Abstract
GroEL, a prototypical member of the chaperonin class of chaperones, is a large supramocular machine that assists protein folding and plays an important role in proteostasis. GroEL comprises two heptameric rings, each of which encloses a large cavity that provides a folding chamber for protein substrates. Many questions remain regarding the mechanistic details of GroEL facilitated protein folding. Thus, data at atomic resolution of the type provided by NMR and EPR are invaluable. Such studies often require complete deuteration of GroEL, uniform or residue specific 13C and 15N isotope labeling, and the introduction of selective cysteine mutations for site-specific spin labeling. In addition, high purity GroEL is essential for detailed studies of substrate-GroEL interactions as quantitative interpretation is impossible if the cavities are already occupied and blocked by other protein substrates present in the bacterial expression system. Here we present a new purification protocol designed to provide highly pure GroEL devoid of non-specific protein substrate contamination.
Keywords: GroEL, supramolecular machine, disassembly/reassembly, purification for biophysical studies
1. Introduction
GroEL, the prototypical member of the chaperonin class of chaperones [1], is a large tetradecameric complex comprising 14 identical subunits (GroEL14) that form two double-ringed cylinders (7 subunits per cylinder) stacked back to back [2]. The resulting two large cavities constitute the protein substrate folding chambers that are capped upon binding the co-chaperone GroES. It has been proposed that confinement of a single protein substrate molecule within the protected cage facilitates folding by preventing aggregation [3, 4]. There are still many unanswered questions regarding the mechanistic basis of GroEL action. For example, GroEL displays positive intra-ring cooperativity with respect to ATP and K+ binding but negative inter-ring cooperativity for GroES and ATP binding [5], yet little is known about how the different subunits communicate with each other. Similarly, each GroEL subunit has a long 23-residue unstructured C-terminal tail, comprising several Gly-Gly-Met repeats located at the base of the cavity, that is essential for the folding of protein substrates [6, 7]; whether the tail is actively involved in the incorporation of protein substrates within the cavity or simply facilitates retention within the cavity is still a matter of debate. Recent developments in NMR and EPR hold promise for addressing such questions, to name only a few, at atomic resolution. For example, selective-isotope labeling [8], TROSY-based pulse sequences [9], and dark state exchange saturation transfer [10] have expanded the field of NMR spectroscopy to the study of megadalton complexes, while recent progress in pulsed double electron-electron resonance (DEER) EPR spectroscopy has extended the range of accessible distances between spin labels up to 170 Å in deuterated proteins [11]. A pre-requisite for quantitative NMR and EPR studies is a GroEL purification protocol that (i) results in high purity GroEL in which the cavities are devoid of any contaminating substrate proteins present in the bacterial expression system, (ii) is reproducible from construct to construct, (iii) allows full deuteration, and (iv) produces sufficiently high yields to permit solution-state NMR studies.
GroEL is essential for E. coli growth [12] and has been shown to interact both in vivo and in vitro with several hundred proteins [13]. Many of these protein substrates bind with micro- to picomolar affinity, and, as a result, the substrate-GroEL complexes survive many passages through various columns, rendering GroEL purification challenging. The general GroEL purification protocol consists of two steps: a standard purification in which GroEL is separated from other unbound proteins, and a final clean-up to remove proteins bound within the GroEL cavity (Fig. 1). The most commonly used final clean-up procedure, introduced by Lorimer’s group [14], consists of an acetone precipitation step in which protein solubility is reduced by lowering the dielectric constant of the solution. GroEL with all bound protein impurities precipitates in the presence of acetone, and for unknown reasons only GroEL can be resolubilized. However, this protocol is not applicable for deuterated proteins and, in our hands, also failed for several GroEL mutants. Although deuterated GroEL has been used by Horwich and Wüthrich in various qualitative solution-state NMR studies, the purification protocol employed did not make use of a final clean-up procedure as this wasn’t required for the particular questions being addressed [15]. Other protocols treat GroEL with 20% methanol [16], salting out the major contaminant, β-galactosidase [17], or make use of disassembly/reassembly methods [18–20]. Unfortunately, in our hands all published clean-up procedures either failed to produce pure enough GroEL (we aimed for less than 10% impurities expressed in terms of mol per mol GroEL14) or the yield was too low to meet the sample requirements for NMR and EPR.
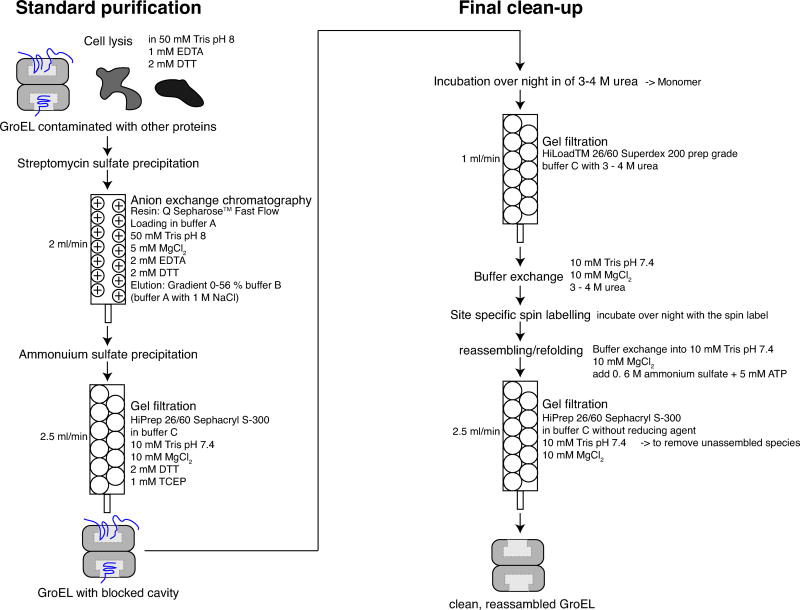
Figure 1. Summary of the GroEL purification protocol.
The protocol consists of two parts: a standard purification protocol (left panel) and a final clean-up (right panel). The standard purification comprises streptomycin sulfate precipitation, ion exchange column chromatography, (NH4)2SO4 precipitation, and gel filtration (Sephacryl S-300). During the first purification phase, GroEL is separated from other proteins produced during protein expression in E. coli that do not bind to GroEL. Tightly binding protein substrates of GroEL are still present in the cavities (illustrated schematically in blue). The second purification phase comprises the final clean-up in which intrinsic protein substrates are removed from the cavities of GroEL. Phase 2 consists of disassembly into monomers by addition of 3–4 M urea (depending on the mutant), gel filtration to purify the monomer (Superdex 200 column), site-specific nitroxide spin labeling (for surface engineered cysteine mutants), reassembly (by the addition of (NH4)2SO4 and ATP), and finally gel filtration (Sephacryl S-300 column) to remove the unassembled GroEL fraction. For each step, the respective columns, buffers, and flow rates are specified.
Here we introduce a novel purification protocol that was successful employed for various mutations located around and within the GroEL cavity (specifically, E315C, A138C, and R268C). We optimized a previously published standard purification protocol [14] and developed a new protocol for the final clean-up phase. The latter consists of disassembly into monomers in 3–4 M urea, purification of the resulting monomer by gel filtration, and finally reassembly back to GroEL14 by the addition of ammonium sulfate and ATP.
2. Materials and Methods
2.1 Materials
Luria Bertani (LB) medium (CAPSULES) was purchased from MP Biomedicals (Solon, OH, USA) and LB agar plates with 100 μg/ml ampicilin from IPMscientific (MD, USA). Ampicillin sodium salt, isopropyl β-D-1-thiogalactopyranoside (IPTG) (OmniPur, Calbiochem), DL-dithiothreitol (DTT), tris(2-carboxyethyl)phosphine (TCEP), sodium chloride (NaCl), albumin from bovine serum (BSA), adenosine 5’ diphosphate disodium salt (ADP), adenosine 5’ triphosphate disodium salt (ATP), ammonium sulfate ((NH4)2SO4) for molecular biology, and streptomycin sulfate salt were obtained from Millipore Sigma (St. Louis, MO, USA). cOmplete tablets EDTA-free (Roche, Germany) were used as a protease inhibitor. 1M Tris-HCl at pH 7.4 or 8, and ethylenediaminetetraacetic acid (EDTA) were purchased from K-D Medicals (Columbia, MD, USA). Molecular biology grade magnesium chloride (MgCl2) was obtained from Quality Biological (MD, USA), and Urea (UltraPure™) and guanidinium hydrochloride (GuHCl) from Invitrogen (ThermoFisher Scientific, NY, USA). Nitroxide spin-labels (MTSL, S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl) methyl methanesulfonothioate, and the diamagnetic control MTS (1-acetoxy-2,2,5,5-tetramethyl-δ-3-pyrroline-3-methyl) methanethiosulfonate) were obtained from Toronto Research Chemicals (Canada).
2.2 Point mutations
One Shot™ MAX Efficiency™ DH5ɑ™−T1R Competent Cells (Invitrogen, ThermoFisher) were used as the bacterial host for DNA cloning. The GroELE315C DNA sequence, containing the wild type sequence with all 3 natural cysteines mutated to alanines and a cysteine introduced at position 315 (C138A, C458A, C519A, E315C), was synthesized by GenScript (NJ, USA) using the OptimumGene™ algorithm for codon optimization, and cloned into a pET-21(a+) vector. A point mutation was subsequently introduced by PCR at position 315 (C315E) to obtain the GroEL sequence without any cysteines (Cys0). Two residues at positions 268 (R268C) and 138 (A138C) of the Cys0 sequence were mutated to cysteines resulting in 3 GroEL constructs, each containing only one cysteine residue: GroELE315C, GroELR268C, and GroELA138C. The primers are shown in Table T1 and were obtained from Integrated DNA Technologies (IA, USA).
Table T1.
Primers used to introduce point mutations into GroELE315C
| GroEL-Cys0 | forward: 5’- CTG GAA AAA GCG ACG CTG GAA GAT CTG GGT CAA GCC AAA -3’ |
| reverse: 5’- TTT GGC TTG ACC CAG ATC TTC CAG CGT CGC TTT TTC CAG -3’ | |
|
| |
| GroEL-A138C | forward: 5’- CTG TCC GTT CCG TGC AGC GAT TCT AAG -3’ |
| reverse: 5’- CTT AGA ATC GCT GCA CGG AAC GGA CAG -3’ | |
|
| |
| GroEL-R268C | forward: 5’- GTT GTC AAC ACG ATG TGT GGT ATT GTC AAA GTG -3’ |
| reverse: 5’- CAC TTT GAC AAT ACC ACA CAT CGT GTT GAC AAC -3’ | |
The PCR reactions were conducted using 0.625 units DNA polymerase, 0.2 mM dNTPs (Taq DNA polymerase master mix, Apex™, Biocompare, CA, USA), 1 μl each, forward and reverse primers at a concentration of 5 μM (Integrated DNA Technologies, IA, USA), and 1 μl DNA plasmid at approximately 5 ng/μl. 17 cycles of 30 seconds denaturation at 95 °C, annealing for 1 minute at 55 °C, selectively denaturation of the mutation for 14 minutes at 68 °C, and finally extension for 20 minutes at 72 °C w as carried out. The template vector was removed by adding 2 μl DpnI restriction endonuclease (New England BioLabs, MA, USA) at 20,000 U/ml at 37 °C for 1h. The plasmid was purified using the plasmid purification kit (QIAprep spin Miniprep Kit, Quiagen, Germany), cloned into DH5ɑ T1R competent cells, and plated on LB agar plates with ampicillin at 100 μg/ml. On the next day the colonies were amplified in 3 ml LB with 100 μg/ml ampicillin overnight and again purified with the plasmid purification kit. All mutations were sequenced by QuintaraBio (DC, USA).
2.3 Protein expression
The various plasmids were transformed into One Shot™ BL21 Star™ (DE3) Chemically Competent E. coli cells (Invitrogen, ThermoFisher). A pre-culture of 3 ml LB was grown overnight at 37 °C and transferred into 1 L LB the next morning. Gene expression was induced with 0.5 mM IPTG at an optical density at 600 nm of 0.8. After overnight expression at 20 °C the cells were harvested by centrifugation at 4500 g (Avanti™ J-20 XP, Beckman Coulter, Inc.) at 4 °C for 30 minutes and stored at −80 °C.
2.4 Cell lysis and fast protein liquid chromatography
Cells were lysed using the Constant System cell disruptor, TS Series 0.75 kW Version 4 (Constant Systems Limited, UK). Fast protein liquid chromatography (FPLC) was performed using an ÄKTA Explorer (GE, Healthcare, MA, USA) with a Frac-950 fraction collector for ion exchange chromatography and a Frac-920 for size exclusion chromatography. Elution of protein was monitored by UV absorption at 254 and 280 nm. A Biologic LP system with a BioFrac fraction collector (BioRAD, CA, USA) was used for all urea containing purification steps and the elution profiles were monitored by UV absorption at 280 nm. For ion exchange chromatography a column was self-packed with 60 ml Q Sepharose™ Fast Flow resin (GE, Healthcare, MA, USA). For gel filtration, a HiPrep 26/60 Sephacryl S-300 column (GE, Healthcare, MA, USA) was employed for assembled GroEL14, and a HiLoad™ 26/60 Superdex 200 prep grade column (GE, Healthcare, MA, USA) for disassembled GroEL monomer. For ion exchange column in urea a HiTrap™ QFF (2 × 5 ml) (GE, Healthcare, MA, USA) column was used; for hydrophobic interaction chromatography (HIC) a HiTrap™ Phenyl FF (1 × 5 ml) (GE, Healthcare, MA, USA) column was used; and for buffer exchange a disposable PD-10 desalting column was used with Sephadex G-25 resin, 2.5 mL samples (GE, Healthcare, MA, USA).
For the purification of GroEL the following buffers were employed:
Lysis buffer: 50 mM Tris, pH 8, 1 mM EDTA, and 2 mM DTT
Buffer A: 50 mM Tris pH 8, 5 mM MgCl2, 2 mM EDTA, and 2 mM DTT
Buffer B: 50 mM Tris pH 8, 5 mM MgCl2, 2 mM EDTA, 1 M NaCl, and 2 mM DTT
Buffer C: 10 mM Tris, pH 7.4, 10 mM MgCl2, 2 mM DTT, and 1 mM TCEP
Buffer D: 10 mM Tris, pH 7.4, 10 mM MgCl2, 2 mM DTT, 1 mM TCEP, and 3 M urea
Buffer E: 10 mM Tris, pH 7.4, 10 mM MgCl2
Centrifugation we carried out at 50,000 g for 25 minutes unless otherwise indicated.
2.5 Gel electrophoresis
For SDS-PAGE gel electrophoresis, 15 μl sample was mixed with 5 μl SDS protein gel loading solution (2x) (Quality Biological, MD, USA) and loaded onto a NuPAGE™ 4–12% Bis-Tris gel with 1.5 mm × 10 or 1.5 mm × 15 wells (Novex, ThermoFisher Scientific, NY, USA). The PageRuler™ Plus prestained protein ladder (ThermoFisher Scientific, NY, USA) was used as a molecular marker. The running buffer was NuPAGE MES SDS buffer (20x) (Novex, ThermoFisher Scientific, NY, USA). Gel electrophoresis was performed according to the manufacturer’s instructions. Gels were stained for 1h or overnight with PageBlue™ protein staining solution (ThermoFisher Scientific, NY, USA) and destained for several hours with deionized water.
Assembly and disassembly of GroEL was monitored using blue native PAGE (BN-PAGE; Native PAGE™ 3–12% Bis-Tris gel) with 1 mm × 15 wells. 7.5 μl sample was mixed with 2.5 μl loading solution (NativePAGE 4x sample buffer). The gel, cathode buffer (20x), running buffer (20x), and loading solution were obtained from Novex, ThermoFisher Scientific (NY, USA). Gel electrophoresis was performed in the cold room following the manufacturer’s instructions, followed by staining for 1h or overnight with PageBlue™ protein staining solution (ThermoFisher Scientific, NY, USA) and destaining for several hours with deionized water.
2.6 Tryptophan fluorescence
The final purity of GroEL can be judged by measuring the intrinsic fluorescence based on the emission of the aromatic amino acid tryptophan [21]. Since GroEL does not contain any tryptophan residues, any residual tryptophan fluorescence must arise from co-purified protein impurities.
An infinite M200 Pro plate reader, in conjunction with the Magellan™ V 7.2 data analysis software from Tecan (Switzerland), was used to measure fluorescence. 96 well plates (PP, FBottom, chimney well black, and greiner bio-one; NC, USA) were used. The measurement was performed at 25 °C, starting with orbital shaking with an amplitude of 1 mm for 10 seconds prior to the actual fluorescence measurement. An emission scan from 325–450 nm was employed, with an excitation wavelength at 295 nm and a top readout. GroEL samples were denaturated in 8 M GuHCl and loaded in the wells at a concentration of 10 μM. All measurements were performed in triplicate and the accurate concentration of GroEL was measured from the A280 of each well. The amount of impurities was estimated using a standard curve for BSA from 0 to 2 μM. BSA contains two tryptophans per molecule, and, assuming the average molecular weight of the protein impurities is around 40 kDa with an average 3–4 tryptophans per protein, the molar amount of impurities in the sample can be estimated [21].
2.7 Expression and purification of FynSH3
The triple A39V/N53P/V55L (SH3vpl) mutant of the Gallus gallus FynSH3 domain was isotopically labelled with 15N and purified using a hexahistidine purification tag according to a previously published protocol [22]. Tobacco etch virus (TEV) protease, required to cleave the N-terminal His tag of FynSH3, was purified according to a published procedure [23].
2.8 Expression and purification of GroES
GroES was expressed in E. coli (BL21 DE3) using LB medium and purified as previously reported [24]. Briefly, a hexahistidine containing construct was purified with a nickel Sepharose column, the affinity tag was cleaved with thrombin, and further purified with a second nickel Sepharose column. A final passage through a benzamidine column removed thrombin from the final sample.
2.9. GroEL/GroES assembly
GroEL was complexed with GroES as previously described [24]. In brief, GroES was added in excess (400 μM in subunits) to 100 μM (in subunits) MTSL-labeled GroEL(A138C) in 50 mM Tris, pH 7.4, 100 mM KCl, and 10 mM MgCl2. Approximately 10 mM ATP was added and left for at least 15 minutes at room temperature. The ATPase reaction was inhibited by the addition of 2 mM AlF3, and by adding either ATP or ADP bullet- (or football-shaped complexes were formed, corresponding to one or two GroES molecules, respectively, bound per GroEL-14mer. [25].
2.10 NMR spectroscopy
GroEL sample quality was assessed by measuring lifetime broadening of SH3vpl resulting from the interaction of SH3vpl with GroEL. This was carried out by measuring 15N-R1?? and 15N-R1 relaxation rates for SH3vpl in the presence and absence of GroELR268C. The entire yield of 1 L cell culture of GroELR268C provided one 250 μl NMR sample at a concentration of 105 μM per subunit. The buffer employed comprised 50 mM sodium phosphate pH 7.0, 0.2 mM EDTA, 0.05% NaN3, 90% H2O/10% D2O (v/v); SH3vpl was added to the sample to a final concentration of 100 μM. Solution-state NMR experiments were performed at 283 K in Shigemi tubes in a volume of 250 μl on a Bruker 600 MHz spectrometer equipped with a triple resonance z axis gradient cryoprobe. Data processing was performed using NMRPipe [26] and spectral analysis was performed with Topspin3.5 (Bruker Biospin) and ccpNMR [27]. 15N-R1?? and 15N-R1 relaxation rates were measured on SH3vpl in the absence or presence of GroELR268C-MTS as described previously [28]. An effective spin-lock field of 1.8 kHz was employed for the 15N-R1?? to suppress chemical exchange. Relaxation rates were obtained by fitting the signal intensity decays to a single exponential. For the 15N-R1?? experiments, relaxation delays of 1, 21, 41, 71, 111 and 150 ms were used; for the 15N-R1 experiments the relation delays were 40, 120, 200, 320, 520 and 720 ms. R1?? values were corrected for off-resonance effects to obtain R2 values using the equation R2 = (R1ρ – R1cos2θ)/sin2, where θ is the angle between the effective spin-lock field and the external magnetic field (where 90° represents a resonance exactly on-resonance with the spin-lock field). θ = tan−1(ω1/Ω), where ω1 is the spin-lock radiofrequency field strength and Ω the 15N carrier offset.
2.11. Electron microscopy
GroEL in the apo state or in complex with GroES was diluted to a final concentration of 1 μM (in subunits), blotted onto carbon-coated copper EM grids (Ultrathin Carbon Film/Holey Carbon; Ted Pella) for 1 min, and stained with 2% uranyl acetate for 30 s. Images were taken with an FEI Tecnai T12 electron microscope (at 120 kV) using a Gatan US1000 CCD camera.
3. Results and Discussion
Purification of recombinant GroEL presents a challenging problem as GroEL binds a wide range of naturally produced proteins in E. coli [3]. Further, since both the N- and C-termini are buried within the cavity, GroEL is not suitable for purification using an affinity tag.
The standard purification of GroEL followed a previously published protocol with minor modifications [29]. Cells from a 1 L culture were resuspended in 25 ml lysis buffer and stirred at 4 °C with a protease inhibitor tablet for 30 minutes. The cells were lysed by one passage through a cell disruptor, and the suspension cleared by centrifugation. 4 ml (100x) streptomycin sulfate was added and stirred at 4 °C for 10 minutes. The suspension was cleared again by centrifugation and the supernatant loaded onto an ion exchange column. Initially a standard 20 ml column was used leading to an insufficient separation; further due to the low capacity of the column not all protein from the 1 L cell culture was bound within a single run. Therefore, the flow through from the first run was reloaded for a second run to recover some sample (Supplementary Fig. S1). Since some protein was lost even after two runs and there was no obvious path for obtaining better separation with the 20 ml column, subsequent work was carried out with a 60 ml self-packed ion exchange column. The sample was loaded in buffer A at 2 ml/min, and GroEL was eluted with buffer B and a 0 to 0.56 M NaCl gradient. The GroEL containing fractions (peak between 0.4 to 0.45 M NaCl) were pooled, and (NH4)2SO4 was added to 66% (w/v) and stirred overnight at 4 °C. The solution was centrifuged for 35 minutes at 10,000 g, and the pellet was resuspended in 7–8 ml buffer C and applied onto a HiPrep 26/60 Sephacryl S-300 column at 2.5 ml/min. The GroEL containing fractions eluted between 105 and 125 ml, and at this stage the resulting GroEL sample is free from unbound protein contaminations. However, according to tryptophan fluorescence, around 1.5–3.5 mol of bound protein impurities per mol GroEL14 remain with large sample to sample variability (Supplementary Fig. S2).
3.1 Initial trials for the final GroEL clean-up
Acetone precipitation is often used to clean the cavities of protonated wild type GroEL resulting in less than 10% protein impurities per GroEL14[29]. Unfortunately, addition of acetone to deuterated GroEL and several GroEL cysteine mutants results in aggregates that are refractory to resolubilization. In addition to the standard solvent (buffer C), stronger solvents such as 3 M urea or 50 mM sodium acetate at pH 4 were also tried in an attempt to dissolve the precipitate. Note, the lower pH maintains in solvent exposed cysteines in a reduced state and prevents the mutants from aggregating. Unfortunately, either GroEL could not be resolubilized or the protein impurities resolubilized simultaneously with GroEL, resulting in no benefit.
Further, addition of 20% (v/v) methanol to the buffers, as previously proposed [16, 21] and re-running GroEL through an ion-exchange column did not improve the purity of GroEL. Slight improvement was achieved by adding 200 mM MgCl2 to salt out β-galactosidase, as suggested earlier [17]: specifically the protein impurity level was decreased from 1.5 mol impurities per mol GroEL14 to about 0.25 mol/mol GroEL14 in the case of the GroELE315C mutant. However, a level of 25% for residual protein impurities is inadequate for our purposes and this approach was therefore discontinued.
We therefore decided to proceed with protocols in which GroEL is disassembled into monomers, purified, and subsequently reassembled back to GroEL14. We started with a previously published protocol, where no chaotropic agents are needed [19]. In this protocol intersubunit interactions within GroEL oligomers are weakened by the addition of MgADP (or MgATP) and then disassembled to monomers using a solid-phase ion exchange medium. Reassembly is achieved by the addition of 1 M (NH4)2SO4. Unfortunately, we were unsuccessful in disassembling GroELE315C under these conditions and consequently the purity did not improve (Supplementary Fig. S3).
Finally, we moved on to a urea-based disassembly/reassemble protocol. GroEL can be disassembled into monomers by the addition of urea and reassembled back to functional GroEL14 by the addition of 0.6 to 1 M (NH4)2SO4 and ATP (or ADP) (Supplementary Fig. S4) [20, 30]. Since it is crucial to use only the minimum amount of chaotropic agent to regain the structure and function of GroEL [31], the minimum amount of urea (between 3–4 M) needed to disassemble GroEL14 was determined for each mutant individually.
Using the latter approach, the GroELE315C monomer was successfully purified with an ion exchange column (2 × 5 ml QFF column, Supplementary Fig. S5). The protein was injected immediately after disassembly in 3 M urea onto the column and washed with 70 ml running buffer D at 2.5 ml/min. GroEL was eluted with a gradient from 0 to 0.56 M NaCl in 100 ml at 5 ml/min with the GroEL containing peak between 0.26 to 0.31 M NaCl. GroELE315C was further reassembled and the final yield from 1 L cell culture was around 42 μM (in subunits) in 1 ml (i.e. 42 nmoles) with less than 0.1 mol protein impurities per mol GroEL14. This purification protocol, however, was not successful for the other two GroEL mutants. The yield for GroELA138C was 240 μM (in subunits) in 1 ml (i.e. 240 nmoles) with 1.6 - 3 mol protein impurities per mol GroEL14. Similarly, the yield for GroELR268C was 190 μM in 1 ml (i.e. 190 nmoles) but had more than 3 mol protein impurities per mol GroEL14. Optimization of the ion exchange purification by the addition of 4 M urea, slowing down the flow rate, or addition of 20% methanol did not result in any increase in purity.
We further tried to purify GroEL monomer with a HIC column equilibrated in buffer C with 1.5 M (NH4)2SO4. GroEL was eluted with a gradient against buffer C at 5 ml/min. However, GroEL bound very tightly to the column and could not be eluted with the buffers mentioned here (data not shown).
3.2 Final protocol for final clean-up
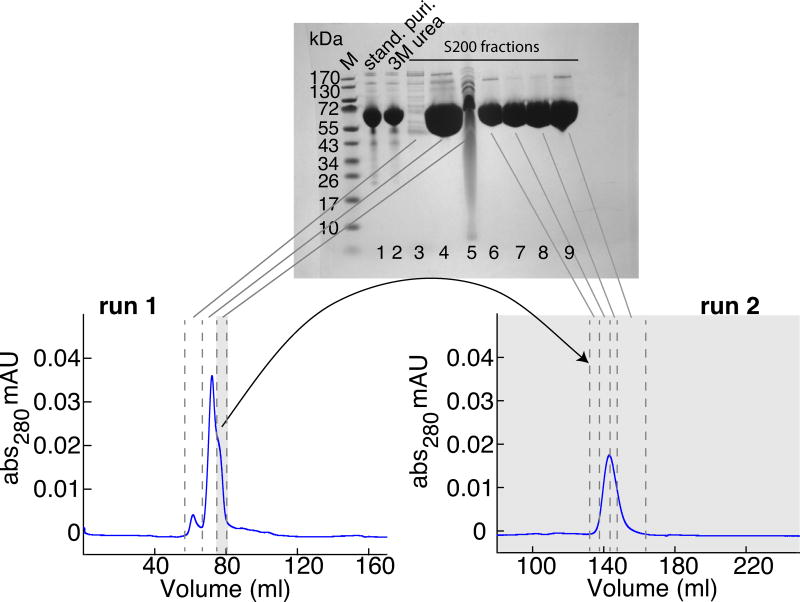
We finally succeeded in purifying all three mutants (GroELE315C, GroELA138C, GroELR268C) by gel filtration chromatography (Superdex 200) (Fig. 1 right panel). GroEL, after the standard purification, was disassembled into monomers by incubation at 4 °C overnight in 3–4 M urea. The sample was loaded onto the gel filtration column at 1 ml/min in buffer C with 3 M urea (Fig. 2). The protein impurities can be successfully separated from the main peak. As the shoulder on the higher elution volume side of the main peak contains too many impurities, only the central portion of the main peak was collected. However, the shoulder can be reloaded onto the column, and by collecting only the middle portion of the resulting main peak and combining it with the fraction collected from the first run, less than 10% impurities remain in the sample. Reassembly was achieved in several steps as follows: (1) Initial dialysis was carried out against buffer E including 3–4 M urea overnight at 4 °C. (2) The GroEL cysteine mutants were spin-labeled with MTSL (or its diamagnetic equivalent MTS) by incubation with a 10-fold excess of spin-label overnight at room temperature. Note, nitroxide spin-labeling of GroEL prior to reassembly was required to avoid aggregation (Supplementary Fig. S6). (3) GroEL was concentrated with a 100 kDa cut off filter to 2.5 ml, and the buffer exchanged with a PD10 column to buffer E following the manufacturer’s spin protocol. (4) 1.7 ml of a buffer containing buffer E and 2 M (NH4)2SO4 was added to the sample, and immediate GroEL reassembly was obtained after addition of 5 mM ATP (Fig. 3). The amount of GroEL14 did not change over time (data not shown). (5) Incorrectly folded or unassembled species were removed with a final gel filtration chromatography step (HiPrep 26/60 Sephacryl S-300) at 2.5 ml/min, in buffer E. For all three GroEL mutants, the yield from 1 L cell culture was between 100–500 μM (in subunits) protein in 500 μl (i.e. 50–250 nmoles), which corresponds to at least one NMR sample per liter. Reassembly was verified with a BN-PAGE gel and purity assessed by SDS-PAGE, mass spectrometry and tryptophan fluorescence (Supplementary Fig. S2).
Figure 2. Analysis of GroEL monomer obtained after disassembly and gel filtration.
The lower left panel shows the elution profile of the GroELE315C monomer from run 1 through the gel filtration column (Superdex 200). The lower right panel shows the elution profile for run 2 in which the shoulder on the side of the main peak from run 1 (indicated in grey) is re-injected. Buffer C with 3 M urea was used at a flow rate of 1 ml/min. SDS-PAGE (4–12% w/v) analysis of the relevant fractions is shown in the top panel. Lane 1: sample after standard purification (stand. puri.); lane 2: after addition of 3 M urea; lanes 3–5: fractions from gel filtration run 1; and lanes 6–9: fractions from gel filtration run 2. During the first gel filtration run only the main peak (lane 4) results in a sample with sufficient purity. When the shoulder of the peak (lane 5) is re-injected for a second gel filtration run, the middle part of that peak (lanes 7 and 8) have less than 10% impurities. Lane M is the molecular weight standard with masses indicated in kDa.
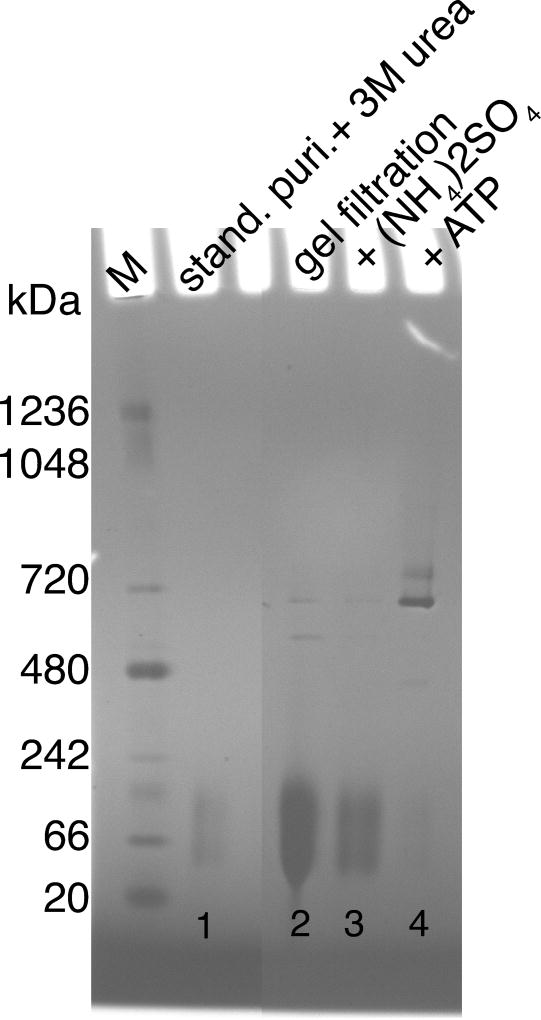
Figure 3. BN-PAGE native gel analysis of GroEL reassembly.
GroELE315C is disassembled into monomers upon addition of 3 M urea to the sample obtained after the standard purification (stand. puri.; lane 1). During the gel filtration step GroEL remains mostly monomeric (lane 2). For reassembly back into GroEL14 both (NH4)2SO4 (lane 3) and ATP (lane 4) are needed (see results section for details). Lane M is the molecular weight standard with masses indicated in kDa.
3.3 Sample quality
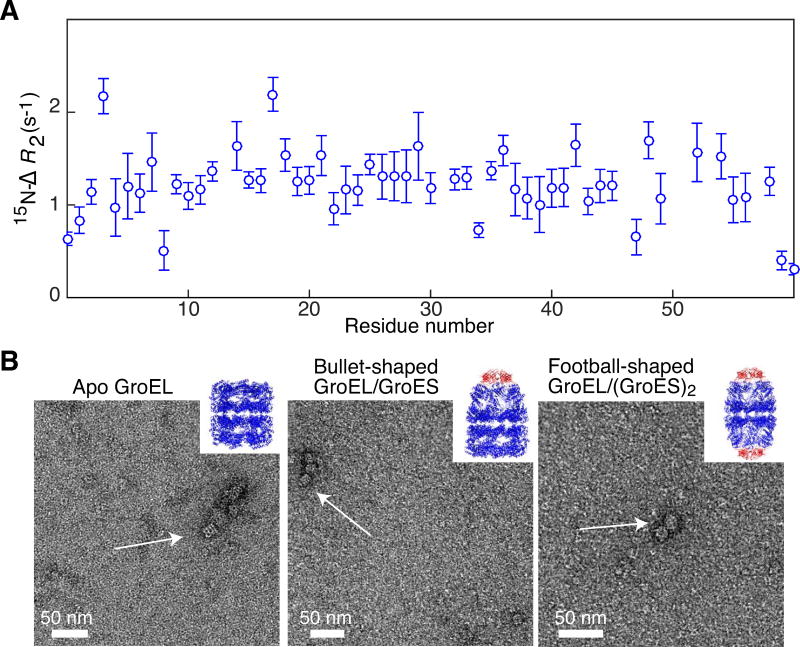
To assess the quality of the sample we made use of 15N lifetime broadening (15N-ΔR2) measurements. Specifically, we measured residue-specific 15N-ΔR2 values for a protein substrate SH3vpl in the presence of GroELR268C and compared the resulting profiles to those previously published for the interaction of SH3vpl with wild type GroEL [32]. 15N-ΔR2 values are obtained by taking the difference in 15N transverse relaxation rates (15N-R2) of uniformly 15N-labeled SH3vpl in the presence and absence of MTS spin-labeled GroELR268C. The ΔR2 profile is similar to that published previously (Fig. 4A), indicating that SH3vpl interacts in the same manner with spin-labeled GroELR268C and wild type GroEL. The slightly lower ΔR2 values (~1.5 s−1 here vs. ~4 s−1 with wild type GroEL) is probably due to the fact that the concentration of GroEL used here (105 μM per subunit) is lower than that used previously (120 μM per subunit). In addition, the R268C mutation is located within the substrate binding site; it is therefore not surprising that the binding affinity for SH3vpl may be reduced slightly, especially when taking into account the presence of seven hydrophobic MTS labels per heptameric ring.
Figure 4. Biophysical characterization of reassembled GroEL14.
(A) 15N lifetime line broadening (15N-ΔR2) of SH3vpl in the presence of reassembled diamagnetic MTS-labeled GroELR268C. 15N-ΔR2 profile of 100 μM SH3vpl obtained in the presence of 105 μM GroELR268C (in subunits) measured at 600 MHz and 283 K. The close to uniform 15N-ΔR2 profile indicates that SH3vpl binds to GroEL as a rigid body. The buffer employed was 50 mM sodium phosphate pH 7.0, 0.2 mM EDTA, 0.05% NaN3, and 90% H2O(v/v)/10% D2O (v/v). (B) Negative stain electron micrographs of reassembled MTSL-labeled GroELA138C in different conformational states: left panel, apo GroEL as seen after the purification; middle panel, bullet-shaped GroEL14/GroES complex formed upon addition of ADP and the co-chaperone GroES, where one end is capped by GroES; right panel, football-shaped GroEL14/(GroES)2 formed upon addition of ATP and GroES with both ends of GroEL capped with GroES. The inserts depict ribbon diagrams of the respective X-ray structures with GroEL in blue and GroES in red (left, 1XCK [34]; middlem 1SX4 [35]; and right, 4PKN [36]). The complexes were formed at 100 μM (in subunits) GroEL with an excess of GroES and diluted 100 times to blot on the grid. For further details see Materials and Methods
Further evidence that the purified GroEL is correctly reassembled and functional is provided by electron microscopy showing that GroEL can adopt different conformational states needed for folding of protein substrates. Binding of the co-chaperonin GroES is only possible when GroEL undergoes a conformational change from the apo to the open conformation. ATP binding triggers tilting and twisting of the domains within each subunit, burying the hydrophobic substrate binding sites and opening up the GroES binding site [33]. Electron micrographs of purified and reassembled MTSL-labeled A138C mutant of GroEL reveal a two-ring structure that is capable of binding GroES, either on one side of GroEL (forming the so-called bullet-shaped complex), or on both sides (forming the football-shaped complex) (Fig. 4B). Thus, the purification protocol described here results in GroEL constructs that are fully capable of adopting the various conformational states present in the natural GroEL cycle.
4. Concluding Remarks
In the present work we describe a new purification protocol to obtain high purity GroEL containing less than 10% of GroEL cavities blocked by substrate proteins present in the bacterial expression system. This method was developed to specifically address the yield and high-sample quality required for quantitative NMR and EPR studies. The method is suitable for a range of mutations, illustrated here by three mutations within the GroEL cavity (E315C, A138C, R268C), and should also be suitable for deuteration and complex isotopic labeling of GroEL. These constructs, with only one cysteine in the sequence, allow site-specific spin labeling needed for fluorescence, EPR double electron-electron resonance and solution NMR paramagnetic relaxation enhancement measurements, and will hopefully contribute to a better understanding of the functioning of GroEL.
Supplementary Material
Highlights.
A straightforward purification protocol involving disassembly and reassembly of GroEL, a dual-ringed tetradecamer, is described
Effectively removes naturally occurring, tightly bound proteins present in the bacterial expression system
Suitable for isotope labeling, deuteration and introduction of site-specific paramagnetic labels via surface engineered cysteine mutations
Provides yields in sufficient quantities suitable for both NMR and EPR studies.
Acknowledgments
We thank John Louis and David Libich for useful discussions. M.A.W. was supported by an Early Postdoc.Mobility Fellowship from the Swiss National Science Foundation. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health and by the AIDS-Targeted Antiviral Program of the Office of the Director of the National Institutes of Health (to G.M.C.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data related to this paper can be found at ………
References
- 1.Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–69. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 2.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, et al. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371:578–86. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 3.Hayer-Hartl M, Bracher A, Hartl FU. The GroEL-GroES Chaperonin Machine: A Nano-Cage for Protein Folding. Trends Biochem Sci. 2016;41:62–76. doi: 10.1016/j.tibs.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Horwich AL, Fenton WA. Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys. 2009;42:83–116. doi: 10.1017/S0033583509004764. [DOI] [PubMed] [Google Scholar]
- 5.Gruber R, Horovitz A. Allosteric Mechanisms in Chaperonin Machines. Chem Rev. 2016;116:6588–606. doi: 10.1021/acs.chemrev.5b00556. [DOI] [PubMed] [Google Scholar]
- 6.Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, et al. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–14. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 7.Weaver J, Rye HS. The C-terminal tails of the bacterial chaperonin GroEL stimulate protein folding by directly altering the conformation of a substrate protein. J Biol Chem. 2014;289:23219–32. doi: 10.1074/jbc.M114.577205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tugarinov V, Kay LE. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. Chembiochem. 2005;6:1567–77. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- 9.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–71. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Atomic-resolution dynamics on the surface of amyloid-beta protofibrils probed by solution NMR. Nature. 2011;480:268–72. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt T, Walti MA, Baber JL, Hustedt EJ, Clore GM. Long Distance Measurements up to 160 A in the GroEL Tetradecamer Using Q-Band DEER EPR Spectroscopy. Angew Chem Int Ed Engl. 2016;55:15905–9. doi: 10.1002/anie.201609617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgopoulos C. Toothpicks, serendipity and the emergence of the Escherichia coli DnaK (Hsp70) and GroEL (Hsp60) chaperone machines. Genetics. 2006;174:1699–707. doi: 10.1534/genetics.104.68262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–20. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 14.Grason JP, Gresham JS, Lorimer GH. Setting the chaperonin timer: a two-stroke, two-speed, protein machine. Proc Natl Acad Sci U S A. 2008;105:17339–44. doi: 10.1073/pnas.0807418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. NMR analysis of a 900K GroEL GroES complex. Nature. 2002;418:207–11. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 16.Weissman JS, Hohl CM, Kovalenko O, Kashi Y, Chen S, Braig K, et al. Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell. 1995;83:577–87. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 17.Molugu SK, Li J, Bernal RA. Separation of E. coli chaperonin groEL from beta-galactosidase without denaturation. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;1007:93–9. doi: 10.1016/j.jchromb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Lissin NM, Venyaminov S, Girshovich AS. (Mg-ATP)-dependent self-assembly of molecular chaperone GroEL. Nature. 1990;348:339–42. doi: 10.1038/348339a0. [DOI] [PubMed] [Google Scholar]
- 19.Ybarra J, Horowitz PM. Inactive GroEL monomers can be isolated and reassembled to functional tetradecamers that contain few bound peptides. J Biol Chem. 1995;270:22962–7. doi: 10.1074/jbc.270.39.22962. [DOI] [PubMed] [Google Scholar]
- 20.Ybarra J, Horowitz PM. Refolding and reassembly of active chaperonin GroEL after denaturation. J Biol Chem. 1995;270:22113–5. doi: 10.1074/jbc.270.38.22113. [DOI] [PubMed] [Google Scholar]
- 21.Todd MJ, Lorimer GH. Criteria for assessing the purity and quality of GroEL. Methods Enzymol. 1998;290:135–41. doi: 10.1016/s0076-6879(98)90012-x. [DOI] [PubMed] [Google Scholar]
- 22.Maxwell KL, Davidson AR. Mutagenesis of a buried polar interaction in an SH3 domain: sequence conservation provides the best prediction of stability effects. Biochemistry. 1998;37:16172–82. doi: 10.1021/bi981788p. [DOI] [PubMed] [Google Scholar]
- 23.Tropea JE, Cherry S, Waugh DS. Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol Biol. 2009;498:297–307. doi: 10.1007/978-1-59745-196-3_19. [DOI] [PubMed] [Google Scholar]
- 24.Wälti MA, Schmidt T, Murray DT, Wang H, Hinshaw JE, Clore GM. Chaperonin GroEL accelerates protofibril formation and decorates fibrils of the Het-s prion protein. Proc Natl Acad Sci U S A. 2017;114:9104–9. doi: 10.1073/pnas.1711645114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taguchi H, Tsukuda K, Motojima F, Koike-Takeshita A, Yoshida M. BeF(x) stops the chaperonin cycle of GroEL-GroES and generates a complex with double folding chambers. J Biol Chem. 2004;279:45737–43. doi: 10.1074/jbc.M406795200. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 28.Lakomek NA, Ying J, Bax A. Measurement of (1)(5)N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR. 2012;53:209–21. doi: 10.1007/s10858-012-9626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grason JP, Gresham JS, Widjaja L, Wehri SC, Lorimer GH. Setting the chaperonin timer: the effects of K+ and substrate protein on ATP hydrolysis. Proc Natl Acad Sci U S A. 2008;105:17334–8. doi: 10.1073/pnas.0807429105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arai M, Inobe T, Maki K, Ikura T, Kihara H, Amemiya Y, et al. Denaturation and reassembly of chaperonin GroEL studied by solution X-ray scattering. Protein Sci. 2003;12:672–80. doi: 10.1110/ps.0233603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Price NC, Kelly SM, Thomson GJ, Coggins JR, Wood S, auf der Mauer A. The unfolding and attempted refolding of the bacterial chaperone protein groEL (cpn60) Biochim Biophys Acta. 1993;1161:52–8. doi: 10.1016/0167-4838(93)90195-w. [DOI] [PubMed] [Google Scholar]
- 32.Libich DS, Tugarinov V, Clore GM. Intrinsic unfoldase/foldase activity of the chaperonin GroEL directly demonstrated using multinuclear relaxation-based NMR. Proc Natl Acad Sci U S A. 2015;112:8817–23. doi: 10.1073/pnas.1510083112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and allostery of the chaperonin GroEL. J Mol Biol. 2013;425:1476–87. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 34.Bartolucci C, Lamba D, Grazulis S, Manakova E, Heumann H. Crystal structure of wild-type chaperonin GroEL. J Mol Biol. 2005;354:940–51. doi: 10.1016/j.jmb.2005.09.096. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhry C, Horwich AL, Brunger AT, Adams PD. Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states. J Mol Biol. 2004;342:229–45. doi: 10.1016/j.jmb.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Fei X, Ye X, LaRonde NA, Lorimer GH. Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form. Proc Natl Acad Sci U S A. 2014;111:12775–80. doi: 10.1073/pnas.1412922111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.