Abstract
The Matchmaker Exchange (MME) connects rare disease clinicians and researchers to facilitate the sharing of data from undiagnosed patients for the purpose of novel gene discovery. Such sharing raises the odds that two or more similar patients with candidate genes in common may be found, thereby allowing their condition to be more readily studied and understood. Consent considerations for data sharing in MME included both the ethical and legal differences between clinical and research settings and the level of privacy risk involved in sharing varying amounts of rare disease patient data to enable patient matches. In this Commentary, we discuss these consent considerations and the resulting MME Consent Policy as they may be relevant to other international data sharing initiatives.
Keywords: Consent, privacy, data sharing, data access, personalized medicine, precision medicine
The Matchmaker Exchange (MME)
The Matchmaker Exchange (MME) was created to aid in discovering the causes of rare disease through sharing data from exome and genome sequencing performed in research and clinical care. As the majority of patients lack a clear diagnosis after initial analysis, finding additional patients with a deleterious variant in the same gene and overlapping phenotype may provide sufficient evidence to causally implicate the gene, enabling a diagnosis for the patient. Several rare disease consortia had established genomic services to facilitate such matching, but fragmentation of efforts made the aggregation of similar cases difficult. An international collaboration consisting of representatives from rare disease consortia and existing matchmaking services therefore launched Matchmaker Exchange in 2013 to unify these efforts and facilitate the discovery of novel Mendelian disease genes through the international sharing of patients' genetic and clinical data (Philippakis, et al., 2015). The Global Alliance for Genomics and Health (Global Alliance; (Global Alliance for Genomics and Health, 2016)) is an international coalition dedicated to improving human health and maximizing the potential of genomic medicine via its Framework for Responsible Sharing of Genomic and Health-Related Data (Knoppers, 2014). The International Rare Diseases Research Consortium (IRDiRC) brings together researchers and organizations investing in rare diseases research with two main objectives: to deliver 200 new therapies for rare diseases and a means to diagnose most rare diseases, by 2020 (Lochmuller, et al., 2016); (Boycott, et al., 2017)). The MME was recognized and supported by both the Global Alliance and IRDiRC.
The MME platform went live in September 2015. MME databases connect to each other through a federated network allowing queries to be exchanged through standardized application programming interfaces (APIs) and procedural conventions (Buske, et al., 2015b). Data from the patient to be matched are submitted to one of the participating matchmaker services and cases with similar phenotypic and genotypic profiles are matched to build evidence for disease-causality. The MME enables queries of multiple matchmaker services without having to separately query all of them, or deposit data in each one. Matches are made based on pairing candidate genes (genes suspected of causing disease) and/or disease phenotype. Each matchmaker uses an algorithm to score the validity of a match. One or multiple candidate genes can be submitted for each case. The strength of the match increases with fewer candidate genes and more detailed phenotypic features.
To promote responsible data sharing, the founding members of the MME have established a set of requirements for participating matchmaking services (www.matchmakerexchange.org/assets/files/MatchmakerExchangeServiceRequirements.pdf), a user agreement for those wishing to use the MME (www.matchmakerexchange.org/assets/files/MatchmakerExchangeEndUserAgreement.pdf), and a steering committee (SC) to govern the program. The SC is composed of a representative from each approved MME service, as well as program organizers and representation from the Global Alliance and IRDiRC. The SC is charged with maintaining the service requirements, user agreement, and oversight of the API to ensure the MME meets the needs of the rare disease community and reflects consensus standards and best practices as set forth by the Global Alliance and IRDiRC. The MME also supports a monthly conference call and periodic in-person meetings, most of which are open to the community to encourage active participation by all stakeholders and the continued expansion of international data sharing so as to elucidate the genetic causes of rare disease.
One of the critical ethical, legal and social issues (ELSI) confronted by MME was understanding the consent requirements for such data sharing. Standard practices interpreting the ethical cornerstone of free and informed consent usually involves a consent process in the context of a particular research study - not an approach attuned to the expansion of genomic research databases in which data may often be collected and stored for future unanticipated research purposes (Greely, 2007; McGuire and Beskow, 2010). Clinical applications of genomic medicine (Manolio, et al., 2013) are placing further strain on the ethical tenets of free and informed consent in ongoing database development via data sharing. In this context, we sought to provide practical guidance to assist the large data sharing initiatives participating in MME, which span countries with different regulatory requirements, in order to produce a consent policy for MME (Philippakis, et al., 2015). The matchmakers currently connected through the MME API are located in four countries: GeneMatcher (Baylor-Hopkins, USA)(Sobreira, et al., 2015), PhenomeCentral (Hospital for Sick Children, Canada)(Buske, et al., 2015a), DECIPHER (Sanger Centre, UK)(Chatzimichali, et al., 2015), MyGene2 (University of Washington, USA)(Chong, et al., 2016), Patient Archive (Australia)(patientarchive.org) and matchbox (Broad Institute, USA). All of the connected nodes handle data from multiple countries and jurisdictions. In addition, the Monarch Initiative API allows queries on model organism databases for interspecies matching (McMurry, et al., 2016).
Bridging Clinical Care and Research
The future of healthcare is geared towards greater integration of research practices with clinical care. Precision medicine in particular, along with improvements to evidence-based medicine and ‘learning healthcare systems’ in the digital era, depend on research and its results making an impact on clinical care. This intertwining of research and care creates challenges for standard consent processes in either setting. One of the main differences between these two spheres is that while consent for clinical care is usually simply implied in the act of seeking care, research consent typically relies on written consent materials as a demonstration of proper awareness and agreement to a specific project (McGuire and Beskow, 2010). Sharing one's data for ongoing research, however, usually takes the form of consent to donate data to a research project or to a biobank or a database, for use in a range of further studies.
MME serves the rare disease clinical and research community by bringing together extensive patient data for analysis to further understand the causes and manifestation of rare diseases. These findings may have a direct impact on patient care, perhaps bringing a diagnosis to undiagnosed patients (Au, et al., 2015; Loucks, et al., 2015). As such, it is both a rich research resource and a valuable clinical tool.
Sharing of a rare disease patient's data can be considered under two different scenarios: 1) to assist the clinician in trying to find a match with a similar case elsewhere in the world to facilitate interpretation of findings and ultimately provide a diagnosis (and similarly to enable their discovery by other clinicians with similar patients); and 2) to enable exploratory research into the causes, development or treatment of rare diseases. The first situation has a reasonable chance of improving the patient's care through a diagnosis or improved management. The second, although building the foundation for evidence-based medicine, is much less likely to be of direct benefit to the patient contributing data. This lack of expected direct, personal, immediate “care” benefit to the patient whose data are shared for research is part of why detailed consent information and signed consent forms are usually deemed appropriate for research consent in this context (as it is when recruiting research participants in settings that do not involve care). Similarly, an ethics framework proposed for learning healthcare systems highlights the importance of avoiding non-clinical risks and burdens on patients (Faden, et al., 2013).
In addition, there is the potential for inducement to participate in research, which is greater when it takes place alongside clinical care. This is due both to the risk of therapeutic misconception (that patients misconstrue research as treatment) and to the imbalance of power in the patient-physician relationship, especially in situations involving sick children (de Vries, et al., 2011). While the risk of misconception is real, it is also true that rare disease patients and their families stand to benefit greatly from a potential match.
Further complicating the picture, data shared through MME may be contributed by doctors on behalf of their patients in a clinical setting, or it may be contributed by researchers who have recruited rare disease patients to a research study or are using pre-existing research datasets. For the latter, in the context of a growing number of “precision medicine” research initiatives, standard research consents are being adapted to suit more frequent expectations that clinical information arising from research analysis could be returned to research participants when so desired and possible and where it could benefit their healthcare (so-called return of research results or of secondary findings) (Knoppers, et al., 2006; Knoppers, et al., 2015), and to then enable future recruitment to suitable clinical studies. Likewise, access to existing clinical data and samples to be mined for eligibility for recruitment into clinical trials is increasing (e.g., through pharmacogenomic selection), so implicit consent to medical care may not be sufficient for data use in these situations (Kris, et al., 2014). Nevertheless, as evidenced by the recognition by the NIH of the use of a broad consent in such initiatives, the ethical chasm between consent in research and the clinic may have been bridged (Us Government, 2017). Yet, as diagnostic testing laboratories come to play a greater role in data sharing (Harrison, et al., 2017), it is not just research consents but also standard diagnostic testing consent forms that will need to be adapted.
From Patient Matches to Further Investigation
The first step of matchmaking involves a search being conducted by a data requester to establish the existence of similar patients (e.g., matching candidate genes) in collections of patient records elsewhere (Buske, et al., 2015b). Once a match has occurred, it is at that stage that it is typically desirable for more detailed patient data (e.g., phenotypic data) to be exchanged between the data depositor and the data requester. This will depend on contact with and approval by both parties. MME data exchange can also be considered at two distinct data levels: Level 1 for matchmaking based on sharing candidate genes and clinical features represented as disease names or as a short list of Human Phenotype Ontology (HPO) terms (Köhler, et al., 2016), and Level 2 for matchmaking based on more detailed phenotypic information and/or DNA or protein sequence information including genomic variant datasets. These two data levels present different levels of privacy risk for individuals, which also informs MME consent policy.
MME “Tiered” Consent Policy
The MME consent policy we propose is “tiered” in that it entails different consent requirements at the different levels of matchmaking or data sharing tiers (see MME Consent Policy in supplementary information S1). The first level addresses matchmaking based on disease name, structured phenotype descriptions and names of genes of interest, and is considered to be of minimal risk for re-identification of individuals (except by their own physician and care community). Sharing this level of information for matchmaking data discovery (i.e., to find similar cases elsewhere among colleagues around the world) would usually not require additional patient consent to data sharing and queries if being done in a clinical context, as it is both expected as part of professional consultation and is necessary to provide the best clinical care for patients. Consent to medical care would therefore entail consent to matchmaking. Along similar lines, guidance in the US on clinical genetics laboratory submissions to public databases, such as ClinVar (Harrison, et al., 2001), including the submission of basic variant and pathogenicity assertion information, does not require explicit patient consent (van El, et al., 2013), www.clinicalgenome.org/site/assets/files/2780/clinvar_data_submission_proposal_7_27_15_final.pdf). As these practices become more widely adopted in the emerging era of genomic medicine, we nonetheless recommend that clinicians raise awareness of them by discussing such data sharing with patients. We also note that, sometimes, local and national policy or regulation will require that specific consent for these purposes be obtained.
In contrast, if rare disease patients are included in MME solely as research participants – whether from a current or past study – consent to genetic analysis and data sharing at level 1 may often be required. This will depend on local/national requirements and research ethics committee assessment.
The second level of matchmaking involves sharing more detailed genotypic and/or phenotypic data that may be unique or sensitive. The risk of re-identification and potential harm can be lowered through pooling sources of data and by removing some phenotype information based on its sensitivity (Dyke, et al., 2016b; The Beacon Project). However, this may not be suitable for the purposes of some scientific research and would potentially prevent matches and follow-up for MME. Use of this level of information carries a possible risk of re-identification and as such requires appropriate patient consent. It should be noted, however, that if a patient has previously consented to genetic analysis and inclusion in an open or registered access database whose declared purpose involves data sharing for purposes consistent with those of this matchmaking, an ethics committee may decide that no further consent would be required.
In short, as MME bridges clinical care and research, the need for consent to share data in the MME depends on the context and objectives of the matching and on the probability of occurrence and seriousness of potential harm introduced by sharing possibly re-identifying information for matchmaking (see figure 1).
Figure 1.
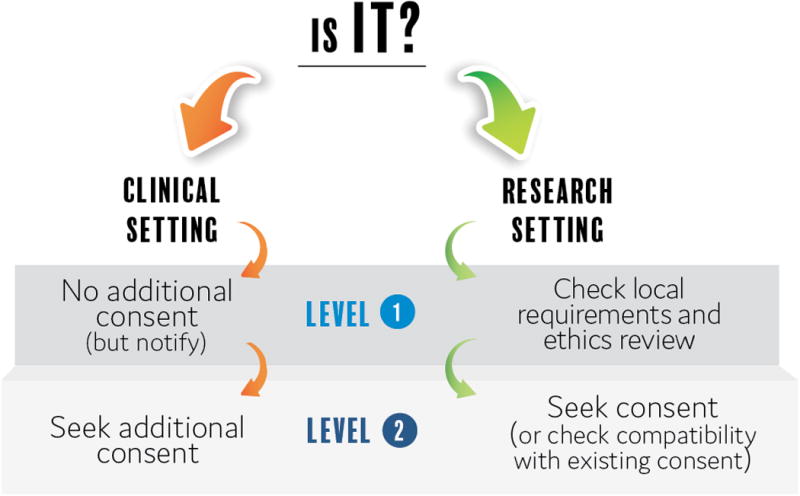
MME Consent Policy considers the setting (clinical or research) and the level of privacy risk associated with shared data.
We drew up further guidance on the content of consent materials for services participating in MME. This consisted of a list of points to consider and information to include when preparing consent materials for data sharing via MME, based on the Global Alliance for Genomics and Health Consent Policy (https://genomicsandhealth.org/files/public/Consent Policy %28Final - 27 May 2015%29.pdf). For example, the consent should make clear that genomic and health-related data (including data from the medical record) may be shared internationally and used by many clinicians and researchers to try to better understand rare diseases. Secondly, consent materials should also specify how confidential data will be protected, such as through coding in accordance with applicable laws and/or guidelines. Thirdly, consent materials should specify that commercialization of discoveries may occur in the future as a result of the development of products, tests, devices, etc. Finally, consent materials must specify how data donors can withdraw from the sharing of clinical or research data, but state that if data have already been shared it may be impossible to retrieve and/or destroy that data, particularly if it has already been used for scientific analysis.
Discussion
The MME consent policy was adopted to provide guidance for projects coming together with a common purpose but operating in different settings, often with different laws and ethics guidance. It was not meant to determine how consent would be sought by participating projects, but rather to assist them in drawing up protocols for sharing via MME. While we believe it may be a helpful consent model for similar data sharing initiatives, several aspects deserve further clarification.
Firstly, the distinction between the two levels of data sharing and consent policy is based on a somewhat crude estimate of the magnitude of the risk of re-identification of shared data as well as of its sensitivity (i.e., the risk of causing harm if re-identified). While in many situations this distinction will be sufficient, the MME Consent Policy acknowledges that it may not always be: “Clinical judgment should be used to assess the potential for re-identification and possible harm depending on the level of phenotypic detail provided”. Ongoing research into data sensitivity aims to provide greater clarity in this area and to further develop useful guidance such as the Data Sharing Privacy Test to distinguish levels of data sensitivity (Dyke, et al., 2016b).
Secondly, to date MME Policy has largely treated consent as “all or nothing” in the project, albeit with the two distinct stages of data deposition and matching and then follow-up data sharing. In practice, consent-based permissions and restrictions, particularly stemming from research protocols, often differentiate between data uses in different fields of research (e.g., for pediatric only or for cancer-related research). This additional layer of complexity may become more relevant as MME expands its research functions, moving beyond just candidate gene discovery to other uses for sharing. Tools such as the Global Alliance Consent Codes, an initiative to simplify the communication of standard consent-based data use conditions in research, are being developed to facilitate the use of data that still include such restrictions (Dyke, et al., 2016a).
Finally, consent will need to be carefully structured as patients contribute to both the clinical and research settings simultaneously, and considered further as they become more directly involved in data sharing and even contribute data directly to MME (MyGene2 and GeneMatcher). Indeed, the ethical and legal distinctions underpinning “research vs. clinical” policies are becoming increasingly blurred.
Uncovering the causes of all rare genetic diseases is a global challenge and there are few better examples of the need for international data sharing. The success of the MME, and the policies it has developed, serve as a model for innovative data sharing that will help guide the community in continually launching new and innovative data sharing platforms in pursuit of the interests and needs of rare disease patients.
Supplementary Material
Acknowledgments
We would like to thank Danielle Azzariti and all other members of the MME for their commitment and thoughtful insights and acknowledge the contributions of the Global Alliance for Genomics and Health (GA4GH) and the International Rare Diseases Research Consortium (IRDiRC) in advancing this collaborative initiative.
Funding: SD is supported by the Canadian Institutes of Health Research (grants EP1-120608; EP2-120609), Genome Quebec, Genome Canada, the Government of Canada, and the Ministère de l'Économie, Innovation et Exportation du Québec (Can-SHARE grant 141210), and the Canada Research Chair in Law and Medicine. BK would like to acknowledge the funding support of the Canada Research Chairs Program (Canada Research Chair in Law and Medicine). AH is supported by NIH grants 1U54HG006542 (Baylor-Hopkins Center for Mendelian Genomics) and 1U41HG006627 (OMIM). HF is supported by the Wellcome Trust award 200990/Z/16/Z (Designing, developing and delivering integrated foundations for genomic medicine). KB is supported by the Care4Rare Canada Consortium funded by Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation. HR was supported by NIH grants UM1HG008900 (Broad Center for Mendelian Genomics) and U41HG006834 (ClinGen).
References
- 1.Federal Policy for the Protection of Human Subjects (‘Common Rule’) US Government; 2017. [Google Scholar]
- 2.Au PY, You J, Caluseriu O, Schwartzentruber J, Majewski J, Bernier FP, Ferguson M, Valle D, Parboosingh JS, Sobreira N, Innes AM, Kline AD. GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue abnormalities caused by de novo variants in HNRNPK. Hum Mutat. 2015;36(10):1009–14. doi: 10.1002/humu.22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boycott KM, Rath A, Chong JX, Hartley T, Alkuraya FS, Baynam G, Brookes AJ, Brudno M, Carracedo A, den Dunnen JT, Dyke SOM, Estivill X, Goldblatt J, Gonthier C, Groft SC, Gut I, Hamosh A, Hieter P, Hohn S, Hurles ME, Kaufmann P, Knoppers BM, Krischer JP, Macek M, Jr, Matthijs G, Olry A, Parker S, Paschall J, Philippakis AA, Rehm HL, Robinson PN, Sham PC, Stefanov R, Taruscio D, Unni D, Vanstone MR, Zhang F, Brunner H, Bamshad MJ, Lochmuller H. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am J Hum Genet. 2017;100(5):695–705. doi: 10.1016/j.ajhg.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buske OJ, Girdea M, Dumitriu S, Gallinger B, Hartley T, Trang H, Misyura A, Friedman T, Beaulieu C, Bone WP, Links AE, Washington NL, Haendel MA, Robinson PN, Boerkoel CF, Adams D, Gahl WA, Boycott KM, Brudno M. PhenomeCentral: a portal for phenotypic and genotypic matchmaking of patients with rare genetic diseases. Hum Mutat. 2015a;36(10):931–40. doi: 10.1002/humu.22851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buske OJ, Schiettecatte F, Hutton B, Dumitriu S, Misyura A, Huang L, Hartley T, Girdea M, Sobreira N, Mungall C, Brudno M. The Matchmaker Exchange API: automating patient matching through the exchange of structured phenotypic and genotypic profiles. Hum Mutat. 2015b;36(10):922–7. doi: 10.1002/humu.22850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatzimichali EA, Brent S, Hutton B, Perrett D, Wright CF, Bevan AP, Hurles ME, Firth HV, Swaminathan GJ. Facilitating collaboration in rare genetic disorders through effective matchmaking in DECIPHER. Hum Mutat. 2015;36(10):941–9. doi: 10.1002/humu.22842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chong JX, Yu JH, Lorentzen P, Park KM, Jamal SM, Tabor HK, Rauch A, Saenz MS, Boltshauser E, Patterson KE, Nickerson DA, Bamshad MJ. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet Med. 2016;18(8):788–95. doi: 10.1038/gim.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries MC, Houtlosser M, Wit JM, Engberts DP, Bresters D, Kaspers GJ, van Leeuwen E. Ethical issues at the interface of clinical care and research practice in pediatric oncology: a narrative review of parents' and physicians' experiences. BMC Med Ethics. 2011;12:18. doi: 10.1186/1472-6939-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyke SO, Philippakis AA, Rambla De Argila J, Paltoo DN, Luetkemeier ES, Knoppers BM, Brookes AJ, Spalding JD, Thompson M, Roos M, Boycott KM, Brudno M, Hurles M, Rehm HL, Matern A, Fiume M, Sherry ST. Consent Codes: Upholding Standard Data Use Conditions. PLoS Genet. 2016a;12(1):e1005772. doi: 10.1371/journal.pgen.1005772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyke SOM, Dove ES, Knoppers BM. Sharing health-related data: a privacy test? Npj Genomic Medicine. 2016b;1:16024. doi: 10.1038/npjgenmed.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faden RR, Kass NE, Goodman SN, Pronovost P, Tunis S, Beauchamp TL. An ethics framework for a learning health care system: a departure from traditional research ethics and clinical ethics. Hastings Cent Rep. 2013:S16–27. doi: 10.1002/hast.134. Spec No: [DOI] [PubMed] [Google Scholar]
- 12.Global Alliance for Genomics and Health. GENOMICS. A federated ecosystem for sharing genomic, clinical data. Science. 2016;352(6291):1278–80. doi: 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]
- 13.Greely HT. The Uneasy Ethical and Legal Underpinnings of Large-Scale Genomic Biobanks. Annual Review of Genomics and Human Genetics. 2007;8(1):343–364. doi: 10.1146/annurev.genom.7.080505.115721. [DOI] [PubMed] [Google Scholar]
- 14.Harrison SM, Dolinsky JS, Knight Johnson AE, Pesaran T, Azzariti DR, Bale S, Chao EC, Das S, Vincent L, Rehm HL. Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genet Med. 2017 doi: 10.1038/gim.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison SM, Riggs ER, Maglott DR, Lee JM, Azzariti DR, Niehaus A, Ramos EM, Martin CL, Landrum MJ, Rehm HL. Current Protocols in Human Genetics. John Wiley & Sons, Inc; 2001. Using ClinVar as a Resource to Support Variant Interpretation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoppers BM. Framework for responsible sharing of genomic and health-related data. The HUGO Journal. 2014;8(3) doi: 10.1186/s11568-014-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knoppers BM, Joly Y, Simard J, Durocher F. The emergence of an ethical duty to disclose genetic research results: international perspectives. Eur J Hum Genet. 2006;14(11):1170–8. doi: 10.1038/sj.ejhg.5201690. [DOI] [PubMed] [Google Scholar]
- 18.Knoppers BM, Zawati MnH, Senecal K. Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet. 2015;16(9):553–559. doi: 10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 19.Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, Aymé S, Baynam G, Bello SM, Boerkoel CF, Boycott KM, Brudno M, Buske OJ, Chinnery PF, Cipriani V, Connell LE, Dawkins HJS, DeMare LE, Devereau AD, de Vries Bert BA, Firth HV, Freson K, Greene D, Hamosh A, Helbig I, Hum C, Jähn JA, James R, Krause R, Laulederkind SJF, Lochmüller H, Lyon GJ, Ogishima S, Olry A, Ouwehand WH, Pontikos N, Rath A, Schaefer F, Scott RH, Segal M, Sergouniotis PI, Sever R, Smith CL, Straub V, Thompson R, Turner C, Turro E, Veltman Marijcke WM, Vulliamy T, Yu J, von Ziegenweidt J, Zankl A, Züchner S, Zemojtel T, Jacobsen Julius OB, Groza T, Smedley D, Mungall CJ, Haendel M, Robinson PN. The Human Phenotype Ontology in 2017. Nucleic Acids Research. 2016 doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson SL, Su PF, Shyr Y, Camidge DR, Sequist LV, Glisson BS, Khuri FR, Garon EB, Pao W, Rudin C, Schiller J, Haura EB, Socinski M, Shirai K, Chen H, Giaccone G, Ladanyi M, Kugler K, Minna JD, Bunn PA. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311(19):1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lochmuller H, Le Cam Y, Jonker AH, Lau LP, Baynam G, Kaufmann P, Lasko P, Dawkins HJ, Austin CP, Boycott KM. ‘IRDiRC Recognized Resources’: a new mechanism to support scientists to conduct efficient, high-quality research for rare diseases. Eur J Hum Genet. 2016 doi: 10.1038/ejhg.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loucks CM, Parboosingh JS, Shaheen R, Bernier FP, McLeod DR, Seidahmed MZ, Puffenberger EG, Ober C, Hegele RA, Boycott KM, Alkuraya FS, Innes AM. Matching two independent cohorts validates DPH1 as a gene responsible for autosomal recessive intellectual disability with short stature, craniofacial, and ectodermal anomalies. Hum Mutat. 2015;36(10):1015–9. doi: 10.1002/humu.22843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, Bick D, Bottinger EP, Brilliant MH, Eng C, Frazer KA, Korf B, Ledbetter DH, Lupski JR, Marsh C, Mrazek D, Murray MF, O/'Donnell PH, Rader DJ, Relling MV, Shuldiner AR, Valle D, Weinshilboum R, Green ED, Ginsburg GS. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15(4):258–267. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire AL, Beskow LM. INFORMED CONSENT IN GENOMICS AND GENETIC RESEARCH. Annual review of genomics and human genetics. 2010;11:361–381. doi: 10.1146/annurev-genom-082509-141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMurry JA, Kohler S, Washington NL, Balhoff JP, Borromeo C, Brush M, Carbon S, Conlin T, Dunn N, Engelstad M, Foster E, Gourdine JP, Jacobsen JO, Keith D, Laraway B, Xuan JN, Shefchek K, Vasilevsky NA, Yuan Z, Lewis SE, Hochheiser H, Groza T, Smedley D, Robinson PN, Mungall CJ, Haendel MA. Navigating the Phenotype Frontier: The Monarch Initiative. Genetics. 2016;203(4):1491–5. doi: 10.1534/genetics.116.188870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, Brunner HG, Buske OJ, Carey K, Doll C, Dumitriu S, Dyke SO, den Dunnen JT, Firth HV, Gibbs RA, Girdea M, Gonzalez M, Haendel MA, Hamosh A, Holm IA, Huang L, Hurles ME, Hutton B, Krier JB, Misyura A, Mungall CJ, Paschall J, Paten B, Robinson PN, Schiettecatte F, Sobreira NL, Swaminathan GJ, Taschner PE, Terry SF, Washington NL, Zuchner S, Boycott KM, Rehm HL. The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Hum Mutat. 2015;36(10):915–21. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Beacon Project. Beacon: A Protocol for Federated Genomic Data Sharing. Submitted. [Google Scholar]
- 28.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van El CG, Cornel MC, Borry P, Hastings RJ, Fellmann F, Hodgson SV, Howard HC, Cambon-Thomsen A, Knoppers BM, Meijers-Heijboer H, Scheffer H, Tranebjaerg L, Dondorp W, de Wert GMWR. Whole-genome sequencing in health care. Eur J Hum Genet. 2013;21(6):580–584. doi: 10.1038/ejhg.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


