Abstract
The fungus Candida albicans can grow as either yeast or filaments, which include hyphae and pseudohyphae, depending on environmental conditions. Filamentous growth is of particular interest because it is required for biofilm formation and for pathogenesis. Environmentally induced filamentous growth is associated with expression of filamentation-associated genes, and both filamentous growth and associated gene expression depend upon several well characterized transcription factors. Surprisingly, strains with reduced expression of many essential genes display filamentous growth under non-inducing conditions – those in which the wild type grows as yeast. We found recently that diminished expression of several essential protein kinase genes leads to both filamentous cell morphology and filamentation-associated gene expression under non-inducing conditions. Reduced expression of the essential protein kinase gene CAK1 promoted filamentation-associated gene expression and biofilm formation in strains that lacked key transcriptional activators of these processes, thus indicating that CAK1 expression is critical for both environmental and genetic control of filamentation. In this study we extend our genetic interaction analysis to a second essential protein kinase gene, KIN28. Reduced expression of KIN28 also permits filamentation-associated gene expression, though not biofilm formation, in the absence of several key transcriptional activators. Our results argue that impairment of several essential cellular processes can alter the regulatory requirements for filamentation-associated gene expression. Our results also indicate that levels of filamentation-associated gene expression are not fully predictive of biofilm formation ability.
Keywords: Candida albicans, EPI induced filamentation, Gene Expression, Mutant Strains, Protein Kinases
Introduction
C. albicans is a natural fungal inhabitant of mammalian mucosal surfaces. It can cause infection in susceptible individuals, whose risk factors include presence of an implanted medical device, immune system defects, or use of broad spectrum antibacterial antibiotics (Macphail et al. 2002; Perlroth et al. 2007; Pfaller and Diekema 2007). Therefore, C. albicans infection seems to arise when the organism reaches an unprotected niche, such as a venous catheter, or when its proliferation is unchecked by host immune defenses or by competition with other commensal microbes.
Environmental signals influence the morphological form in which the fungus grows. Two of the most well studied forms are yeast cells and filamentous cells, which include hyphae and pseudohyphae (Sudbery et al. 2004). These growth forms are associated with gene expression programs that seem much broader than required for cellular morphogenesis alone; for example, mutations in many filamentation-associated genes do not measurably impair the physical appearance of hyphae or pseudohyphae ((Naglik et al. 2003; Nobile et al. 2006); reviewed in (Mayer et al. 2013)).
The infection capability of C. albicans is tied to its filamentous growth program. This connection is illustrated by the fact that many filamentation-associated genes promote infectivity or pathogenesis. For example, ALS3 and HWP1 specify cell surface adhesins that are required for host cell interaction and biofilm formation ((Nobile et al. 2006; Phan et al. 2007; Staab et al. 1999) reviewed in (Mayer et al. 2013)); ECE1 specifies a secreted toxin that causes epithelial cell damage (Moyes et al. 2016). The connection is also supported by the observation that impaired expression of filamentation-associated genes, through deletion mutations of transcriptional activators or overexpression of transcriptional repressors, generally results in prominent pathogenicity defects (Banerjee et al. 2008; Bernstein et al. 2012; Braun and Johnson 1997; Braun et al. 2001; Murad et al. 2001; Nobile and Mitchell 2005). There are filamentation-competent mutants that are defective in pathogenicity, so filamentation is not sufficient for pathogenicity (Moyes et al. 2016). However, the filamentous growth program is clearly one determinant of C. albicans pathogenic potential (Lo et al. 1997).
Many of the environmental signals that induce filamentous growth seem to be relevant to an infection site, including 37° temperature, presence of serum, and limitation for iron or oxygen (reviewed in (Biswas et al. 2007; Du and Huang 2016; Huang 2012; Mayer et al. 2013; Shapiro et al. 2012; Sudbery 2011; Whiteway and Bachewich 2007)). However, many genetic determinants of filamentous growth have a less obvious connection to infection biology. For example, an engineered reduction in expression of many essential genes leads to a filamentous cell morphology in the absence of normal inducing signals (Berman 2006; O'Meara et al. 2015; Sudbery 2011). In many cases, impairment of essential processes induces filamentation-associated gene expression along with filamentous morphogenesis (Bachewich et al. 2005; Woolford et al. 2016). We refer to the filamentation program that is induced by Essential Process Impairment as EPI-induced filamentation. Whether environmentally-induced filamentation and EPI-induced filamentation are governed by identical regulatory mechanisms is uncertain.
We recently explored the regulatory requirements for EPI-induced filamentation that resulted from reduced expression of the protein kinase gene CAK1 (Woolford et al. 2016). We focused on five transcription factors – Bcr1, Brg1, Efg1, Tec1, and Ume6 – that are required for full expression of filamentation-associated genes in otherwise wild-type cells. Surprisingly, reduced expression of CAK1 promoted considerable expression of filamentation-associated genes in the absence of any one of these transcription factors. Elevated expression of filamentation-associated genes was associated with biological impact: reduced CAK1 expression enabled biofilm formation in the absence of Bcr1, Brg1, Tec1, or Ume6, and enabled filamentation in the absence of Efg1. These results suggested that regulation of EPI-induced filamentation may have features of genetic control that are distinct from those of environmentally induced filamentation.
We have thus far explored the genetic control of EPI-induced filamentation only with CAK1. Here we turn our attention to the impact of reduced expression of protein kinase gene KIN28. The cellular functions of Cak1 and Kin28 are distinct, based upon studies of their orthologs in Saccharomyces cerevisiae. Cak1 is inferred to function as a cell cycle regulator through its phosphorylation of cyclin-dependent kinase Cdc28 (Tsakraklides and Solomon 2002). Kin28 is inferred to function to promote transcription initiation through its phosphorylation of the RNA polymerase II C-terminal domain (Akhtar et al. 2009). Through comparison of Cak1 and Kin28 deficiencies, we seek to determine whether there may be general principles that underlie EPI-induced filamentation.
Material and Methods
Media
C. albicans strains were grown at 30° or 37°in YPD. Transformants were selected on synthetic medium (2% dextrose, 1.7% Difco yeast nitrogen base with ammonium sulfate and auxotrophic supplements).
Construction of Mutants
The kin28 DX mutant was constructed as described in Woolford et al. (2016). Briefly, one allele of the gene was replaced with the URA3 marker in the BWP17 background, and the other allele of KIN28 had the promoter of ORF19.7606 (950 base pairs upstream sequence), marked by the ARG4 gene, replace its own promoter. To construct a complement of the kin28 DX mutant, the WT allele was amplified from genomic DNA (SC5314), including 350bp upstream (the neighboring gene ends 358 bp upstream of the KIN28 ATG) and 100 bp downstream, and gap repaired into the plasmid pDDB78, as described in Woolford et al. (2016). The resulting complementing plasmid was digested with NruI to direct integration to the his1 locus of the DX mutant strains. A marker matched prototrophic strain was constructed by inserting NruI digested pDDB78 into the DX mutant.
Construction of transcription factor kin28 DX double mutants was performed as described in Woolford et al. (2016). First an unmarked deletion of the transcription factor gene was obtained using the URA3 mini-blaster protocol (Ganguly and Mitchell 2012). This strain was used as the parent for the kin28 DX mutant. This strain was made prototrophic as described above. All strains are listed in Table S1.
RNA extraction and NanoString Analysis
RNA extractions were performed using the Qiagen RNeasy Mini Kit as described in Woolford et al. (2016). Gene expression levels were measured for 181 genes using the NanoString nCounter platform as described in Woolford et al. (2016). The heat maps were generated using Multiexperimental Viewer 4.9.0 (Saeed et al. 2003)
Cell Microscopy
Cells grown overnight were freshly diluted into YPD and grown for 4 hours at 37° before harvesting and prepared for calcofluor staining as described in Woolford et al. (2016).
Biofilm Imaging
Cells were allowed to adhere for 90 min and biofilms grown for 48 hours in YPD before imaging as described in Woolford et al. (2016).
Results
The protein kinase gene KIN28 is formally a negative regulator of filamentous growth and filamentation associated gene expression. This conclusion is based on the properties of strains with reduced KIN28 expression, which we refer to as kin28 DX strains (Woolford et al. 2016). In these strains, one KIN28 allele has been deleted and the second allele has been fused to a weak constitutive promoter. We reported previously (Woolford et al. 2016) that kin28 DX strains grow as filaments and express several core filamentation-associated genes (Martin et al. 2013) under noninducing conditions (YPD medium at 30°). We also observed that complementation of a kin28 DX mutant with a wild-type copy of KIN28 restored wild-type growth and gene expression properties. We concluded from these observations that wild-type levels of KIN28 expression are necessary to maintain environmental control of the filamentous growth program (Woolford et al. 2016).
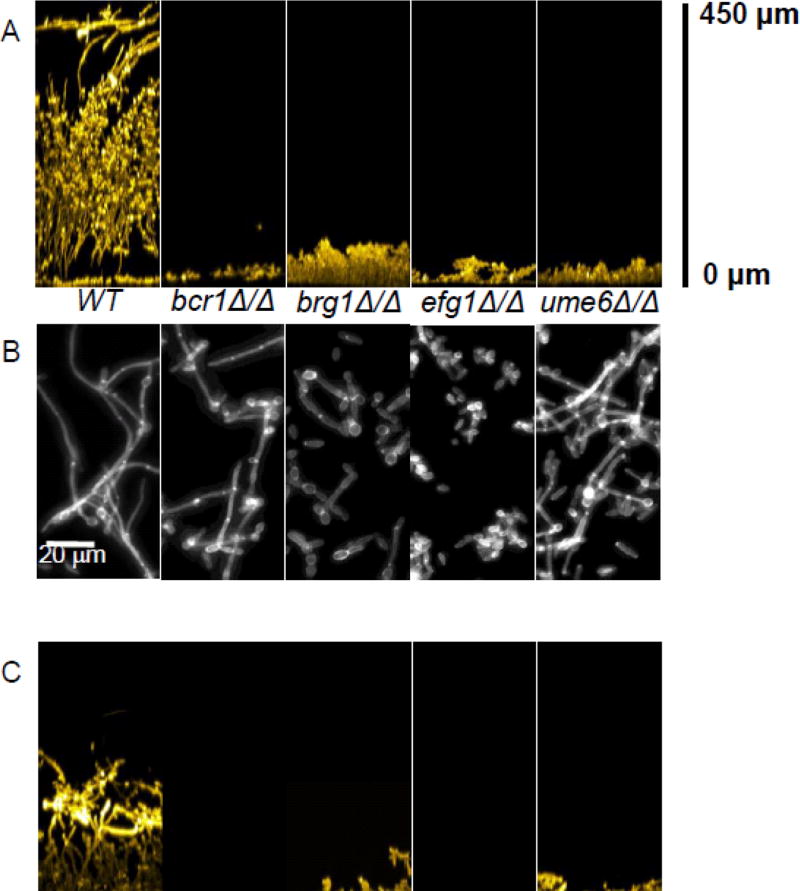
In order to determine whether wild-type levels of KIN28 expression are necessary to maintain genetic control of filamentation, we examined deletions of each of four key activators of filamentation-associated genes in the kin28 DX background. Deletion mutations in the genes BCR1, BRG1, EFG1 and UME6 are known to cause defects in filamentation, biofilm formation, and expression of filamentation-associated genes (Banerjee et al. 2008; Du et al. 2012; Nobile et al. 2012; Nobile and Mitchell 2005; Ramage et al. 2002). Under the growth conditions we use (YPD medium at 37° with no added inducer), filamentous cell morphogenesis was severely defective in the efg1Δ/Δ mutant, partially defective in the brg1Δ/Δ mutant, and only subtly defective in the bcr1Δ/Δ and ume6Δ/Δ mutants (Fig. 1B), as we reported previously (Woolford et al. 2016). The kin28 DX strain yielded shorter and more heterogeneous filamentous cells than the wild-type strain under these growth conditions (Fig. 1D), as reported previously (Woolford et al. 2016). In the kin28 DX background, the defect of the efg1Δ/Δ mutant in filamentous cell morphogenesis remained severe (Fig. 1D). Strikingly, in the kin28 DX background, the defect of the brg1Δ/Δ mutant in filamentous cell morphogenesis increased in severity, and defects of the bcr1Δ/Δ or ume6Δ/Δ mutants became evident (Fig. 1D). These results indicate that the kin28 DX defect causes a mild filamentation defect under inducing conditions, and seems to augment rather than suppress the phenotypes of filamentation activator mutations.
Fig 1.
Phenotypes of filamentation/biofilm activator mutants in KIN28 and kin28 DX backgrounds. Panels A and C. In vitro biofilms. Cells were grown under in vitro biofilm conditions for 48 hr, then visualized by confocal microscopy. Cross-sectional views are shown. Panels B and D. Cell morphology. Cell cultures were grown for 4 hr at 37° in YPD, then fixed and stained with Calcofluor White prior to visualization. Strains in panels A and B include WT (DAY185), bcr1Δ/Δ (CW1627), brg1Δ/Δ (CW1639), efg1Δ/Δ (CW1651), and ume6Δ/Δ (CW1633). Strains in panels C and D include kin28 DX (CW1202), kin28 DX bcr1Δ/Δ (CW1549), kin28 DX brg1Δ/Δ (CW1561), kin28 DX efg1Δ/Δ (CW1609), and kin28 DX ume6Δ/Δ (CW1540). Images shown in panels A and B were originally published in Woolford et al. (2016) and have been reprinted to aid comparison of results.
We also examined effects of the kin28 DX genotype on genetic control of filamentation through biofilm formation assays, which provide an appraisal of filamentous cell function. In these assays, we examined cells at 37° to help relate our findings to growth under infection or colonization conditions, and to ensure that each transcription factor mutant examined had a clear phenotype. The kin28 DX strain produced a biofilm under our growth conditions, though its depth was not as great as the wild-type strain's biofilm (Fig. 1A,C). In both the wild-type and kin28 DX backgrounds, the bcr1Δ/Δ, brg1Δ/Δ, efg1Δ/Δ and ume6Δ/Δ mutations each caused severe biofilm defects (Fig. 1A,C). This result stands in contrast to our finding that, in the cak1 DX background, the bcr1Δ/Δ, brg1Δ/Δ, and ume6Δ/Δ mutations failed to cause a biofilm defect (Woolford et al. 2016). Our results indicate that the kin28 DX genotype does not bypass the genetic control of filamentation or biofilm formation.
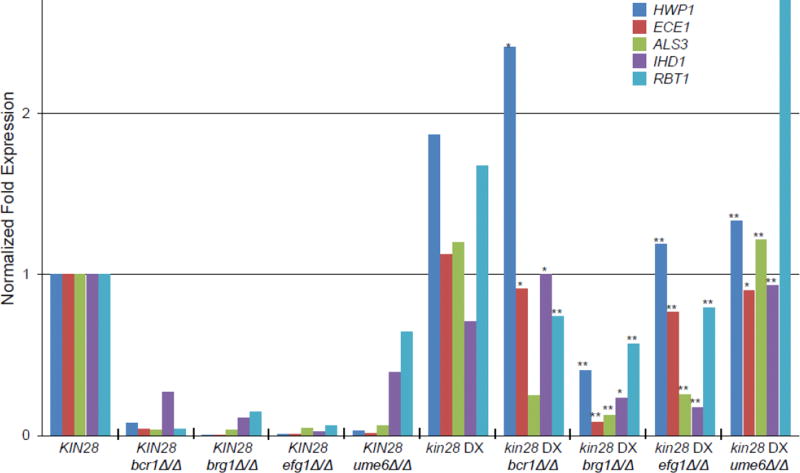
We used expression of filamentation-associated genes as a third assay for genetic control of the filamentation program in the kin28 DX background. We first focused on expression of core filamentation genes HWP1, ECE1, ALS3, IHD1, and RBT1 (Martin et al. 2013). Our published data (Woolford et al. 2016) were renormalized to facilitate comparison to new results; the data are presented in Fig. 2 and Table S2. In a wild-type background, deletion of any of the four activator genes caused a reduction of ~2-fold to over 20-fold in each core gene expression level under inducing conditions. The kin28 DX strain expressed the core filamentation-associated genes at similar levels to the wild-type under inducing conditions (Fig. 2). However, in the kin28 DX background, the impact of each activator gene deletion mutation was much more limited than in the wild-type background (Fig. 2). For example, the activator gene mutations caused ALS3 RNA levels to be reduced by 15- to 30-fold in the wild-type background, but by no more than 8-fold in the kin28 DX background. In fact, the deletion of the activator gene UME6 caused only a marginal gene expression defect in kin28 DX background. These results indicate that wild-type levels of KIN28 expression are necessary to maintain genetic control of filamentation-associated gene expression.
Fig 2.
Core filamentation gene expression of filamentation/biofilm activator mutants in KIN28 and kin28 DX backgrounds. RNA levels for environmental response genes were determined by nanoString for strains grown for 4 hr at 37° in YPD (Table S2). Averaged normalized expression levels are shown for core filamentation genes in the strain indicated. Normalized expression levels in bcr1Δ/Δ, brg1Δ/Δ, efg1Δ/Δ, and ume6Δ/Δ strains originally presented in Woolford et al. 2016 and were renormalized with new WT and kin28 DX background strains. All expression ratios were calculated using mean values of three independent isolates and statistical significance was determined by two-tailed Student’s t-test. Symbols: * = p<0.05; ** = p<0.01 for comparison of KIN28 and kin28 DX strains carrying the same activator mutant.
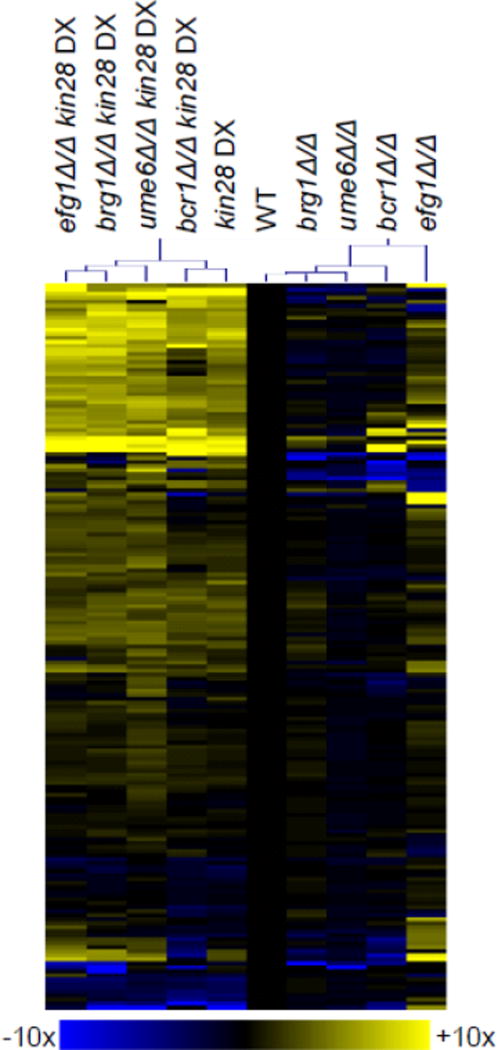
A broader view of the kin28 DX gene expression impact is consistent with its overriding effects on filamentation-associated genes. We assayed expression levels of 181 genes through triplicate nanoString determinations, and the results are presented as a sample-clustered heat map (Fig. 3; Table S2). The kin28 DX genotype had a profound effect on gene expression, and caused a significant change in expression of 55 of the 181 genes (≥2-fold up/downregulation, p<0.05). The broad-ranging effects of the kin28 DX genotype were evident regardless of deletion of any of the four filamentation-associated gene activators. These results indicate that reduced KIN28 expression has global gene expression effects, and that they are manifested for the most part independently of the filamentation-associated gene activators Bcr1, Brg1, Efg1, and Ume6.
Fig 3.
Gene expression profile of filamentation/biofilm activator mutants in KIN28 and kin28 DX backgrounds. RNA was extracted from cells grown for 4 hr at 37° in YPD and used for nanoString analysis (Table S2). Hierarchal clustering of gene expression data was performed from the average of three isolates using MeV software. Fold change values were obtained by dividing normalized expression values for each strain genotype by the wild type strain for each of the probes. The color scale represents Log2 fold change compared to wild type.
Discussion
It is well established that inhibition of the C. albicans cell cycle or cell growth can induce filamentation (reviewed in (Berman 2006; Sudbery 2011)), a phenomenon that we have called EPI-induced filamentation here to distinguish it from environmentally induced filamentation. Activation of filamentation-associated gene expression accompanies EPI-induced filamentation in the majority of cases examined ((Bachewich et al. 2003; O'Meara et al. 2015; Umeyama et al. 2006; Wightman et al. 2004), reviewed in (Berman 2006)). We have sought to determine whether the same transcriptional regulators that govern environmentally induced filamentation also govern EPI-induced filamentation. We found here that EPI-induced filamentation, brought about by diminished expression of KIN28, partially relieves dependence of filamentation-associated genes on the key transcription factors Bcr1, Brg1, Efg1, and Ume6. This result mirrors our previous findings from diminished expression of CAK1 (Woolford et al. 2016). However, biological assays of filamentous cell morphology and biofilm formation ability give opposite results in the cases of KIN28 and CAK1. Below we discuss our interpretation of these assay results and some additional thoughts about EPI-induced filamentation.
Our most important biological conclusion is that EPI-induced filamentation has distinct genetic requirements from environmentally induced filamentation. In the case of core filamentation-associated gene expression, the results for both kin28 DX and cak1 DX backgrounds are similar: dependence of the response on several well characterized transcriptional activators of filamentation-associated genes is less severe than it is in the wild-type background. This observation may be explained by either of two simple models: the novel regulator model, or the redundant activation model. The novel regulator model explains the result by proposing that EPI signals alter activity of a filamentation gene regulator for which no mutation was tested in our epistasis analysis. For example, EPI signals might activate Ndt80 (Nobile et al. 2012), a positive regulator, or might inhibit Tup1 (Braun and Johnson 1997), a negative regulator. The redundant activation model explains the result by proposing that EPI signals lead to concerted stimulation of expression or activity of several filamentation gene activators, for example Brg1 and Efg1 and Ume6. These activators share multiple target genes (Nobile et al. 2012), so inactivation of any one may have limited impact if several are stimulated. We argued previously in favor of the redundant activation model (Woolford et al. 2016), but we have no direct evidence that either model explains our kin28 DX observations. However, the fact that so many known regulators converge to affect expression of filamentation-associated genes makes the two models quite challenging to distinguish experimentally.
Whereas gene expression assays for kin28 DX and cak1 DX strains yield similar conclusions about genetic control of EPI-induced filamentation, our biological assays yield opposite conclusions. Specifically, the cak1 DX genotype was epistatic to most filamentation activator defects for both filamentation and biofilm formation ability; the kin28 DX genotype was hypostatic or, in some cases, even aggravated mild filamentation-defective phenotypes. The difference in these results may reflect in part the morphology of each DX strain: cak1 DX strains make fairly uniform hyphae; kin28 DX strains produce heterogeneous and slightly aberrant hyphae and pseudohyphae. Our gene expression assays argue that the difference in morphology is not simply a consequence of the strength of the filamentation-inducing signal, because core filamentation-associated gene expression levels are comparable in kin28 DX and cak1 DX strains. We suggest that the biological differences between the two DX strains may arise from the phenotypic impact of non-filamentation-associated genes that are expressed in each strain. For example, the kin28 DX strain may express genes that disrupt proper morphogenesis and adherence, leading to defects in hyphae and biofilms. We note that the cell wall protein genes PGA17, PGA25, PGA34, and PGA37 are up-regulated in the kin28 DX strain, not in the cak1 DX strain. One or several of those gene products may disrupt organization of the filamentous cell wall. The fact that biologically-based and gene-expression-based epistasis tests give different outcomes reflects the fact that many features of C. albicans biological processes have yet to be worked out in detail.
Do the mechanisms that underlie EPI-induced filamentation have relevance to the biology of natural C. albicans isolates? We hypothesize that they do, particularly in the context of biofilm growth. First, internal biofilm cells are relatively insulated from nutrients in the surrounding environment (Stewart and Franklin 2008), and their protracted starvation may inhibit several essential processes. It seems possible that a resulting induction of filamentation-associated genes may increase cell-cell adherence and thus reinforce biofilm integrity. Second, it is known that biofilms are notoriously tolerant to antimicrobial treatment, and many mechanisms contribute to this phenomenon. One mechanism may be that growth inhibition from antimicrobial treatment may cause EPI-induced filamentation, once again leading to reinforcement of biofilm integrity. In that context, we note a recent study that showed that the growth inhibitor Staurosporine induces filamentation, even in an efg1Δ/Δ background (Xie et al. 2017). That outcome is similar to our finding that filamentation-associated genes are induced by kin28 DX or cak1 DX defects in an efg1Δ/Δ background. These parallels suggest that an understanding of EPI-induced filamentation may reveal general aspects of antimicrobial drug responses and thus point toward approaches that minimize potential resistance.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R21 AI100270 (APM) and R01 AI067703 (APM).
References
- Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich C, Nantel A, Whiteway M. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol Microbiol. 2005;57:942–959. doi: 10.1111/j.1365-2958.2005.04727.x. [DOI] [PubMed] [Google Scholar]
- Bachewich C, Thomas DY, Whiteway M. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol Biol Cell. 2003;14:2163–2180. doi: 10.1091/mbc.02-05-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee M, et al. UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell. 2008;19:1354–1365. doi: 10.1091/mbc.E07-11-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DA, Vyas VK, Weinberg DE, Drinnenberg IA, Bartel DP, Fink GR. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proc Natl Acad Sci U S A. 2012;109:523–528. doi: 10.1073/pnas.1118859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Guan G, Xie J, Sun Y, Tong Y, Zhang L, Huang G. Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One. 2012;7:e29707. doi: 10.1371/journal.pone.0029707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Huang G. Environmental pH adaption and morphological transitions in Candida albicans. Curr Genet. 2016;62:283–286. doi: 10.1007/s00294-015-0540-8. [DOI] [PubMed] [Google Scholar]
- Ganguly S, Mitchell AP. Mini-blaster-mediated targeted gene disruption and marker complementation in Candida albicans. Methods Mol Biol. 2012;845:19–39. doi: 10.1007/978-1-61779-539-8_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence. 2012;3:251–261. doi: 10.4161/viru.20010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Macphail GL, Taylor GD, Buchanan-Chell M, Ross C, Wilson S, Kureishi A. Epidemiology, treatment and outcome of candidemia: a five-year review at three Canadian hospitals. Mycoses. 2002;45:141–145. doi: 10.1046/j.1439-0507.2002.00741.x. [DOI] [PubMed] [Google Scholar]
- Martin R, Albrecht-Eckardt D, Brunke S, Hube B, Hunniger K, Kurzai O. A core filamentation response network in Candida albicans is restricted to eight genes. PLoS One. 2013;8:e58613. doi: 10.1371/journal.pone.0058613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes DL, et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 2016;532:64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad AM, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor. Bcr1p Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- O'Meara TR, Veri AO, Ketela T, Jiang B, Roemer T, Cowen LE. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat Commun. 2015;6:6741. doi: 10.1038/ncomms7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QT, et al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007;5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage G, VandeWalle K, Lopez-Ribot JL, Wickes BL. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Shapiro RS, Ryan O, Boone C, Cowen LE. Regulatory circuitry governing morphogenesis in Saccharomyces cerevisiae and Candida albicans. Cell Cycle. 2012;11:4294–4295. doi: 10.4161/cc.22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Sudbery PE. Growth of Candida albicans hyphae. Nat Rev Microbiol. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Tsakraklides V, Solomon MJ. Comparison of Cak1p-like cyclin-dependent kinase-activating kinases. J Biol Chem. 2002;277:33482–33489. doi: 10.1074/jbc.M205537200. [DOI] [PubMed] [Google Scholar]
- Umeyama T, Kaneko A, Niimi M, Uehara Y. Repression of CDC28 reduces the expression of the morphology-related transcription factors, Efg1p, Nrg1p, Rbf1p, Rim101p, Fkh2p and Tec1p and induces cell elongation in Candida albicans. Yeast. 2006;23:537–552. doi: 10.1002/yea.1373. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R, Bates S, Amornrrattanapan P, Sudbery P. In Candida albicans, the Nim1 kinases Gin4 and Hsl1 negatively regulate pseudohypha formation and Gin4 also controls septin organization. J Cell Biol. 2004;164:581–591. doi: 10.1083/jcb.200307176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford CA, et al. Bypass of Candida albicans Filamentation/Biofilm Regulators through Diminished Expression of Protein Kinase Cak1. PLoS Genet. 2016;12:e1006487. doi: 10.1371/journal.pgen.1006487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie JL, O'Meara TR, Polvi EJ, Robbins N, Cowen LE. Staurosporine Induces Filamentation in the Human Fungal Pathogen Candida albicans via. Signaling through Cyr1 and Protein Kinase A mSphere. 2017;2 doi: 10.1128/mSphere.00056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.