Abstract
Purpose
To implement a fully automated atlas based method for prescribing 3D PRESS MR spectroscopic imaging (MRSI).
Methods
The PRESS selected volume and outer-volume suppression bands were predefined on the MNI152 standard template image. The template image was aligned to the subject T1-weighted image during a scan, and the resulting transformation was then applied to the predefined prescription. To evaluate the method, H-1 MRSI data were obtained in repeat scan sessions from 20 healthy volunteers. In each session, datasets were acquired twice without repositioning. The overlap ratio of the prescribed volume in the two sessions was calculated and the reproducibility of inter- and intra-session metabolite peak height and area ratios was measured by the coefficient of variation (CoV). The CoVs from intra- and inter-session were compared by a paired t-test.
Results
The average overlap ratio of the automatically prescribed selection volumes between two sessions was 97.8%. The average voxel-based inter-session CoVs were less than 0.124 and 0.163 for peak height and area ratios, respectively. Paired t-test showed no significant difference between the intra- and inter-session CoVs.
Conclusion
The proposed method provides a time efficient method to prescribe 3D PRESS MRSI with reproducible imaging positioning and metabolite measurements.
Keywords: MRSI, Automated, Prescription, Atlas-based, Reproducibility
Introduction
Magnetic resonance spectroscopic imaging (MRSI) is able to non-invasively provide information about the spatial distribution of tissue metabolites that cannot be obtained by conventional anatomical MRI. Techniques such as point-resolved spectroscopy (PRESS)1 have been implemented and used to assist in the diagnosis and evaluation of a number of different human neurological disorders2.
The widespread clinical application of MRSI in human brain is currently limited by the need to prescribe both the volume of interest and outer-volume suppression (OVS) bands that reduce signals from subcutaneous lipid. Manual prescription is time-consuming and requires experienced operators to perform. Another level of complexity is that the prescription of the selected region may be difficult to replicate in serial scans. For example, because the subject is positioned differently at each scan, it may be necessary to define an oblique volume for follow-up, which may not be supported on commercial scanners. Inconsistent prescriptions result in scan-to-scan differences in MRSI and variations in measured metabolite levels. Li et al.3 have demonstrated that even a 1-mm shift of a 1-cm3 voxel in all canonical directions could cause 10~30% variation in estimated metabolite concentration due to the variation in the percentage of gray, white matter and cerebrospinal fluid in each voxel. Although the problem can be mitigated by the retrospective registration of images from different scans4, the interpolation required by the registration can make it impractical for metabolite quantification in the voxels at the edge of the volume of interest. A more promising strategy is to perform a prospective registration. This has been done indirectly by either using a stereotactic frame to position the subject’s head in the same place5 or by acquiring uniformly oriented reference images for prescribing the MRSI volume6. Even though these methods simplify the definition of the triple oblique prescription, the volume of interest still has to be prescribed manually. Hancu et al.7 and Ratai et al.8 used uniformly oriented background images and the transformation matrix between them to propagate the center of the selected volume from baseline to the follow-up scans. While this does enforce consistency between time points, the prescription for the baseline scan is still made manually and does not adjust for differences between subjects.
The prescription of OVS bands is even more complex and time consuming. Although previous studies9–12 have considered the possibility of automating OVS band prescription, few of them have provided a fully integrated solution. In this study, we propose an atlas-based registration method for automatic prescription of both the selection volume and OVS bands. Our method predefines the prescription on a standard image atlas and transforms both the selected volume and OVS bands to individual subject space during each scan session. This allows us to provide a fully automated prescription that covers the desired anatomy consistently not only for different time points in a single subject, but also across a group of subjects. Although a similar method was previously implemented for automatic placement of OVS bands9, including the definition of the selection volume is critical for studies where reproducible anatomic localization is required. The purpose of this study was to implement this method for prescribing 3D PRESS MRSI and to evaluate its reliability by assessing the reproducibility of both selection volume/OVS prescription and metabolite measurements.
Methods
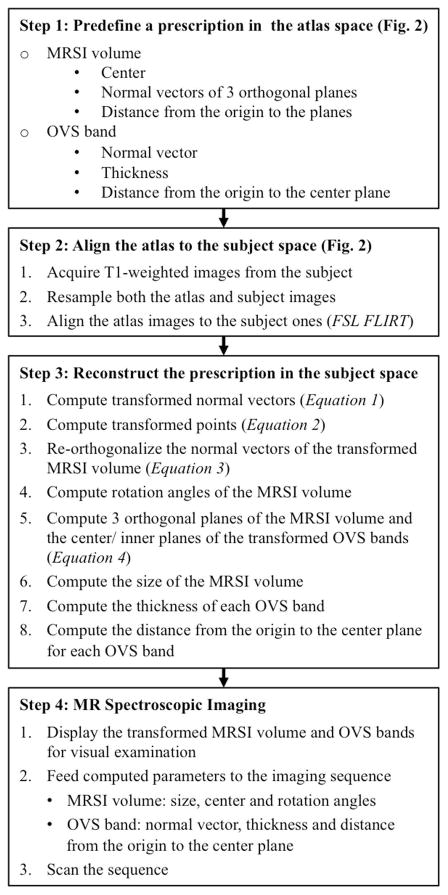
The overall process of defining and transforming the PRESS volume and OVS bands is summarized in Figure 1, with the details described in the following sessions.
Figure 1.
Summary of the steps required for the automatic MRSI prescription.
Predefinition of PRESS Volume and OVS Bands
The Montreal Neurological Institute MNI152 standard-space 1mm T1-weighted images (152 nonlinear 6th generation)13 provided with the FSL14 distribution were used as the atlas, on which a 3D rectangular PRESS box and 16 OVS bands were prescribed (Figure 2). The box had a size of 8(RL)×10(AP)×5(SI) cm3 and covered most of the cingulate cortex and deep gray matter structures in the brain. The OVS bands had a thickness of 40mm and were placed to cover the brain skull and suppress signals from subcutaneous lipid. Parameters that define the box and OVS bands in LPS coordinates included the center point of the box p⃗c = (pl,pp,ps)T, where T represents vector transpose; the normal vectors of the left, posterior and superior planes of the box n⃗L = (nLl,nLp,nLs)T, n⃗P = (nPl,nPp,nPs)T, and n⃗S = (nSl,nSp,nSs)T; the distance from the coordinate origin to the 3 planes dL, dP, and dS; the normal vector of the center plane of each OVS band n⃗i = (nil,nip,nis)T (i=1…16); the distance form the coordinate origin to the center plane of each OVS band di (i=1…16).
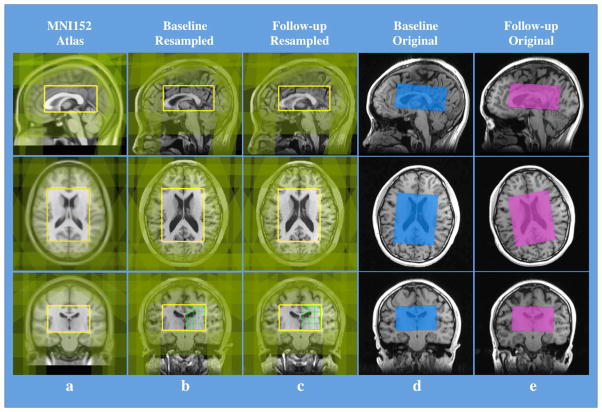
Figure 2.
Intra-subject reproducibility of the placement of MRSI PRESS volume/OVS bands and the spectra in serial scans. (a) Predefined MRSI PRESS volume (yellow rectangular boxes) and OVS bands (green-shaded bands) overlaid on the MNI152 standard 1mm T1-weighted atlas image. The size of the volume box is 80mm in RL (R51 to L130 in the atlas space coordinates), 100mm in AP (A157 to P58) and 50mm in SI (S114 to I65). (b) & (c) The PRESS volume and OVS bands transformed to a subject space in the baseline and follow-up scans, respectively. Note the PRESS volume is overlaid on the T1-weighted images that have been resampled and aligned to the PRESS volume. (d) & (e) The transformed PRESS volume and OVS bands overlaid on the subject’s original baseline and follow-up T1-weighted images, respectively. In (a), (b) and (c) the 3 orthogonal background image slices were chosen such that they all transverse the center of the PRESS volume box. It can be seen that although the orientation of the subject’s head was different between its baseline and follow-up scans, the locations of both PRESS volumes and OVS bands from both scans were still matched, with a Dice overlap ratio of 0.977. Also from (d) &(e), it is clear that the triple oblique prescription of PRESS volume was achieved.
Transformation of the predefined prescription
The PRESS volume and OVS band locations were transformed from the MNI152 atlas space to the subject space in each scan session. Firstly, the atlas T1-weighted image was affinely registered to the subject’s T1-weighted image on the scanner console using FSL FLIRT (FMRIB’s Linear Image Registration Tool)14 with 12 degrees of freedom. To accelerate computation, both T1-weighted images were down-sampled to an isotropic resolution of 2mm, and no skull stripping or image intensity corrections were applied. Once the registration was completed, the 4×4 output transformation matrix M was applied to each normal vector n⃗ = (nl,np,ns)T and point p⃗ = (pl,pp,ps)T defined in the atlas space in order to obtain their counterparts and in the subject space, using following two equations in the homogeneous coordinate system, respectively,
| (1) |
and
| (2) |
Because of shear, normal vectors calculated by (1) are in general no longer orthogonal. Therefore, a correction was made to recreate orthogonal vectors through the following operations
| (3) |
where × represents the vector product operator. Knowing the normal vectors, the equations that define the left, superior and posterior box planes can be formulated as follows
| (4) |
where · represents the inner product operation, , and are any points on the individual planes, and , and are fixed points transformed from P⃗L0 = dLn⃗L, P⃗P0 = dPn⃗P, and P⃗S0 = dSn⃗S, respectively, using the equation (2). Then the size of the transformed box was calculated as twice the distance from its center to each of the 3 orthogonal planes. Similarly, the equations that defined the center and inner planes of a transformed OVS band were formulated such that the thickness of the band was calculated as twice the distance between the two planes. In addition, by constructing a 3×3 rotation matrix , 3 rotation angles α, β, and γ of the box in the subject space were calculated.
The process was implemented and performed on the scanner console, with the output prescription being displayed on the background of the subject’s T1-weighted images for visual examination. If an unexpected prescription occurs, manual prescription can be performed to override the automated prescription. The MRSI sequence was tailored to accept parameters generated after the registration10, including the size, center and rotation angles of the box; the normal vector, thickness and distance from coordinate origin to the center plane of each OVS band.
Subjects and MR Imaging
Ten healthy male and ten female volunteers were recruited with similar age ranges: 27.8±5.1 years (mean±stdev) and 25.1±4.5 years, respectively. None of them had a history of neurological disorders and all of them signed an informed consent form approved by our Institutional Review Board before their MR scans.
All scans were performed on a GE Discovery 750 3T scanner (GE Healthcare, Waukesha, WI) with a 32-channel head coil (Nova Medical, Wilmington, MA). In each scan session, an axial 3D inversion recovery spoiled gradient recalled (IR-SPGR) whole brain T1-weighted image was acquired using TI/TE/TR = 450/2.5/7ms, flip angle = 12°, FOV = 25×25×18cm3 and resolution = 0.5×0.5×1.5mm3. This image volume defined the subject space, to which the atlas T1-weighted image was aligned and the predefined MRSI volume and OVS bands were transformed. High-order shimming was performed to reduce the degree of magnetic field inhomogeneity. Immediately after the field shimming, the 3D multi-voxel H-1 MRSI was acquired with PRESS volume selection, CHESS water suppression, phase encoding in two dimensions and flyback encoding in the superior-inferior direction15. The sequence parameters were as follows: TE/TR = 80/1800ms, phase encoding grid = 16×16×16, nominal resolution = 1×1×1cm3, total acquisition time = 7.8 minutes. An over prescription factor of PRESS = 1.2 was used to reduce chemical shift artifacts16. In each scan session, the MRSI was repeated without repositioning to evaluate the intra-session variation. To evaluate the inter-session variation in metabolite quantification, every volunteer was scanned twice in two sessions separated by one week. The study comprised a total of 80 scans, which were completed within 5 months, during which time there was no software or hardware upgrade on the scanner.
Data analysis
The reproducibility of the automated MRSI volume prescription was assessed by the Dice ratio17,
| (5) |
where A and B are volumes from two separate scans and A ∩ B is the volume covered by both. A ratio of 1 indicates a perfect overlap. The computation was implemented by first rigidly co-registering a subject’s follow-up T1-weighted image to its baseline one using FSL FLIRT14, followed by sampling at an isotropic 0.1mm interval inside the both baseline and aligned follow-up volumes7, and finally counting the number of samples in each individual volume and the number of samples in the both volumes. The reproducibility of the prescription of OVS bands was assessed by measuring the inter-scan differences in the band’s normal vector, thickness and distance (from the coordinate origin to the center plane) after transforming the follow-up OVS band prescription to the baseline image space.
The MRSI data were processed as described previously18 using in-house software packages19, which perform the following operations: zero filling, spectra apodization, phase/frequency correction, baseline subtraction, and calculation of peak heights and areas of N-acetyl aspartate (NAA), Choline (Cho), and Creatine (Cre).
The intra-subject reproducibility of the ratios of the peak heights and peak areas for NAA/Cre, Cho/Cre and NAA/Cho in a voxel were evaluated by the coefficient of variance (CoV), which is calculated as follows20:
| (6) |
where Rbase and Rfollow are ratios from the baseline and follow-up, respectively. The CoVs from all voxels of a subject were averaged, and the final results were reported as the group means and standard deviations calculated across subjects. To exclude regions with no metabolites or extremely low SNR, only voxels with ratios >0 for all 4 scans from each subject were included for the analysis. The CoVs were calculated for both intra-session scans and one set of inter-session scans (between the very first scans from both scan sessions). The CoVs from intra- and inter-session scans were compared using a paired t-test with the significant level being set as p < 0.05 after the Bonferroni correction for multiple comparisons.
Results
Reproducibility of the automated MRSI prescription
The automated MRSI prescription took only 50 seconds to complete on the scanner console and was successfully applied for all scans without any manual modifications. Visual inspection of the prescription found little difference in its anatomy coverage between the transformed and predefined PRESS volume/OVS bands Figure 2. The intra-subject Dice overlap ratio of the MRSI volume between the baseline and follow-up scans was 0.978±0.005; the average total shift of its center in all 3 orthogonal dimensions of the volume was 0.84±0.22mm; the average relative volume variation was 0.43%±0.43%, and the average variation in its 3 rotation angles relative to the physical space was less than 0.4 degree (Table 1a). For the 320 (16×20) OVS bands, the average angle between their baseline and follow-up (transformed to the baseline space) normal vectors was 0.86±0.82 degrees, and the average variations in their thickness and distance were 0.14±0.15mm and 0.11±0.10mm, respectively (Table 1b).
Table 1.
Reproducibility of the Automated MRSI Prescription
| (a) PRESS Volume
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dice Ratio | Center Shift1 (mm) | Rotation Angle Variation (Degree) | Size Variation (mm) | Relative Volume Variation2 (%) | ||||||||
| RL | AP | SI | Total3 | Alpha | Beta | Gamma | RL | AP | SI | |||
| Mean (N=20) | 0.978 | 0.49 | 0.34 | 0.46 | 0.84 | 0.37 | 0.16 | 0.19 | 0.20 | 0.24 | 0.14 | 0.43 |
| Standard Deviation | 0.005 | 0.29 | 0.26 | 0.22 | 0.22 | 0.26 | 0.14 | 0.16 | 0.16 | 0.18 | 0.14 | 0.43 |
| Min | 0.968 | 0.03 | 0.00 | 0.18 | 0.50 | 0.02 | 0.00 | 0.01 | 0.00 | 0.06 | 0.00 | 0.05 |
| Max | 0.989 | 0.90 | 0.94 | 1.08 | 1.35 | 0.83 | 0.58 | 0.68 | 0.77 | 0.79 | 0.58 | 1.89 |
| (b) OVS Bands
| |||
|---|---|---|---|
| Angle4 between Normal Vectors (Degree) | Distance5 Variation (mm) | Thickness Variation (mm) | |
| Mean (N=320) | 0.86 | 0.14 | 0.11 |
| Standard Deviation | 0.82 | 0.15 | 0.10 |
| Min | 0.00 | 0.00 | 0.00 |
| Max | 2.80 | 1.08 | 0.58 |
The shifts and variations in the table are absolute differences between the baseline and follow-up scans.
The relative volume variation is calculated as the absolute volume difference between the baseline and follow-up scans divided by the volume of the baseline.
The total variation in volume center is calculated by:
An angle of 0 degrees indicates a perfect alignment between the normal vectors of the baseline and follow-up (transformed to the baseline space) OVS bands.
Distance is measured from the coordinate origin to the center plane of an OVS Band.
Reproducibility of metabolite ratio from MRSI
The quality of metabolite spectra was sufficient for data analysis for all 80 scans, with the mean Cre SNR and linewidth being 12.3±1.4 and 7.0±0.4Hz, respectively. The appearance of spectra from two inter-session scans of each subject was similar, with an example shown in Figure 3. The mean inter-session CoVs of peak height ratios were 0.102, 0.124 and 0.115 for NAA/Cre, Cho/Cre and NAA/Cho, respectively. The CoVs of peak area ratio were 0.142, 0.153 and 0.163 for NAA/Cre, Cho/Cre and NAA/Cho, respectively (Table 2). On average each subject’s PRESS volume had 327 voxels, and 92% of them were included for analyzing the CoVs. When the analysis was performed separately for the top and bottom slices, the CoVs of both peak height and peak area ratios was approximately 20% smaller in the top slice and 20% larger in the bottom slice. Paired t-tests showed no significant difference in CoVs between intra- and inter-sessions after the Bonferroni correction for multiple comparison (Table 2). Visual inspection of spectra from different subjects showed similar metabolite intensities and spatial variations (See the supporting Figure S1).
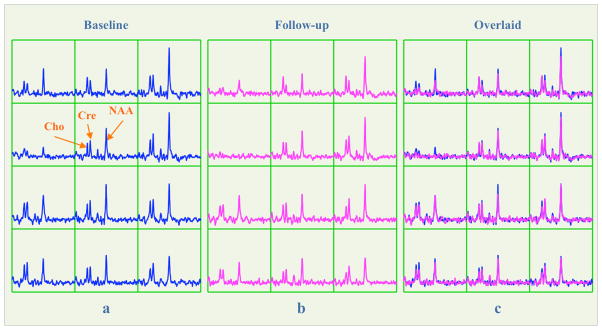
Figure 3.
(a) and (b) The baseline and follow-up spectra in the green grid on the coronal slices in Figure 2(b) and (c), respectively. (c) Overlay of the follow-up spectra on the top of the baseline ones. Note all spectra were displayed with the same intensity range, but the SNR and line width of Cre were similar between the two scans (Baseline/follow-up SNR: 13.4/12.2; Baseline/follow-up line width: 6.8/6.6Hz).
Table 2.
Reproducibility Evaluation for Metabolite Measurements from MRSI
| Ratio | Metabolite | Location | Inter-Session CoV1 | Intra-Session1 CoV | Intra-Session2 CoV | p value2 of paired t-test | |
|---|---|---|---|---|---|---|---|
| Inter vs. Intra-session1 | Inter vs. Intra-session2 | ||||||
| Peak Height | Whole Brain | 0.102±0.011 | 0.101±0.011 | 0.099±0.011 | 0.320 | 0.128 | |
| NAA/Cre | Top Slice | 0.081±0.010 | 0.078±0.009 | 0.078±0.009 | 0.142 | 0.212 | |
| Bottom Slice | 0.123±0.021 | 0.124±0.021 | 0.124±0.025 | 0.752 | 0.670 | ||
| Whole Brain | 0.124±0.015 | 0.118±0.013 | 0.118±0.011 | 0.037 | 0.014 | ||
| Cho/Cre | Top Slice | 0.102±0.013 | 0.095±0.011 | 0.096±0.010 | 0.031 | 0.040 | |
| Bottom Slice | 0.145±0.019 | 0.143±0.022 | 0.143±0.023 | 0.592 | 0.643 | ||
| Whole Brain | 0.115±0.010 | 0.111±0.012 | 0.110±0.012 | 0.027 | 0.005 | ||
| NAA/Cho | Top Slice | 0.089±0.009 | 0.086±0.010 | 0.086±0.012 | 0.108 | 0.233 | |
| Bottom Slice | 0.140±0.019 | 0.142±0.023 | 0.140±0.020 | 0.700 | 0.864 | ||
| Peak Area | Whole Brain | 0.142±0.015 | 0.134±0.028 | 0.139±0.0169 | 0.172 | 0.175 | |
| NAA/Cre | Top Slice | 0.116±0.015 | 0.115±0.014 | 0.115±0.015 | 0.691 | 0.727 | |
| Bottom Slice | 0.165±0.039 | 0.168±0.029 | 0.161±0.038 | 0.639 | 0.328 | ||
| Whole Brain | 0.153±0.018 | 0.149±0.015 | 0.146±0.016 | 0.156 | 0.046 | ||
| Cho/Cre | Top Slice | 0.127±0.018 | 0.123±0.012 | 0.123±0.013 | 0.171 | 0.146 | |
| Bottom Slice | 0.173±0.022 | 0.176±0.027 | 0.164±0.023 | 0.528 | 0.150 | ||
| Whole Brain | 0.163±0.017 | 0.153±0.046 | 0.162±0.024 | 0.338 | 0.841 | ||
| NAA/Cho | Top Slice | 0.131±0.014 | 0.127±0.014 | 0.131±0.019 | 0.380 | 0.987 | |
| Bottom Slice | 0.199±0.035 | 0.206±0.030 | 0.200±0.034 | 0.347 | 0.893 | ||
The CoV values are reported as mean ± standard deviation.
The p values are the ones before the Bonferroni correction, with the numbers in bold indicating the tests that showed significant differences before the correction.
Discussion
In this study we implemented an atlas-based fully automated 3D PRESS MRSI prescription method, which extends the previous implementation of automatic placement of OVS bands9–12 to include the PRESS volume. We achieved an intra-subject Dice overlap ratio with smaller variance than Ratai et al8 (97.8%±0.5% vs. 94.9%%±6.6%) for the PRESS volume. The intra-subject variation of parameters that determine the location of OVS bands was also reproducible. Our atlas-based method maintains a consistent prescription of the PRESS volume and OVS bands both within and between subjects, facilitating both intra- and inter-subject group analyses. This is an advantage compared with the segmentation-based prescription method10, which, as implemented, prescribes only a single oblique angle. The atlas-based method also supports a broader range of acquisitions including single voxel studies. In addition, our implementation provides a time efficient method (50 seconds) for transforming the predefined prescription from atlas to subject space by subsampling both the atlas and subject T1-weighted images. Being able to reduce the time interval between the completion of the T1-weighted imaging and the start of the MRSI acquisition is crucial, as it minimizes the likelihood of head movement.
To assess the reproducibility of the method, repeat MRSI acquisitions were preformed with (inter-session) and without (intra-session) repositioning. The reproducibility of the intra-session scans is likely to be the best achievable, as the only source of variation in this case would be system noise provided no subject motion occurs between the scans. The reproducibility of inter-session scans was influenced not only by the system noise but also the error from repositioning and physiological variations. Since there was no system upgrade and the inter-session interval was only a week, the system noise level should be similar and the physiological variations can be ignored, making the repositioning error the major source of variation for these scans. However, the paired comparisons showed no significant difference between inter- and intra-session CoVs, indicating that the variance introduced by repositioning was also small and could be ignored. Six comparisons showed slightly higher inter-session CoVs before correcting multiple comparisons. Interestingly, all these CoVs included measurements of Cho. This may be explained by a larger scan-to-scan variation in the level of Cho compared to others metabolites, which have been observed in previous studies4.
The CoVs of metabolite levels reported in previous studies3–5, 8, 21 varied widely, from less than 10% to more than 20%. This variation may be explained by differences in experimental designs, which included the scanner field strength, echo time, voxel size, metrics of metabolite level, and the regions that are included in the data analysis. In a similar study to ours, Ratai et al.8 obtained median concentration ratio CoVs of 11.6% for NAA/Cre and 13.8% Cho/Cre, using an automated selection volume prescription. Our measurements are comparable to theirs, but obtained in a more challenging setting. First, our 3D selection volume covered a larger region than their 2D volume, especially in the deep gray matter, which often has low SNR and poor shimming. Our study clearly shows that the reproducibility in this region is lower than that in gray or white matter located above the lateral ventricles. In addition, our analysis included 92% voxels in the selection volume, with the excluded ones being mainly in the ventricles where no relevant metabolite signals are available, while only 76% voxels were analyzed in the Ratai study. By including regions in deep grey matter, our assessment of reproducibility should be representative of values observed in regions of interest for studying neurodegenerative disorders such as multiple sclerosis22, Alzheimer’s23 and Parkinson’s24 diseases.
There were several limitations in our study that should be mentioned. First, the success of prospective registration requires there to be no motion during the interval between T1-weighted imaging and MRSI. Second, we did not attempt evaluate the accuracy of the atlas-based prescription in pathological situations where dramatic tissue displacements may occur between examinations. Finally, our study focused on the intra-subject reproducibility and did not attempt to examine inter-subject biological variability. This would require a more extensive study on a broader population.
Conclusions
The proposed atlas-based MRSI prescription method provides a time efficient and reproducible prescription for both selection volume and OVS bands, which results in robust estimation of metabolite levels.
Supplementary Material
Inter-subject reproducibility of the placement of MRSI PRESS volume/OVS bands and spectra. (a) and (b) The PRESS volume and OVS bands transformed to the spaces of subject 1 and 2 (who had a similar head size, hence a similar size of PRESS volume), respectively, in their baseline scans. Similar to Figure 2, the 3 orthogonal background image slices were chosen such that they all transverse the center of the PRESS volume box in (a) and (b). (c) and (d) Spectra of subject 1 and 2 in the green grid on the axial slices in (a) and (b), respectively. Good inter-subject reproducibility was achieved for the two subjects, which is evident by the visual inspection of the similarity between: 1) the anatomy covered by the OVS bands, PRESS volume, and voxels in the grid; 2) the spatial variation of spectra, e.g., the decrease of absolute metabolite intensity from the right to the left columns; higher Cre and lower Cho levels in cortical gray matter (voxels in the right column) vs. lower Cre and higher Cho levels in cortical white matter (voxels in the middle column), which is in line with the observation from a previous study.
Acknowledgments
The authors would like to thank Dr. Eugene Ozhinsky for his insightful discussion of this study and the participation of all volunteers. This study was supported by the funding from National Institutes of Health NIHR01 CA127612 and the National Center for Advancing Translational Sciences, National Institutes of Health, through UCSF-CTSI Grant Number UL1 TR000004.
References
- 1.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–48. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 2.Oz G, Alger JR, Barker PB, et al. Clinical proton MR spectroscopy in central nervous system disorders. Radiology. 2014;270:658–79. doi: 10.1148/radiol.13130531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li BS, Babb JS, Soher BJ, Maudsley AA, Gonen O. Reproducibility of 3D proton spectroscopy in the human brain. Magn Reson Med. 2002;47:439–46. doi: 10.1002/mrm.10081. [DOI] [PubMed] [Google Scholar]
- 4.Maudsley AA, Domenig C, Sheriff S. Reproducibility of serial whole-brain MR spectroscopic imaging. NMR Biomed. 2010;23:251–6. doi: 10.1002/nbm.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Langer DL, Rakaric P, Kirilova A, Jaffray DA, Damyanovich AZ. Assessment of metabolite quantitation reproducibility in serial 3D-(1)H-MR spectroscopic imaging of human brain using stereotactic repositioning. Magn Reson Med. 2007;58:666–73. doi: 10.1002/mrm.21351. [DOI] [PubMed] [Google Scholar]
- 6.Van Cauter S, Sima DM, Luts J, et al. Reproducibility of rapid short echo time CSI at 3 tesla for clinical applications. J Magn Reson Imaging. 2013;37:445–56. doi: 10.1002/jmri.23820. [DOI] [PubMed] [Google Scholar]
- 7.Hancu I, Blezek DJ, Dumoulin MC. Automatic repositioning of single voxels in longitudinal 1H MRS studies. NMR Biomed. 2005;18:352–61. doi: 10.1002/nbm.965. [DOI] [PubMed] [Google Scholar]
- 8.Ratai EM, Hancu I, Blezek DJ, Turk KW, Halpern E, Gonzalez RG. Automatic repositioning of MRSI voxels in longitudinal studies: impact on reproducibility of metabolite concentration measurements. J Magn Reson Imaging. 2008;27:1188–93. doi: 10.1002/jmri.21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yung KT, Zheng W, Zhao C, Martinez-Ramon M, van der Kouwe A, Posse S. Atlas-based automated positioning of outer volume suppression slices in short-echo time 3D MR spectroscopic imaging of the human brain. Magn Reson Med. 2011;66:911–22. doi: 10.1002/mrm.22887. [DOI] [PubMed] [Google Scholar]
- 10.Ozhinsky E, Vigneron DB, Chang SM, Nelson SJ. Automated prescription of oblique brain 3D magnetic resonance spectroscopic imaging. Magn Reson Med. 2013;69:920–30. doi: 10.1002/mrm.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozhinsky E, Vigneron DB, Nelson SJ. Improved spatial coverage for brain 3D PRESS MRSI by automatic placement of outer-volume suppression saturation bands. J Magn Reson Imaging. 2011;33:792–802. doi: 10.1002/jmri.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Ramon M, Gallardo-Antolin A, Cid-Sueiro J, et al. Automatic Placement of Outer Volume Suppression Slices in MR Spectroscopic Imaging of the Human Brain. Magnetic Resonance in Medicine. 2010;63:592–600. doi: 10.1002/mrm.22275. [DOI] [PubMed] [Google Scholar]
- 13.Grabner G, Janke AL, Budge MM, Smith D, Pruessner J, Collins DL. Symmetric atlasing and model based segmentation: an application to the hippocampus in older adults. Med Image Comput Comput Assist Interv. 2006;9:58–66. doi: 10.1007/11866763_8. [DOI] [PubMed] [Google Scholar]
- 14.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–41. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 15.Cunningham CH, Vigneron DB, Chen AP, et al. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54:1286–9. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Osorio JA, Ozturk-Isik E, et al. Considerations in applying 3D PRESS H-1 brain MRSI with an eight-channel phased-array coil at 3 T. Magn Reson Imaging. 2006;24:1295–302. doi: 10.1016/j.mri.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 17.Dice LR. Measures of the Amount of Ecologic Association between Species. Ecology. 1945;26:297–302. [Google Scholar]
- 18.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46:228–39. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 19.Crane JC, Olson MP, Nelson SJ. SIVIC: Open-Source, Standards-Based Software for DICOM MR Spectroscopy Workflows. Int J Biomed Imaging. 2013;2013:169526. doi: 10.1155/2013/169526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu P, Xu F, Lu H. Test-retest reproducibility of a rapid method to measure brain oxygen metabolism. Magn Reson Med. 2013;69:675–81. doi: 10.1002/mrm.24295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tedeschi G, Bertolino A, Campbell G, et al. Reproducibility of proton MR spectroscopic imaging findings. AJNR Am J Neuroradiol. 1996;17:1871–9. [PMC free article] [PubMed] [Google Scholar]
- 22.Geurts JJ, Reuling IE, Vrenken H, et al. MR spectroscopic evidence for thalamic and hippocampal, but not cortical, damage in multiple sclerosis. Magn Reson Med. 2006;55:478–83. doi: 10.1002/mrm.20792. [DOI] [PubMed] [Google Scholar]
- 23.Foy CM, Daly EM, Glover A, et al. Hippocampal proton MR spectroscopy in early Alzheimer’s disease and mild cognitive impairment. Brain Topogr. 2011;24:316–22. doi: 10.1007/s10548-011-0170-5. [DOI] [PubMed] [Google Scholar]
- 24.Groger A, Chadzynski G, Godau J, Berg D, Klose U. Three-dimensional magnetic resonance spectroscopic imaging in the substantia nigra of healthy controls and patients with Parkinson’s disease. Eur Radiol. 2011;21:1962–9. doi: 10.1007/s00330-011-2123-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inter-subject reproducibility of the placement of MRSI PRESS volume/OVS bands and spectra. (a) and (b) The PRESS volume and OVS bands transformed to the spaces of subject 1 and 2 (who had a similar head size, hence a similar size of PRESS volume), respectively, in their baseline scans. Similar to Figure 2, the 3 orthogonal background image slices were chosen such that they all transverse the center of the PRESS volume box in (a) and (b). (c) and (d) Spectra of subject 1 and 2 in the green grid on the axial slices in (a) and (b), respectively. Good inter-subject reproducibility was achieved for the two subjects, which is evident by the visual inspection of the similarity between: 1) the anatomy covered by the OVS bands, PRESS volume, and voxels in the grid; 2) the spatial variation of spectra, e.g., the decrease of absolute metabolite intensity from the right to the left columns; higher Cre and lower Cho levels in cortical gray matter (voxels in the right column) vs. lower Cre and higher Cho levels in cortical white matter (voxels in the middle column), which is in line with the observation from a previous study.