Abstract
Clinical narratives (the text notes found in patients’ medical records) are important information sources for secondary use in research. However, in order to protect patient privacy, they must be de-identified prior to use. Manual de-identification is considered to be the gold standard approach but is tedious, expensive, slow, and impractical for use with large-scale clinical data. Automated or semi-automated de-identification using computer algorithms is a potentially promising alternative. The Informatics Institute of the University of Alabama at Birmingham is applying de-identification to clinical data drawn from the UAB hospital’s electronic medical records system before releasing them for research. We participated in a shared task challenge by the Centers of Excellence in Genomic Science (CEGS) Neuropsychiatric Genome-Scale and RDoC Individualized Domains (N-GRID) at the de-identification regular track to gain experience developing our own automatic de-identification tool. We focused on the popular and successful methods from previous challenges: rule-based, dictionary-matching, and machine-learning approaches. We also explored new techniques such as disambiguation rules, term ambiguity measurement, and used multi-pass sieve framework at a micro level. For the challenge’s primary measure (strict entity), our submissions achieved competitive results (f-measures: 87.3%, 87.1%, and 86.7%). For our preferred measure (binary token HIPAA), our submissions achieved superior results (f-measures: 93.7%, 93.6%, and 93%). With those encouraging results, we gain the confidence to improve and use the tool for the real de-identification task at the UAB Informatics Institute.
Keywords: Automatic de-identification, clinical natural language processing, shared task, machine learning
Graphical abstract
1. Introduction
Clinical narratives are a rich resource for clinical research that contain individual identifiable information (e.g patient names, contact, and address). Removal of those patient identifiers (anonymization) is necessary to protect patient privacy. According to the HIPAA Privacy Rule, 18 “safe harbor” identifiers need to be removed in order for a data set to qualify as de-identified (1). Using de-identified data for research allows researchers to be exempt from oversight by Institutional Review Boards (IRB).
Manual de-identification (de-id) can be expensive and is subject to human errors(2). Automatic de-id using computer algorithms is a promising alternative. At the UAB Informatics Institute, we are applying automatic de-id as a cost-effective approach for anonymizing clinical data on a large scale. We participated in the CEGS N-GRID 2016 shared task challenge to compare the performance of our de-id system to others. We participated in the late phase of the challenge and were able to submit results to the regular track (Track 1B). The total development time since starting the project to result submissions was about 2 months. Our main strategy was to prioritize implementation of successful methods used in other challenges, and combine them with our own techniques developed in previous research.
2. Background
In a restricted domain, medical text exhibits distinguishable characteristics and constitutes a medical sublanguage(3, 4). Different note types may use different templates and use specialized language (e.g acronyms, notations). Different hospitals might use different EHR vendors, each vendor might have localized implementations. NLP systems optimized to one note type or EHR implementation might not generalize well to the others. Therefore, local optimization of de-id method is still required.
The 2016 CEGS N-GRID is the third de-id challenge organized by Uzuner et al. The first challenge (2006) created a reference standard from discharge summaries(5). The second challenge (2014) used a mix of discharge summaries, admission notes, and physician correspondences(6). The third challenge (2016) used notes from psychiatric initial evaluations(7). The psychiatric intake notes contain significantly more personal information about patients, their relatives and other social relationships (e.g, friends, pets, employers, etc). Psychiatric notes used in the current challenge also have the highest density of PHI, averaging 34.4 instances of PHI per records, a 55.3% increase over the 2014 challenge. These characteristics make development of an automatic de-id solution more challenging in psychiatric notes. The performance of the best team was slightly lower when compared with the previous challenge (from 93.6% to 91.4%).
Many de-id systems and methods were developed and evaluated both inside and outside the scope of the shared task challenge(8–20). Successful systems used the machine-learning algorithm Conditional Random Field (CRF) for labeling a sequence of tokens(11, 18, 19). The popular rule-based method was pattern-matching using a formal language such as regular expressions. De-id systems might or might not use the dictionary-matching method. In the 2014 i2b2/UTHealth challenge, four teams used the dictionary method including the winning team(11).
2. Methods
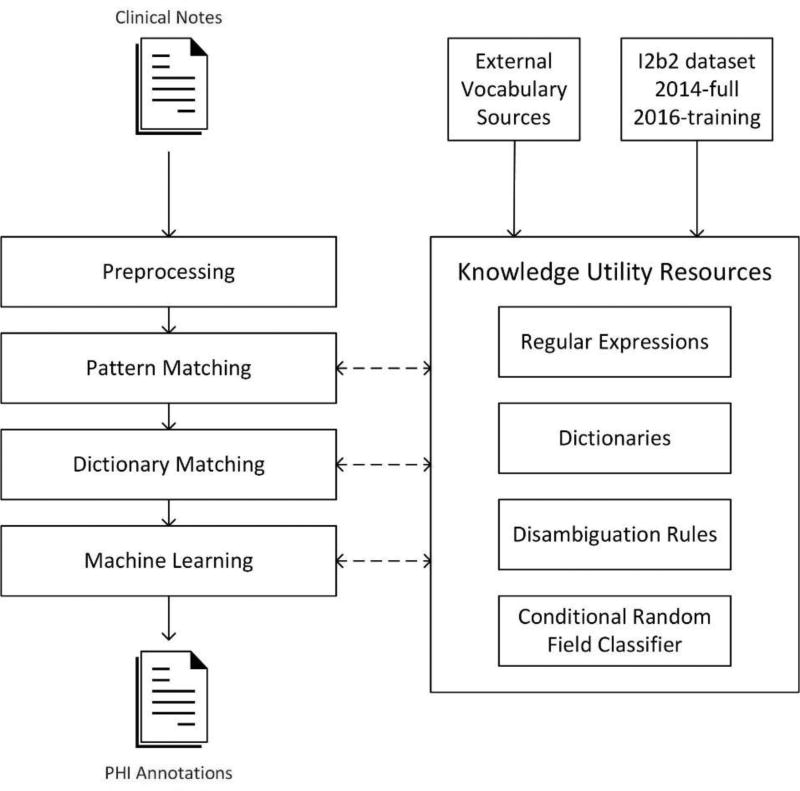
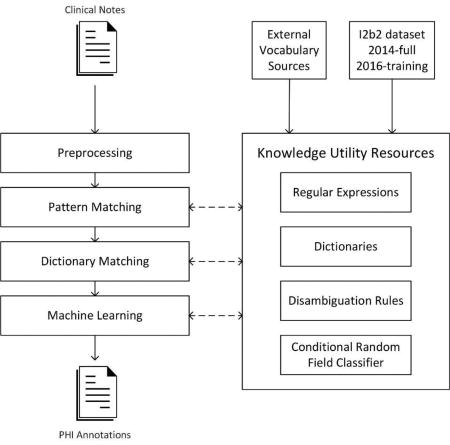
Our de-id system architecture comprises a preprocessing stage, three main processing stages and knowledge utility resources (See Figure 1). Each processing stage represents an underlining computational approach and contains a stack of modules specialized to identify specific types of PHI elements. Knowledge utilities such as pattern matchers, rules, machine-learning models, and dictionaries were created during the development phase to aid the extraction process in the test phase. The system overview and the composition of each stage are described below.
Figure 1.
An overview of the system architecture.
2.1 Overview of system architecture
Our general design uses a multi-pass sieve framework to organize our de-id methods. The multi-pass sieve framework has been applied to resolving co-reference resolution(21, 22) and successfully generalized to other NLP tasks(23, 24). The framework favors breaking a complex task into independent sub-tasks (or sieves) and iteratively passing through the document. Each sieve is specialized to identify a specific type of annotation (i.e PHI sub-category). We used the Interval Tree data structure(25) as a central repository to store and query annotations generated from sieves. This data structure does not require traversing the whole collection to match annotations within a given range, providing faster query times when compared to arrays or list structures. Different sieves’ annotations may cause conflicts by claiming the same span in a document. To resolve those conflicts, we applied a simple rule that first-come annotations are permanent and cannot be overridden by subsequent annotations. This rule expects top-ranked sieves to generate the most trustworthy annotations.
The system is composed of 25 sieves organized into three groups: pattern-matching (15 sieves), dictionary-matching (9 sieves), and machine-learning (1 sieve). The composition of each group is shown in Table 1. The pattern-matching method was only applied to restrictive PHI types such as email, url, fax, phone, and Ids. In our development phase, using the pattern-matching method alone could achieve nearly perfect performance on the training set. For other PHI types, we used the combination of two or three approaches, given certain approaches are applicable and effective on some types but not the others. The ordering of sieves was left-to-right and top-down of Table 1. The pattern-matching sieves could be extremely accurate and reliable in our development phase, therefore we assigned them into the top to take the priority over other methods. The machine-learning sieve was not expected to generalize well on an unseen test set and was assigned to the end. The ordering of sieves within each group was the result of iterative testing and adjustment on the training set.
Table 1.
The composition of three sieve group.
| PHI type | Pattern matching |
Dictionary matching |
Machine learning |
|---|---|---|---|
| x | |||
| URL | x | ||
| FAX | x | ||
| PHONE | x | ||
| MEDICALRECORD | x | ||
| LICENSE | x | ||
| USERNAME | x | ||
| IDNUM | x | ||
| DATE | x | x | |
| AGE | x | x | |
| COUNTRY | x | x | |
| STATE | x | x | |
| CITY | x | x | |
| STREET | x | x | x |
| HOSPITAL | x | x | x |
| ORANIZATION | x | x | x |
| DOCTOR | x | x | x |
| PATIENT | x | x | x |
| PROFESSION | x | x | |
| LOCATION_OTHER | x |
2.2. Preprocessing
In the preprocessing stage, the system performed basic natural langue processing steps including section detection, sentence splitting, and tokenization. We constructed a list of common section headings by analyzing the provided training set. The system first matches the headings in the document and partitions it into separate sections. It then uses the Stanford NLP tool(26) to split text into sentences and tokens. Performing section partition before sentence splitting and tokenization can partially solve the merged word problem. Whitespaces are occasionally missing after pulling texts from the EHRs, which sometime causes two or three words or be merged into a single, incorrect word. Merged words affect the accuracy of tokenization, which leads to unrecognizable patterns. For an example, the text fragment “Age: 51Sex: Female Primary Language (If not English): SpanishChief Complaint” has two merged words “51Sex” and “SpanishChief”. The preprocessing stage in this case first detects the span for headings “Age:”,” Sex:”,” Primary Language (If not English):”, and “Chief Complaint”, therefore conserves the span for two other PHI tokens.
2.3 Pattern-matching approach
We used regular expressions (regex), a formal language, to describe a text pattern. Regex is supported by most programming languages (e.g, .NET, Java, Python, Perl, Ruby, etc). We used regex in the Java programming language for easy integration with our development environment although the use of regex syntax is similar across other supported languages. We used regex’s capturing group feature to divide regex into the core and the optional context parts. The core parts are the PHI texts to be captured, while the context parts are primarily used for disambiguation. For example, the regex pattern “patient|she|pt|he was (\d+ (months|weeks)?)” was used to match AGE values. In this case, the core age values have the numerical pattern with optional time unit (\d+ (months| weeks)?). The context part “patient|she|pt|he was” restricts the matching to those values preceding by “patient was” or “he was”. We also complemented regex with disambiguation rules to further enhance accuracy of regex matching. In the above example, when testing the age pattern against the training set, we found two incorrect matchings related to patient’s duration of pregnancy and weight. To address this, we added a disambiguation rule that invalidates the matching if the system detects stop words (“pounds”, “pregnant”) within 5 words to the right of the matching.
We analyzed the training set to develop patterns for 15 PHI sub-categories. Our patterns achieved nearly perfect precision on the training set while recall was maximized as much as possible. In some cases, we needed to adjust the training annotations if they are annotation errors or inconsistencies. For instance, a curator may treat “Monday, 9/17” as a single annotation, while others may treat it as two separate annotations “Monday” and “9/17”. We merged such adjacent annotations into a single annotation. Regular expressions by default are “greedy”, matching the longest possible span. Sometimes, the pattern found correct PHI that was missed by curators. We added those missed annotations to the training set to assist pattern development and testing. Table 2 shows a snapshot of regular expressions used in 15 pattern-matching sieves. Some patterns are complemented with disambiguation rules derived from our iterative testing. We reviewed the false-positive matches and added more restrictive conditions to reduce them. Common disambiguation techniques are based on neighboring words, section headings, and previously accepted annotations. Disambiguation rules implemented in programming logic are too long to fit in this paper. Figure 2 shows an example pseudo-code of the disambiguation rules for the dictionary approach. The disambiguation rules for regex patterns have a similar form but apply to a single pattern or group of patterns.
Table 2.
Examples of regular expression pattern and matched instances. The context parts are colored blue.
| PHI subcategory |
Regular expressions | Example of matched instances |
|---|---|---|
| ([A-Za-z0-9][A-Za-z0-9._%+-]*@[A-Za-z0-9.-]+\.[A-Za-z]{2,6}) | gzumwait@mailhot.com hcuutaj@bdd.com | |
| URL | ((https?:)?[a-zA-Z0-9-\.\/]+\.(com|org|net|mil|edu|gov|us)([/\][a-zA-Z0-9-\.\/]+)?) | www.sssppsqeslctfuksdo.com |
| (https?:[a-zA-Z0-9-\.\/]+\.([a-z]{3}|us)(?:[/\][a-zA-Z0-9-\.\/]*)?) | http://amzn.com/0195189183 | |
| FAX | Fax.{1,30}(\d{3}[-\.]\d{2,3}[-\.]\d{4,5}) | Fax: 700-126-0424 |
| PHONE | Phone:? ((\d [-\.])?\d{3}[-\.]\d{2,3}[-\.](\d{4,5}|TALK)) | Phone 870-355-1031 Phone: 083-380-6789 |
| MEDICALRECORD | (MMRN|MRN|MR|Patient ID|Medical Record Number|OCH) *[:#]+\s+(\d+) | MRN: 2418195 MMRN: 3161614 |
| LICENSE | (DEA Number|DEA) *[:#]?\s+([A-Z][A-Z0-9] *[-]? *[0-9-]+) | DEA NH9787347 DEA KR 2870315 |
| USERNAME | \[([a-z]{2,3}\d{1,3})\] *\n | [mpl12] |
| IDNUM | (ID) *[:#]? *([A-Z]?\d{5,}) | Pharm ID: 50970046433 ID 8345948 |
| DATE | DAYWEEK=Monday|Tuesday|Wednesday|Thursday|Friday|Saturday|Sunday|Mon|Tue|Wed|Thu|Fri|Sat|Sun | |
| TERM=Spring|Fall|Summer|Winter | ||
| MONTH=January|February|March|April|May|June|July|August|September|October|November|December| Jan|Feb|Mar|Apr|May|Jun|Jul|Aug|Sep|Oct|Nov|Dec | ||
| YEAR= (16|17|18|19|20|21|22|00|01|02)\d{2} | ||
| DATE= [1-9]|0[1-9]|[12]\d|3[01] | ||
| (TERM *(of| *,)? *YEAR) | Spring 2005 Fall of 2016 | |
| (DAYWEEK *,? *" DATE/\d{1,2}) | Mon 06/17 | |
| ((DAYWEEK *,?)? MONTH *[\.,]? *DATE *[,~/] *YEAR) | Wed, Nov 19, 2014 June 10, 2009 | |
| AGE | AGE=1[0-3][0-9]|[1-9][0-9]|[0-9] | |
| RELATIVE= brother|boyfriend|bride|brother|cousin|dad|daughter|father|…. | ||
| AGE *[+-]? *((years?|yrs?)[-]old|y\/?o|y\.o|y o) | 18 year old;36 y.o | |
| age:?(?: +of)? AGE | age of 42 | |
| (?:patient|she|pt|he|I) was AGE | Patient was 56 | |
| RELATIVE *(?:who is|is|was)? AGE | Brother who is 7 | |
| STREET | (\d+) (?:[A-Z][A-Z'’a-z]+){1,2}(?i)(?:Road|Rd|Drive|Lane|Street|St|Avenue|Court) | 607 Cornell Road |
| (on|in|at) (the +)?(([A-Z][A-Z'’a-z]){1,2}(Road|Rd|Drive|Lane|Street|Avenue) | at Piermont Street | |
| NAME=[A-Z][a-z]*(?: [A-Z][a-z]*){0,4} | ||
| HOSPITAL | NAME (Hospital Center|Hospital|Medical Center|Health Center|Clinic|Sanitarium|Sanatorium|Care Center|Family Care|Care Tower|Physician Center…) | Saint Christophers Hospital;Minden Medical Center; Kinston Clinic |
| ORGANIZATION | NAME (College|University|Restaurant|Pharmacy|Corporation|Academy|Company|Agency|Technologies|Airport|Studio|Industries|Bank|Entertainment|Solutions|High School|…) | Houghton Community College;Salem University;Albemarle Corporation |
| DOCTOR | [Dd]r\.? NAME | Dr. Harlan Oneil; Dr. Dailey |
| PATIENT | (Mrs?\.?) NAME is a | Mr. Yager is a |
| NAME 's RELATIVE | Russell’s husband |
Figure 2.
Pseudocode to disambiguate COUNTRY terms
2.4 Dictionary-matching approach
This approach generates annotations by searching terms or key words contained in PHI-specific dictionaries. We used the Aho Corasick algorithm(27) to quickly find occurrence of terms in the documents. We collected PHI terms from the 2014 i2b2/UTHealth (full set), the 2016 CEGS N-GRID (training set), and those from external vocabulary sources (Table 3) and applied the dictionary-matching approach to nine types of PHI (Table 1), given those types use proper names, and their occurrences are noticeably repeated.
Table 3.
Complementary vocabularies sources.
| PHI category | Description | Source |
|---|---|---|
| COUNTRY | List of country names and nationalities | http://www.state.gov/misc/list/ |
| https://en.wikipedia.org/wiki/Lists_of_people_by_nationality | ||
| STATE | List of US states and acronyms | https://simple.wikipedia.org/wiki/List_of_U.S._states |
| CITY | List of US cities and world capitals | http://simplemaps.com/data/us-zips |
| http://geographyfieldwork.com/WorldCapitalCities.htm | ||
| ORANIZATION | List of US notable companies | https://en.wikipedia.org/wiki/List_of_companies_of_the_United_States |
We evaluated dictionary terms to measure their ambiguity level and determine the best matching strategy. Matching a term can be influenced by case-sensitivity and word-boundary. If case-sensitivity is set to “false”, the term “Birmingham” will match “Birmingham”, “BIRMINGHAM” and “birmingham”. If case-sensitivity is set to “true”, only the “Birmingham” instances are found. Word-boundary controls partial vs. exact matching. If word-boundary is set to “false”, the term “India” can find a partial matching inside the word “Indian”. If word-boundary is set to “true”, only exact word matches are counted. For a given term, we tested all four variants of those two control parameters to determine the best matching strategy.
Highly ambiguous terms such as short words and common words may inflate the number of false positives, and need to be controlled. Therefore, measuring term ambiguity level is a useful practice to control ambiguous matching. We used a simple approach to measure term ambiguity. First, any given term is tested against the training set in four matching strategies. Then, the highest precision is determined and that is used as the best matching strategy for that term. Term ambiguity is defined as 1 minus the highest precision of four matching strategies. The system filters terms whose term ambiguities are above a certain system configurable threshold: maxAmbiguity. Ideally, the maxAmbiguity of zero means the system achieved 100% precision on the training set. Increasing maxAmbiguity results in more false-positive matches. We improved precision by analyzing those false matches and developing disambiguation rules. Figure 2 provides an algorithm to disambiguate terms in the COUNTRY dictionary. In this example, a term is not assigned a PHI label if the term ambiguity is greater than the system’s maxAmbiguity. Otherwise, it uses the local context and section heading clues to classify the annotation to COUNTRY, PROFESSION, or unassigned.
2.5 Machine-learning approach
We developed a machine-learning classifier using the conditional random fields (CRFs) algorithm. The Stanford Named Entity Recognizer (NER) implementation of CRF was selected given its easy integration with our Java implementation. The Stanford NER(28) was originally designed to recognize 7 entities (Location, Person, Organization, Money, Percent, Date, and Time). We re-trained Stanford NER to recognize 12 PHI categories as shown in Table 1. From a sequence of input tokens, Stanford NER automatically generates linguistic features such as word features, orthographic features, prefixes/suffixes, label sequences, conjunction features, and distributional similarity. We used the enhanced tokenizer in the pre-processing stage to generate the sequence of tokens and keep other CRF configurations at default parameters. The training data were the 600 documents of the 2016 CEGS N-GRID training set released in the training phase.
2.6 Challenge submission and evaluation
In our submissions, we examined the impacts of external vocabulary sources to the de-id performance. We submitted 3 runs to the challenge based on the variation of the dictionary-based approach. The configuration used for each run is summarized in Table 4. In the first run, we configured maxAmbiguity to 30% and developed disambiguation rules to improve precision to nearly perfect. In the second run, we adjusted the maxAmbiguity to 50% without extending disambiguation rules further. In the third run, we completely excluded the dictionary approach. It was our prediction that run 1 would perform best among three.
Table 4.
Summary of three run configurations.
| Methods | Run 1 | Run 2 | Run 3 |
|---|---|---|---|
| Pattern-matching | Yes | Yes | Yes |
| Dictionary-matching |
|
|
No |
| Machine-learning | Yes | Yes | Yes |
We reported recall, precision, and f-measure for different measurement types defined by the challenge. The “entity” measures requires matching each PHI entity exactly including all words in multi-word PHI. The “token” measures relax the all-words condition and gives credit to recognition of composition words. Most measures follow the “strict” criterion that requires matching exactly at beginning and ending offsets. The “relaxed” measures allow the end part to be flexible by 2 characters. Most measures require correctly classifying PHI to one of seven categories. The “binary” measures simplifies the multi-class classification to binary classification (PHI vs. non-PHI). Lastly, the “HIPAA” measures evaluate only sub-categories that are considered PHI under HIPAA definition(1).
3. Results
The challenge organizer selected the “entity strict” measure as the primary outcome. Among our submissions, Run 1 achieved the best f-measure (87.3%) as we predicted, followed by Run 2 (87.1%) and Run 3 (86.7%). The use of the dictionary approach in Runs 1 and 2 improved performance 0.65% and 0.46% respectively when compared with the model that did without them (Run 3). In comparison with other submissions, our best submission scored 9.4% above the mean of the challenge participants and 4.1% below the best submission. Our team ranked 4th among 13 participants in the challenge.
Other evaluation measures are summarized in Table 5. Not surprisingly, the performance is increased for more relaxed measures. For the binary (PHI vs. non-PHI) token measure, our best f-measure was 92.4%. The ranking remains the same irrespective of which measure. Table 6 showed measures focused only on HIPAA elements. With HIPAA elements, the system scored 91% in with the entity strict measure and 93.7% for the token binary measure. The HIPAA binary token best reflects our system design goal. Classifying PHI to sub-categories and missing non-PHI is not as important as identifying and redacting true PHIs in texts.
Table 5.
The aggregated performance for all PHI on the unseen test set.
| Run 1 | Run 2 | Run 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | |
| Token | 0.9484 | 0.8597 | 0.9019 | 0.9455 | 0.8596 | 0.9005 | 0.9463 | 0.8494 | 0.8952 |
| Token Binary | 0.9721 | 0.8811 | 0.9244 | 0.9696 | 0.8815 | 0.9235 | 0.972 | 0.8725 | 0.9196 |
| Entity Strict | 0.9162 | 0.8338 | 0.8731 | 0.9118 | 0.8342 | 0.8712 | 0.9164 | 0.822 | 0.8666 |
| Entity Relaxed | 0.9194 | 0.8367 | 0.8761 | 0.915 | 0.8371 | 0.8743 | 0.9195 | 0.8248 | 0.8696 |
| Entity Binary Strict | 0.934 | 0.85 | 0.89 | 0.9292 | 0.8501 | 0.8879 | 0.9362 | 0.8398 | 0.8854 |
The challenge’s primary measures are marked in bold.
Table 6.
The aggregated performance for HIPAA-only PHI on the unseen test set.
| Run 1 | Run 2 | Run 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | |
| Token | 0.956 | 0.9053 | 0.93 | 0.9547 | 0.9054 | 0.9294 | 0.9534 | 0.8941 | 0.9228 |
| Binary Token | 0.9629 | 0.9118 | 0.9366 | 0.9615 | 0.9119 | 0.936 | 0.9605 | 0.9008 | 0.9297 |
| Entity Strict | 0.9394 | 0.8827 | 0.9102 | 0.9378 | 0.8828 | 0.9095 | 0.9367 | 0.869 | 0.9016 |
| Entity Relaxed | 0.9411 | 0.8843 | 0.9118 | 0.9396 | 0.8845 | 0.9112 | 0.9383 | 0.8705 | 0.9031 |
| Entity Binary Strict | 0.9435 | 0.8866 | 0.9142 | 0.9421 | 0.8868 | 0.9136 | 0.9411 | 0.8732 | 0.9059 |
Measures most useful for de-id of HIPAA-compliant PHI are marked in bold.
The system’s precision is significantly better than recall (about 5%). Since our method used a pipeline of high precision sieves, the high precision rate is anticipated. The precision for HIPAA binary token measure was satisfactory (96.3%). The recall 91.2% can be improved further by stacking additional sieves. Table 7 summarized the category-specific performance of our three submissions. Overall, Run 1 generally outperformed other runs on all categories except the PROFESSION category. The dictionary-matching approach was not effective for the identification of profession categories. Our system performed best on AGE (95.9%) and DATE (96.3%) categories. Since those are the most frequent instances of PHI and exhibit restrictive patterns, we dedicated time to carefully craft patterns and rules for those categories and obtained superior results. The worst performances were the PROFESSION (64.6%) and ID (65.6%) categories. De-id of the PROFESSION category is not required by HIPAA. For the ID category, the training and testing sets are small and not representative, which challenges both system development and evaluation. Performances of CONTACT (90.8%), NAME (88.5%), LOCATION (76.8%) are promising for future improvement.
Table 7.
The category-specific entity strict performance on the unseen test set.
| Run 1 | Run 2 | Run 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | |
| CONTACT | 0.9187 | 0.8968 | 0.9076 | 0.9187 | 0.8968 | 0.9076 | 0.9187 | 0.8968 | 0.9076 |
| NAME | 0.9619 | 0.8191 | 0.8847 | 0.9619 | 0.8191 | 0.8847 | 0.9596 | 0.7808 | 0.861 |
| DATE | 0.9656 | 0.961 | 0.9633 | 0.9656 | 0.961 | 0.9633 | 0.9656 | 0.961 | 0.9633 |
| AGE | 0.9692 | 0.9482 | 0.9586 | 0.9692 | 0.9482 | 0.9586 | 0.9692 | 0.9482 | 0.9586 |
| PROFESSION | 0.7645 | 0.5594 | 0.6461 | 0.7454 | 0.5564 | 0.6372 | 0.8052 | 0.5812 | 0.6751 |
| ID | 0.7143 | 0.6061 | 0.6557 | 0.7143 | 0.6061 | 0.6557 | 0.7143 | 0.6061 | 0.6557 |
| LOCATION | 0.8285 | 0.7163 | 0.7683 | 0.8184 | 0.7184 | 0.7651 | 0.8201 | 0.6927 | 0.751 |
4. Discussion
The UAB Informatics Institute is optimizing our de-id software for use with UAB Hospital medical records. We participated in the CEGS N-GRID 2016 de-id shared task challenge to compare our software with other state-of-art de-id systems. The challenge organizer selected the entity strict measure as the primary outcome, raising the difficulty bar for an NLP challenge. We achieved a 4th-place performance (87.3%); however, we consider the HIPAA token binary measure to be sufficient for our goal, to identify and redact PHI defined by the HIPAA Privacy Rule, while classification of PHI is secondary and optional. With that practical measure, our system achieved a superior performance (93.7%), which ranked 3rd place (1.3% below the best team). The precision is satisfactory (96.3%), which indicates our method can be easily extended or combined with other methods. The recall rate (91.2%) is a promising baseline performance. Incorporating additional rules, training data, or external terminological resources should improve recall performance further.
We used the multi-pass sieve framework to help organize our NLP processing modules. Multi-pass systems traverse the document several times while a single-pass system process the document only once. Prior de-id studies used multi-pass architectures at the macro level. For instance, Dehghan et al. used a two-pass approach, where the second pass was a post-processing phase to filter low-quality annotations from the first pass(16). Our system applied a multi-pass framework at a micro level: by methodology and PHI type. Each of 24 sieves was dedicated to annotate a specific PHI type using a uniform approach. Exceptionally, the machine-learning sieve handled 12 types of PHI at once to benefit multi-class learning. In our development experience, having more sieves promotes better code maintenance, unit testing, debugging, and extension. In our current version, we applied a simple first-come first-serve rule to resolve the conflicted annotations. Future work can explore complex rules such as inter-sieve agreement, context, and document features to combine annotations.
In the pattern-matching and dictionary-matching methods, we introduced the use of disambiguation rules to control false-positive matches using heuristics with contextual and pragmatic features. These rules increase the system coverage by permitting the uses of more relaxed patterns and ambiguous terms. Researchers can combine the pattern and dictionary methods in the machine-learning model by using matching results as features (11, 19). This method is simple and likely optimal within the scope of the challenge. However, applying it to our real scenario is difficult due to its “black-box” nature and its dependency on labeled data. PHI in UAB Hospital medical records might have different characteristics (e.g, PHI distribution, document templates, local language, etc,) than those in the shared task corpora. Creating a large and representative de-identification corpus at UAB will be resource-intensive and time-consuming. We prefer a hybrid deterministic and probabilistic model that allows us incorporate human knowledge such as rules and terminological resources.
In our submissions, we evaluated the effect of the dictionary method on overall de-id performance. The presence of the dictionary method improved the f-measure by 0.65% over the baseline (without dictionary). The method was more helpful for name categories such as NAME (+2.4%) and LOCATION (+1.7%), but was not effective for the PROFESSION category (−2.9%). In runs 1 and 2, we raised the maxAmbiguity 30% to 50% without adding more disambiguation rules and witnessed a performance drop of 0.19%. Therefore, the use of ambiguous terms must be coupled with appropriate disambiguation strategies. Overall, dictionary-matching remains a useful de-id approach, especially for recognition of popular named entities. Term ambiguity level and disambiguation rules are useful tools to control ambiguous term matches.
4.1. Error analysis
We analyzed the system errors generated from our best submission (Run 1). The errors can be classified into one of in following categories:
Classification errors
The system correctly identifies the span of PHI but classifies it into an incorrect sub-category. Many entities such as STATE, CITY, STREET, HOSPITAL, ORANIZATION, DOCTOR, PATIENT share the same pool of vocabularies. Improving disambiguation rules can reduce classification errors; however, there are challenging cases that are impossible to resolve using simple contextual features. In the sentence “He was sent to Guatemala for 3 weeks”, the system classified “Guatemala” as a city, but it is actually a country. Guatemala is the capital city of the country Guatemala, and the text does not give sufficient clues to make the distinction. The classification errors are unimportant in the binary measure (PHI vs. non-PHI).
Acronyms and misspelled words
Our system did not have a dedicated acronym normalization module and therefore failed to recognize a large number of named entities based on their acronyms. Recognizing and classifying abbreviated PHI (e.g NAME: “NC”, ”NG”, “EU”,“L”, “KT”; DATE: “Su”, “Tu”, “Th”, “We”) is challenging since they are easily confused with abbreviated medical terms having the same pattern of characters. The acronyms themselves are ambiguous terms that trouble any re-identification efforts. The system failed to recognize a number of PHI instances that were misspelled words (e.g noverber 2067, Aprill 2094). Future integration of spell-correction techniques(29) should attenuate this problem.
Named entities (Person and Location)
Identifying the names of people and locations relies on the dictionary and machine-learning approaches. The system sensitivity errors can be attributed to the prevalence of unseen terms and unfamiliar contexts. In the software version evaluated in this challenge, we did we used location vocabularies that were mostly US-centric and did not incorporate person name vocabularies at all. Future integration of additional vocabularies (e.g common first name and last name complied by U.S. Census Bureau) and balancing term ambiguity level will improve the recall performance, while most precision errors are simply due to incorrect classification, as mentioned earlier.
Corpora and annotation problems
Missed PHI related to merging words (e.g “Lewis ClarkIf,” “Hancevillewith,” “inCoffeyville,” etc.) are still present. Section recognition can detect merged words related to section heading but cannot detect those in the content parts. Future work needs to employ lexicon based tokenization(30) or character-level machine-learning techniques(11). The inter-rater agreement of the dataset was about 80% for entity and 90% for token, which indicates there is a small number of human errors or inconsistencies in the evaluation set. Errors include false negative annotations (e.g., “DATE: December (0033_gs)”, “DOCTOR: Carr (0147_gs)”, and “DOCTOR: Zahn Gabapentin (0568_gs)”). Inconsistent annotations are not considered as errors but caused difficulty in counting true positive annotations. For example, the temporal phrase “November of 2090” can be treated as two annotations “November” and “2090” or a single annotation. When development and testing new rules, developers should be aware of those issues and actively adjust the gold standard to measure the system more accurately.
5. Conclusion
We present an automatic de-id system that can identify protected health information from free-text notes. Our system achieved a competitive performance in the CEGS N-GRID 2016 shared task challenge, de-id regular track. We considered HIPAA binary token measures best reflect how we intent to use the system in real de-id task (i.e identification of HIPAA-mandated PHI without the need for classification). We deem the precision rates to be satisfactory, while recall rates can be improved further. The high precision rates indicate that our method can be easily extended or used in combination with other approaches. Our innovative contributions are the use of multi-pass sieve framework at micro level (by methodology and PHI type), measuring term ambiguity level, and the use of disambiguation rules. We emphasized more on creating effective rules and keywords, rather than optimizing the machine-learning model. When applying to new document types, machine-learning approaches often lacks generalizability and requires additional labeled data. Iterative testing and debugging rule and keyword give developers a better insight of clinical text, and can quickly adapt to new note types. We are adapting the tool to the clinical texts at UAB Hospital’s medical records, and will use the tool in real de-id tasks.
Highlights.
We described an automatic de-identification (de-id) system for clinical texts.
We used three de-id methods: pattern-matching, dictionary-matching, and machine-learning.
Dictionary-matching with disambiguation remained a useful de-id approach.
We also explored multi-pass sieve framework, term ambiguity measurement and disambiguation rule.
The system achieved competitive results in CEGS N-GRID 2016 challenge, de-id regular track.
Acknowledgments
This project was supported by internal research funds of the UAB Informatics Institute. We are also thankful to NIH grants (P50MH106933 and 4R13LM011411) which made the organization of the challenge possible.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest.
The authors have no conflicts of interest to declare.
References
- 1.Guidance regarding methods for de-identification of protected health information in accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. US Department of Health Human Services. 2012 [Google Scholar]
- 2.Douglass M, Clifford G, Reisner A, Moody G, Mark R, editors. Computers in Cardiology. Vol. 2004. IEEE; 2004. Computer-assisted de-identification of free text in the MIMIC II database. [Google Scholar]
- 3.Grishman R, Kittredge R. Analyzing language in restricted domains: sublanguage description and processing. Psychology Press; 2014. [Google Scholar]
- 4.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform. 2002;35(4):222–35. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 5.Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc. 2007;14(5):550–63. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stubbs A, Kotfila C, Uzuner Ö. Automated systems for the de-identification of longitudinal clinical narratives: Overview of 2014 i2b2/UTHealth shared task Track 1. J Biomed Inform. 2015;58:S11–S9. doi: 10.1016/j.jbi.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stubbs A, Filannino M, Uzuner O. De-identification of psychiatric intake records: Overview of 2016 CEGS N-GRID Shared Tasks Track 1. J Biomed Inform. 2017 doi: 10.1016/j.jbi.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC medical research methodology. 2010;10(1):70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrandez O, South BR, Shen S, Friedlin FJ, Samore MH, Meystre SM. BoB, a best-of-breed automated text de-identification system for VHA clinical documents. Journal of the American Medical Informatics Association : JAMIA. 2013;20(1):77–83. doi: 10.1136/amiajnl-2012-001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neamatullah I, Douglass MM, Li-wei HL, Reisner A, Villarroel M, Long WJ, et al. Automated de-identification of free-text medical records. BMC Med Inform Decis Mak. 2008;8(1):32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Garibaldi JM. Automatic detection of protected health information from clinic narratives. J Biomed Inform. 2015;58:S30–S8. doi: 10.1016/j.jbi.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dernoncourt F, Lee JY, Uzuner O, Szolovits P. De-identification of patient notes with recurrent neural networks. Journal of the American Medical Informatics Association. 2016:ocw156. doi: 10.1093/jamia/ocw156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scaiano M, Middleton G, Arbuckle L, Kolhatkar V, Peyton L, Dowling M, et al. A unified framework for evaluating the risk of re-identification of text de-identification tools. J Biomed Inform. 2016;63:174–83. doi: 10.1016/j.jbi.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 14.Salloway MK, Deng X, Ning Y, Kao SL, Chen Y, Schaefer GO, et al., editors. Biomedical and Health Informatics (BHI), 2016 IEEEE-MBS International Conference on. IEEE; 2016. A de-identification tool for users in medical operations and public health. [Google Scholar]
- 15.Phuong ND, Chau VTN, editors. Computing & Communication Technologies, Research, Innovation, and Vision for the Future (RIVF), 2016 IEEE RIVF International Conference on. IEEE; 2016. Automatic de-identification of medical records with a multilevel hybrid semi-supervised learning approach. [Google Scholar]
- 16.Dehghan A, Kovacevic A, Karystianis G, Keane JA, Nenadic G. Combining knowledge-and data-driven methods for de-identification of clinical narratives. J Biomed Inform. 2015;58:S53–S9. doi: 10.1016/j.jbi.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szarvas G, Farkas R, Busa-Fekete R. State-of-the-art anonymization of medical records using an iterative machine learning framework. Journal of the American Medical Informatics Association. 2007;14(5):574–80. doi: 10.1197/j.jamia.M2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellner B, Huyck M, Mardis S, Aberdeen J, Morgan A, Peshkin L, et al. Rapidly retargetable approaches to de-identification in medical records. Journal of the American Medical Informatics Association. 2007;14(5):564–73. doi: 10.1197/jamia.M2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Chen Y, Tang B, Wang X, Chen Q, Li H, et al. Automatic de-identification of electronic medical records using token-level and character-level conditional random fields. J Biomed Inform. 2015;58:S47–S52. doi: 10.1016/j.jbi.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kayaalp M, Browne AC, Dodd ZA, Sagan P, McDonald CJ, editors. AMIA Annual Symposium Proceedings. American Medical Informatics Association; 2014. De-identification of address, date, and alphanumeric identifiers in narrative clinical reports. [PMC free article] [PubMed] [Google Scholar]
- 21.Jonnalagadda SR, Li D, Sohn S, Wu ST, Wagholikar K, Torii M, et al. Coreference analysis in clinical notes: a multi-pass sieve with alternate anaphora resolution modules. J Am Med Inform Assoc. 2012;19(5):867–74. doi: 10.1136/amiajnl-2011-000766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raghunathan K, Lee H, Rangarajan S, Chambers N, Surdeanu M, Jurafsky D, et al., editors. Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing. Association for Computational Linguistics; 2010. A multi-pass sieve for coreference resolution. [Google Scholar]
- 23.Bui DDA, Del Fiol G, Jonnalagadda S. PDF text classification to leverage information extraction from publication reports. J Biomed Inform. 2016 doi: 10.1016/j.jbi.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambers N, Cassidy T, McDowell B, Bethard S. Dense event ordering with a multi-pass architecture. Transactions of the Association for Computational Linguistics. 2014;2:273–84. [Google Scholar]
- 25.Samet H. The design and analysis of spatial data structures. Addison-Wesley Reading, MA: 1990. [Google Scholar]
- 26.Manning CD, Surdeanu M, Bauer J, Finkel JR, Bethard S, McClosky D, editors. The Stanford CoreNLP Natural Language Processing Toolkit. ACL (System Demonstrations); 2014. [Google Scholar]
- 27.Aho AV, Corasick MJ. Efficient string matching: an aid to bibliographic search. Communications of the ACM. 1975;18(6):333–40. [Google Scholar]
- 28.Finkel JR, Grenager T, Manning C, editors. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics. Association for Computational Linguistics; 2005. Incorporating non-local information into information extraction systems by gibbs sampling. [Google Scholar]
- 29.Lai KH, Topaz M, Goss FR, Zhou L. Automated misspelling detection and correction in clinical free-text records. J Biomed Inform. 2015;55:188–95. doi: 10.1016/j.jbi.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Carus AB. Method and apparatus for improved tokenization of natural language text. Google Patents. 1999 [Google Scholar]