Abstract
Antibiotics started to be used almost 90 years ago to eradicate life-threatening infections. The urgency of the problem required rapid, broad-spectrum elimination of infectious agents. Since their initial discovery, these antimicrobials have saved millions of lives. However, they are not exempt from side effects, which include the indiscriminate disruption of the beneficial microbiota. Recent technological advances have enabled the development of antimicrobials that can selectively target a gene, a cellular process, or a microbe of choice. These strategies bring us a step closer to developing personalized therapies that exclusively remove disease-causing infectious agents. Here, we advocate the preservation of our beneficial microbes and provide an overview of promising alternatives to broad-spectrum antimicrobials. Specifically, we emphasize nucleic acid and peptide-based systems as a foundation for next-generation alternatives to antibiotics that do not challenge our microbiota and may help to mitigate the spread of resistance.
Introduction
Currently, there is a serious global health problem as increasing numbers of multidrug-resistant bacteria are continuously being isolated from hospitals. According to a recent report published by the United Kingdom government, antibiotic-resistant infections are predicted to result in the death of 10 million people worldwide per year by 2050 if new antimicrobial strategies are not discovered [1]. In fact, the rise in antibiotic resistance has led to a post-antibiotic era in which many standard-of-care antimicrobials are no longer effective. This is partly a result of the decline in antibiotic innovation: no new class of antibiotics has been approved for Gram-negative infections in >45 years, and only 37 antibiotic drugs are in either Phase II or III clinical trials as of 2016 [2]. Therefore, there is an urgent need to develop alternative approaches to treat drug-resistant bacterial infections.
Besides leading to emergence of resistant bacteria, antibiotics may have unintended consequences, such as triggering hyper-inflammatory responses or, most notably, displaying numerous off-target effects (i.e., killing) that disturb beneficial microbiota [3,4]. This is a very significant side effect, as these microbes are associated with our health, and their perturbation can lead to disease. Preserving our microbiota is thus an additional critical aspect to be considered when developing next-generation antimicrobials (Figure 1). Several technologies have been developed for the selective targeting of microbes. In this review, we discuss nontraditional strategies, exemplified in Figure 2, to treat bacterial infections, emphasizing precision approaches that serve as a framework for the design of next-generation antimicrobials.
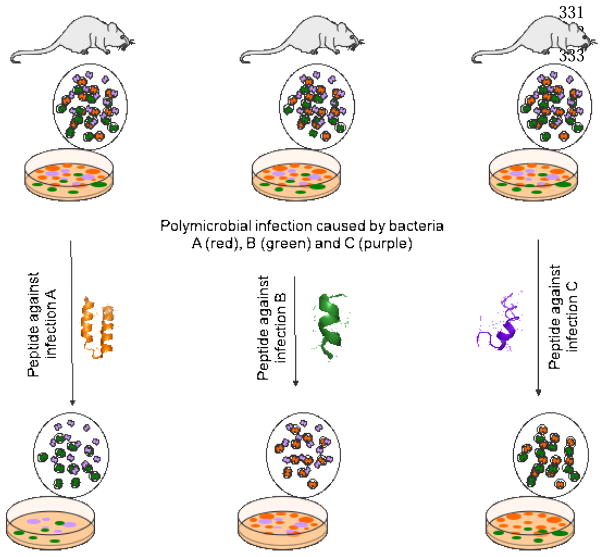
Figure 1. Precision antibiotics.
Hypothetical graphic showing treatment of specific disease causing bacterial infections with precision antimicrobials. For example, in the left side of the figure, we have an infection caused by the orange bacteria, which are surrounded by beneficial microbiota bacteria (colored in green and purple). In this scenario, a precision peptide designed to target the green pathogen will exclusively target this organism. The same applies to the other two examples, where peptides against green (center image) and purple bacteria (right image) would selectively remove these organisms.
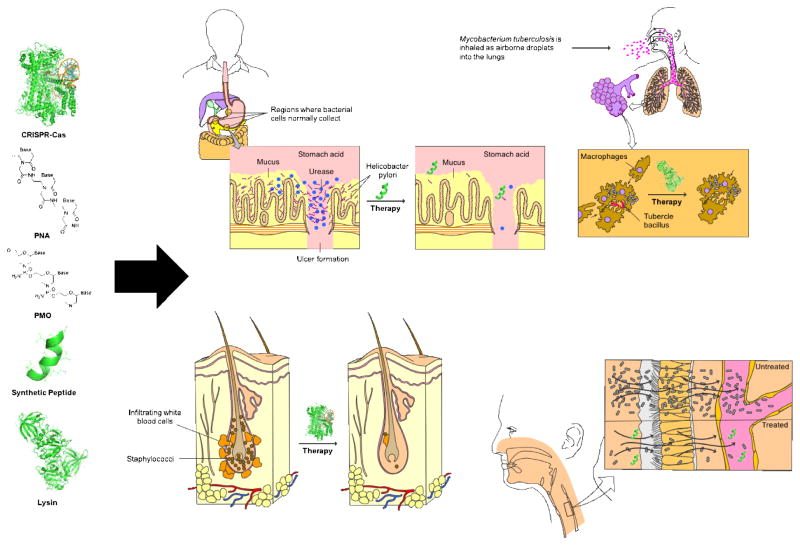
Figure 2. Personalized approach to infectious diseases.
Potential of precision antimicrobial strategies such as the ones described in this review (shown on the left of the figure) for personalized treatment of infectious diseases. In the future, early diagnostic methods may enable rapid identification of disease-causing pathogens, and precision antimicrobial agents may allow specific treatment of infection, while leaving non-causative, beneficial microbial communities unaffected. These advances, coupled with early-diagnostic tools for rapid detection of infectious agents, will constitute the foundation of personalized treatment of infectious diseases.
A call to action: the need to preserve the microbiota
The microbiome plays a key role in human health and disease. Indeed, the composition of the indigenous microbial communities living in the skin, mouth, urinary tract, and especially the gastrointestinal tract profoundly affects many aspects of human health, including immunity and metabolism [5–7]. For instance, a disrupted or unbalanced gut microbiota (i.e., dysbiosis) has been linked to autoimmune disorders (e.g., irritable bowel syndrome and inflammatory bowel disease), obesity, higher propensity to infection, cardiovascular disease, colon cancer, rheumatoid arthritis, depression, Parkinson’s disease, multiple sclerosis, and autism spectrum disorder [8]. Furthermore, dysbiosis can lead to infection with opportunistic pathogens such as Enterococcus faecium, carbapenem-resistant Enterobacteriaceae, and Clostridium difficile [9,10], the latter being responsible for 14,000 deaths per year only in the US. Consistent with the relationship between microbiota disruption and C. difficile infection (CDI), fecal transplantations, which give rise to a healthy human gut microbiota and colonization resistance, are known to prevent the onset of CDI [11].
Microbiome perturbations are of particular concern in children because antibiotics are widely used to treat childhood infections. To put this into perspective, the average child in the US receives three courses of antibiotics by age two, and ten courses by age ten [12].
Therefore, next-generation precision antimicrobials should exhibit high specificity towards target species. We briefly outline nucleic-acid- and peptide-based strategies which may constitute the basis for new types of precision antimicrobials. Additional emerging approaches for the precision killing of bacteria, not covered in this review due to space are the use of bacteriocins, antibodies or anti-virulence compounds [13–21].
Precision antimicrobials: bringing precision medicine to infectious diseases
Nucleic acid-based systems
CRISPR-Cas
The clustered regularly interspersed short palindromic repeats (CRISPR)-CRISPR-associated (Cas) system is widespread in the microbial world [22], as it is found in about 40% of known bacterial species and 90% of archaeal ones. In many prokaryotes, CRISPR-Cas operates as an adaptive immune mechanism that protects them from invading infectious viruses and plasmids. In recent years, apart from its application for precise RNA-guided genome editing, epigenetic modification, and gene regulation in numerous eukaryotic and prokaryotic organisms, CRISPR-Cas tools have been harnessed to generate sequence-specific antimicrobials [23–26]. In these studies, CRISPR-Cas constructs directed towards specific antibiotic resistance genes were delivered via bacteriophages or phagemids serving as specific and high-capacity vectors that allow host-specific targeting and efficient delivery of DNA into bacteria. The approach was very effective in vitro, leading to the selective killing of target microbes carrying antibiotic-resistance genes. This strategy was shown to work in an invertebrate model in which the wax moth Galleria mellonella was infected with Escherichia coli [23], as well as in a mouse model of Staphylococcus aureus skin infection [24].
Virulence factors such as toxin-encoding genes could be targeted instead of antibiotic resistant loci. This opens the door for selective gene therapies that exclusively target pathogenic bacteria, while leaving the beneficial microbiota intact in humans and other animals. However, genetic barriers of the targeted bacterium, notably resident CRISPR-Cas systems, might result in the generation, and further selection upon treatment, of resistant cells. Anti-CRISPR proteins recently identified in some bacteriophages [27] or potentially targeting anti-cas mechanisms [28] could help to circumvent this resistance. Therapeutic strategies combining both CRISPR-based antimicrobials and anti-CRISPR factors offer a promising line of attack against infectious diseases.
Peptide Nucleic Acids (PNAs)
Peptide nucleic acids (PNAs) are synthetic polymers composed of a backbone of repeating N-(2-aminoethyl)-glycine units (and associated with purine and pyrimidine bases) linked by peptide bonds [29]. PNAs operate as antisense molecules by inhibiting the translation of target genes. These agents have traditionally shown promise as therapies for in vivo applications and as alternatives to classical antibiotics, as they exhibit resistance to degradation by nucleases, proteases, and other enzymes. However, PNAs have limited activity in the host due to their inherent high hydrophobicity resulting from the lack of phosphate groups within their backbone, which leads to unselective interactions, and consequently cytotoxicity and hemolytic activity that still need to be addressed. Overcoming this physicochemical barrier will be key to reinstating these agents as eventual therapies.
An example of a peptide-PNA antimicrobial with selective binding properties in mixed bacterial cultures was reported by Mondhe et al. [30]. By designing PNAs aimed at different translational initiation regions, these authors could target several essential genes. This strategy produced a tunable spectrum of activity against Bacillus subtilis, Klebsiella pneumoniae, Salmonella Typhimurium and E. coli. PNAs were designed accordingly to achieve species selectivity based on the presence of 20–25 bases within their sequence that confer selectivity: the target gene (and homologs) were present in the four bacterial species tested, the translation initiation region of the mRNA had at least two base-pair differences between the different bacteria and was amenable to the design of the peptides, and there was evidence that gene silencing of the target and/or inhibition of its cognate protein inhibited growth. The peptides were tested against target and non-target species and exhibited great selectivity. For example, the BS0001 peptide was designed to kill B. subtilis and achieved a minimal inhibitory concentration (MIC) of 4.0 μM but no activity against the other bacteria in the concentration range analyzed. The KS0001 peptide exhibited a MIC of 2.5 μM against K. pneumoniae but was not active against the other bacteria tested [30].
Phosphorodiamidate morpholino-oligomers (PMOs)
Like DNA, phosphorodiamidate morpholino-oligomers (PMOs) are synthetic oligomers composed of the base pairs A, G, C and T; however, they contain a synthetic morpholino and phosphorodiamidate backbone within their sequence [29]. These molecules can be used as antisense strategies as their oligomer sequences are complementary to their target mRNAs. PMOs can be delivered into living cells through different means, for example via conjugation with cell-penetrating peptides. PMOs have shown efficacy both in vitro and in vivo against clinically relevant pathogens such as Acinetobacter baumannii, Burkholderia cepacia complex, and E. coli [31–33] and represent a promising new class of precision antimicrobials. Indeed, a study was recently published in which PMOs targeting essential genes such as acpP, lpxC, and rpsJ were conjugated with the cationic antibiotic polymyxin B leading to 2–8-fold increased anti-P. aeruginosa activity [34]. The compound targeting acpP also inhibited P. aeruginosa biofilm formation. Biofilms are surface-associated communities of microorganisms that are associated with numerous infections in humans and that exhibit increased resistance to antibiotic therapy. The compound targeting rpsJ synergized with the aminoglycoside antibiotic tobramycin. Importantly, these therapies led to 3-log reductions in bacterial loads in the lungs of mice. PMOs are currently limited mostly by delivery efficiency and in vivo stability.
Peptide-based systems
Synthetic peptides
Although still at an early stage, synthetic peptides have been engineered, by tuning their primary structure, for the targeted killing of specific microbes. A range of approaches have been developed to design targeted peptides, including the ab initio method based on database filtering applied by Misha et al., which led to the generation of synthetic peptides with anti-methicillin-resistant S. aureus (MRSA) antimicrobial function [35]. Another strategy to reach this level of specificity was reported by Wang et al. in the identification of a peptide variant of cathelicidin LL-37 containing three D-amino acids (residues 20, 24 and 28) that was resistant to degradation by the protease chymotrypsin, and was able to kill E. coli but did not affect MRSA growth [36]. The selectivity observed may be due to the difference in the NMR structure of the synthetic peptide (unstructured) compared to its parent (alpha-helical) whose amphipathic structure facilitates optimal hydrophobic interactions with the outer membrane of Gram-negative organisms. Further modification of the hydrophobicity of the parent peptide broadened its host range, likely resulting from increased hydrophobic interactions with target bacterial membranes, so that it killed other pathogens such as E. faecium, K. pneumonia, and P. aeruginosa. Other peptides have been designed to target bacterial biofilms [37]. These synthetic peptides act as prophylactic therapies to prevent biofilm colonization on surfaces, and eradicate pre-formed biofilms formed by the most dangerous pathogens, including those highlighted as highest risk by the World Health Organization in February 2017. So far, a number of peptides have been identified that target biofilms formed by various drug-resistant bacteria [38–41]. Interestingly, one of these anti-biofilm peptides has been turned into a dimer by incorporation of a disulfide bond, which led to increased antimicrobial and anti-biofilm activities [42].
Additional studies have also successfully incorporated precision engineering capabilities into peptides. One approach consists of a hybrid peptide (called C16G2) containing a targeting sequence fused to an antimicrobial peptide [43]. This peptide selectively killed the oral bacterial pathogen Streptococcus mutans. The target sequence of C16G2 is composed of 16 amino acids derived from a S. mutans pheromone (also known as a competence-stimulating peptide) and 16 amino acids corresponding to an antimicrobial peptide called G2. The authors demonstrated the specificity of C16G2 towards S. mutans using a saliva-derived in vitro biofilm consortia model containing >100 species of the human oral microbiota. These experiments confirmed the selectivity of peptide C16G2 towards S. mutans, as treatment led to decreased average relative abundance of this bacterium from 24 to 0.1%. Interestingly, upon peptide treatment, a community-level shift in species composition and relative abundance was observed, likely as an ecological response to eliminating S. mutans, a key member of the microbial biofilm community.
Another example is bacaucin-1, a small and stable linear derivative of bacaucin, a natural biosurfactant broad-spectrum antimicrobial peptide originally isolated from Bacillus subtilis strain CAU21. To suppress the hemolytic and cytotoxic effects of the natural peptide bacaucin, the authors removed the lipid portion and the ring opening of the parent peptide, thus exposing its hydrophilic portion and thereby decreasing the overall hydrophobicity of the structure. Contrary to its predecessor, bacaucin-1 displayed selective activity against MRSA and no detectable cytotoxicity. Bacaucin-1 selectively targeted S. aureus antibiotic-resistant strains by a membrane-disruptive mechanism even though it has no lipid portion within its sequence. The authors reported a gradual decrease of cell wall integrity, observed by fluorescence intensity of propidium iodide uptake and DNA binding in a dose-dependent manner. In addition, they described dissipation of the membrane potential, evidencing membrane disruption. Bacaucin-1 has been used as a preventive therapeutic in mice and as a meat preservative [44].
Peptides, including those with selective activity, may also be immobilized on surfaces such as medical devices to prevent colonization with pathogenic organisms [45,46]. Numerous techniques have been described to make peptides adhere to surfaces and materials; these techniques are mostly dependent on the surface and the application desired. For example, cathelicidin LL-37 has been grafted on silanized titanium surfaces, and the final material presents selective activity against E. coli strain K12 [47]. Another example is the lipopeptide C16-KKFF anchored in hybrid biomaterial membranes that generate capsules that are active against Listeria monocytogenes, S. aureus, and E. faecalis [48]. Immobilized antimicrobial peptides with antibiofilm activity have also been described, including peptide CWR11 immobilized on catheters via a polydopamine-peptide strategy, which exhibits activity against E. coli, S. aureus, and P. aeruginosa [49].
A study by Yadavalli and colleagues [50] demonstrated that certain antimicrobial peptides at sublethal concentrations cause filamentation via interference with cell division in E. coli. The authors found that this effect was dependent on the PhoP/PhoQ system, and resulted from peptide-mediated increased expression of the enzyme QueE, which was shown by the authors to be involved in cell division (it localizes to the division septum in filamentous bacterial cells) apart from its known role in tRNA modification. These observations may be exploited in the future to generate peptide antimicrobials that target cell division in bacteria.
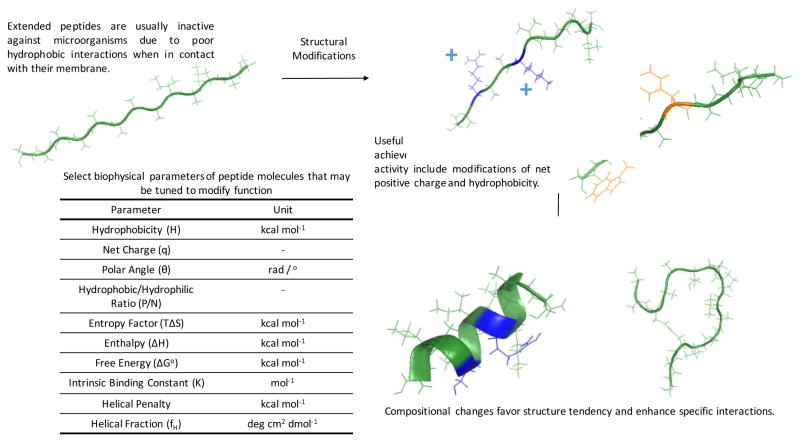
Overall, the studies outlined in this section illustrate the feasibility of incorporating functional specificity into peptide molecules through peptide engineering approaches to precisely manipulate microbial communities. Some of these strategies are shown in Figure 3. Future work will have to address issues with delivery, stability in vivo and potential off-target killing effects of precision peptides to prevent unintended removal of microbes.
Figure 3. Design principles for selective peptides.
Hypothetical images of modifications that can be made to generate precision peptides.
Lysins
Lysins, also known as endolysins or murein (peptidoglycan) hydrolases, are hydrolytic enzymes produced by bacteriophages (or phages) that break down the cell wall of target bacteria during the final stage of the lytic phage cycle. Phage lysins are generally species- or subspecies-specific; however broad-spectrum lysins have also been identified [51]. Lysins, exploited as antimicrobial agents delivered from the outside, are effective primarily against Gram-positive bacteria, which is not surprising as the presence of an outer membrane in Gram-negative bacteria prevents extracellular lysin molecules from digesting peptidoglycan. Still, lysins with a high content of cationic residues on their C-terminal domain have been found to possess activity against Gram-negative pathogens, likely due to positive charges enabling the penetration of outer membranes. For example, certain endolysins (e.g., OBPgp279) have been recently reported to have activity against bacteria such as P. aeruginosa and A. baumannii [52].
In another study, lysins were engineered to kill A. baumannii. The authors identified the C-terminal amino acids 108 to 138 of phage lysin PlyF307 (termed P307) to be sufficient to kill A. baumannii [53]. Lysin P307 was further engineered to generate improved variants such as P307SQ-8C, which had activity against biofilms, MIC values comparable to those of standard-of-care antibiotics such as levofloxacin and ceftazidime, and synergy with polymyxin B. P307SQ-8C further reduced the bacterial burden in a mouse model of A. baumannii skin infection. This study demonstrates the prospect of using peptide derivatives from bacteriophage lysins to treat topical infections and remove biofilms caused by Gram-negative pathogens.
Overall, phage lysins, owing to their precise mode of action resulting partly from their co-evolution with bacteria, constitute potential alternatives to antibiotics. They may also prove useful for developing adjuvant therapies with currently available antibiotics. However, delivery methods still need to be developed to successfully implement the use of lysins in the clinic.
Conclusions
There is currently an urgent need for a new generation of antimicrobials that mitigates the spread of antibiotic resistance and preserves our beneficial microbiota. Despite the tremendous benefit to human health of traditional broad-spectrum antibiotics, these agents exert selection pressure leading to the emergence of resistance in all targeted microbes and disrupt our microbiota, often causing detrimental effects. Here, we have briefly described both nucleic acid-based and peptide-based approaches that constitute a framework for the design of precision antimicrobials.
Nucleic acid-based approaches represent a new paradigm to achieve the selective killing of microbes. For example, CRISPR-Cas has been used to target specific antibiotic resistance genes to selectively remove drug-resistant organisms in pure cultures and in complex microbial consortia. The recent discovery of anti-CRISPR proteins, which might enable evasion of resident CRISPR-based resistance to therapeutic, exogenous CRISPR-Cas constructs, may now be exploited to increase the efficiency of this antimicrobial strategy. Despite being limited by their hydrophobicity, PNAs have also been used directly or as templates for the design of selective molecules that target the translation of desired genes. PMOs show specific complementarity to their target mRNAs and constitute another category of promising candidates when delivered efficiently into bacterial cells.
Peptide-based systems with enhanced specificity have recently been reported. These molecules can be easily tuned to achieve specific functions in living cells given their vast sequence space (20n, n being the number of amino acids in a given peptide chain). Their numerous biophysical properties, which include hydrophobicity, net charge, and amphiphilicity, can be modified to enhance their antibacterial function. To get around the complexity of this combinatorial problem, template molecules that already exhibit activity have been used to generate optimized synthetic variants based on structure-activity relationship studies. Another example of synthetic peptide-based antibiotics is lysins, which show antimicrobial activity against Gram-positive bacteria and have been engineered to expand their host range to Gram-negative pathogens.
Both nucleic acid and peptide-based systems still have to overcome certain limitations in order to ensure translation into the clinic. Most notably, technologies need to be developed to prevent degradation in the host by nucleases and proteases, novel methods are required to efficiently deliver these agents to target sites in the body, and precision has to be optimized to prevent off-target effects.
These diverse, narrow-spectrum polymer-based antimicrobials have been shown to be effective both in vitro and in animal models. The strategies described here thus represent an excellent framework for designing next-generation precision antibacterials.
Highlights.
Precision antimicrobials represent a novel approach for treating infectious diseases;
The role of the microbiota in health and disease is indisputable and so is the need to preserve these microbial communities by using specific therapies;
Nucleic-acid and peptide based antibacterials represent promising technologies to achieve selective killing of microbes.
Acknowledgments
Some of the figures shown here were prepared using the Motifolio drawing toolkit. Cesar de la Fuente-Nunez is a Ramon Areces Foundation Fellow (Spain). Marcelo Der Torossian Torres is supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (Brazil). Francisco J. M. Mojica is funded by the Spanish Ministerio de Economía y Competitividad (BIO2014-53029P) and the European Commission/Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (291815 Era-Net ANIHWA). Timothy Lu is supported by the National Institutes of Health, National Science Foundation, Defense Threat Reduction Agency, Defense Advanced Research Projects Agency, Center for Microbiome Informatics and Therapeutics, Kenneth Rainin Foundation, Koch Institute for Integrative Cancer Research, and Broad Institute of MIT and Harvard.
Footnotes
Conflict of interest
The authors report no conflict of interests associated with this manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Davies DS, Farrar J, Rex J, White LJ, RM . Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. In: JON, editor. The Review on Antimicrobial Resistance. 2014. [Google Scholar]
- 2.Trusts TPC. Antibiotics Currently in Clinical Development. 2016;2017 [Google Scholar]
- 3.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nature reviews Endocrinology. 2015;11:182–190. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemente JC, Ursell LK, Parfrey LW, Knight R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell. 2012;148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y, Hand Timothy W. Role of the Microbiota in Immunity and Inflammation. Cell. 157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 8.Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. New England Journal of Medicine. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 9.Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA, Kelly C, Khoruts A, Louie T, Martinelli LP, et al. Treating Clostridium difficile Infection With Fecal Microbiota Transplantation. Clinical Gastroenterology and Hepatology. 2011;9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belkaid Y, Hand Timothy W. Role of the Microbiota in Immunity and Inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dy RL, Richter C, Salmond GPC, Fineran PC. Annual Review of Virology. Vol. 1. Enquist LW: Annual Reviews; 2014. Remarkable Mechanisms in Microbes to Resist Phage Infections; pp. 307–331. Annual Review of Virology, vol 1.] [DOI] [PubMed] [Google Scholar]
- 12.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nature Communications. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nature Chemical Biology. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 14.Ilangovan A, Fletcher M, Rampioni G, Pustelny C, Rumbaugh K, Heeb S, Camara M, Truman A, Chhabra SR, Emsley J, et al. Structural Basis for Native Agonist and Synthetic Inhibitor Recognition by the Pseudomonas aeruginosa Quorum Sensing Regulator PqsR (MvfR) Plos Pathogens. 2013:9. doi: 10.1371/journal.ppat.1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein T, Henn C, de Jong JC, Zimmer C, Kirsch B, Maurer CK, Pistorius D, Muller R, Steinbach A, Hartmann RW. Identification of Small-Molecule Antagonists of the Pseudomonas aeruginosa Transcriptional Regulator PqsR: Biophysically Guided Hit Discovery and Optimization. Acs Chemical Biology. 2012;7:1496–1501. doi: 10.1021/cb300208g. [DOI] [PubMed] [Google Scholar]
- 16.Lesic B, Lepine F, Deziel E, Zhang JW, Zhang QH, Padfield K, Castonguay MH, Milot S, Stachel S, Tzika AA, et al. Inhibitors of pathogen intercellular signals as selective anti-infective compounds. Plos Pathogens. 2007;3:1229–1239. doi: 10.1371/journal.ppat.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lidor O, Al-Quntar A, Pesci EC, Steinberg D. Mechanistic analysis of a synthetic inhibitor of the Pseudomonas aeruginosa LasI quorum-sensing signal synthase. Scientific reports. 2015:5. doi: 10.1038/srep16569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Loughlin CT, Miller LC, Siryaporn A, Drescher K, Semmelhack MF, Bassler BL. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonetti O, Cirioni O, Cacciatore I, Baldassarre L, Orlando F, Pierpaoli E, Lucarini G, Orsetti E, Provinciali M, Fornasari E, et al. Efficacy of the Quorum Sensing Inhibitor FS10 Alone and in Combination with Tigecycline in an Animal Model of Staphylococcal Infected Wound. Plos One. 2016:11. doi: 10.1371/journal.pone.0151956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He JX, Kitao T, Righi V, Milot S, Tzika A, et al. Identification of Anti-virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity. Plos Pathogens. 2014:10. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storz MP, Maurer CK, Zimmer C, Wagner N, Brengel C, de Jong JC, Lucas S, Musken M, Haussler S, Steinbach A, et al. Validation of PqsD as an Anti-biofilm Target in Pseudomonas aeruginosa by Development of Small-Molecule Inhibitors. Journal of the American Chemical Society. 2012;134:16143–16146. doi: 10.1021/ja3072397. [DOI] [PubMed] [Google Scholar]
- 22.de la Fuente-Nunez C, Lu TK. CRISPR-Cas9 technology: applications in genome engineering, development of sequence-specific antimicrobials, and future prospects. Integrative Biology. 2017;9:109–122. doi: 10.1039/c6ib00140h. [DOI] [PubMed] [Google Scholar]
- 23**.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotech. 2014;32:1141–1145. doi: 10.1038/nbt.3011. The authors reported the use of CRISPR-Cas technology to create sequence-specific antimicrobials whose spectrum of activity is chosen by design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotech. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable Removal of Bacterial Strains by Use of Genome-Targeting CRISPR-Cas Systems. Mbio. 2014:5. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawluk A, Staals RHJ, Taylor C, Watson BNJ, Saha S, Fineran PC, Maxwell KL, Davidson AR. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nature Microbiology. 2016;1:16085. doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- 28.Almendros C, Guzmán NM, García-Martínez J, Mojica FJM. Anti-cas spacers in orphan CRISPR4 arrays prevent uptake of active CRISPR–Cas I-F systems. Nature Microbiology. 2016;1:16081. doi: 10.1038/nmicrobiol.2016.81. [DOI] [PubMed] [Google Scholar]
- 29.Sully EK, Geller BL. Antisense antimicrobial therapeutics. Current Opinion in Microbiology. 2016;33:47–55. doi: 10.1016/j.mib.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Mondhe M, Chessher A, Goh S, Good L, Stach JEM. Species-Selective Killing of Bacteria by Antimicrobial Peptide-PNAs. Plos One. 2014:9. doi: 10.1371/journal.pone.0089082. Authors report the design and evaluation of peptide nucleic acids against a range of different pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenberg DE, Marshall-Batty KR, Brinster LR, Zarember KA, Shaw PA, Mellbye BL, Iversen PL, Holland SM, Geller BL. Antisense Phosphorodiamidate Morpholino Oligomers Targeted to an Essential Gene Inhibit Burkholderia cepacia Complex. Journal of Infectious Diseases. 2010;201:1822–1830. doi: 10.1086/652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geller BL, Deere JD, Stein DA, Kroeker AD, Moulton HM, Iversen PL. Inhibition of gene expression in Escherichia coli by antisense phosphorodiamidate morpholino oligomers. Antimicrobial Agents and Chemotherapy. 2003;47:3233–3239. doi: 10.1128/AAC.47.10.3233-3239.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geller BL, Marshall-Batty K, Schnell FJ, McKnight MM, Iversen PL, Greenberg DE. Gene-Silencing Antisense Oligomers Inhibit Acinetobacter Growth In Vitro and In Vivo. Journal of Infectious Diseases. 2013;208:1553–1560. doi: 10.1093/infdis/jit460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howard JJ, Sturge CR, Moustafa DA, Daly SM, Marshall-Batty KR, Felder CF, Zamora D, Yabe-Gill M, Labandeira-Rey M, Bailey SM, et al. Inhibition of Pseudomonas aeruginosa by Peptide-conjugated Phosphorodiamidate Morpholino Oligomers. Antimicrobial Agents and Chemotherapy. 2017 doi: 10.1128/AAC.01938-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra B, Wang GS. Ab Initio Design of Potent Anti-MRSA Peptides Based on Database Filtering Technology. Journal of the American Chemical Society. 2012;134:12426–12429. doi: 10.1021/ja305644e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36*.Wang GS, Hanke ML, Mishra B, Lushnikova T, Heim CE, Thomas VC, Bayles KW, Kielian T. Transformation of Human Cathelicidin LL-37 into Selective, Stable, and Potent Antimicrobial Compounds. Acs Chemical Biology. 2014;9:1997–2002. doi: 10.1021/cb500475y. Reported a D-amino acid-containing peptide initially selective against E. coli, whose target range was broadened to kill six pathogenic bacterial strains through re-design of hydrophobic portion of parent peptide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de la Fuente-Núñez C, Cardoso MH, de Souza Cândido E, Franco OL, Hancock REW. Synthetic antibiofilm peptides. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2016;1858:1061–1069. doi: 10.1016/j.bbamem.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de la Fuente-Núñez C, Korolik V, Bains M, Nguyen U, Breidenstein EBM, Horsman S, Lewenza S, Burrows L, Hancock REW. Inhibition of Bacterial Biofilm Formation and Swarming Motility by a Small Synthetic Cationic Peptide. Antimicrobial Agents and Chemotherapy. 2012;56:2696–2704. doi: 10.1128/AAC.00064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribeiro SM, de la Fuente-Núñez C, Baquir B, Faria-Junior C, Franco OL, Hancock REW. Antibiofilm Peptides Increase the Susceptibility of Carbapenemase-Producing Klebsiella pneumoniae Clinical Isolates to β-Lactam Antibiotics. Antimicrobial Agents and Chemotherapy. 2015;59:3906–3912. doi: 10.1128/AAC.00092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva ON, de la Fuente-Núñez C, Haney EF, Fensterseifer ICM, Ribeiro SM, Porto WF, Brown P, Faria-Junior C, Rezende TMB, Moreno SE, et al. An anti-infective synthetic peptide with dual antimicrobial and immunomodulatory activities. Scientific reports. 2016;6:35465. doi: 10.1038/srep35465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dosler S, Karaaslan E. Inhibition and destruction of Pseudomonas aeruginosa biofilms by antibiotics and antimicrobial peptides. Peptides. 2014;62:32–37. doi: 10.1016/j.peptides.2014.09.021. [DOI] [PubMed] [Google Scholar]
- 42.Thamri A, Létourneau M, Djoboulian A, Chatenet D, Déziel E, Castonguay A, Perreault J. Peptide modification results in the formation of a dimer with a 60-fold enhanced antimicrobial activity. Plos One. 2017;12:e0173783. doi: 10.1371/journal.pone.0173783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo L, McLean JS, Yang Y, Eckert R, Kaplan CW, Kyme P, Sheikh O, Varnum B, Lux R, Shi W, et al. Precision-guided antimicrobial peptide as a targeted modulator of human microbial ecology. Proceedings of the National Academy of Sciences. 2015;112:7569–7574. doi: 10.1073/pnas.1506207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44**.Liu Y, Ding SY, Dietrich R, Martlbauer E, Zhu K. A Biosurfactant-Inspired Heptapeptide with Improved Specificity to Kill MRSA. Angewandte Chemie-International Edition. 2017;56:1486–1490. doi: 10.1002/anie.201609277. Reported the design of a linear peptide based on a biosurfactant that shows specific antibacterial activity against MRSA. [DOI] [PubMed] [Google Scholar]
- 45.Mishra B, Lushnikova T, Golla RM, Wang XQ, Wang GS. Design and surface immobilization of short anti-biofilm peptides. Acta Biomaterialia. 2017;49:316–328. doi: 10.1016/j.actbio.2016.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa F, Carvalho IF, Montelaro RC, Gomes P, Martins MCL. Covalent immobilization of antimicrobial peptides (AMPs) onto biomaterial surfaces. Acta Biomaterialia. 2011;7:1431–1440. doi: 10.1016/j.actbio.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Gabriel M, Nazmi K, Veerman EC, Amerongen AVN, Zentner A. Preparation of LL-37-grafted titanium surfaces with bactericidal activity. Bioconjugate Chemistry. 2006;17:548–550. doi: 10.1021/bc050091v. [DOI] [PubMed] [Google Scholar]
- 48.Castelletto V, Kaur A, Hamley IW, Barnes RH, Karatzas KA, Hermida-Merino D, Swioklo S, Connon CJ, Stasiak J, Reza M, et al. Hybrid membrane biomaterials from self-assembly in polysaccharide and peptide amphiphile mixtures: controllable structural and mechanical properties and antimicrobial activity. Rsc Advances. 2017;7:8366–8375. [Google Scholar]
- 49.Lim KY, Chua RRY, Bow H, Tambyah PA, Hadinoto K, Leong SSJ. Development of a catheter functionalized by a polydopamine peptide coating with antimicrobial and antibiofilm properties. Acta Biomaterialia. 2015;15:127–138. doi: 10.1016/j.actbio.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 50.Yadavalli SS, Carey JN, Leibman RS, Chen AI, Stern AM, Roggiani M, Lippa AM, Goulian M. Antimicrobial peptides trigger a division block in Escherichia coli through stimulation of a signalling system. Nature Communications. 2016:7. doi: 10.1038/ncomms12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoong P, Schuch R, Nelson D, Fischetti VA. Identification of a broadly active phage lytic enzyme with lethal activity against antibiotic-resistant Enterococcus faecalis and Enterococcus faecium. Journal of Bacteriology. 2004;186:4808–4812. doi: 10.1128/JB.186.14.4808-4812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay JP, et al. Engineered Endolysin-Based “Artilysins” To Combat Multidrug-Resistant Gram-Negative Pathogens. Mbio. 2014:5. doi: 10.1128/mBio.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53*.Thandar M, Lood R, Winer BY, Deutsch DR, Euler CW, Fischetti VA. Novel Engineered Peptides of a Phage Lysin as Effective Antimicrobials against Multidrug-Resistant Acinetobacter baumannii. Antimicrobial Agents and Chemotherapy. 2016;60:2671–2679. doi: 10.1128/AAC.02972-15. Reported novel engineered peptides from a phage lysin. [DOI] [PMC free article] [PubMed] [Google Scholar]