein interactomes (Figure 7E, Supplemental Figure 9–11, and Supplemental Table 5). This finding was confirmed also by immunoprecipitation–Western blot for mediator 12 (MED12), performed in triplicate (Figure 7F and Supplemental Figure 11).
Figure 7. Mutant GATA4 V267M does not recruit the mediator complex and is transcriptionally less active.
(A) Impaired activation of HNF4A and CEBPD by GATA4 V267M versus GATA4. Cells transiently transfected with expression vector for GATA4, GATA4 V267M, or empty vector control. QRT-PCR, relative to nontransfected cells. (B) Western blot for GATA4, c-MYC, p27/CDKN1B, and actin. (C) Cell proliferation. (D) GATA4 and GATA4 V267M binding to GATA response elements. PLC HCC cells transfected with expression vectors for GATA4 or GATA4 V267M. Nuclear lysates incubated with DNA probes containing GATA4 response elements X3-biotin tag or with control scrambled probes. Bound protein detected by Western blot after streptavidin pull-down. (E) Mediator was absent from the GATA4 V267M interactome. LC-MS/MS analysis of protein interactome of GATA4 and GATA4 V267M immunoprecipitated from transfected PLC cells using anti-Flag antibody. Circle size indicates protein abundance in the coimmunoprecipitate (Supplemental Figure 10). (F) Confirmation by immunoprecipitation/Western blot of absence of mediator, but not other coactivators. Triplicate results are shown in Supplemental Figure 11.
Major coactivators for GATA4 are recurrently inactivated in HCC.
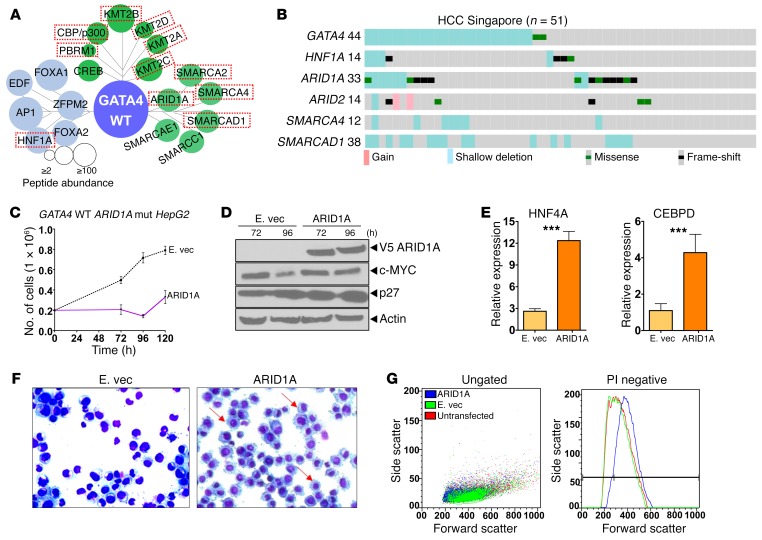
GATA4 V267M loss of interactions with mediator complex suggested that direct inactivation of key coactivators that mediate transcription activation by GATA4 could be another mechanism by which GATA4 function is impaired in HCC. Coactivators for GATA4 were identified by LC-MS/MS analysis of the GATA4 protein interactome (Figure 8A and Supplemental Table 5). Several of the identified coactivators were recurrently inactivated at a high rate in HCC (~40% of cases), observed in both the Singapore and TCGA series, with the highest inactivating mutation rate observed for AT-rich interactive domain 1A (ARID1A) (Figure 8B and Supplemental Figure 12). HNF1A, a differentiation-driving transcription factor found in the GATA4 protein interactome, was also frequently mutated in both the Singapore and TCGA series (Figure 8, A and B, Supplemental Figure 12, and Supplemental Table 5). Consistent with the idea that coactivator inactivation will negatively affect GATA4-mediated gene activation, there was similar suppression of key late-differentiation factors downstream of GATA4 in both GATA4-haploinsufficient and ARID1A/SMARCAD1/ARID2/SMARCA4-deficient HCC; this suppression was greater than that observed in HCC without these alterations in both the Singapore and TCGA series (Supplemental Figure 13). Reintroduction of ARID1A into ARID1A-mutant/GATA4-intact HCC cells (HepG2) by expression vector activated key terminal hepatocyte epithelial-differentiation genes, downregulated MYC, upregulated p27/CDKN1B, induced morphologic changes of epithelial differentiation, and terminated proliferation (Figure 8, C–G). These analyses suggested a model in which inactivation of hepatocyte master transcription factor GATA4 and/or GATA4 coactivators impaired activation of terminal hepatocyte epithelial genes that physiologically antagonize MYC to mediate precursor-to-hepatocyte transition (Figure 9).
Figure 8. Recurrent inactivation of GATA4 coactivators in HCC.
(A) Coactivators highly represented in the GATA4 protein interactome. Circle size indicates protein abundance in the GATA4 coimmunoprecipitate. (B) Coactivators frequently deleted and mutated (inactivating mutations) in primary HCC (n = 51). Similar results in the TCGA series are shown in Supplemental Figure 12. ARID1A reintroduction into ARID1A-mutated, GATA4 WT HCC cells (HepG2) induced terminal epithelial differentiation. Transfection with V5-tagged ARID1A expression vector versus empty vector control. (C) Cell counts were determined using an automated counter. (D) Changes in MYC and p27/CDKN1B protein levels were consistent with terminal differentiation. Western blot. (E) ARID1A reintroduction increased HNF4A and CEBPD expression. QRT-PCR (relative to nontransfected cells). ***P < 0.001, Wilcoxon rank sum test, 2-tailed. (F) Increased cytoplasmic to nuclear ratio of ARID1A transfected cells consistent with epithelial differentiation. Giemsa stain 96 hours after transfection. (G) Increased cell size (forward scatter) and granularity (side scatter) by flow cytometry consistent with epithelial differentiation 96 hours after transfection.
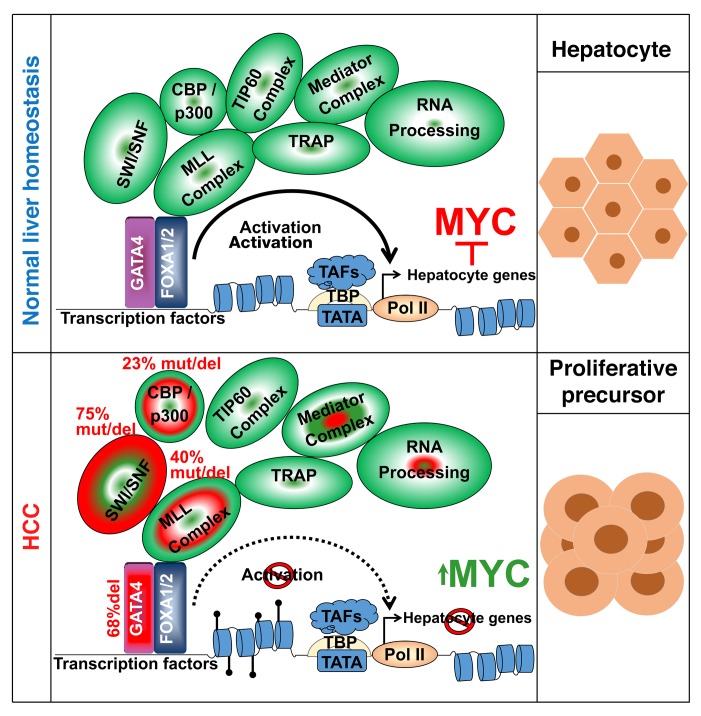
Figure 9. The model: several genetic alterations in HCC impair master transcription factor–mediated (GATA4) chromatin remodeling/transactivation to suppress precursor-to-epithelial transition.
Normal liver cells have intact master transcription factors (e.g., GATA4/FOXA1) that dictate cell fate by recruiting coactivators (e.g., ARID1A) to remodel and activate downstream transcription factors and hundreds of hepatocyte epithelial-differentiation genes, antagonizing MYC in the process for terminal epithelial differentiation. In HCC, haploinsufficiency or loss-of-function mutation in 1 allele of GATA4 and/or inactivation of GATA4 coactivators (e.g., ARID1A) impairs this chromatin remodeling and transcription activation to produce a selective suppression of terminal epithelial-differentiation genes, a selective repression that is facilitated by the inherently closed chromatin state of these versus commitment and MYC proliferation genes.
CTNNB1 mutations were not detected in our HCC series.
Mutations in CTNNB1 are known to be less frequent in HCC in Asian subjects and/or patients with hepatitis B than in HCC in patients of other ethnicities or those without hepatitis B (refs. 39–43; reviewed in ref. 44). To extend these observations, we used targeted next-generation sequencing to evaluate for CTNNB1 mutations in the Singapore HCC series and found no mutations (mean coverage 2,777, range 38–5,645; 56% of the Singapore HCC cases were hepatitis B sAg positive). In contrast, approximately 25% of HCC in the TCGA series from the USA contained CTNNB1 mutations (Supplemental Figure 14). There was no significant difference, however, in the rate of GATA4 deletion, ARID1A mutation or deletion, or TP53 mutation or deletion in the Singapore versus TCGA series.
Discussion
The liver replenishes hepatocytes consumed by metabolic work and insults such as viruses routinely and in controlled fashion. One control on this tissue homeostasis is WNT-CTNNB1 signaling, which initiates tissue precursor proliferation (10), and mutations that constitutively activate CTNNB1 are frequent in HCC in Europe and North America (20%–40% of cases). CTNNB1 mutations are less frequent in Asia and in patients with hepatitis B–associated HCC (39–44), possibly because hepatitis B viral X protein directly increases WNT/CTNNB1 signaling (45, 46). Other homeostatic controls terminate precursor proliferation once it is initiated; advancing differentiation (transition to specialized lineage fate) routinely terminates exponential proliferation of precursors, and degree of differentiation failure correlates with clinical aggressiveness of HCC (47). Even if differentiation derangement is not obvious by light microscopy, as can be the case with HCC classified as well differentiated (grade 1 histologic grade), it is evident by gene expression analyses that precursor genes are elevated and hundreds of hepatocyte genes are suppressed in grade 1 HCC compared with normal liver. That HCC phenocopies proliferating precursors with suspended epithelial differentiation has also been observed by others by (i) detailed immunophenotyping (15); (ii) a near-diploid genome characteristic of liver precursors (instead of the polyploid genome of mature hepatocytes) (10, 16); (iii) capacity of HCC cells to differentiate into hepatocytes (13, 14); and (iv) pericentral vein location of HCC that is similar to that of liver precursors (10, 16).
The mechanisms underlying this precursor phenotype of HCC were not known. A small subset of the hundreds of transcription factors expressed in cells are masters that command and coordinate other transcription factors to drive lineage fates. It is thus notable that a master transcription factor driver of hepatocyte lineage fate, GATA4 (24–26), is located in a minimal deleted segment of chromosome 8p in this and other HCC series (1, 3, 6, 7). Conditional deletion of 1 allele of Gata4 from murine liver, to model the haploinsufficiency observed in human HCC, created an enlarged, proliferative liver phenotype, with a gene expression profile of precursors and failed terminal hepatocyte epithelial differentiation that mimicked that of HCC. Presumably only 1 allele of GATA4 is inactivated in HCC because some GATA4 is needed for existence along the GATA4-dependent liver differentiation continuum; Gata4 complete knockouts have liver agenesis (21, 22).
GATA4 is a known “pioneer” transcription factor, that is, GATA4 has been shown to access compacted chromatin that is inaccessible to lesser transcription factors to initiate the remodeling needed for subsequent gene activation (30). It is notable, therefore, that transcription factor drivers of proliferation, hepatocyte commitment, and early differentiation (Myc, Hnf6, Hhex, Tbx3) that had preserved or even increased expression in Gata4-haploinsufficient livers and HCC versus normal livers had chromatin already poised or open at the earliest baseline of ESC, while in contrast, transcription factor drivers of terminal epithelial differentiation (Hnf4a, Hlf, and Nr1h4) and hundreds of hepatocyte epithelial-differentiation genes that had significantly decreased expression in Gata4-haploinsufficient livers and HCC had closed chromatin at this same baseline. That is, marked baseline differences in the chromatin state of proliferation/commitment versus terminal differentiation genes may interact with pioneer/master transcription factor deficiency to stall differentiation at committed, intrinsically proliferative points in the differentiation continuum.
Also implicating chromatin remodeling failure in HCC genesis is direct inactivation of coactivators (chromatin remodelers) themselves — transcription factors are selective in their coactivator usage (48, 49) — and of the coactivators interacting with GATA4, identified by MS analysis of the GATA4 protein interactome, several (ARID1A, SMARCAD1, ARID2, SMARCA4) are genetically inactivated at a high rate in HCC (>40% inactivating mutations or deletions). Consistent with these loss-of-function events negatively affecting transactivation by GATA4, reintroduction of ARID1A into ARID1A-deficient/GATA4-intact HCC cells activated GATA4 target genes, resumed hepatocyte epithelial differentiation, and terminated proliferation. Also, liver-conditional deletion of 1 Arid1a allele in mice created a phenotype of proliferative liver precursors with impaired hepatocyte differentiation (50), and genetic inactivation of Smarcb1 in mice potently suppressed hepatocyte differentiation and increased proliferation (51).
We also found 2 HCC cases without GATA4 deletion, but with a very rare germline missense mutation in GATA4 (GATA4 V267M) that abrogated its interactions with mediator (MED12, etc.), another complex that mediates transcription activation. Both patients did not have usual HCC risk factors; they were female and did not have hepatitis B, alcohol use, or cirrhosis, and interestingly, neither HCC contained frequent chromosomal structural gains or deletions by SNP array karyotyping. Based on this atypical character of the cases and the enrichment of the mutation in the Singapore HCC series (4% from 0.4% reported in the general ethnic Han Chinese population), this germline mutation could be a risk factor for HCC.
Biochemical actions of coactivators to open chromatin are balanced by corepressors that keep it compacted. A corollary of coactivator loss of function, therefore, is unbalanced activity of opposing corepressors (52). This suggests pharmacologic inhibition of corepressors can potentially compensate for genetic reduction in coactivators to crack the chromatin barrier and resume epithelial differentiation of HCC cells (52). This result is observed in vitro with the clinical drug decitabine, which inhibits a corepressor, DNA methyltransferase 1 (DNMT1), that creates and/or maintains the CpG methylation repression marks strikingly enriched at terminal differentiation versus proliferation/commitment genes (53–58). Unfortunately decitabine has trivial distribution into the liver in vivo, hindering clinical translation to treat HCC (57, 59). Several clinical development efforts are thus directed toward improving distribution of decitabine and related 5-azacytidine into liver (57, 59). A reason to pursue such development is that cell cycle exits by resumed epithelial differentiation do not require p53 — conventional oncotherapy applies stress upstream of p53 to upregulate it and cause apoptosis (cytotoxicity); such intent is undermined when p53 or its key cofactors (e.g., p16/CDKN2A) are absent/nonfunctional, as in HCC (60).
Can MYC gain of function itself inhibit terminal epithelial differentiation (13)? Although we cannot rule this out, consideration of ordered multicellularity and experimental data here and elsewhere indicate that MYC is subordinate to terminal differentiation. For example, HNF4A dominantly antagonizes MYC to terminate proliferation even in cancer cells with MYC amplification (58, 61–70), and GATA4 and ARID1A restoration in the experiments here activated hundreds of epithelial-differentiation genes, suppressed MYC expression, and terminated proliferation despite several genetic alterations in the HCC cells that amplify and stabilize MYC.
GATA4 has pioneer and/or master transcription factor roles in tissues originating from other germ layers besides endoderm. Also, the chromatin landscape of ESCs, the ultimate tissue precursors, suggests that pioneer function is broadly less needed for activation of intrinsically accessible proliferation/commitment genes, but necessary for activation of intrinsically compacted terminal epithelial-differentiation genes. Notably, therefore, chromosome 8p deletion is also highly recurrent in cancers of the lung, colon, breast, prostate, head, neck, bladder, ovary, prostate, and brain, and minimal commonly deleted regions in these cancers also incorporate GATA4 (71–75). GATA4 reintroduction into glioblastoma cell lines promoted differentiation and decreased proliferation (71). It is conceivable, therefore, that GATA4 haploinsufficiency and/or coactivator inactivation is how MYC-antagonizing terminal-differentiation programs are selectively suppressed in multiple cancers.
Missing from our understanding of HCC genesis was the gene(s) and pathways driving selection pressure for highly recurrent chromosome 8p deletions. Here, we showed that GATA4 is a key TSG targeted by this alteration and that GATA4 loss of function explains in large part how HCC cells suspend the epithelial fate that would otherwise suppress replicative precursor phenotype.
Methods
Primary HCC and paired nonmalignant liver tissue.
Fresh primary HCC tumors were surgically removed (therapeutic segmentectomy or hemihepatectomy) at the National Cancer Centre Singapore between 2008 and 2011. Resected liver tissue was analyzed by frozen section to identify neoplastic and nonneoplastic areas. These were macrodissected for analyses as HCC and paired nonmalignant liver tissue.
Liver-conditional knockout of Gata4.
Gata4 double-floxed mice were purchased from the Jackson Laboratory (stock 008194). These mice were crossed to albumin-cre mice, also from the Jackson Laboratory (stock 003574). 100% of F1 progeny were Gata4 haploinsufficient only in hepatocyte cells. Both double-floxed and liver-conditional Gata4-haploinsufficient mice were monitored daily; animals with signs of distress were euthanized by an IUCAC-approved protocol. PCR genotyping primer (Supplemental Table 6) sequences were provided by the Jackson Laboratory. DNA was isolated from mouse tails using the DNA Purification Kit (Promega catalog A1020).
RNA and DNA extraction.
Snap-frozen specimens were equilibrated with a buffer (RNAlater-ICE, Ambion) that preserves RNA integrity. Frozen specimens, no larger than 0.5 cm, were added to RNAlater-ICE that was first cooled to –80°C in polypropylene tubes and equilibrated with the RNAlater-ICE at –20°C overnight. The specimens were separated for (a) tissue homogenization and RNA extraction and (b) proteinase K digestion and DNA extraction. The fragment for RNA extraction was homogenized using a tissue homogenizer. The mirVana Kit (Ambion) was used per the manufacturer’s protocol for extraction of total RNA. The fragment for DNA extraction was minced with a blade into fragments approximately 2 mm in dimension. These fragments were placed in Eppendorf tubes with cold PBS and left on ice for 5 to 10 minutes to leach out the RNAlater-ICE before digestion in proteinase K for DNA extraction. After removing PBS, lysis buffer was added followed by proteinase K digestion as per the manufacturer’s instructions for the DNeasy Blood and Tissue Kit (QIAGEN).
Cell culture and transfection.
Human HCC cell lines HepG2 (ATCC, stock Hb-8065) and PLC (ATCC, stock CRL-8024) were cultured in RPMI media with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin (Mediatech). Cells were incubated at 37°C in a 5% CO2 atmosphere. DNA was isolated from both cell lines for SNP array analysis. PLC cells were transfected with GATA4 WT and GATA4 V267M vector, and approximately 60 × 106 cell pellets were harvested at 72 hours. Harvested cells were resuspended in 1× PBS plus PI plus PMSF (PBSW buffer PH7-9). The pellet was centrifuged at 835 g for 5 minutes. Cells were washed 3 times in PBSW buffer.
GATA4 and ARID1A expression vectors.
WT GATA4 cDNA was cloned into pFlag-CMV4 (OriGene Technologies) using Clontech infusion cloning (Clontech Laboratories Inc.). The primers used for infusion cloning (Supplemental Table 6) were designed to insert WT GATA4 cDNA at the C terminal end of the Flag tag in the pFlagCMV4 vector (OriGene Technologies). Transient transfection of GATA4 WT or empty vector into HCC cell lines was performed using Xfect transfection (Clontech) following the manufacturer’s guidelines. Cell pellets were isolated at 0, 48, and 96 hours after transfection for downstream analysis. pLenti-puro-ARID1A was a gift from Ie-Ming Shih (Johns Hopkins, Baltimore, MD, USA) (Addgene plasmid 39478) (76).
In vitro site-directed mutagenesis.
In vitro site-directed mutagenesis was performed using the Stratagene QuikChange Site-Directed Mutagenesis Kit (catalog 200519). Mutagenic oligonucleotides specific for point mutation 11607635G>A in GATA4 exon 4 (Supplemental Table 6) were designed and ordered from Integrated DNA Technologies (ITD). Both forward and reverse primers annealed to the same sequence on opposite strands of the plasmid expressing GATA4 cDNA. The mutant strand synthesis reaction was performed with the recommended PCR conditions from a Stratagene kit and using PfuTurbo DNA polymerase (Stratagene 200519). After the thermocycler reaction, the PCR product was treated with Dpn I endonuclease reagent to digest the WT parenteral DNA template. The Dpn I–treated PCR product was purified and used to transform XL1 Blue Supercompetent Cells. The cells containing the mutant cDNA were used to transform E. coli at 37°C overnight. Colonies were selected and mini-prep reaction was carried out using the QIAGEN Mini-Prep Kit (catalog 27106). The isolated bacteria DNA was sequenced to confirm the presence of the mutant GATA4 cDNA by both Sanger sequencing and target deep sequencing using primers designed to amplify WT GATA4 cDNA (data not shown) (Supplemental Table 2). Colonies with mutant GATA4 were maxi-prepared using the Promega Pure Yield Plasmid Kit (catalog A2492) and used for transfection experiments.
QRT-PCR using SYBR green.
RNA was isolated using the RNeasy method (QIAGEN), and cDNA was prepared using the iScript cDNA Synthesis Kit (Bio-Rad). QRT-PCR was done using an ABI Prism 7500 Sequence Detection System (Applied Biosystems) and SYBR Premix Ex Taq II (TakaRa). Real-time PCR primers (Supplemental Table 6) were designed with PrimerQuest (https://www.idtdna.com/primerquest/Home/Index). The relative number of copies of mRNA (RQ) was calculated based on the average Ct values using the housekeeping gene GAPDH as internal control and baseline controls for relative expression. Results are shown as mean ± SD of 3 independent experiments.
Sanger and targeted next-generation sequencing.
All coding region exons of GATA4 in genomic DNA from paired primary HCC and nonmalignant liver tissue were Sanger sequenced by ABI 3730×I DNA analyzer (Applied Biosystems). Primers for bidirectional sequencing were designed using PrimerQuest (www.idtdna.com) (Supplemental Table 6). DNA sequences were analyzed using FinchTV DNA analysis software. In patients in whom mutations were identified, DNA isolated from peripheral blood mononuclear cells was similarly sequenced to determine whether the mutation was germline.
In addition, targeted next-generation sequencing was applied to exon 4 of GATA4 (to confirm the mutation identified by Sanger sequencing) and to all coding exons of HNF1A, ARID1A, SMARCA4, ARID2, CTNNB1, and TP53. Primers (Supplemental Table 6) were designed using PrimerQuest (www.idtdna.com). Each target exon was amplified by PCR using 100 ng/μl or more of primary DNA. The PCR amplification protocol included an initial denaturation step (94°C for 5 minutes), followed by 19 cycles of denaturation (94°C for 30 seconds, at 25% ramp cooling temperature), annealing, and extension (60°C for 3 minutes at 40% ramp cooling). After PCR amplification, subsequent purification and sequencing library preparation were according to Illumina pair-end library protocol. Briefly, DNA was purified using magnetic beads. The purified PCR products were end repaired to introduce sticky ends using end-repair enzyme (NEB catalog E6050S). Paired-end adapters were then ligated using T4 DNA Ligase (NEB catalog M0202S) to the amplified PCR fragments of about 250 bp in length. Nick fill reaction was performed using Bst DNA polymerase (NEB catalog MO374S). Library fragments and all PCR amplification were performed using HotStart Applied Biological Material (ABM) Taq DNA polymerase (ABM catalog G011). The library was subjected to deep sequencing on Illumina MiSeq using 2 × 300 paired-end sequencing following standard loading procedures. Coverage information for each sequenced gene for each sample is reported in Supplemental Table 4. Raw sequencing reads were trimmed of Illumina adapters, and low quality reads were discarded. Paired-end alignment was done using BWA 0.6 aligner with hg19 as the reference sequence. Variant extraction was done using the GATK pipeline (GATKv3.3). We prioritized variants after an elimination of sequencing/mapping errors (removal of low-quality reads, sequencing depth of at least 30 reads and 10 mutation reads, removal of variants with directional bias), removal of potential, common, benign polymorphisms (using publicly available databases of common and rare germline variants such as ExAC [http://exac.broadinstitute.org/], ESP6500 [http://evs.gs.washington.edu/EVS/], and 1000 Genomes [http://www.internationalgenome.org/]), and removal of variants with minor allelic frequency of more than 0.0001. Sequences were aligned to the reference genome using NovoAlign and were analyzed using Integrative Genome Viewer (IGV) software.
RNA sequencing.
RNA was extracted from murine livers by the methods discussed above; however, the liver samples were also treated with DNaseI to remove potential DNA contamination. RNA integrity was measured by 1% 2D gel electrophoresis. Samples with 28S and 18S RNA (n = 3 mice per group) were sent for sequencing by ABM. RNA quality check was confirmed using Agilent 2100 Bioanalyzer, with all samples passing quality control. The samples were subjected to poly A enrichment, followed by fragmentation, first- and second-strand synthesis, adenylation of 3′ ends, adapter ligation, DNA fragmentation enrichment, and real-time PCR quantification. Cluster generation and sequencing were carried out in one run on NextSeq 500 (Illumina, cluster generation and 2-channel sequencing), and Bcl files were converted to FastQ data immediately after the run. Over forty million paired-end reads for all 6 samples were recovered from the sequencing. Sequence reads were aligned to murine reference genome mm10 by Bowtie 2, and expression data were generated for analysis. Heat maps were generated using ArrayStarv3 (DNASTAR).
Gene expression analysis by microarray.
The HumanHT-12 v3 gene expression microarray (Illumina) was used to analyze RNA from paired HCC and nonmalignant liver. The array evaluated over 25,000 annotated genes with over 48,000 probes designed using RefSeq (Build 36.2, Rel 22) and UniGene (Build 199). Microarray probe intensity values were subject to average normalization by GenomeStudio software to minimize the effects of variation from nonbiological factors and to calculate expression measures from the raw data. Expression measures of probe sets covering specific genes of interest were exported as a spreadsheet to the SAS System V9.2 (SAS Institute Inc.) for further statistical analysis. Only probe intensity data with detection P values of less than 0.05 (a statistical calculation that provides the probability that the signal from a given probe is greater than the average signal from the negative controls) were used in analyses of differences between groups of samples. Heat maps were generated by ArrayStarv3 (DNASTAR). All original microarray data were deposited in the NCBI’s Gene Expression Omnibus (GEO GSE57958).
High-resolution molecular karyotyping by SNP array.
The Human660W-Quad v1.0 DNA BeadChip Kit was used for high-resolution molecular karyotyping of DNA isolated from primary HCC specimens and a control nonmalignant DNA. The BeadChip analyzed more than 660,000 individual loci. Genome Studio (Illumina), KaryoStudio (Illumina), and IGV (77) software were used to document large chromosome aberrations (e.g., >75 kb), to score these aberrations as loss, gain, or uniparental disomy (UPD), and for crossmatching these aberrations with information from public databases. Affymetrix SNP6 CEL files of 27 liver-derived CCLE samples were converted into A- and B-allele frequencies using crlmm of the Bioconductor R package oligo. log R ratios (LRR) were then computed as log2 of A+B divided by its median. Curves fitted to LRR were obtained using the ggplot2 function stat_smooth with its span parameter set to 0.3. The smooth curve displayed without its data was found similarly, but using data from all 27 CCLE liver cell lines combined.
Cell fractionation and nuclear protein extraction.
Cells were resuspended in 500 μl PBSW followed by addition of 10% NP-40 (1:20 μl). This was incubated on ice for 1 minute followed by centrifugation at 835 g for 10 minutes. Supernatant containing the cytoplasmic fraction was transferred to a separate tube. Nuclear pellets were washed in PBSW buffer and centrifuged at 835 g for 10 minutes. Benzonase was added (1 μl), and this was incubated on ice for 90 minutes and vortexed every 30 minutes. 250 μl of nuclear extraction buffer E1 (250 μl PBSW + 2% NP40 + 500 mM [5 μl] + 5 M NaCl [25 μl]) was added, followed by a 10-minute incubation on ice; mixture was vortexed every 5 minutes. Sample was centrifuged at full speed for 10 minutes. The supernatant, which contained the cytoplasmic fraction, was transferred to a separate tube. 5 μl of 10% SDS was added to the remaining nuclear pellet. Nuclear protein was extracted again by adding 250 μl of nuclear extraction buffer E2 (250 μl PBSW + 1% NP40 + 500 mM [2.5 μl] NaCl) to the remaining pellet. Sample was incubated on ice for 10 minutes and vortexed every 5 minutes as above. This was followed by centrifugation at full speed for 15 minutes. Supernatant was added to the nuclear protein extraction tube. Nuclear extraction buffer (200 μl) was added to the tube containing the remaining pellet. This was incubated on ice for 10 minutes with vortexing every 5 minutes. The sample was centrifuged at full speed for 15 minutes, and supernatant was added to the nuclear pellet tube. Concentration of the total protein extracted was measured using BCA.
Covalent binding of antibody to protein G beads.
25 mg (200 μl) of protein G–sepharose was washed twice with 1× PBS, followed by incubation with 200 μl of Flag antibody for 1 hour at room temperature and pressure. Antibody-bound protein G was incubated in 1% chicken egg albumin for 1 hour. This was washed twice with 1× PBS. 25 mg of dimethylpimelimidate was added to 1 ml of 300 mM NEM, followed by swirling for 30 minutes at room temperature and pressure. This was repeated twice. Glycine-HCl (PH3), was added, followed by spin down. This was washed 3× using 1× PBS. Samples were then washed ×2 using nuclear extraction buffer.
Immunoprecipitation.
200 μl of nuclear protein lysate was precleared using protein G-sepharose (50% slury). This was incubated at 4°C for approximately 60 minutes and spun for 10 minutes at 4°C. Supernatant was transferred to fresh tubes. 30 mg nuclear protein extracts (precleared lysate) was transferred to tubes with antibody-bound protein G beads and rocked gently at 4°C overnight. This mixture was washed 5 times with 1× PBS containing 1% NP-40. Samples were dried using Spin-Dry vacuum centrifugation at –100 on a SpeedVac vapor trap. Immunoprecipitation products were extracted from the protein G beads using Laemmli sample buffer.
Western blot analysis.
Western blot was by standard methods: antibodies used were GATA4 (Abcam, catalog Ab124265), anti-Flag (Sigma-Aldrich, catalog F7425-2MG), c-MYC (Cell Signaling Technology, catalog 5605), p27/CDKN1B (Cell Signaling Technology, catalog 3833) and β-actin (Sigma-Aldrich, catalog a3854), histone 3 (Cell Signaling Technology, catalog 9715S), MED12 (Cell Signaling Technology, catalog 4529S,) SMARCA5/SNF2 (Cell Signaling Technology, catalog 13543s), and anti-V5 (Abcam, catalog Ab27671).
Protein identification by LC-MS/MS.
Anti-Flag and isotype antibody immunoprecipitation products were separated by molecular weight using SDS-polyacrylamide gel electrophoresis and stained with colloidal Coomassie blue (GelCode Blue, Pierce Chemical). All separated proteins on the gel slices were excised (Supplemental Figure 9); proteins were reduced with dithiothreitol (Sigma-Aldrich, D0632, 10 mM), alkylated with iodoacetamide (Sigma-Aldrich, I1149, 55 mM), and digested in situ with trypsin. Peptides were extracted from gel pieces 3 times using 60% acetonitrile and 0.1% formic acid/water. The dried tryptic peptide mixture was redissolved in 20 μl of 1% formic acid for MS analysis. Tryptic peptide mixtures were analyzed by online LC-MS/MS on an Orbitrap Mass Spectrometer (Thermo Fisher Scientific).
Database search and data validation.
Mascot Daemon software (version 2.3.2; Matrix Science) was used to perform database searches using the Extract_msn.exe macro provided with Xcalibur (version 2.0 SR2; Thermo Fisher Scientific) to generate peaklists. The following parameters were set for creation of the peaklists: parent ions in the mass range of 400–4500, no grouping of MS/MS scans, and threshold at 1000. A peaklist was created for each analyzed fraction (i.e., gel slice), and individual Mascot (version 2.3.01) searches were performed for each fraction. The data were searched against Homo sapiens entries in the UniProt protein database (May 2015 release, 151,569 total sequences; http://www.uniprot.org/). Carbamidomethylation of cysteines was set as a fixed modification, and oxidation of methionine was set as a variable modification. Specificity of trypsin digestion was set for cleavage after Lys or Arg, and 2 missed trypsin cleavage sites were allowed. The mass tolerances in MS and MS/MS were set to 10 ppm and 0.6 Da, respectively, and the instrument setting was specified as ESI-Trap. To calculate the FDR, the search was performed using the decoy option in Mascot. The spectral FDR and protein FDR were 0.35% ± 0.17% and 4.28% ± 1.99%, respectively. A minimum Mascot ion score of 25 and peptide rank 1 were used for automatically accepting all peptide MS/MS spectra.
LFQ.
Relative protein quantification was performed using spectral count-based label-free relative protein quantitation (LFQ). For each biological sample, data from the individual gel slices were combined. Statistical analysis was performed on all proteins identified, with average spectral counts of 2 or more for at least 1 of the 3 experiments. The spectral count data were normalized by total spectral counts of the bait protein (GATA4) in each sample to adjust for differences in overall protein levels among samples. Proteins were considered to have a significant difference in abundance if there was a difference of 2-fold or greater in normalized spectral counts between experiments and a P value of 0.01 or less using a 2-tailed t test. Spectral counts for all proteins and peptides identified are provided in Supplemental Table 5.
GATA4 DNA-binding analysis.
A 5′-GATA probe (ATTACTGATAATGGTG-3′ X3) and negative control probe (5′-ATTACTCCCCATGGTG-3′ X3) (underlines indicate GATA4 response element versus scrambled sequence ‘CCCC’) were designed and ordered from ITD. The probes were biotinylated with a biotin-labeling kit following the manufacturer’s guidelines (Thermo Fisher Scientific, catalog K0651). GATA4 WT and GATA4 V267M vector were transfected in HCC cell PLC, cell pellets from 6.0 × 106 cells were isolated at 48 hours, and nuclear fractions were extracted using nuclear extraction buffer: (250 μl PBSW buffer PH7-9) + ( 2% NP40 + 500 mM [5 μl] + 5 M NaCl [25 μl]). Isolated nuclear fractions were incubated with the GATA probe and the scramble control probe at 4°C overnight. Incubated fractions were pulled down using streptavidin beads for 1 hour at room temperature.
Bioinformatic and statistical analysis.
Proteins identified by label-free LC-MS/MS were analyzed by the Ingenuity Pathway Analysis Tool (IPA, Ingenuity Systems). The core analysis function included in IPA was used to interpret the data in the context of biological processes, pathways, and networks. Right-tailed Fisher’s exact test was used to determine a P value indicating that the probability of biological functions, canonical pathways, and diseases associated with the networks was not because of chance alone. Each protein identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base.
For classification of hepatocyte precursor and hepatocyte genes, a public gene expression database (GEO GSE13149) of genes expressed sequentially from early to late stages of liver development was analyzed to identify genes enriched at different stages of development (23). Comparative Marker Selection (V10) in GenePattern (Broad Institute, Massachusetts Institute of Technology, Boston, Massachusetts, USA) (78) was used to find genes whose expression correlated with phenotype. Statistical significance was determined by the 1000 permutations test and an FDR (Benjamini-Hochberg) cutoff of less than 0.01. In a separate approach to identifying genes that were differentially expressed between HCC and paired nonmalignant liver, the average expression of each gene across 46 HCC samples and, separately, across 46 adjacent nonmalignant livers was calculated. Genes with average expression values in HCC (n = 46) that were less than 66% of the average expression value in nonmalignant liver (n = 46) were identified. To determine tissue expression (UNIGENE EST QUARTILE) associations, gene lists were uploaded into DAVID (79) to provide a ranked representation of tissue expression associations that were most saturated or “enriched” with the input gene lists. For analysis of public ChIP sequencing (ChIP-Seq) data, aligned H1 ESC H3K4me3 ChIP-Seq data (ENCFF775QSF) were imported, analyzed, and visualized using EaSeq and its suite of integrated tools; all values were normalized to reads per million per 1 kbp (80).
Wilcoxon rank sum and Student’s t tests were 2-sided and performed at the 0.05 significance level unless stated otherwise. SDs for each set of measurements were calculated and represented as y axis error bars on each graph. JMP Pro 10.0 or SAS statistical software was used to perform statistical analysis (SAS Institute Inc., http://www.jmp.com) including correlation analyses.
Study approval.
Human samples were obtained in the context of clinically indicated surgery and with written informed consent from patients in accordance with the Declaration of Helsinki and using protocols approved by the SingHealth Institutional Review Board at the National Cancer Centre Singapore; human HCC tumors and paired normal samples were surgically removed at the National Cancer Centre Singapore between 2008 and 2011. Animal studies were conducted at the Cleveland Clinic using protocols approved by the Cleveland Clinic IACUC.
Author contributions
All authors reviewed and edited the manuscript. FOE designed and performed experiments. WHS validated sequencing experiments of GATA4 and in vitro functional effects of GATA4 transfection. XG validated GATA4 immunoprecipitation. EQ generated the Gata4 conditional KO mouse model. BP performed bioinformatics and next-generation sequence analysis. HM analyzed GATA4 sequences. JB performed IHC studies. JN, CLC, RB, LSK, JL, RC, and MT isolated DNA and RNA from paired normal versus HCC samples. ZH isolated and analyzed gDNA from HCC and adjacent normal tissue. KPN analyzed ChIP-Seq data. JM reviewed deep sequencing data and edited the manuscript. TR performed biostatistics and analyzed SNPA data. AC, LLO, YMT, PCC, PC, CYC, and KHL isolated and characterized tissues from HCC patients. LY and EH validated IHC studies. HCT and YS conceived hypotheses, designed experiments, obtained funding, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
HCT is supported by the National Medical Research Council (NMRC)/MOHIAFCAT2/001/2014, the Biomedical Research Council (BMRC) EDB-IAF311020, a National Cancer Centre Research Fund (NCCRF) pilot grant, a Terry Fox Foundation Pilot Grant, and block funding from the National Cancer Centre Singapore. YS is supported by case GI SPORE P50CA150964, Comprehensive Cancer Center grant P30 CA043703, and Cleveland Clinic HCC Center of Excellence grants. Financial support was also received from the NIH (RO1 CA204373).
Version 1. 07/31/2017
Electronic publication
Version 2. 09/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(9):3527–3542.https://doi.org/10.1172/JCI93488.
Contributor Information
Wai Ho Shuen, Email: timothy.shuen.w.h@nccs.com.sg.
Bartlomiej Przychodzen, Email: przychb@ccf.org.
Hideki Makishima, Email: makishimah@gmail.com.
Juraj Bodo, Email: bodoj@ccf.org.
Joanna Ng, Email: joannanht@gmail.com.
Chit Lai Chee, Email: chee.chit.lai@nccs.com.sg.
Rebecca Ba, Email: RBA1@e.ntu.edu.sg.
Lip Seng Koh, Email: koh.lip.seng@nccs.com.sg.
Janice Lim, Email: janice.lim.c.w@nccs.com.sg.
Rachael Cheong, Email: rachael.cheong.k.m@nccs.com.sg.
Marissa Teo, Email: marissa_teo@yahoo.com.
Zhenbo Hu, Email: huzhenbo@wfmc.edu.cn.
Kwok Peng Ng, Email: ngk2@ccf.org.
Jaroslaw Maciejewski, Email: maciejj@ccf.org.
Tomas Radivoyevitch, Email: radivot@ccf.org.
Alexander Chung, Email: alexander.chung.y.f@singhealth.com.sg.
Yu Meng Tan, Email: ymtan.surgery@gmail.com.
Peng-Chung Cheow, Email: cheow.peng.chung@singhealth.com.sg.
Pierce Chow, Email: gsupc@sgh.com.sg.
Chung Yip Chan, Email: chan.chung.yip@sgh.com.sg.
Kiat Hon Lim, Email: lim.kiat.hon@sgh.com.sg.
Lisa Yerian, Email: YERIANL@ccf.org.
Eric Hsi, Email: hsie@ccf.org.
Han Chong Toh, Email: dmothc@nccs.com.sg.
Yogen Saunthararajah, Email: saunthy@ccf.org.
References
- 1.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci U S A. 2012;109(21):8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kan Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23(9):1422–1433. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooke SL, Pole JC, Chin SF, Ellis IO, Caldas C, Edwards PA. High-resolution array CGH clarifies events occurring on 8p in carcinogenesis. BMC Cancer. 2008;8:288. doi: 10.1186/1471-2407-8-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan J, et al. LFIRE-1/HFREP-1, a liver-specific gene, is frequently downregulated and has growth suppressor activity in hepatocellular carcinoma. Oncogene. 2004;23(10):1939–1949. doi: 10.1038/sj.onc.1207306. [DOI] [PubMed] [Google Scholar]
- 5.Demchev V, et al. Targeted deletion of fibrinogen like protein 1 reveals a novel role in energy substrate utilization. PLoS ONE. 2013;8(3):e58084. doi: 10.1371/journal.pone.0058084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue W, et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes Dev. 2008;22(11):1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan BZ, Miller MJ, Keck CL, Zimonjic DB, Thorgeirsson SS, Popescu NC. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58(10):2196–2199. [PubMed] [Google Scholar]
- 8.Durkin ME, Avner MR, Huh CG, Yuan BZ, Thorgeirsson SS, Popescu NC. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 2005;579(5):1191–1196. doi: 10.1016/j.febslet.2004.12.090. [DOI] [PubMed] [Google Scholar]
- 9.Boulter L, Lu WY, Forbes SJ. Differentiation of progenitors in the liver: a matter of local choice. J Clin Invest. 2013;123(5):1867–1873. doi: 10.1172/JCI66026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Zhao L, Fish M, Logan CY, Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524(7564):180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardinale V, et al. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology. 2011;54(6):2159–2172. doi: 10.1002/hep.24590. [DOI] [PubMed] [Google Scholar]
- 12.Conigliaro A, et al. Evidence for a common progenitor of epithelial and mesenchymal components of the liver. Cell Death Differ. 2013;20(8):1116–1123. doi: 10.1038/cdd.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 14.Zhang F, et al. Combined hepatocellular cholangiocarcinoma originating from hepatic progenitor cells: immunohistochemical and double-fluorescence immunostaining evidence. Histopathology. 2008;52(2):224–232. doi: 10.1111/j.1365-2559.2007.02929.x. [DOI] [PubMed] [Google Scholar]
- 15.Köhn-Gaone J, Gogoi-Tiwari J, Ramm GA, Olynyk JK, Tirnitz-Parker JE. The role of liver progenitor cells during liver regeneration, fibrogenesis, and carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2016;310(3):G143–G154. doi: 10.1152/ajpgi.00215.2015. [DOI] [PubMed] [Google Scholar]
- 16.Wang R, Ferrell LD, Faouzi S, Maher JJ, Bishop JM. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol. 2001;153(5):1023–1034. doi: 10.1083/jcb.153.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarevich NL, et al. Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology. 2004;39(4):1038–1047. doi: 10.1002/hep.20155. [DOI] [PubMed] [Google Scholar]
- 18.Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem. 2012;287(10):7345–7356. doi: 10.1074/jbc.M111.334599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ning BF, et al. Hepatocyte nuclear factor 4 alpha suppresses the development of hepatocellular carcinoma. Cancer Res. 2010;70(19):7640–7651. doi: 10.1158/0008-5472.CAN-10-0824. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty A, et al. Neil2-null mice accumulate oxidized DNA bases in the transcriptionally active sequences of the genome and are susceptible to innate inflammation. J Biol Chem. 2015;290(41):24636–24648. doi: 10.1074/jbc.M115.658146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watt AJ, Zhao R, Li J, Duncan SA. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol. 2007;7:37. doi: 10.1186/1471-213X-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 23.Li T, et al. Multi-stage analysis of gene expression and transcription regulation in C57/B6 mouse liver development. Genomics. 2009;93(3):235–242. doi: 10.1016/j.ygeno.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang P, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 25.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 26.DeLaForest A, et al. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138(19):4143–4153. doi: 10.1242/dev.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawar SA, et al. C/EBP{delta} targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proc Natl Acad Sci U S A. 2010;107(20):9210–9215. doi: 10.1073/pnas.0913813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang-Verslues WW, Sladek FM. Nuclear receptor hepatocyte nuclear factor 4alpha1 competes with oncoprotein c-Myc for control of the p21/WAF1 promoter. Mol Endocrinol. 2008;22(1):78–90. doi: 10.1210/me.2007-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parviz F, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34(3):292–296. doi: 10.1038/ng1175. [DOI] [PubMed] [Google Scholar]
- 30.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/S1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 31. Amin MB, et al., eds. AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer International Publishing; 2017. [Google Scholar]
- 32.Fero ML, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85(5):733–744. doi: 10.1016/S0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 33.Kiyokawa H, et al. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85(5):721–732. doi: 10.1016/S0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama K, et al. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85(5):707–720. doi: 10.1016/S0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 35.Tang ZH, et al. Two novel missense mutations of GATA4 gene in Chinese patients with sporadic congenital heart defects. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23(2):134–137. [PubMed] [Google Scholar]
- 36.Wang E, et al. Identification of functional mutations in GATA4 in patients with congenital heart disease. PLoS ONE. 2013;8(4):e62138. doi: 10.1371/journal.pone.0062138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teo YY, et al. Singapore Genome Variation Project: a haplotype map of three Southeast Asian populations. Genome Res. 2009;19(11):2154–2162. doi: 10.1101/gr.095000.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(5 Suppl 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Kawai-Kitahata F, et al. Comprehensive analyses of mutations and hepatitis B virus integration in hepatocellular carcinoma with clinicopathological features. J Gastroenterol. 2016;51(5):473–486. doi: 10.1007/s00535-015-1126-4. [DOI] [PubMed] [Google Scholar]
- 40.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyault S, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 42.Kim YD, et al. Genetic alterations of Wnt signaling pathway-associated genes in hepatocellular carcinoma. J Gastroenterol Hepatol. 2008;23(1):110–118. doi: 10.1111/j.1440-1746.2007.05250.x. [DOI] [PubMed] [Google Scholar]
- 43.Imbeaud S, Ladeiro Y, Zucman-Rossi J. Identification of novel oncogenes and tumor suppressors in hepatocellular carcinoma. Semin Liver Dis. 2010;30(1):75–86. doi: 10.1055/s-0030-1247134. [DOI] [PubMed] [Google Scholar]
- 44.Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102(2):74–83. doi: 10.1016/j.ygeno.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Ding Q, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19(2):159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh A, Kim HS, Lim SO, Yu DY, Jung G. Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/β-catenin signaling. Cancer Lett. 2011;300(2):162–172. doi: 10.1016/j.canlet.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 47.Subramaniam S, Kelley RK, Venook AP. A review of hepatocellular carcinoma (HCC) staging systems. Chin Clin Oncol. 2013;2(4):33. doi: 10.3978/j.issn.2304-3865.2013.07.05. [DOI] [PubMed] [Google Scholar]
- 48.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 49.Sikorski TW, Joo YJ, Ficarro SB, Askenazi M, Buratowski S, Marto JA. Proteomic analysis demonstrates activator- and chromatin-specific recruitment to promoters. J Biol Chem. 2012;287(42):35397–35408. doi: 10.1074/jbc.M112.391581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun X, et al. Suppression of the SWI/SNF component arid1a promotes mammalian regeneration. Cell Stem Cell. 2016;18(4):456–466. doi: 10.1016/j.stem.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gresh L, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24(18):3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu X, et al. Runx1 regulation of Pu.1 corepressor/coactivator exchange identifies specific molecular targets for leukemia differentiation therapy. J Biol Chem. 2014;289(21):14881–14895. doi: 10.1074/jbc.M114.562447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venturelli S, et al. Epigenetic combination therapy as a tumor-selective treatment approach for hepatocellular carcinoma. Cancer. 2007;109(10):2132–2141. doi: 10.1002/cncr.22652. [DOI] [PubMed] [Google Scholar]
- 54.Dannenberg LO, Edenberg HJ. Epigenetics of gene expression in human hepatoma cells: expression profiling the response to inhibition of DNA methylation and histone deacetylation. BMC Genomics. 2006;7:181. doi: 10.1186/1471-2164-7-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arai M, et al. Sequential gene expression changes in cancer cell lines after treatment with the demethylation agent 5-Aza-2’-deoxycytidine. Cancer. 2006;106(11):2514–2525. doi: 10.1002/cncr.21905. [DOI] [PubMed] [Google Scholar]
- 56.Mei Q, et al. An open-label, single-arm, phase I/II study of lower-dose decitabine based therapy in patients with advanced hepatocellular carcinoma. Oncotarget. 2015;6(18):16698–16711. doi: 10.18632/oncotarget.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saunthararajah Y. Key clinical observations after 5-azacytidine and decitabine treatment of myelodysplastic syndromes suggest practical solutions for better outcomes. Hematology Am Soc Hematol Educ Program. 2013;2013:511–521. doi: 10.1182/asheducation-2013.1.511. [DOI] [PubMed] [Google Scholar]
- 58.Saunthararajah Y, et al. Evaluation of noncytotoxic DNMT1-depleting therapy in patients with myelodysplastic syndromes. J Clin Invest. 2015;125(3):1043–1055. doi: 10.1172/JCI78789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ebrahem Q, Mahfouz RZ, Ng KP, Saunthararajah Y. High cytidine deaminase expression in the liver provides sanctuary for cancer cells from decitabine treatment effects. Oncotarget. 2012;3(10):1137–1145. doi: 10.18632/oncotarget.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velcheti V, Radivoyevitch T, Saunthararajah Y. Higher-level pathway objectives of epigenetic therapy: A solution to the p53 problem in cancer. Am Soc Clin Oncol Educ Book. 2017;37:812–824. doi: 10.14694/EDBK_174175. [DOI] [PubMed] [Google Scholar]
- 61.Acosta JC, et al. Myc inhibits p27-induced erythroid differentiation of leukemia cells by repressing erythroid master genes without reversing p27-mediated cell cycle arrest. Mol Cell Biol. 2008;28(24):7286–7295. doi: 10.1128/MCB.00752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Negrotto S, et al. Noncytotoxic differentiation treatment of renal cell cancer. Cancer Res. 2011;71(4):1431–1441. doi: 10.1158/0008-5472.CAN-10-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grote D, Souabni A, Busslinger M, Bouchard M. Pax 2/8-regulated Gata 3 expression is necessary for morphogenesis and guidance of the nephric duct in the developing kidney. Development. 2006;133(1):53–61. doi: 10.1242/dev.02184. [DOI] [PubMed] [Google Scholar]
- 64.Green LM, Wagner KJ, Campbell HA, Addison K, Roberts SG. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res. 2009;37(2):431–440. doi: 10.1093/nar/gkn955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lucas B, Grigo K, Erdmann S, Lausen J, Klein-Hitpass L, Ryffel GU. HNF4alpha reduces proliferation of kidney cells and affects genes deregulated in renal cell carcinoma. Oncogene. 2005;24(42):6418–6431. doi: 10.1038/sj.onc.1208794. [DOI] [PubMed] [Google Scholar]
- 66.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell. 2002;2(1):81–91. doi: 10.1016/S1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 67.Aschauer L, et al. Delineation of the key aspects in the regulation of epithelial monolayer formation. Mol Cell Biol. 2013;33(13):2535–2550. doi: 10.1128/MCB.01435-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kojima T, et al. FOXO1 and TCF7L2 genes involved in metastasis and poor prognosis in clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2010;49(4):379–389. doi: 10.1002/gcc.20750. [DOI] [PubMed] [Google Scholar]
- 69.Negrotto S, et al. CpG methylation patterns and decitabine treatment response in acute myeloid leukemia cells and normal hematopoietic precursors. Leukemia. 2012;26(2):244–254. doi: 10.1038/leu.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ng KP, et al. p53 independent epigenetic-differentiation treatment in xenotransplant models of acute myeloid leukemia. Leukemia. 2011;25(11):1739–1750. doi: 10.1038/leu.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Agnihotri S, et al. A GATA4-regulated tumor suppressor network represses formation of malignant human astrocytomas. J Exp Med. 2011;208(4):689–702. doi: 10.1084/jem.20102099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akiyama Y, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23(23):8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo M, et al. Hypermethylation of the GATA gene family in esophageal cancer. Int J Cancer. 2006;119(9):2078–2083. doi: 10.1002/ijc.22092. [DOI] [PubMed] [Google Scholar]
- 74.Cai KQ, et al. Loss of GATA4 and GATA6 expression specifies ovarian cancer histological subtypes and precedes neoplastic transformation of ovarian surface epithelia. PLoS ONE. 2009;4(7):e6454. doi: 10.1371/journal.pone.0006454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hua G, et al. A negative feedback regulatory loop associates the tyrosine kinase receptor ERBB2 and the transcription factor GATA4 in breast cancer cells. Mol Cancer Res. 2009;7(3):402–414. doi: 10.1158/1541-7786.MCR-08-0175. [DOI] [PubMed] [Google Scholar]
- 76.Guan B, Wang TL, Shih IeM ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res. 2011;71(21):6718–6727. doi: 10.1158/0008-5472.CAN-11-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 79.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 80.Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol. 2016;23(4):349–357. doi: 10.1038/nsmb.3180. [DOI] [PubMed] [Google Scholar]
- 81.Nazor KL, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10(5):620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.