Precision medicine has the potential to profoundly improve the practice of medicine. However, the advances required will take time to implement. Genetics is already being used to direct clinical decision-making and its contribution is likely to increase. To accelerate these advances, fundamental changes are needed in the infrastructure and mechanisms for data collection, storage and sharing. This will create a continuously learning health-care system with seamless cycling between clinical care and research. Patients must be educated about the benefits of sharing data. The building blocks for such a system are already forming and they will accelerate the adoption of precision medicine.
The practice of medicine is an inexact science. The clinician assesses the patient’s symptoms and decides which tests to perform to gather more data. They must determine the cause of the symptoms and the patient’s prognosis, whether clinical intervention is warranted and, if so, which intervention to prescribe. To do this effectively, the clinician might need to assess several potential courses of action and incorporate all that is known about the patient into his or her decision. Human physiology is complex. In some cases, the cause of the patient’s symptoms cannot be ascertained. In other cases, clinicians cannot gather enough data to make a fully informed decision. The guesswork inherent in the practice of medicine reduces the efficacy of the interventions that are prescribed.
Genetics is an important contributor to this complexity. Distinct genetic variants cause conditions that respond to different treatments yet share a similar set of symptoms. Without a mechanism to determine the underlying genetic cause of a set of symptoms, it might not be possible to determine which treatment will be most effective a priori. For instance, although there are many causes of lung cancer, only people who have an alteration in the gene EGFR respond to treatment with tyrosine kinase inhibitors1,2. Similarly, many genetic lesions lead to a thickened heart and an increased risk of sudden cardiac death, but only people with mutations in the gene GLA respond to enzyme replacement therapy3. Even when the cause of a condition is known, unrelated genetic variants can affect treatment efficacy by altering the way in which drugs are metabolized or by increasing the likelihood of adverse events. For example, patients who are treated with conventional doses of the immunosuppressive drug azathioprine for an extended period are at risk of developing life-threatening myelosuppression if they harbour genetic variants that prevent the drug from being properly metabolized4. And approximately 6% of European populations carry HLA-B alleles that predispose them to potentially life-threatening hypersensitivity reactions if they are treated with the antiretroviral drug abacavir5.
Understanding the patient’s genetic make-up is crucial for providing optimal care for many diseases. Clinicians now have access to an increasing array of tests that allow them to determine which genetic variants exist in their patients. These include: genotyping tests that look at variants in a patient’s DNA sequence that are known to associate strongly with important clinical effects; panel-based gene sequencing, which looks at many genes related to a specific indication to detect known and new variants; sequencing of the exome — all known protein-coding genes; and whole-genome analysis that attempts to sequence a patient’s genome. However, simply determining which variants are present is insufficient. The implications of these variants must also be determined for each clinical indication. This genetic understanding must then be considered in conjunction with other clinical data to decide the path that will produce the best results for the patient.
The precision-medicine ecosystem
The goal of precision medicine is to enable clinicians to quickly, efficiently and accurately predict the most appropriate course of action for a patient. To achieve this, clinicians are given tools — in the form of tests and information-technology support — that are both compatible with their clinical workflow and economically feasible to deploy in the modern health-care environment. These tools help to simplify the process of managing the extreme biological complexity that underlies human disease. To support the creation and refinement of these tools, a precision-medicine ‘ecosystem’ is developing. This ecosystem is beginning to link clinicians, laboratories, research enterprises and clinical-information-system developers together in new ways. There is increasing hope that these efforts will create the foundation of a continuously learning health-care system that is capable of fundamentally accelerating the advance of precision-medicine techniques.
Interpretation is key to the precision-medicine ecosystem. It occurs at several levels. Individual variants can be interpreted in relation to specific indications. Sets of variants can be assessed in relation to their collective impact on patients. Genetic and clinical data can be combined to determine the best course of action for a patient. The quality of these interpretations is highly dependent on the data on which they are based. For this reason, research and clinical databases provide the foundation for precision medicine. Continuous learning in health care is in many ways driven by improvements to the content and structure of these resources. For example, the highly collaborative Clinical and Functional Translation of CFTR (CFTR2) project brought together researchers from around the world to share extensive patient data on CFTR variants to distinguish between pathogenic and benign lesions. This work is leading to more effective treatments for patients with cystic fibrosis6. Similarly, the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium has engaged a large collaborative network to share data on the BRCA1 and BRCA2 genes that predispose patients to breast and ovarian cancer7. Patients with pathogenic variants in these genes can now take preventive action through monitoring and prophylactic surgical procedures8 and those with active breast cancer are candidates for targeted treatments9,10. In addition, large collaborative programmes led by the US National Institutes of Health (NIH)-supported Clinical Genome (ClinGen) Resource11 and the Global Alliance for Genomics and Health have begun to tackle the development of reliable resources for systematically defining the pathogenicity of all human variation through broad and targeted efforts. When optimized, the infrastructure that supports the precision-medicine ecosystem efficiently manages and integrates the flow of material, knowledge and data needed to generate, validate, store, refine and apply clinical interpretations (Fig. 1). Biobanks link samples with patient data to support discovery. Research databases record the data, calculations and results that provide evidence for clinical interpretations. Clinical-knowledge-sharing networks enable the refinement of interpretations. Clinical laboratories and their information systems facilitate the consolidation of interpretations into reports and alerts. Electronic health records (EHRs) and associated systems help clinicians to apply results, both when they are received and as the patient’s condition and knowledge of the variants evolve. Patient-facing infrastructure or ‘portals’ provide individuals with access to their genetic data and — if appropriate — the ability to decide how they should be used, including whether to participate in research. At present, much of this infrastructure is at a very early stage of development. However, the infrastructural foundation for precision medicine is beginning to emerge. In this Review, we explore its crucial components.
Figure 1.
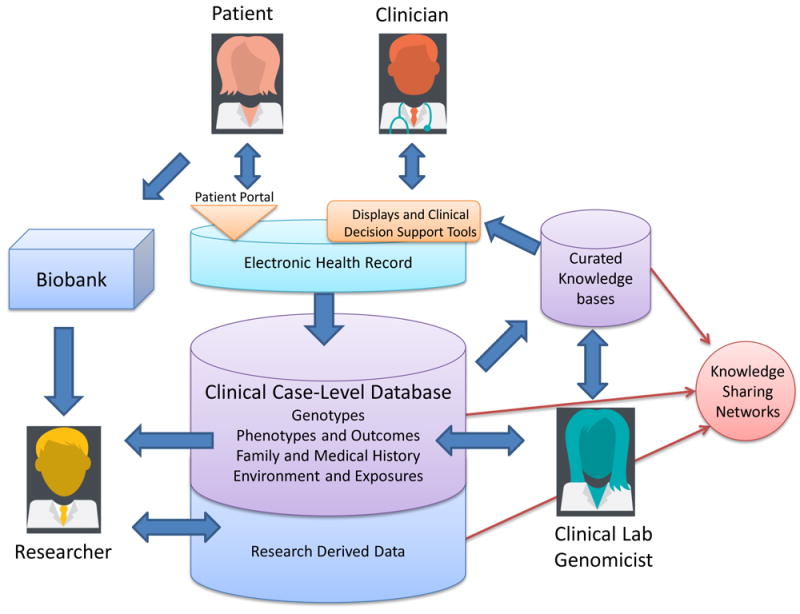
The precision-medicine ecosystem. The precision-medicine ecosystem contains building blocks that optimally connect patients, clinicians, researchers and clinical laboratories to one another. Patients and clinicians access information through portals or EHRs. The ecosystem can include displays or CDS augmented by curated knowledge that is supplied and shared by multiple stakeholders. Case-level databases and biobanks receive case data and samples from clinical and research workflows. Researchers benefit from all of these information sources and also contribute to knowledge sources. Clinical laboratories leverage data and inform the clinical community as they assess genomic variation and its impact on human health
The patient viewpoint
The role of the patient in supporting precision medicine is becoming increasingly important. Patients are obtaining a growing number of genetic results in the course of their care. Typically, clinicians involved in their treatment order such tests for them. However, patients are also now able to access direct-to-consumer testing, sometimes through the help of someone who is not directly involved in their care. To ensure that precision medicine is tailored to the unique genetic make-up of each patient, we must gather as much information as possible from individual patients. Yet there are risks associated with widespread sharing of patient data. To gain access to these data, researchers must actively engage patients, teach them about the benefits of data sharing and help them to weigh up the risks and benefits. This can be done by making the process of obtaining consent more effective.
There are two major forms of consent that are relevant: consent for receiving medical treatment or procedures; and consent for releasing data or samples for use in research. In both cases, the risks and benefits must be conveyed to the patient. However, the conventional distinction is that obtaining consent for treatment focuses on benefits to the individual whereas obtaining consent for research focuses on generalizable knowledge12. Increasingly, the line between clinical care and research is blurring; participation in research studies can lead to a direct improvement in outcome for the patient13,14, and the continuous capture of clinical-care data has been proved an effective way to inform generalizable knowledge15. As a result, efforts are under way to ask all patients who enter the clinical-care setting to sign a form that permits their data to be used in research16-19. In addition, those signing clinical genetic-testing consent forms now commonly agree to share their data broadly to help advance knowledge11. Nevertheless, there is still a need for more uniform consenting processes. It is difficult to generate consent forms in language that is both easy to understand and robustly conveys the main issues associated with genetic testing. Sharing such language across institutions could be helpful in this context. Harmonizing consent language across providers, laboratories and biobanks would make it easier to administer and adhere to those agreements. Recently, the Regulatory and Ethics Working Group of the Global Alliance for Genomics and Health published a framework for the responsible sharing of genomic and health-related data20. The group has also created consent tools and policies to aid the development of standardized approaches to obtaining consent and that support data sharing in the global community. Consistent with the Global Alliance for Genomics and Health framework, ClinGen has developed standardized consenting approaches (http://clinicalgenome.org/data-sharing/) for use in the clinical-care setting, which will enable sharing of genetic-test results and accompanying phenotypic data in the absence of research-study enrolment.
Some patients are extremely interested in supporting research and are willing to take proactive steps to facilitate the sharing of genetic information. The Global Network of Personal Genome Projects recruits volunteers who are prepared to share their genomic data and medical histories publicly. ClinGen manages the GenomeConnect patient portal, built on the Patient Crossroads platform, which allows individuals to share health and genetic information to form communities. The Platform for Engaging Everyone Responsibly (PEER), supported by the Genetic Alliance, enables individuals to control sharing, privacy and access preferences for their health and genomic data with a high degree of precision.
The clinician viewpoint
Clinicians gain access to patients’ genetic information through tests. Tests have two components: a technical component that focuses on identifying which variants are present in the patient; and an interpretive component in which the implications of identified variants are assessed. In most scenarios, genetic testing is performed to determine either the cause of a specific indication or the most appropriate treatment21. However, exome and genome data can be reused to perform multiple assessments over time. This opens up the possibility of obtaining and storing genome and exome sequences before disease manifests, with the intention that they will be interpreted and reinterpreted as indications arise. Irrespective of when the sequence is obtained, the interpretation step is crucial.
Clinical interpretation is a multiple-component process (Fig. 2). After a pool of variants have been identified, step 1 determines which of those variants should be assessed further. In step 2, the clinical impact of each of those variants is assessed. In step 3, the relevance of the combination of variants identified in step 2 are considered in relation to the patient’s indication. Finally, in step 4, the test results are placed within the context of all known information about the patient to determine clinical care. Many laboratories base their reports on steps 1–3, although some simply report the variants they have identified. The patient’s clinician often reviews the laboratory analysis and completes any remaining steps. These processes can be time consuming and therefore expensive. They also involve a considerable amount of professional judgement, which makes them subject to human error and differences of opinion. The quality and efficiency of these processes is highly dependent on the data available in clinical and research databases. For this reason, these databases are in many ways the core of the ecosystem that is needed to advance the practice of precision medicine. In addition, more standardized ways to evaluate evidence, such as those released by the American College of Medical Genetics and Genomics22, will be crucial for interpreting human genetic variation in a more consistent way. Finally, the open sharing of clinical interpretations to distribute the labour of variant assessment, to identify and resolve differences in interpretation, and to catalogue variation for research studies is essential for improving the care of patients with genetic-based conditions11.
Figure 2.
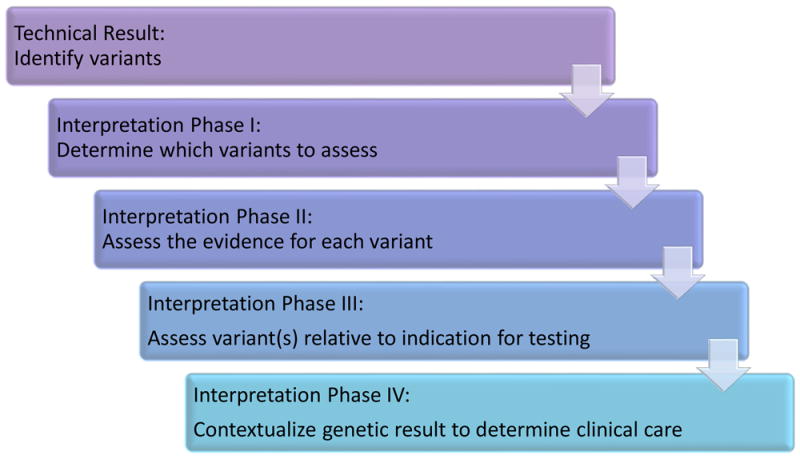
Stages of the genetic interpretation process. Once genetic variants have been identified, they are filtered to select those of interest (step 1). Next, the evidence for each variant is assessed to determine the variant’s clinical impact (step 2). One or more assessed variants are then interpreted with respect to the specific condition for which the patient is being investigated (step 3). Last, the overall genetic assessment is placed into the patient’s clinical and personal context to inform the clinical-care decision-making process (step 4)
In each patient encounter, clinicians must address several questions that relate to precision medicine. First, they must assess whether genetics could be relevant — and if so, order the appropriate tests. Once the test results are received, the clinician must determine how to apply them. Then they must manage the results over time. Information-technology support is needed to manage the large amount of patient data and other information that are required to execute these processes optimally. EHRs and their associated systems are the main means of providing such support to clinicians.
Electronic health records
EHRs are well positioned to be the apex of genetic information-technology support. They should serve as the clinician’s gateway to all of the patient’s information, including any genetic data. Information should be organized and displayed in a way that integrates with the clinician’s workflow and facilitates diagnostic and treatment decisions. EHR and related systems can also provide clinicians with electronic clinical-decision support (CDS) that provides extra information about a genetic test or result through an e-resource or InfoButton23,24 that links to electronic resources such as websites or databases. They can also issue pre-test and post-test pharmacogenomic warnings that highlight potentially adverse interactions between drugs and specific genetic variants. Pre-test warnings are triggered when a clinician takes an action that should be informed by a genetic assessment but there is no record of the assessment being performed. Post-test alerts are triggered when an action is taken that may be contraindicated by a patient’s genetic profile. An example is ordering a high dose of azathioprine for a patient with a thiopurine methyltransferase deficiency4. CDS systems can also alert clinicians when important information emerges on a patient’s previously reported variant25. In the future, CDS systems might be able to guide clinicians through complex scenarios that take into account multiple types of patient data, including genetics. Evolving such CDS is essential for the formation of a learning health-care system.
Genetic information and CDS do not necessarily have to be implemented directly into the EHR — it is possible to integrate EHRs with external systems25,26. Such integration can be seamless so that clinicians need never know that they are working with multiple systems. Providing genetic support through the EHR is complex27 and it is currently unclear how much genetic functionality EHR vendors will build into their systems. Some have indicated that they are unlikely to store full genomic sequencing in the EHR, instead choosing to link to external genomic data stores and focus their internal functionality on managing test results that have been interpreted at a higher level. Irrespective of whether patient genetic profiles are stored in the EHR, a constellation of systems will need to be tightly integrated with the EHR to provide optimal support to clinicians.
Displays of genetic information and CDS are often impossible to provide without robust access to the patient’s genetic results and reports. The EHR and related clinician-facing systems must obtain genetic results from laboratory systems. This requires interfaces between the EHR provider and the laboratory. Creating these interfaces often involves establishing electronic connections that span multiple organizations and integrate systems from competing vendors. Relatively few such interfaces exist, largely because of the expense associated with creating them. Generally, results are transmitted from the laboratory to the provider by fax, which makes it difficult to keep the results organized in the EHR. CDS usually relies on access to structured electronic data that cannot be reliably extracted from a fax. Even if a genetic result is recorded in a structured format, this structure is often lost when the result is transferred to clinicians involved in the patient’s care who operate out of different institutions. Any results from direct-to-consumer testing are also unlikely to be transferred in a structured format.
Several groups are working to promote interconnectivity that would enable CDS systems that incorporate genetic information. The Institute of Medicine Roundtable on Translating Genomic-Based Research for Health established the Displaying and Integrating Genetics Information Through the EHR Action Collaborative (DIGITizE AC). DIGITizE AC brings together clinicians, laboratories, vendors, standards organizations, government agencies and patient representatives to increase support for genetics in the EHR. The group has defined a set of genetics-based CDS rules that it seeks to roll out widely. This will involve leveraging the frameworks of standards bodies, such as Health Level Seven (HL7) International, and the Logical Observation Identifiers Names and Codes (LOINC) database, as well as ontology and rule creators such as the Clinical Pharmacogenomics Implementation Consortium (CPIC). The National Human Genome Research Institute (NHGRI) has also established several consortia with EHR working groups to investigate how genetics can be supported in the EHR. These include: the Electronic Medical Records and Genomics (eMERGE) Network28, Clinical Sequencing Exploratory Research (CSER) and ClinGen11.
However, the problem that these organizations are trying to solve is very difficult. Standardized message formats and ontologies are the best way to reduce the cost of establishing the laboratory–provider and provider–provider interfaces needed to underlie precision medicine. However, these standards are helpful only if they robustly account for the different real-world scenarios they are intended to support and are broadly implemented by the vendor community (Fig. 3). Developing such standards requires an enormous amount of input from groups that combine deep clinical, laboratory, vendor and information-technology expertise. The DIGITizE AC has found that even defining the specific requirements for its initial set of narrow-use cases entailed a considerable amount of interdisciplinary effort. Much more work is needed to build truly robust, general-purpose standards.
Figure 3.
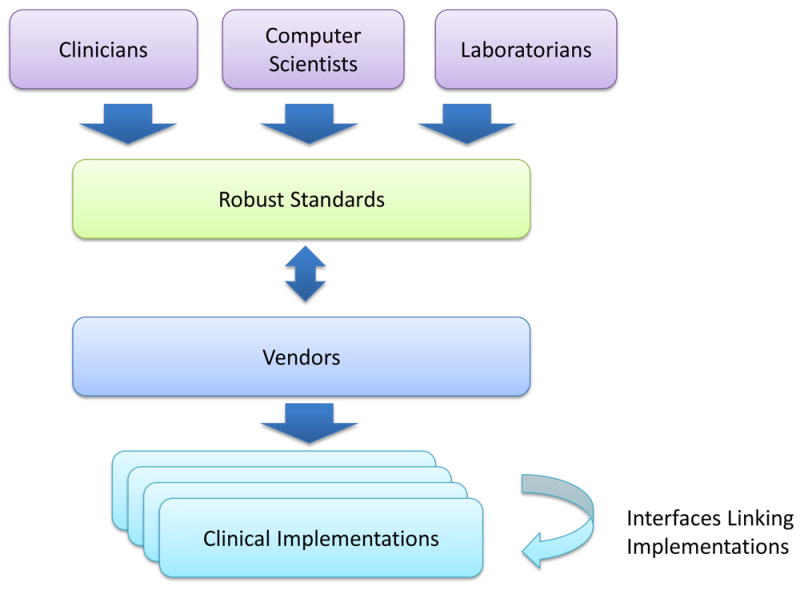
Creating and implementing robust standards for the description and structuring of data in laboratory processing and patient-care systems. Professionals with diverse expertise interact with vendors of laboratory-information systems and EHR systems to iteratively design and implement standards that effectively enable techniques to be used in the clinic.
The clinical laboratory viewpoint
Clinical laboratories sit at the core of the interpretative process. Ideally, they provide both the evidence for individual variants as well as a case-level report that places all potentially relevant variants in the context of the patient’s presentation. Laboratories that perform genome sequencing often discover variants that they have never seen before, which must then be assessed. Similarly, variants that have been seen before might need to be reassessed as new knowledge emerges. Variant assessment is becoming an important factor in the cost of genetic tests. It must be performed by skilled individuals because errors could result in inappropriate patient care. Yet we know that variants can be interpreted differently. As of 11 September 2015, 369 organizations had submitted a total of 158,668 variants to ClinVar, a National Center for Biotechnology Information (NCBI) database that acts as a single centralized public repository to which institutions can submit their interpreted variants as well as retrieve data from others29. At least 2,000 of these have been interpreted differently by submitters11.
Laboratories and clinicians can be assisted in two ways: better access to variant assessments performed by other institutions using consistent approaches, and tools to improve and standardize the variant assessment process.
Building clinical genomic knowledge
Sharing variant- and gene-level assessments between laboratories and clinicians can increase the quality and efficiency of the variant assessment process. Multiple efforts are under way to increase the sharing of such knowledge30-34. The ClinGen programme is building an authoritative central resource that defines the clinical relevance of genomic variants for use in precision medicine and research. The programme aims are to increase the rate of submission to ClinVar and to improve the content of ClinVar and other genomic resources through expert curation. ClinGen has worked together with ClinVar to create a ‘star system’ that defines the level of review for each variant that is submitted to ClinVar11. ClinGen working groups have been established in multiple clinical domains to curate gene–disease relationships and to interpret variants through expert consensus.
Centralized knowledge repositories can also be created by linking together the infrastructure that supports different laboratories. For example, laboratories that use the GeneInsight Lab application35 are able to use the system to communicate and share knowledge in real time. This functionality has been used to create a network called VariantWire and also supports the Canadian Open Genetics Repository (COGR)36 network of Canadian labs. Importantly, an organization can both participate in a knowledge-sharing network and contribute their data to ClinVar. By adopting a standardized infrastructure that helps to structure data for submission to ClinVar, public sharing becomes cheaper, more efficient and more comprehensive with respect to supplying the supporting evidence.
Case repositories and biobanks
An important driver of improvements to variant assessment processes is the collection and analysis of case data. Clinical and research laboratories often develop case repositories. The power of these repositories is a function of the number of cases that they contain. Therefore sharing cases across institutions is beneficial. However, it is difficult to combine data that have been stored in information systems developed by different groups. Trade-offs must be made when deciding what data to capture and how deeply to standardize and structure them. The amount of data in a case repository can be increased by allowing contributors to deposit heterogeneous data that are incomplete or inconsistently validated and may therefore be difficult to process downstream37. If repository developers insist on the submission of complete, validated and consistent data, many cases will have to be excluded.
Several databases have been launched that share case-level data across broad disease areas. The NCBI’s database of Genotypes and Phenotypes (dbGaP)38 places minimal restrictions on the types of case data that can be submitted and therefore serves as a generalized repository. However, because phenotypic data are often limited, making informative use of the information is difficult. Similarly, the European Bioinformatics Institute (EBI) maintains the European Genome-phenome Archive for storing case-level genomic data. The International Cancer Genome Consortium (ICGC)39 and The Cancer Genome Atlas (TCGA)40 have each set up large repositories of somatic cancer sequencing data. The American Society of Clinical Oncology (ASCO) is looking to incorporate the tracking of patient outcomes to enable a learning health-care system in its CancerLinQ platform41. Repositories have also been developed through direct patient participation and span non-profit, academic and commercial activities.
Access to clinical specimens associated with patient data is often necessary to fully inform discovery and continuous learning. The costs associated with collecting samples prospectively for research studies are enormous. However, when samples are collected in bulk and then placed into biobanks, which allows their reuse across studies, the costs decrease considerably. One of the keys to success is ensuring that strong consent processes are in place. Biobanks are moving from repositories of de-identified, unconsented specimens towards fully consented models that allow association with longitudinal health records. This is another area in which universal consent language would simplify the infrastructure development process. Direct engagement of patients in this type of sharing can help participants to balance the risks and benefits. Patients must fully understand the scope of their consent and be actively informed and engaged as participants in the advancement of knowledge. The return to patients of clinically relevant results from research studies should also be supported because this allows patients to benefit directly from their data sharing.
In addition to supporting individual case-level repositories and biobanks, efforts are being made to connect divergent databases. Launched in 2015, the Matchmaker Exchange is a centralized network for sharing case-level data within an international set of case-level repositories focused on gene discovery42. Although each database has its own data schema, the development of a common application programming interface43 means that users can query genomic and phenotypic data across multiple systems. This has encouraged the member databases to move towards implementing a common set of fields to facilitate effective data exchange for gene discovery.
There is no doubt that other such efforts will emerge, particularly as the Genomics England and Precision Medicine Initiative programmes develop. Furthermore, the data-standardization efforts that help to establish interfaces between laboratories and providers could assist the development of these case repositories. Organizations that supply patient data to these efforts must develop mechanisms for collecting data more uniformly and for sharing them consistently. By enabling patients to contribute their data directly, the collection of phenotype data can be accelerated and broadened44.
Knowledge resources and tools
In addition to clinical-knowledge and case-sharing networks, many laboratories and clinicians use research-grade knowledge resources and tools. Many types of tools and resources are used daily in the clinical workflow, even if they are not intended for direct clinical usage. In silico assessment tools use computational algorithms to assess the likely effects of DNA variation45-47. Genome browsers can display multiple tracks of information, including species conservation data, the location of gene transcripts and regulatory elements, and population genetic variation. A number of allele-frequency databases, such as dbSNP Short Genetic Variations, the National Heart, Lung, and Blood Institute (NHLBI) Grand Opportunity Exome Sequencing Project’s Exome Variant Server, the Exome Aggregation Consortium (ExAC) Browser and the 1000 Genomes Project48, provide data that are used by clinical laboratories to define variation that is unlikely to cause Mendelian disorders. The NCBI’s PubMed database provides access to published biomedical literature, and public and commercialized efforts exist to curate and present the data contained in such literature in a more useful format, such as Online Mendelian Inheritance in Man (OMIM) and The Human Gene Mutation Database49. To define the role of these knowledge resources, users must assess which resources are useful, how their quality is controlled and how best they can be integrated into clinical workflows50. ClinGen maintains a list (http://clinicalgenome.org/tools/web-resources/) of web-based tools that members of its community have found useful. This resource has also been designed to serve as an e-resource that can be accessed through EHRs.
The researcher viewpoint
The advent of precision medicine and its supporting infrastructure has given researchers the ability to influence clinical care directly. The release of innovative research tools and the addition of new information in the form of knowledge bases can immediately influence patient care by changing how genomic variants are assessed. Although data obtained through clinical settings typically require processing in specific ways, those obtained from research tend to be more flexible. This means that researchers can often tolerate more variability and inconsistency in their data sources than clinicians. For this reason, new infrastructure is usually released to the research community before it is optimized for clinical use.
Clinical knowledge-sharing infrastructure and case repositories, especially when combined with EHR-derived content, can provide clinicians and clinical laboratories not only with unprecedented access to clinical data, but also make this information accessible to researchers. New models are emerging for the broad sharing of data for discovery purposes. For example, the crowd-sourced approach to solving complex biological problems taken by the DREAM Challenges is now being applied to clinical-trial data in the hope of advancing precision medicine51,52. Such challenges pair the brightest computer scientists with unprecedented open access to data to allow the development of highly informative models for predicting patient outcomes. To support this era of open data and discovery, it is crucial that appropriate consent approaches are established to allow clinical data to be used in these ways. Opportunities for discovery are also being created as the cost of sequencing falls and the processing capabilities of ‘big data’ become increasingly accessible53. All these factors could contribute further to the formation of a continuously learning health-care system that simultaneously engages clinical-care providers and researchers and is necessary to support the development of precision medicine.
Realizing continuously learning health care
Ideally, continuous learning in health care would involve the capture of all incremental data, knowledge and experience gained through each patient interaction. This information would then be used in real time to improve the care of current and future patients. The ability to stratify patients, understand scenarios and optimize decision-making would consistently improve based on the myriad data obtained during the care-delivery process. This would be the ultimate expression of precision medicine. The infrastructure we discuss in this Review represents initial steps in this direction. We have already seen evidence to show that continuous-learning processes are achievable. Figure 4 depicts an example of a continuous-learning system in which hypothetical historical patient data from breast-cancer testing are accessed to determine the pathogenicity of a new BRCA1 variant as ‘likely benign’. The variant would otherwise be considered of ‘uncertain significance’. Clinical laboratories often classify variants on the basis of historical clinical case histories. Because each laboratory has access to only a fraction of patients tested, optimal learning can only happen when data are shared broadly between organizations. ClinGen has made advances in clinical-laboratory genetic-data sharing through the use of the ClinVar database11. However, this level of sharing is more likely to lead to therapeutic development and improved outcomes if the results of genetic testing are accompanied by greater amounts of patient health data. This will require the emerging genetic infrastructure to be extended, such that it can integrate with as many other forms of patient data as possible.
Figure 4.
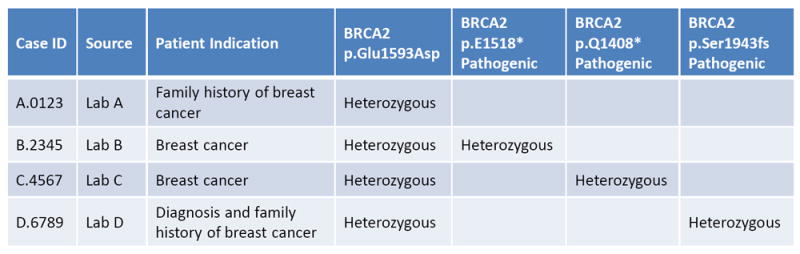
Example of a learning health-care system. Case data can be shared between laboratories to support variant assessment. In this example, the BRCA2 p.Glu1593Asp variant in case D is classified initially as being of ‘uncertain significance’. After accessing genetic and phenotypic patient data from cases A, B and C, in which there are other genetic explanations for the clinical phenotype, the necessary evidence becomes available to classify the BRCA2 p.Glu1593Asp variant as ‘likely benign’
Addressing barriers to precision medicine
Multiple issues must be overcome for personalized medicine to reach its potential, as summarized by Joyner and Paneth in seven key questions54. Although some doubt has been expressed that personalized medicine will reach its full potential for common diseases, the recent shift in emphasis to studies of the genetic basis of rare diseases and somatic cancer could provide tangible success in this field. For example, mechanistic understanding of rare disease and cancer pathways might inform the understanding of common diseases and approaches to reducing risk more effectively than has been achieved through genome-wide association studies. However, to ensure that we can learn from our evolving experience in the diagnosis and treatment of all types of disease, continuously learning health-care systems and broad data-sharing approaches must be supported. The absence of such systems is likely to be responsible for the limited success of personalized medicine to date. The continuous-learning infrastructure could be used to add a testing methodology for new hypotheses, in which real-time evaluation is repeatedly conducted against a limited set of treatment decisions for a given condition to determine which treatments provide the best results for different patient subgroups. Improved decision-making — at present, based on access to more up-to-date knowledge, and in the future, based on real-time evaluation techniques — has the potential to partially offset cost concerns by reducing expenditures associated with unnecessary or ineffective care. These improvements are also likely to generate public-health benefits. Improved infrastructure to capture both test results and patient outcomes should enable the measurement of such benefits.
The type and quality of patient data stored in EHRs are clearly issues that need to be addressed to support a continuously learning health-care system. In our experience, the investment required to capture higher-quality and more clinically relevant data is made only when a near-term financial return on those investments can be established. An improved foundational infrastructure provides an expanded basis for innovation and thereby facilitates the development of tools and analysis that are capable of justifying these foundational investments.
Open data-sharing resources, as well as the principle of open data itself, can help to reduce the cost of conducting genetic research. They can also limit the number of conflict-of-interest problems that occur as academic medical centres increasingly partner with commercial activities. Although this infrastructure does not help to solve generalized funding issues, it does set a precedent for sharing data rather than keeping it proprietary. In doing so, it reduces the scope and impact of potential conflicts and helps to ensure that commercial relationships are based on open principles.
Future directions
Despite compelling examples of the use of genomics to support precision medicine, the core building blocks that will be necessary to scale up the field are still in a very primitive state. However, as the community works to improve these building blocks and link them up, transformations are beginning to occur. Clinicians, researchers, laboratories and vendors are working together to build the tools that will close the distance between each stakeholder. It is becoming easier to move, compare, apply and reproduce knowledge, data and samples. The basic infrastructure required to support a continuously learning health-care system has started to evolve spontaneously in many different areas. Furthermore, a cultural change is emerging as researchers, clinicians and patients embrace the open sharing of data to facilitate scientific advancement. Although it is unclear how long it will take to build an infrastructure that fully supports the widespread sharing and effective use of genomic and health data, the ultimate result will be a transformation of health care that allows continuous advances in medicine to occur within a clinical-care system that is less dependent on externally funded research endeavours.
Acknowledgments
H.L.R. was supported in part by NIH grants U41HG006834, U01HG006500 and U19HD077671. S.J.A. was supported in part by U41HG006834.
Footnotes
The authors declare competing financial interests: see go.nature.com/7jayhk for details.
References
- 1.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Morel CF, Clarke JT. The use of agalsidase alfa enzyme replacement therapy in the treatment of Fabry disease. Expert Opin Biol Ther. 2009;9:631–639. doi: 10.1517/14712590902902296. [DOI] [PubMed] [Google Scholar]
- 4.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther. 2013;93:324–325. doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin MA, et al. Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing: 2014 update. Clin Pharmacol Ther. 2014;95:499–500. doi: 10.1038/clpt.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutting GR. Cystic fibrosis genetics: from molecular understanding to clinical application. Nature Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spurdle AB, et al. ENIGMA—evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat. 2012;33:2–7. doi: 10.1002/humu.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domchek SM, et al. Association of risk-reducing surgery in BRCA1 or BRCA2 mutation carriers with cancer risk and mortality. J Am Med Assoc. 2010;304:967–975. doi: 10.1001/jama.2010.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 10.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 11.Rehm HL, et al. ClinGen — The Clinical Genome Resource. N Engl J Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. This article describes ClinGen, an NIH-supported programme to build an authoritative central resource that defines the clinical relevance of genomic variants for use in precision medicine and research, employing systematic sharing of clinical knowledge and expert curation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.US Department of Veterans Affairs Office of Research & Development. Informed Consent for Human Subjects Research: a Primer. VA Boston Health Care System. 2002 http://www.research.va.gov/resources/pubs/docs/consent_primer_final.pdf.
- 13.Jameson E, Jones S, Wraith JE. Enzyme replacement therapy with laronidase (Aldurazyme®) for treating mucopolysaccharidosis type I. Cochrane Database Syst Rev. 2013;11 doi: 10.1002/14651858.CD009354.pub2. CD009354. [DOI] [PubMed] [Google Scholar]
- 14.Hacein-Bey Abina S, et al. Outcomes following gene therapy in patients with severe Wiskott–Aldrich syndrome. J Am Med Assoc. 2015;313:1550–1563. doi: 10.1001/jama.2015.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy SN, et al. High throughput tools to access images from clinical archives for research. J Digit Imaging. 2015;28:194–204. doi: 10.1007/s10278-014-9733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCarty CA, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen NL, et al. Biobank participants’ preferences for disclosure of genetic research results: perspectives from the OurGenes, OurHealth, OurCommunity project. Mayo Clin Proc. 2014;89:738–746. doi: 10.1016/j.mayocp.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toledo JB, et al. A platform for discovery: The University of Pennsylvania Integrated Neurodegenerative Disease Biobank. Alzheimers Dement. 2014;10:477–484. doi: 10.1016/j.jalz.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milani L, Leitsalu L, Metspalu A. An epidemiological perspective of personalized medicine: the Estonian experience. J Intern Med. 2015;277:188–200. doi: 10.1111/joim.12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoppers BM. Framework for responsible sharing of genomic and health-related data. HUGO J. 2014;8:3. doi: 10.1186/s11568-014-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korf BR, Rehm HL. New approaches to molecular diagnosis. J Am Med Assoc. 2013;309:1511–1521. doi: 10.1001/jama.2013.3239. [DOI] [PubMed] [Google Scholar]
- 22.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. These guidelines provide a standardized approach to the interpretation of genetic variants for monogenic disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman MA, Williams MS. Electronic medical records and personalized medicine. Hum Genet. 2011;130:33–39. doi: 10.1007/s00439-011-0992-y. [DOI] [PubMed] [Google Scholar]
- 24.Del Fiol G, et al. Integrating genetic information resources with an EHR. AMIA Annu Symp Proc 2006. 2006:904. [PMC free article] [PubMed] [Google Scholar]
- 25.Aronson SJ, et al. Communicating new knowledge on previously reported genetic variants. Genet Med. 2012;14:713–719. doi: 10.1038/gim.2012.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starren J, Williams MS, Bottinger EP. Crossing the omic chasm: a time for omic ancillary systems. J Am Med Assoc. 2013;309:1237–1238. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kho AN, et al. Practical challenges in integrating genomic data into the electronic health record. Genet Med. 2013;15:772–778. doi: 10.1038/gim.2013.131. This review summarizes challenges that the eMERGE consortium has encountered when integrating genetics into the EHR and suggests approaches for addressing these challenges. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottesman O, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Béroud C, Collod-Béroud G, Boileau C, Soussi T, Junien C. UMD (Universal Mutation Database): a generic software to build and analyze locus-specific databases. Hum Mutat. 2000;15:86–94. doi: 10.1002/(SICI)1098-1004(200001)15:1<86::AID-HUMU16>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Sosnay PR, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nature Genet. 2013;45:1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Firth HV, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller DT, et al. Consensus statement: chromosomal microarray is a first tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson BA, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nature Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aronson SJ, et al. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat. 2011;32:532–536. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerner-Ellis J, Wang M, White S, Lebo MS Canadian Open Genetics Repository Group. Canadian Open Genetics Repository (COGR): a unified clinical genomics database as a community resource for standardising and sharing genetic interpretations. J Med Genet. 2015;52:438–445. doi: 10.1136/jmedgenet-2014-102933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riggs ER, Jackson L, Miller DT, Van Vooren S. Phenotypic information in genomic variant databases enhances clinical care and research: the International Standards for Cytogenomic Arrays Consortium experience. Hum Mutat. 2012;33:787–796. doi: 10.1002/humu.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tryka KA, et al. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, et al. International Cancer Genome Consortium Data Portal—a one-stop shop for cancer genomics data. Database (Oxford) 2011;2011:bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schilsky RL, Michels DL, Kearbey AH, Yu PP, Hudis CA. Building a rapid learning health care system for oncology: the regulatory framework of CancerLinQ. J Clin Oncol. 2014;32:2373–2379. doi: 10.1200/JCO.2014.56.2124. This article provides an overview of the challenges of applying precision medicine techniques to cancer and then describes the CancerLinQ system and the regulatory framework under which it operates. [DOI] [PubMed] [Google Scholar]
- 42.Philippakis AA, et al. The matchmaker exchange: a platform for rare disease gene discovery. Hum Mutat. 2015 doi: 10.1002/humu.22858. http://dx.doi.org/10.1002/humu.22858. This paper describes an international system for sharing genomic cases to aid in gene discovery. [DOI] [PMC free article] [PubMed]
- 43.Buske OJ, et al. The matchmaker exchange API: automating patient matching through the exchange of structured phenotypic and genotypic profiles. Hum Mutat. 2015 doi: 10.1002/humu.22850. http://dx.doi.org/10.1002/humu.22850. [DOI] [PMC free article] [PubMed]
- 44.Almalki M, Gray K, Sanchez FM. The use of self-quantification systems for personal health information: big data management activities and prospects. Health Inf Sci Syst. 2015;3(suppl):S1. doi: 10.1186/2047-2501-3-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 46.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jian X, Boerwinkle E, Liu X. In silico tools for splicing defect prediction: a survey from the viewpoint of end users. Genet Med. 2014;16:497–503. doi: 10.1038/gim.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. erratum 473, 544 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenson PD, et al. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gargis AS, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nature Biotechnol. 2012;30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarchum I, Jones S. DREAMing of benchmarks. Nature Biotechnol. 2015;33:49–50. doi: 10.1038/nbt.3115. [DOI] [PubMed] [Google Scholar]
- 52.Abdallah K, Hugh-Jones C, Norman T, Friend S, Stolovitzky G. The Prostate Cancer DREAM Challenge: A community-wide effort to use open clinical trial data for the quantitative prediction of outcomes in metastatic prostate cancer. Oncologist. 2015;20:459–460. doi: 10.1634/theoncologist.2015-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O’Driscoll A, Daugelaite J, Sleator RD. ‘Big data’, Hadoop and cloud computing in genomics. J Biomed Inform. 2013;46:774–781. doi: 10.1016/j.jbi.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 54.Joyner MJ, Paneth N. Seven questions for personalized medicine. J Am Med Assoc. 2015 doi: 10.1001/jama.2015.7725. http://dx.doi.org/10.1001/jama.2015.7725. This review discusses cloud computing and big data concepts and their application to the field of genomics. [DOI] [PubMed]


