Summary
RNA viruses pose a threat to public health that is exacerbated by the dearth of antiviral therapeutics. The RNA-dependent RNA polymerase (RdRp) holds promise as a broad-spectrum, therapeutic target because of the conserved nature of the nucleotide-substrate-binding and catalytic sites. Conventional, quantitative, kinetic analysis of antiviral ribonucleotides monitors one or a few incorporation events. Here, we use a high-throughput magnetic tweezers platform to monitor the elongation dynamics of a prototypical RdRp over thousands of nucleotide-addition cycles in the absence and presence of a suite of nucleotide analog inhibitors. We observe multiple RdRp-RNA elongation complexes; only a subset of which are competent for analog utilization. Incorporation of a pyrazine-carboxamide nucleotide analog, T-1106, leads to RdRp backtracking. This analysis reveals a mechanism of action for this antiviral ribonucleotide that is corroborated by cellular studies. We propose that induced backtracking represents a distinct mechanistic class of antiviral ribonucleotides.
Keywords: RNA virus, RNA polymerase, magnetic tweezers, inhibitor, nucleoside analog, backtracking, pyrazine carboxamide, T-1106, T-705
Graphical Abstract
Highlights
-
•
Several unique conformational states of an elongating RdRp exist
-
•
Only one conformation incorporates nucleotide analogs with therapeutic potential
-
•
An analog thought to be a chain terminator actually promotes RdRp backtracking
-
•
Distinctive behavior of backtrack-inducing analog on virus variants in cell culture
Dulin et al. find that a prototypical RNA-dependent RNA polymerase (RdRp) visits several states during nucleotide synthesis, of which only one incorporates nucleotide analogs with therapeutic potential. Different analogs exhibit distinct kinetic signatures, with an analog thought to induce chain termination actually promoting RdRp backtracking.
Introduction
RNA viruses represent a threat to global public health, and this has been confirmed time and time again over the past 20 years. During this period, the world has witnessed outbreaks of West Nile virus, severe acute respiratory syndrome coronavirus, chikungunya virus, Middle East respiratory syndrome coronavirus, and currently Zika virus. These events highlight our inability to predict the next outbreak as well as our inability to treat or prevent infections once the outbreak is underway (Bekerman and Einav, 2015, Narasimhan, 2014). Antiviral therapeutics with broad-spectrum activity would greatly improve our readiness for the inevitable outbreaks of the future, but such therapeutics are few, at best.
All RNA viruses encode an RNA-dependent RNA polymerase (RdRp) to produce viral mRNA and progeny genomes (Ng et al., 2008). While the mechanisms for initiation can vary (Ng et al., 2008), once initiated, all RdRps transfer a ribonucleotide, selected in a template-dependent manner, to the 3′ end of a primer. Consistent with this conserved mechanism is the conserved nature of the RdRp active site (Ng et al., 2008). This conservation makes the viral RdRp a very attractive target for developing broad-spectrum antiviral therapeutics. Indeed, ribavirin, a nucleoside analog, is one of the few antiviral therapeutics with broad-spectrum activity. For more than 30 years, however, the mechanism of action was thought to be inhibition of a cellular enzyme involved in purine nucleotide biosynthesis (Streeter et al., 1973), while it is now known that the antiviral activity is mediated by the RdRp in the case of poliovirus (PV) and likely other RNA viruses (Crotty et al., 2000, Crotty et al., 2001).
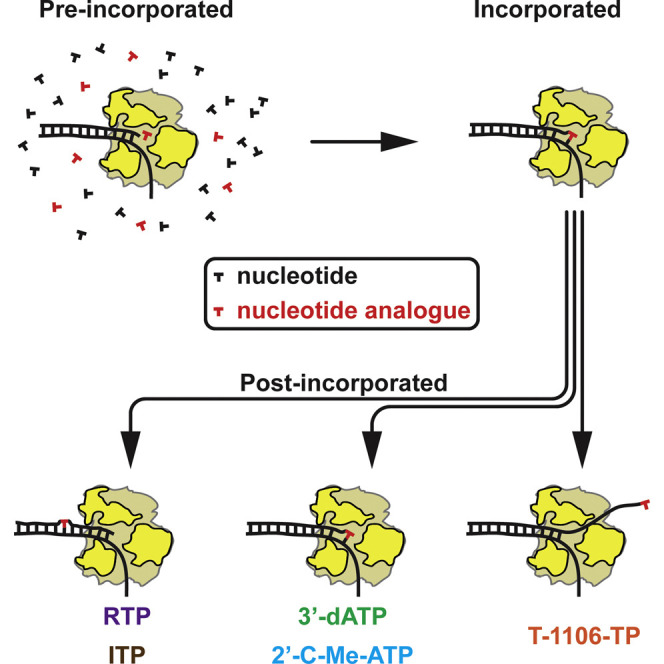
Since the mechanism of ribavirin action was discovered, efforts to develop antiviral nucleosides against myriad RNA viruses have ensued (Deval et al., 2014). To date, antiviral nucleosides fall into three categories: mutagen, obligate chain terminators, and non-obligate chain terminators. Ribavirin is an example of a mutagen because its triazole carboxamide pseudobase can hydrogen bond to both uracil and cytosine, leading to ambiguous incorporation (Crotty et al., 2000). Cordycepin is the classic obligate chain terminator. This adenosine analog contains a 3′-deoxyribose, thus precluding extension once incorporated (Maale et al., 1975). Non-obligate chain terminators produced to date have a natural base and a 3′-hydroxyl moiety on the sugar but have a substituent appended to the ribose ring that interferes with a post-incorporation translocation event by some still-unknown mechanism (Carroll et al., 2003, Sofia et al., 2010). Among the first member of this class is the 2′-C-methyl substituent (Carroll et al., 2003). This substituent is a critical component of the anti-hepatitis C virus drug sofosbuvir (Sofia et al., 2010).
Recently, a new class of antiviral base/ribonucleoside has been approved for use in the treatment of influenza virus infection (Furuta et al., 2009). These drugs contain a pyrazine-carboxamide pseudobase, which is similar to the pseudobase of ribavirin in that both contain a rotationally mobile carboxamide moiety as a determinant for base-pairing. The approved drug is just the pseudobase and has been named favipiravir (also known as T-705). Favipiravir and a related nucleoside, T-1106, exhibit broad-spectrum activity (Furuta et al., 2009). However, the mechanism of action of this class of antiviral agents is unclear. Some have suggested that it is a non-obligate chain terminator (Sangawa et al., 2013), others have suggested that it is a mutagen (de Ávila et al., 2016, de Avila et al., 2017), and others have suggested that it is both a non-obligate chain terminator and a mutagen (Jin et al., 2013b).
The inability to define the mechanism of action of favipiravir reflects the lack of tools available to study nucleic acid polymerases and their inhibitors. Experiments employing long templates that reveal inhibition cannot distinguish among nucleotide competition, decreased elongation efficiency, and chain termination (Jin et al., 2013b). Experiments employing short templates that reveal specificity and efficiency of addition do not offer varied sequence context that could be essential to observe perturbed elongation efficiency or even chain termination (Sangawa et al., 2013). We have recently developed a multiplexed magnetic tweezers apparatus in which hundreds of magnetic beads are tethered to a flow chamber surface by individual double-stranded RNA (dsRNA) molecules (Cnossen et al., 2014, Dulin et al., 2015a, Dulin et al., 2015c, Berghuis et al., 2015), all of which experience the same force (Yu et al., 2014, De Vlaminck et al., 2012). This high-throughput instrument enables probing of rare events that interrupt polymerase kinetics (e.g., error or nucleotide analog incorporation). This approach was established using the RdRp from bacteriophage Φ6 RdRp (Dulin et al., 2015c), where we followed the activity of tens of polymerases on RNA templates thousands of nucleotides in length and provided sufficient statistics to quantitatively examine nucleotide (analog) incorporation in the presence of all four natural nucleotides. This provides the ability to assess not only the consequences of nucleotide analog incorporation but also its mechanistic basis.
The RdRp from PV is the most extensively characterized human RNA virus polymerase known (Cameron et al., 2016) and was used to elucidate the mechanism of action of ribavirin (Crotty et al., 2000). Here, we apply our high-throughput magnetic tweezers approach to the PV RdRp with the goal of obtaining insight into the mechanism of action of the pyrazine-carboxamide class of antiviral therapeutics. The basic mechanics of nucleotide-addition observed for the Φ6 RdRp is consistent with our PV RdRp data. We exploited the existence of a mutator PV RdRp to prove the existence of a low-fidelity elongation complex and examine its dynamics. While prokaryotic and nuclear RNA polymerases have a tendency to backtrack when forward motion is obstructed (e.g., by binding of inhibitors), incorporation of neither class of chain terminator induced backtracking. In contrast, incorporation of a pyrazine-carboxamide nucleotide caused backtracking of the PV RdRp, an event from which the enzyme was able to recover given sufficient time. Ensemble experiments showed that this backtracking behavior manifests as inhibition and that incorporation exhibited a very relaxed template specificity. Studies in cells were consistent with this unique mechanism of action, as PV mutants exhibiting enhanced or diminished sensitivity to mutagens and chain terminators did not exhibit a corresponding change in sensitivity to the pyrazine-carboxamide nucleotide. We conclude that the active pyrazine-carboxamide nucleoside triphosphate metabolite defines a distinct mechanistic class of antiviral (deoxy)ribonucleotides.
Results
Interpreting Pauses of Polymerase Activity Observed in High-Throughput Magnetic Tweezers Experiments Using PV RdRp and a Mutator Derivative
Since the elucidation of the first kinetic mechanism for a nucleic acid polymerase nearly 30 years ago (Kuchta et al., 1987, Kuchta et al., 1988), the approaches used to interrogate the kinetics, mechanism, and fidelity of nucleic acid polymerases have remained essentially the same. A single cycle of nucleotide addition is monitored. When processive incorporation is monitored, quantitative analysis is only possible when measuring ten or fewer incorporation events. How ensemble measurements made on a short primed template, and corresponding inferences, inform the real-world scenario in which thousands or even millions of incorporation events must occur remains to be determined. Here, we study polymerase kinetics by monitoring elongation for thousands of nucleotide-incorporation events with near-single-nucleotide resolution in relative position. (As absolute position is established at lower resolution, we do not report on the sequence context.) The initial study employing this method used the RdRp from bacteriophage Φ6, an enzyme for which extensive kinetic data and corresponding mechanistic models had been lacking (Dulin et al., 2015c).
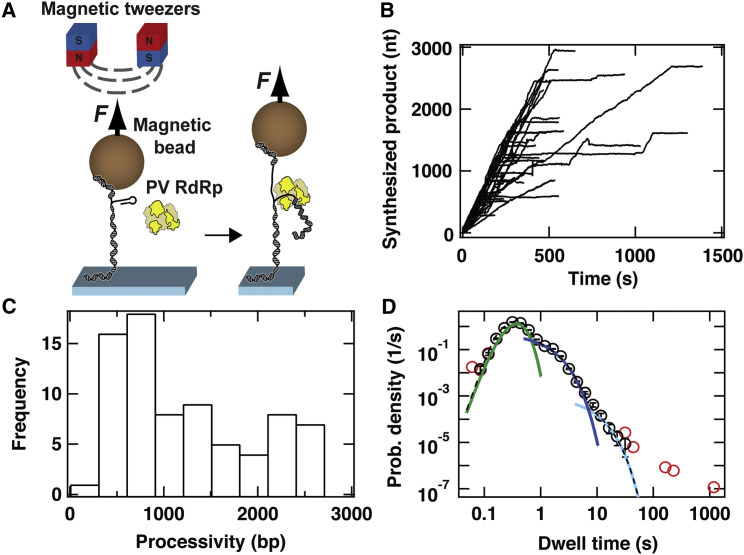
Much of what is known about the kinetics, mechanism, and fidelity of viral RdRps has been learned from studies of picornaviral RdRps, in particular PV RdRp (Cameron et al., 2016). The PV enzyme therefore permits comparisons between the magnetic tweezers experiment and classical approaches. In addition, PV RdRp derivatives exist to permit kinetic and mechanistic parameters determined by using the magnetic tweezers experiment to be correlated to rate and equilibrium constants obtained by using classical approaches. The experimental design is shown in Figure 1 . Following successful initiation on a short hairpin located at the 3′ end of the template strand of the dsRNA tether, RNA synthesis by PV RdRp displaces the templating strand from the tethering strand, lengthening the bead-tethered RNA because of its conversion from dsRNA to single-stranded RNA (ssRNA) (Figure 1A) (Dulin et al., 2015c). Single, elongating molecules of PV RdRp examined in high-throughput mode (Figure S1A) exhibited stochastic dynamics in which bursts of nucleotide addition were interrupted by pauses lasting <10 s (Figure 1B). From these time courses, we first extract coarse-grained information pertaining to the processivity of PV RdRp on the dsRNA template. The median processivity was found to equal 1,390 ± 160 bp (Figure 1C; unless mentioned otherwise, see Experimental Procedures for descriptions of error determination) at a saturating concentration of nucleotide triphosphates (NTPs; 1 mM of each NTP). Second, we collect more fine-grained information that allows us to extract mechanistic insight from the dataset by recording the dwell times of PV RdRp in consecutive windows of 5-nt-addition cycles (Dulin et al., 2015a, Dulin et al., 2015c).
Figure 1.
Characterizing PV RdRp Activity Using High-Throughput Magnetic Tweezers
(A) Schematic of the experimental assay to monitor RNA synthesis. F represents the constant force applied to the (predominantly) dsRNA tether. Apart from a short hairpin to the 3′ end of the 2.8-kb template strand to enable primed initiation by PV RdR, the dsRNA tether is as described in Dulin et al. (2015c). During RNA synthesis, the RdRp displaces the template strand from the tethering strand, increasing the end-to-end distance of the magnetic bead from the coverslip surface.
(B) 40 time courses of individual RdRp-elongation complexes acquired in a single experiment at 30 pN applied force and 100 μM NTP concentration. The traces are acquired at an acquisition frequency of 25 Hz and are subsequently low-pass filtered at 0.5 Hz and synchronized, resulting in the representation shown. Collectively, these traces illustrate the variation in the dynamics and processivity of PV RdRp, with the leftmost traces including almost no pauses and the rightmost traces including many short pauses.
(C) Histogram of the processivity of 77 PV RdRps under the conditions described in (B).
(D) The dwell-time distribution is extracted from 77 time courses of RNA synthesis by PV RdRp acquired at 30 pN applied force and 100 μM NTP concentration. The red circles represent all the data, whereas the black circles represent the data that remain after filtering out the shortest and longest dwell times (0.05% of the total data collected). We fit this distribution to a stochastic-pausing model (Supplemental Experimental Procedures) using MLE (dashed black line). For clarity, we individually plot each contribution to the dwell-time distribution: the gamma distribution (green), the first short pause (pause 1; dark blue), and the second short pause (pause 2; light blue). Error bars are determined as described in Experimental Procedures.
We fit a general stochastic-pausing model to the data using maximum-likelihood estimation (MLE) (Figures S1B and S1C; Supplemental Experimental Procedures) and illustrate the results by showing empirical dwell-time distributions together with the fits (Figure 1D). Three distinct kinetic behaviors were observed in the dwell-time analysis: a fast phase of sub-second duration and two slow phases of durations ∼1 s (termed pause 1) and ∼5 s (termed pause 2) (Figure 1D). The component of the distribution in the time regime less than 1 s is dominated by polymerases synthesizing RNA through the dwell-time window without pausing. In the absence of noise, we expect this portion of the curve to be described by a gamma distribution of an order equal to the number of nucleotides in a dwell-time window (5 nt in this case). Our fits do not consistently yield a number of sub-steps agreeing with the 5-nt dwell-time window; this suggests that noise influences the short dwell times, preventing us from reporting the nucleotide addition rate. For longer dwell times, noise can be suppressed by filtering, and we can determine the fraction of dwell times where RNA synthesis was momentarily paused. We can extract several useful parameters relating to the pause dynamics: the apparent pause exit rates of pauses 1 and 2 ( and , respectively) and the pause probabilities ( and , respectively). At saturating nucleotide concentrations (100 μM per NTP), we find that elongating polymerases enter the fast phase with a probability of 83.0% (Figure 1D; calculated using the integral under the green solid line as detailed in Supplemental Experimental Procedures), enter pause 1 with a probability of 16.7% (Figure 1D; calculated using the integral under the dark blue solid line), and enter pause 2 with a probability of 0.3% (Figure 1D; calculated using the integral under the light blue solid line).
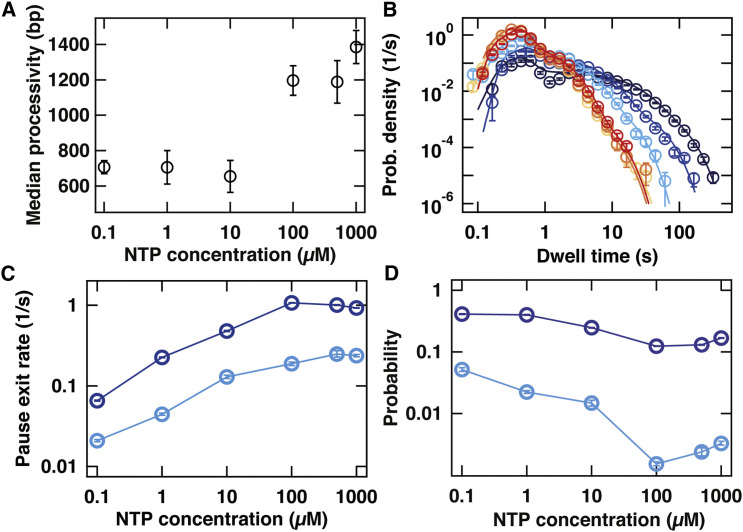
To obtain a baseline understanding of PV dynamics, we examine the nucleotide concentration dependence of the overall processivity and the dwell-time distributions. As expected, values of the median processivity decrease with decreasing nucleotide concentration (Figure 2 A). The largest effect of decreasing nucleotide concentration on the dwell-time distribution is observed in the time regime greater than 10 s (Figure 2B). Pause exit rate is directly correlated with nucleotide concentration, ranging from 0.9/s to 0.06/s (Figure 2C, dark blue points). Probability is inversely correlated with nucleotide concentration, ranging from 17% to 41% (Figure 2D, dark blue points). Pause exit rate also correlated directly with nucleotide concentration, ranging from 0.2/s to 0.02/s. (Figure 2C, light blue points). Probability also correlated inversely with nucleotide concentration, ranging from 0.3% to 5% (Figure 2D, light blue points). As the probability of PV RdRp to engage in only nucleotide addition equals one minus the sum of and , this approximates unity at saturating concentrations.
Figure 2.
Influence of Nucleotide Concentration on the Pausing Behavior of PV RdRp
(A) Median processivity of PV RdRp at 30 pN applied force for different concentrations of NTPs in the reaction.
(B) The dwell-time distributions (circles) of PV RdRp activity at 30 pN applied force and 1 mM (red), 500 μM (orange), 100 μM (light orange), 10 μM (cyan), 1 μM (blue), and 0.1 μM (dark blue) nucleotide concentrations. The solid lines are the MLE fits to a scenario where rapid elongation competes with two long-lived pause states, as illustrated in Figure 3G. The kinetic parameters extracted from these fits are shown in (C) and (D).
(C) Exit rates out of pause 1 (, dark blue) and pause 2 (, light blue) as a function of NTP concentration. As the NTP concentration is decreased from 1 mM to 0.1 μM, decreases from to and decreases from to .
(D) Probabilities of finding PV RdRp in pause 1 (, dark blue) or pause 2 (, light blue) as a function of NTP concentration. As the NTP concentration is decreased from 1 mM to 0.1 μM, increases from to and increases from to .
In all panels, error bars are determined as described in Experimental Procedures.
Since pause 1 is quite probable, we cannot rule out that the fitted exit rate simply inherits its dependence on nucleotide concentration (Figure 2C) from the facts that repeated entries into the pause would extend its apparent lifetime and the probability of repeated entry is determined through a kinetic competition between the pause entry rate and a nucleotide-dependent catalytic step. Since pause 2 will only have been entered multiple times for a few percent of the scored long pauses (Figure 2D), repeated pause entry cannot affect the order of magnitude change seen in the exit rate of pause 2 (Figure 2C).
We consider whether nucleotide misincorporation by PV underlies the observed pausing behavior. Misincorporation of nucleotides by PV RdRp in vitro and in cells occurs at a frequency no greater than 0.0001 per nucleotide-incorporation event at rates much less than 0.1/s (Arnold and Cameron, 2004). Therefore, it is impossible to conclude that pause 1 is the result of misincorporation. A state capable of correct nucleotide incorporation, but with low efficiency, is most probable. If pause 2 is a signature of nucleotide misincorporation (Dulin et al., 2015c), then one must account for the higher frequency and rate observed here relative to classical methods (Arnold and Cameron, 2004). However, such methods generally only evaluate utilization of a single, incorrect nucleotide (Arnold and Cameron, 2004), and the experimental configuration employed differs in the use of a dsRNA template under applied force. To explore the potential impact of these differences, we examined the dependence of PV kinetics on applied force (Figure S2A). When the applied force is lowered, the exit rates and both decrease by a factor of ∼2 (Figure S2B, dark blue and light blue points), showing that our experimental configuration impacts the measured rates. However, the probabilities to enter each state are maintained constant over the experimentally accessible force range (Figure S2C, dark and light blue points).
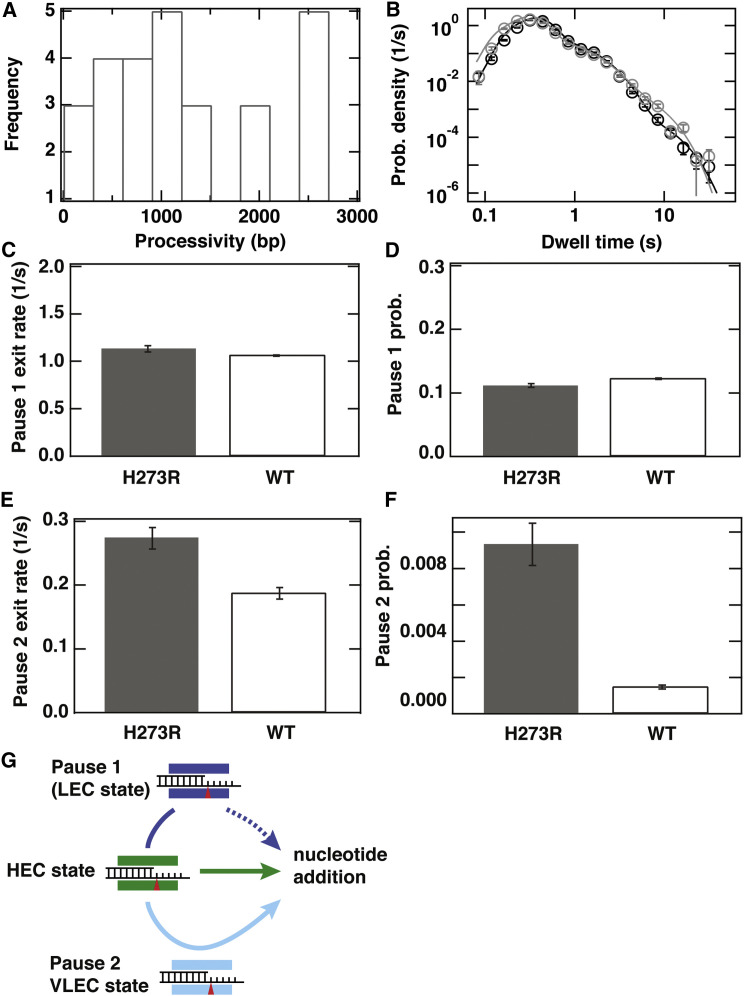
To rule in or rule out nucleotide misincorporation as a contributor to pause 1 and/or pause 2, we exploited the existence of a PV derivative with a mutator phenotype (Korboukh et al., 2014). The H273R PV RdRp increases the frequency of all types of mutations by relaxing one of the fidelity checkpoints used by PV RdRp (Korboukh et al., 2014). When examined at the single-molecule level, H273R PV RdRp exhibited a processivity of bp (Figure 3 A), unaltered within experimental error compared to wild-type (WT) PV RdRp. We also examined the corresponding dwell-time distributions (Figure 3B), reasoning that an increased pause probability would be observed if the pause reflected, even in part, nucleotide misincorporation. When the H273R PV RdRp derivative was employed, it appeared that the parameters related to pause 1 (Figures 3C and 3D) do not change relative to WT. The probability increased by ∼6.3-fold (Figure 3F); a 3- to 5-fold increase in mutation frequency was observed for H273R PV RdRp using classical methods and in cells (Korboukh et al., 2014). The exit rate increased by ∼50% (Figure 3E); a 20-fold increase in G:U mispair extension is observed for H273R PV RdRp using classical methods (Moustafa et al., 2014). These observations with H273R PV RdRp point to nucleotide misincorporation as the origin of pause 2 during the elongation reaction observed in the magnetic tweezers experiments. If this is the case, then the magnetic tweezers data suggest that the frequency of mutation may be underestimated by classical methods, perhaps a reflection of evaluating one nucleotide at a time (see probability at concentrations ≥100 μM in Figure 2D). In addition, the magnetic tweezers data reveal a higher probability of misincorporation at lower nucleotide concentration (see probability at concentrations ≤100 μM in Figure 2D). Based on these findings, we construct a working kinetic model for elongation by PV RdRp that invokes a high-efficiency state favoring nucleotide addition and two paused states, of which at least the longest-lived state is elongation competent, albeit error-prone (Figure 3G; Discussion).
Figure 3.
A Mutator PV RdRp (H273R) Displays Altered Pausing Behavior
(A) Histogram of the processivities of H273R PV RdRp at 30 pN applied force and 100 μM NTP concentration.
(B) Dwell-time distribution for H273R PV RdRp (gray) compared to WT PV RdRp (black). The solid lines are the MLE fits to a scenario where rapid elongation competes with two long-lived pause states, as illustrated in (G). The kinetic parameters extracted from these fits are shown in (C)–(F).
(C) Exit rate for H273R PV RdRp (gray) compared to WT PV RdRp (white rectangle with black border). (D) Probability for H273R (gray) compared to WT (white rectangle with black border).
(E) Exit rate for H273R PV RdRp (gray, ) compared to WT PV RdRp (white rectangle with black border, ).
(F) Probability for H273R (gray, ) compared to WT (white rectangle with black border, ).
(G) Schematic illustration of the kinetic pathway proposed to underlie elongation by PV RdRp. In this model, the incorporation of nucleotides can occur via one of several catalytically competent pathways. (Center, green) a principal pathway emanating from a high-efficiency catalytic (HEC) state in which PV RdRp adds nucleotides at a high rate with high fidelity. Presence in the HEC state is associated with bursts of nucleotide addition. (Top, dark blue) A second pathway emanating from a low-efficiency catalytic (LEC) state, in which PV RdRp adds nucleotides at a reduced rate. Presence in the LEC is associated with pause 1. The possibility that the fitted exit rate inherits its nucleotide dependence from repeated entry into pause 1 competing with a nucleotide-dependent catalytic step as shown in the center is indicated by the dashed arrow. (Bottom) A third pathway emanating from an error-prone, very low-efficiency catalytic (VLEC) state. Presence in the VLC is associated with pause 2.
In (B)–(F), error bars are determined as described in Experimental Procedures.
Lessons on RdRp Misincorporation and Mismatch Bypass Learned from Magnetic Tweezers Experiments Performed Using Imbalanced Nucleotide Pools
Classical methods studying polymerase error frequencies do so in the absence of other nucleotides. Our ability to link pause 2, at least in part, to nucleotide misincorporation provides a unique opportunity to study polymerase error frequencies in the presence of all four nucleotides. Here, we determine the impact of nucleotide-pool bias on the misincorporation frequency of PV RdRp and the extent to which a misincorporation event can be bypassed by the PV RdRp. In the control experiment, all four nucleotides were present at equimolar concentrations of 100 μM, for which we observed a median processivity of bp (Figure 2A) and a probability of % (Figure 2D). Subsequent experiments elevated the concentration of one of the four nucleotides by 10-fold to 1 mM (Figure S3). We find no change in the median processivity when cytidine triphosphate (CTP) is in excess bp; Figure S3A), whereas the presence of adenosine-, uridine-, or guanosine triphosphate (ATP, UTP, or GTP, respectively) in excess results in substantial decreases in the median processivity by ∼24%, ∼30%, and ∼45% to , bp, and bp, respectively; Figure S3A). Potentially, following misincorporation of cytidine monophosphate (CMP), PV RdRp is able to recover and continue elongation as before; conversely, following misincorporation of adenosine-, uridine-, or guanosine monophosphate (AMP, UMP, or GMP, respectively), PV RdRp may have more difficulty bypassing these mispairs, an increased tendency to stall, or an increased propensity to dissociate from the template. Analysis of the dwell-time distributions (Figure S3B) shows that the parameters of pause 1 were slightly altered by an imbalanced nucleotide pool, with the value of being consistently lowered by 10%–40% for all NTPs with the exception of ATP (Figure S3C) and a slight increase of probability under all conditions (Figure S3D). The changes in probability were found to be more significant and ranged from no change for ATP or a moderate 2- to 3-fold change for UTP and GTP to a substantial 6-fold change for CTP (Figure S3F). The frequency of each nucleotide in the template is essentially equal; therefore, these differences cannot be attributed to templating bias. The misincorporation events caused by the skewed nucleotide pool were not all bypassed equivalently; exit rate for GTP was significantly reduced (Figure S3E).
Characterization of a Lethal Mutagen, Obligate Chain Terminator, and a Non-obligate Chain Terminator Using Magnetic Tweezers
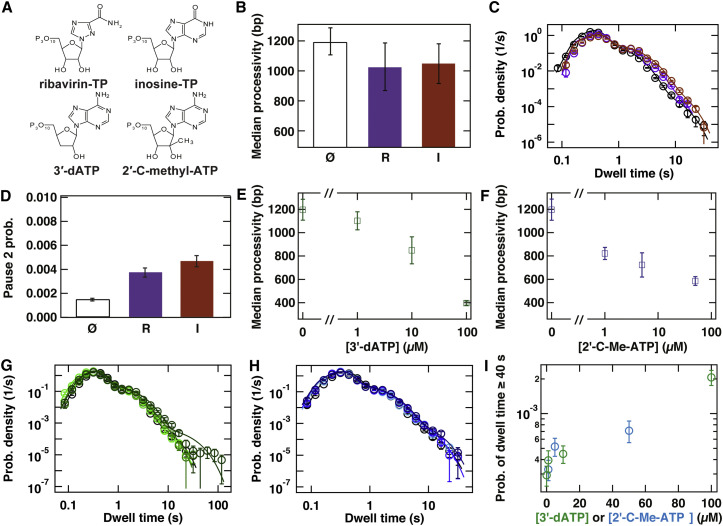
We next characterize the dynamic response of PV RdRp to two nucleotide analogs known to be substrates for a variety of RdRps, including PV RdRp, inosine triphosphate (ITP), and ribavirin triphosphate (RTP) (Figure 4 A). ITP is known to increase for bacteriophage Φ6 RdRp (Dulin et al., 2015c), and RTP has been described as a mutagen because of its ability to template by both uridine and cytidine (Crotty et al., 2000, Crotty et al., 2001). To assess how PV RdRp dynamics change in the presence of these nucleotide analogs, we compare experiments at saturating NTP concentrations (100 μM per NTP) to experiments at these NTP concentrations supplemented with 1 mM of either ITP or RTP). In the latter case, we observe that median values for processivity are not significantly affected (Figure 4B). The corresponding dwell-time distributions are shown in Figure 4C; analysis thereof shows that the parameters associated with pause 1 are slightly affected, with exit rate being lowered by 10%–40% for ITP and RTP (Figure S4A) and a slight increase of probability (Figure S4B). The presence of both ITP and RTP leads to increases in probability relative to standard conditions (3-fold for ITP and 2-fold for RTP; Figure 4D).
Figure 4.
The Presence of Nucleotide Analogs Increases Misincorporation and Stalling by PV RdRp
(A) Chemical structure of the different antiviral nucleoside triphosphate analogs tested.
(B) Processivity at 100 μM NTPs (Ø) and supplemented with 1 mM RTP (R, purple) or 1 mM ITP (I, crimson).
(C) The dwell-time distributions acquired at 30 pN applied force with 100 μM NTPs (black circles) supplemented with 1 mM RTP (purple circles) or 1 mM ITP (crimson circles). The solid lines are the MLE fits to a scenario where rapid elongation competes with two long-lived pause states, as illustrated in Figure 3G. The kinetic parameters extracted from these fits are shown in (D) and Figure S4.
(D) The probability of pause 2 under the conditions described in (B) and (C): ), , and .
(E) The median processivity of PV RdRp at 30 pN applied force and the standard concentration of 100 μM NTPs in the absence or presence of the indicated concentration of 3′-dATP.
(F) The median processivity of PV RdRp at 30 pN applied force and the standard concentration of 100 μM NTPs in the absence or presence of the indicated concentration of 2′-C-Me-ATP.
(G and H) The dwell-time distributions at 30 pN applied force in the presence of 100 μM NTPs (black circles) supplemented with 1 μM 3′-dATP (light green circles), 10 μM 3′-dATP (green circles), or 100 μM 3′-dATP (dark green circles) (G) or 1 μM 2′-C-Me-ATP (light blue circles), 5 μM 2′-C-Me-ATP (blue circles), and 50 μM 2′-C-Me-ATP (dark blue circles) (H). The solid lines are the MLE fits to a scenario where rapid elongation competes with two long-lived pause states, as illustrated in Figure 3G. The kinetic parameters extracted from these fits are shown in Figure S4.
(I) Probability of dwell times longer than 40 s as a function of 3′-dATP or 2′-C-Me-ATP concentration from the distributions presented in (G) and (H).
In (B)–(I), error bars are determined as described in Experimental Procedures.
To extend our study of the effect of nucleotide analogs on PV RdRp, we also characterize two ATP analogs that act as chain terminators, namely the obligate terminator 3′-deoxyadenosine-TP (3′-dATP)(Arnold et al., 2012) and the non-obligate terminator 2′-C-methyl-adenosine-TP (2′-C-Me-ATP) (Carroll et al., 2003) (Figure 4A). 3′-dATP terminates RNA extension because of the absence of a 3′-OH to serve as the nucleophile during the next cycle of nucleotide addition (Steitz, 1998). For 2′-C-Me-ATP, the reason for termination is unknown. One suggestion is that a methyl group above the ribose ring may hinder translocation (Carroll and Olsen, 2006). To test the effect of these chain terminators, we supplement our standard NTP concentration with 1, 10, and 100 μM 3′-dATP or 1, 5, and 50 μM 2′-C-Me-ATP, respectively. Under these conditions, both terminators compete with the 100 μM ATP present in the reaction buffer. When the concentrations of 3′-dATP or 2′-C-Me-ATP are increased, we observe the median values of the processivity to consistently decrease, reaching minimum values of bp for 100 μM 3′-dATP and bp for 50 μM 2′-C-Me-ATP (Figures 4E and 4F). While these observations are consistent with the proposed mode of action of these terminators (Arnold et al., 2012), they also suggest that these nucleotide analogs compete relatively poorly with ATP for incorporation; a median value for processivity of ∼400 bp in the presence of 100 μM 3′-dATP is expected, taking into account the uracil content of our template, to correspond to the incorporation of ∼100 ATP prior to termination. Similarly, ensemble experiments show a 17-fold higher selectivity for correct ATP than for 3′-dATP (Arnold et al., 2012).
The dwell-time distributions (Figures 4G and 4H) of PV RdRp in the presence of chain terminators do not show any difference on short timescales (<10 s), indicating that the pauses 1 and 2 are unperturbed by 3′-dATP or 2′-C-Me-ATP (Figures S4E–S4H and Figures S4I–S4L, respectively). However, examination of the dwell-time distributions on longer timescales reveals the existence of a new type of long pause (>40 s; Figures 4G and 4H) in the presence of both 3′-dATP (at 10 and 100 μM; Figure 4I, green points) and 2′-C-Me-ATP (Figure 4I, blue points). While these long pauses may affect the fit parameters used to characterize pause 2 kinetics, they are clearly separate from the pause 2 distribution.
Pyrazine-Carboxamide Nucleotide Induces Polymerase Backtracking, Defining a Distinct Mechanistic Class of Antiviral Nucleotide
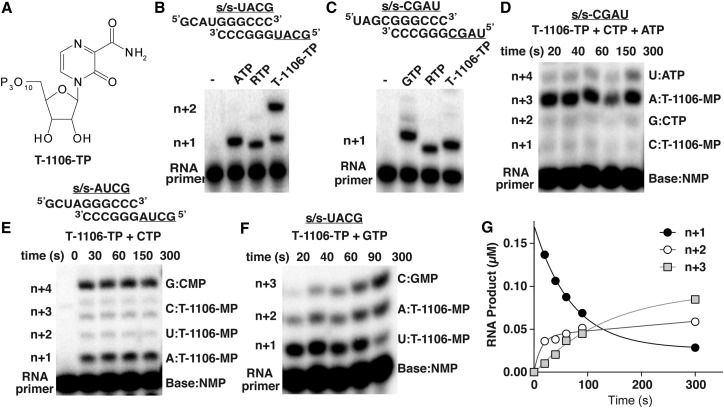
To illustrate the utility of the molecular tweezers approach in providing insight into the mechanism of action of an antiviral nucleotide, we chose to study a member of the pyrazine-carboxamide family. The best-known member of this family is T-705, which has been approved for treatment of influenza virus infection, but its mechanism of action is actively debated (Furuta et al., 2009). Here, we studied T-1106, which is closely related to T-705, but its nucleosides and nucleotides are more synthetically tractable (Figure 5 A). Because there is no mechanistic information for this class of antiviral nucleotide with PV RdRp, we first evaluated the specificity and efficiency of T-1106-TP utilization using our standard, bulk biochemical assays (Arnold and Cameron, 2004). PV RdRp incorporates T-1106-MP opposite uridine, adenosine, or cytidine in the template, but not opposite guanosine (Figures 5B and 5C). As a control, we show that PV RdRp incorporates RMP opposite uridine and cytidine, as shown previously (Crotty et al., 2000). PV RdRp efficiently bypasses T-1106:C pair, but the enzyme stalls at a T-1106:A pair (strong band at line ; Figure 5D). The stalling occurs for durations as long as 300 s. PV RdRp exhibits a context dependence for T-1106-MP incorporation opposite uridine, ranging from very inefficient (Figure 5D) to quite efficient (Figure 5E). The context dependence for incorporation and/or bypass also extends to T-1106:A pairs (Figures 5E and 5F). These experiments suggest that incorporation of T-1106-MP opposite adenosine creates an impediment to RNA synthesis. Interestingly, when T-1106-MP is in the RNA template, PV RdRp only incorporates UMP or CMP (Figures S5A–S5D). Only a fraction of elongation complexes are competent for efficient incorporation and bypass opposite T-1106-MP (Figures S5A–S5D). Thus, from these bulk biochemical experiments, we conclude that incorporation of T-1106-MP by PV RdRp opposite adenosine substantially reduces the nucleotide-addition rate of subsequent nucleotides (on timescales from tens to hundreds of seconds; Figure 5G), and its presence in the template strand hinders nucleotide addition. PV RdRp exhibits the same properties described above when using T-705-TP (Figure S6). These observations are consistent with studies in other viral systems, suggesting that pyrazine-carboxamide analogs are chain terminators (Sangawa et al., 2013, Jin et al., 2013b).
Figure 5.
Ambiguous Templating of a Pyrazine-Carboxamide Nucleotide Analog, T-1106-TP, Induces Context-Dependent Stalling of PV RdRp
(A) Chemical structure of T-1106-triphosphate (T-1106-TP).
(B and C) The symmetrical primed templates employed are referred to as sym/sub-UACG (B) or sym/sub-CGAU (C). The first and subsequent templating bases are underlined. PV RdRp incorporates T-1106-MP opposite uridine, adenosine, or cytidine, but not guanosine. For comparison, the utilization of RTP and the first correct NTP substrate (ATP or GTP) are shown as controls. Each reaction was quenched after 60 s.
(D–F) The elongation reaction products from PV RdRp-catalyzed T-1106-MP incorporation in the presence of additional correct nucleotide substrates. Reactions contained the indicated symmetrical primed templates and nucleotides. The incorporation of T-1106-MP opposite adenosine (A:T-1106-MP) (D) results in the production of terminated products or in a substantial reduction in the efficiency of utilization of the next correct nucleotide substrate. Comparable delays are not observed for incorporation opposite cytidine (C:T-1106-MP) or uridine (U:T-1106-MP) (E). (F) Stalling occurs opposite adenosine at n + 2.
(G) Kinetics of RNA synthesis for the reaction products (n + 1, n + 2, and n + 3) shown in (F). Incorporation of T-1106-MP opposite adenosine at n+2 slows down the rate of utilization of the next correct nucleotide substrate (GTP, n+3) consistent with the presence of a stalled polymerase.
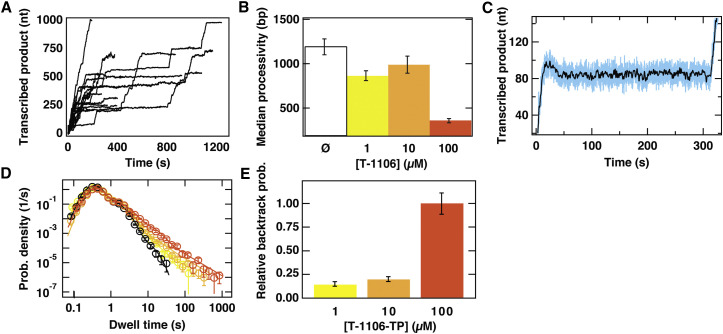
We now determine the impact of T-1106-TP utilization on the elongation dynamics of PV RdRp in the presence of standard NTP concentrations using the magnetic tweezers platform. From the time courses, it is immediately evident the presence of T-1106-TP causes the appearance of previously unobserved long pauses (>20 s; Figure 6 A; compare to Figure 1B), even at low concentrations of T-1106-TP (1 μM). Following such pauses induced by T-1106-TP, PV RdRp resumes elongation; the apparent decrease in the median processivity to (at 100 μM T-1106-TP; Figure 6B) results from temporal limits on the length of our data acquisitions. Based on these observations, we conclude that the effect of T-1106-MP incorporation is to stall RdRp elongation for long periods of time, but not to terminate elongation. Zooming in on the corresponding traces, we observe that several of them show significant extension decreases (Figure 6C) that likely correspond to backtracking by PV RdRp. In addition, the lifetime of this new pause appears distributed in a way consistent with backtracking, in contrast to the pause introduced by chain terminators (compare Figure 6D to Figures 4H and 4I). Backtracking has previously been reported for Φ6 P2 RdRp (Dulin et al., 2015d) and for multi-subunit RNA polymerases (Galburt et al., 2007, Shaevitz et al., 2003). We include the contribution from backtracked pauses in our model (Depken et al., 2009, Dulin et al., 2015c, Dulin et al., 2015d), but as we cannot extract the position of shoulder corresponding to the basic hopping rate in the backtrack (Depken et al., 2009) from under the distribution of pause 1 and pause 2, we cannot deduce the total fraction of backtracking events. However, since the basic hopping rate is unlikely to depend on T-1106-TP concentration, we can still determine the relative change in the probability to enter a backtrack as the T-1106-TP concentration is varied (Figure 6E). In the absence of T-1106-TP, we find no evidence of backtracking in the dwell-time distributions.
Figure 6.
T-1106-TP Induces Backtracking of PV RdRp
(A) Sixteen time courses of RNA synthesis acquired at 30 pN applied force and the standard concentration of 100 μM NTPs supplemented with 100 μM of T-1106-TP. These time courses are acquired at an acquisition frequency of 25 Hz.
(B) Processivity of PV RdRp in the absence of T-1106 (white with black border) and in the presence of 1 μM (light yellow), 10 μM (light orange), or 100 μM (dark orange) T-1106-TP.
(C) A zoom-in onto a single time course illustrating a decrease in the length of the RNA tether, corresponding to the rehybrization of ∼20 nt, followed by a long pause prior to resumption of elongation. This phenomenon is consistent with backtracking. The time course acquired at 25 Hz is shown in turquoise and, following low-pass filtering at 0.5 Hz, in black.
(D) Dwell-time distributions assembled from the PV RdRp time courses acquired at 30 pN applied force with 100 μM NTPs (black circles) complemented with 1 μM T-1106TP (yellow circles), 10 μM T-1106TP (light orange circles), or 100 μM T-1106-TP (dark orange circles). The solid lines are the MLE fits to a scenario where rapid elongation competes with two long-lived pause states, as illustrated in Figure 3G, and extended to include a backtracked state in the presence of T-1106-TP as described in Dulin et al. (2015c). The kinetic parameters extracted from these fits are shown in (E) and Figure S7.
(E) The relative probability of finding PV RdRp in the backtracked state (as deduced from the fits in D) in the presence of 1 μM (light yellow), 10 μM (light orange), or 100 μM (orange) T-1106-TP.
In (B), (D), and (E), error bars are determined as described in Experimental Procedures.
A Unique Mechanism of Action of T-1106 against PV in Cell Culture
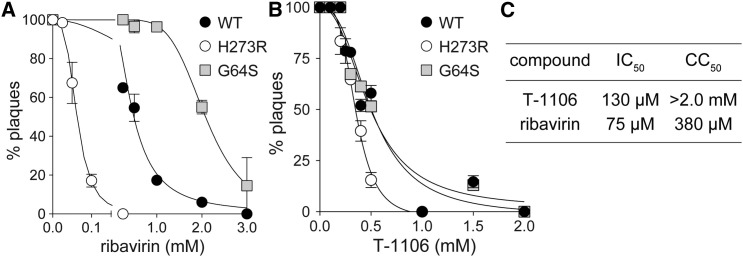
The biochemical studies suggest that primary cause of the inhibitory activity of T-1106 relates to the backtracking induced by T-1106-MP incorporation into viral RNA. We have described PV mutants exhibiting increased sensitivity (H273R PV) or decreased sensitivity (G64S PV) to ribavirin caused by decreased or increased fidelity of the encoded RdRp (Arnold et al., 2005, Korboukh et al., 2014). As shown in Figure 7 A, plating virus in the presence of increasing concentrations of ribavirin yields a dose-dependent reduction in the yield of plaques. H273R PV is more sensitive to ribavirin than WT PV, and G64S PV is less sensitive to ribavirin than WT PV. In contrast, the response of the PV strains to T-1106 is identical (Figure 7B). This observation is consistent with an event post-incorporation causing the antiviral outcome of T-1106 (e.g., backtracking). Finally, we performed a direct comparison of the efficacy and toxicity of T-1106 to ribavirin as described in Experimental Procedures. The efficacy of both compounds is within twofold of each other (half maximal inhibitory concentration [IC50] in Figure 7C). However, T-1106 is fivefold less toxic to HeLa cells than ribavirin (50% cytotoxic concentration [CC50] in Figure 7C).
Figure 7.
Unique Mechanism of Action of a Pyrazine-Carboxamide Nucleoside Analog Observed in Virus-Infected Cells
(A and B) Inhibition of virus production by ribavirin (A) or T-1106 (B). We infected drug-treated cells with PV expressing WT, high-fidelity (G64S) RdRps, or low-fidelity (H273R) RdRps. Sensitivity to ribavirin correlates directly to RdRp fidelity, but sensitivity to T-1106 does not, consistent with a mechanism of action different from that of ribavirin. The solid lines represent the fit of the data to a sigmoidal dose-response equation (four-parameter logistic model). The IC50 values for ribavirin with WT, G64S, and H273R are 0.55 ± 0.03 mM, 2.05 ± 0.10 mM, and 0.062 ± 0.003 mM, respectively. The IC50 values for T-1106 with WT, G64S and H273R are 0.51 ± 0.03 mM, 0.49 ± 0.03 mM, and 0.35 ± 0.01 mM, respectively. Error bars represent ±SEM.
(C) Anti-poliovirus activity (IC50) and HeLa cell toxicity (CC50) of T-1106 and ribavirin.
Discussion
Nucleic acid polymerases can transcribe or replicate genomes with both speed and accuracy over thousands, millions, or even billions of nucleotide-addition cycles. For decades, experimental systems suitable for quantitative kinetic analysis of nucleotide addition have been capable of evaluating on the order of ten events, with emphasis often placed on one or two incorporation events (Arnold and Cameron, 2004, Johnson, 1995). When one or two incorporation events are monitored, only one or two NTPs are present in the reaction, which is an unnatural circumstance. Notwithstanding this limitation, the approach has yielded copious insight into polymerase mechanism and has helped to explain and predict myriad biological phenotypes (Estep and Johnson, 2011, Arnold et al., 2005, Arnold et al., 2012, Batabyal et al., 2010, Korboukh et al., 2014, Lee et al., 2003). Nevertheless, two questions remain: (1) how does incorporation of thousands of nucleotides compare to incorporation of a few, and (2) what new insights can be gleaned from monitoring many, many incorporation events?
This study reports the second use of our magnetic tweezers instrument, which was designed to simultaneously measure hundreds of polymerases performing thousands of cycles of nucleotide addition (Cnossen et al., 2014, Dulin et al., 2015a, Dulin et al., 2015c), in the examination of RdRp kinetics. Our first study characterized the RdRp from bacteriophage Φ6 (Dulin et al., 2015c), a system for which the initiation phase is well characterized (Butcher et al., 2001, Makeyev and Bamford, 2000, Sarin et al., 2009, Wright et al., 2012) but which lacks quantitative, kinetic, and thermodynamic information describing the behavior of the ensemble during elongation. We here alter that circumstance by studying the RdRp from PV, an enzyme for which substantial kinetic and thermodynamic data exist, as well as a deep understanding of the mechanisms governing enzyme speed and accuracy (Cameron et al., 2016). Studies of the Φ6 RdRp identified three apparently catalytic states and one non-catalytic, backtracked state of an elongating polymerase, and most of these states are also observed for PV RdRp (Figure 1D). Nucleotide addition from the state with the shortest lifetime accounts for high-fidelity nucleotide incorporation and can be captured by a gamma distribution (Figure 1D). Because our data-analysis pipeline is unreliable for events occurring at rates faster than ∼1 s−1 due to filtering, it is not possible to extract rate constants for nucleotide addition. However, high-efficiency incorporation clearly occurs at rates of 10 s−1 and above, comparable to ensemble measurements (Arnold and Cameron, 2004). The population of dwell times greater than ∼50 s (Figure 1D) occurs at such a low probability that we consider it an insignificant pause species. In contrast, this population of dwell times was more prevalent for Φ6 RdRp, where it was shown to represent polymerases that have undergone backtracking (Dulin et al., 2015d). The states bracketed in time by the high-efficiency state and backtracked state are referred to as pause 1 and pause 2 (Figure 1D). The nature of these states is discussed below.
Our data analysis shows that the probability of entering pause 1 stabilizes at ∼10% during each nucleotide addition step at nucleotide concentrations higher than 100 μM, while the probability of entering this state increases when reducing the nucleotide concentration (Figure 2D). Because of the low probability of formation of pause 1, observing this elongating species in typical quench-flow ensemble experiments is unlikely. The 1 s−1 rate constant is too fast to represent misincorporation by PV RdRp based on ensemble experiments (Arnold and Cameron, 2004). As suggested in the Φ6 RdRp study, it is possible that pause 1 reflects incorporation of correct nucleotides by a low-efficiency catalytic state of the elongation complex (Dulin et al., 2015c). Molecular dynamics simulations of a PV RdRp elongation complex reveal numerous possibilities for the low-efficiency catalytic state, all of which use residues conserved in the catalytic sites of all RdRps with a PV RdRp fold (Moustafa et al., 2014). The catalytic site toggles between a nucleotide-binding-competent state and a nucleotide-binding-occluded state (Moustafa et al., 2014). Two primary conformations exist for interaction of the enzyme with the nascent base pair (Moustafa et al., 2014). The motif-B loop of the enzyme can adopt four conformations, each with a unique outcome on the stability of the nascent base pair (Moustafa et al., 2014). PV RdRp derivatives with substitutions in these conserved motifs analyzed using the magnetic tweezers platform should facilitate connection of pause 1 to a structural state. The exit rate is on par with observed rates of mispair extension measured by ensemble experiments (Moustafa et al., 2014, Garriga et al., 2013, Sholders and Peersen, 2014), and hence one could postulate that pause 1 reflects extension past a terminal mismatch. However, below we present evidence that associates the longer-lived pause 2 with nucleotide misincorporation, rendering this interpretation of the data unlikely.
Pause 2 must represent misincorporation. Ensemble experiments show that this event occurs on a timescale of minutes at a frequency of 10−4 or less (Arnold and Cameron, 2004). The single-molecule experiments predict a timescale of seconds (Figure 2C) and probabilities of 10−2 (Figure 2D), both of which are substantially higher than ensemble measurements. Before considering these quantitative differences, we wanted to ensure that pause 2 reported on misincorporation at least in part. To do this, we used a PV RdRp derivative known to exhibit an elevated frequency of misincorporation, H273R (Korboukh et al., 2014). This derivative exhibits an increase in exit rate (Figure 3E) and probability (Figure 3F); its 4-fold increase in probability relative to WT matches the magnitude of the increase in observed rate constants for nucleotide misincorporation measured in bulk (Moustafa et al., 2014, Korboukh et al., 2014). In terms of the absolute differences in rate constants, it is possible that, like pause 1, pause 2 represents a very low-efficiency catalytic state competent for correct nucleotide incorporation and reflecting nucleotide misincorporation. As suggested above, there are sufficient conformations of the PV RdRp catalytic site to make this plausible. These correct incorporation events would add to the complexity of the pause 2 exit rate and probability, but there is no doubt that a component of pause 2 derives from misincorporation. An alternative possibility for the differences in exit rate measured in the single-molecule experiment and for misincorporation measured in bulk is that the single-molecule experiment measures the activity of the pause 2 conformational state and/or its attenuation of the next incorporation event, while the bulk experiment measures the composite rate of, first, formation of the pause 2 conformational state and, subsequently, misincorporation. Regardless of the precise mechanism, the data support the notion that particular states of Φ6 and PV RdRps exist that are competent for misincorporation and that binding of an incorrect nucleotide does not induce this conformation. Identification of this state kinetically motivates its identification structurally, as this conformation has myriad practical applications. Perturbing the kinetic partitioning between these states either chemically or genetically underpins strategies for antiviral therapy and viral attenuation, respectively.
The ability of pause 2 to report on misincorporation permits the analysis of misincorporation of nucleotides harboring natural nucleobases, synthetic bases, and/or modified ribose sugars, all in the presence of all four natural nucleotides. This attribute of our magnetic tweezers approach is unique among the single-molecule and ensemble approaches used to study polymerase fidelity and nucleotide analog utilization, including its consequences for nucleotide incorporation. The interruptions are likely termination events as the mean processivity declines for most nucleotides (Figure S3A). Imbalanced NTP pools decrease the probability of uninterrupted cycles of nucleotide addition, as probabilities and both increase (Figures S3D and S3F). Probability is the most affected parameter, as expected for a parameter responsive to misincorporation (Figure S3F). Two outliers exist. Excessive ATP is well tolerated (Figure S3F). A concentration of 1 mM ATP is lower than the cellular concentration of ATP; thus, the RdRp may have evolved a mechanism to diminish misincorporation of AMP. The ability to incorporate a nucleotide after AMP misincorporation is quite efficient (Figure S3E), again suggesting that the RdRp may have evolved mechanisms to prevent this misincorporation event from having a significant effect on viral RNA synthesis. Levels of CTP in the cell are far below 1 mM, but CMP misincorporation occurs with the highest probability of all nucleotides analyzed (Figure S3F), is easily bypassed (Figure S3E), and has absolutely no impact on processivity (Figure S3A). U-to-C transition mutations are the most frequent RdRp-dependent mutation observed in cells during PV infection (Korboukh et al., 2014). Interestingly, C-to-U transition mutations are the most frequent RdRp-independent mutation in cells, suggested to be the result of a member of the APOBEC family of cytidine deaminases (Korboukh et al., 2014). We speculate that the propensity to misincorporate CMP may help to mitigate the impact of cytidine deamination. Finally, GMP misincorporation (Figure S3F) has the greatest negative impact on processivity (Figure S3A), which is likely a reflection of the impediment GMP mispairs pose to RdRp bypass (Figure S3E).
Ribonucleotide analogs represent a class of antiviral agent with the greatest potential for broad-spectrum activity (Debing et al., 2015). Analogs like ribavirin function by increasing the apparent mutation frequency of the virus because of the ambiguous base-pairing capacity of its pseudobase (Crotty et al., 2000). Efficacy of this class of antiviral ribonucleotide requires multiple incorporation events. Our studies here show that magnetic tweezers can be used to study incorporation of this analog in the presence of other NTPs. Probability increases (Figure 4D) without any impact on median processivity (Figure 4B). As a result, ribonucleotide analogs with chain-terminating activity are favored. A single incorporation event per RNA is sufficient to render the RNA biologically useless. Compounds with a 3′-deoxyribose, for example, 3′-dATP, are obligate chain terminators because of the loss of the nucleophile for nucleotidyl transfer. Obligate chain terminators are often substrates for cellular polymerases, leading to substantial toxicity (Arnold et al., 2012). In contrast, non-obligate chain terminators, those that retain a 3′-hydroxyl but inhibit subsequent rounds of nucleotide addition as a result of substituents added to the ribose ring, can exhibit higher selectivity and therefore lower cytotoxicity (Arnold et al., 2012). One of the first non-obligate chain terminators developed was 2′-C-Me-ATP (Carroll et al., 2003). The magnetic tweezers experiment reveals quite nicely the negative impact of both of these analogs on the median processivity, consistent with the chain-termination activity of these compounds. While termination activity is easily monitored in bulk experiments, the magnetic tweezers experiment also revealed restart of a few percent of the terminated traces on a timescale greater than 40 s (Figure 4I). Excision of chain terminators has been reported for several viral RdRps, including the RdRp from hepatitis C virus (D’Abramo et al., 2004, Deval et al., 2007, Jin et al., 2013a). Demonstration of excision in bulk experiments often requires a contrived experimental setup. The magnetic tweezers experiment reveals this outcome under normal experimental conditions.
Our ability to evaluate the above antiviral ribonucleotides to obtain new information on these compounds using the magnetic tweezers motivated us to investigate a class of antiviral ribonucleotides for which the mechanism of action was unclear. This class of compounds contains a pyrazine-carboxamide pseudobase; we use the T-1106 nucleotide for our studies (Figure 5A) (Furuta et al., 2009). T-1106 is a synthetically more tractable alternative to T-705, which is currently approved to treat influenza virus infection but exhibits activity against numerous viruses (Furuta et al., 2009). The debate in the literature about this class compound is whether it is a mutagen, like ribavirin, or a non-obligate chain terminator. The structure of the compound fails to support the latter, as there is absolutely no modification of the ribose. How does a base induce termination? Ensemble experiments with PV RdRp clearly reveal the mutagenic activity and reveal a context-dependent failure of elongation, the definition of chain termination using this assay format (Figures 5B–5G). The failure to extend occurs primarily when T-1106-MP is incorporated opposite A in the template (Figures 5D–5F). What the magnetic tweezers experiment reveals is that incorporation of T-1106-MP induces long pauses (Figure 6A). Close inspection of the traces shows reannealing of the displaced strand to the tethered RNA (Figure 6C), an event identical to the backtracking events observed for Φ6 RdRp. Furthermore, the distribution of the long dwell times is also consistent with a backtracking process (Dulin et al., 2015d, Depken et al., 2009) upon T-1106 addition (Figure 6D). Backtracking of an RdRp elongation complex in response to incorporation of a nucleotide analog defines a distinct mechanistic class of antiviral ribonucleotide, thus providing an explanation for the controversy. Is backtracking the mechanism of action in a cellular context? To address this question, we used PV mutants that exhibited decreased (G64S) or increased (H273R) sensitivity to nucleotide analogs with mutagenic or chain-terminating activity (Figure 7A) (Arnold et al., 2005, Korboukh et al., 2014). The response of these strains to T-1106 does not resemble the response to ribavirin (Figure 7B), consistent with a unique mechanism of action. Galidesivir (BCX4430), a ribonucleoside with broad-spectrum antiviral activity, was recently described (Warren et al., 2014). This analog has a modified glycosidic bond without any other changes to the ribose or adenine base and has been suggested to be a non-obligate chain terminator (Warren et al., 2014). The backtracking activity of this drug should be evaluated in the future.
Although we use an RdRp here, our approach and conclusions likely apply to all classes of nucleic acid polymerases. Combining traditional ensemble approaches with the magnetic tweezers approach produces a comprehensive understanding of polymerase speed, accuracy, and inhibition. Measuring rate constants for nucleotide incorporation with confidence is best done one nucleotide at a time using chemical quench-flow or stopped-flow instrumentation. However, as we demonstrate here, insight into processivity, strand-displacement synthesis, and rare asynchronous events such as misincorporation and incorporation of nucleotide analogs in the presence of a pool of natural nucleotides, excision of chain terminators and backtracking by polymerase are all best described by using magnetic tweezers instrumentation with high-throughput capabilities. Our ability to unmask these steps of the PV RdRp nucleotide-addition cycle has revealed unique, nucleotide-specific signatures in the pause kinetics of the enzyme and the discovery that some analogs can result in extreme pauses by inducing backtracking of the enzyme. We concluded that RdRp backtracking represents a viable mechanism for antiviral therapy. Studies of the physical and structural basis of backtracking are warranted.
Experimental Procedures
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
dsRNA Tethers
Each (predominantly) dsRNA molecule consists of a template strand for PV RdRp hybridized to a complementary tethering strand that is linked at its two extremities to the flow chamber and the beads, respectively (Dulin et al., 2015c). Apart from a small hairpin that terminates the 3′ end of the template strand (sequence of the initial 24 bases of the template strand: 3′-GGGGAGCUCCCCUUUUUUUUUUUU…-5′, where the hairpin sequence is underlined), the dsRNA construct used in this study is identical to that employed in our preceding studies of Φ6 P2 RdRp (Dulin et al., 2015c, Dulin et al., 2015d) (Figure 1A). This hairpin forms an efficient way to mimic a primer hybridized to the template strand and promotes primer-dependent initiation (Arnold and Cameron, 1999, Arnold and Cameron, 2000, Arnold and Cameron, 2004).
Magnetic Tweezers Instrumentation
The magnetic tweezers instrumentation employed here has been described previously (Berghuis et al., 2015, Cnossen et al., 2014, Dulin et al., 2015c, Dulin et al., 2015b), with specifics of assays for detecting the activity of RdRps detailed in Dulin et al. (2015c). The magnetic beads are each tethered to the flow chamber surface by a single dsRNA molecule. Hundreds of magnetic beads are tethered to a flow chamber surface by individual dsRNA molecules. The magnetic beads experience a constant force from a pair of permanent magnets placed above the flow chamber (Figures 1A and S1A). This force is then transmitted directly to the RNA tether.
Reaction Conditions in the Magnetic Tweezers
Once the RNA construct length is calibrated inside a flow cell containing PV RdRp reaction buffer (50 mM HEPES [pH 7.9], 5 mM MgCl2, 0.01% Triton X-100, and 5% Superase RNase inhibitor [Life Technologies]), 500 nM PV RdRp is flushed in. We perform experiments at 21°C for 1 hr at 30 pN applied force and fixed NTP concentration. We follow the (x, y, z) positions of up to 800 tethered magnetic beads in real time at 25 Hz (Berghuis et al., 2015, Cnossen et al., 2014, Dulin et al., 2015c). Distinct traces are low-pass filtered at 0.5 Hz, providing an optical resolution of 0.3 nm along the optical axis (z axis) (Cnossen et al., 2014), and aligned with respect to their starting position. The changes in extension are converted into the numbers of nucleotides transcribed using the known force-extension relationships for dsRNA and ssRNA molecules under the employed buffer conditions (Dulin et al., 2015c).
To set up the stalling reaction with PV RdRp, we perform a 10-min incubation of PV RdRp with the dsRNA tethers in the flow cell in PV RdRp initiation buffer (PV RdRp reaction buffer supplemented with 1 mM rATP, 1 mM rGTP). We subsequently rinse the reaction chamber with an excess amount of PV RdRp reaction buffer containing 0.5% Superase RNase inhibitor (Life Technologies) in the absence of NTPs. We then trigger elongation by adding PV RdRp reaction buffer supplemented with NTPs and/or nucleotide analogs, as indicated.
Determination of Uncertainties in Measured and Fitted Parameter Values
We compute a statistical error estimation for the median processivity (e.g., Figure 2A) by bootstrapping the median estimation 10,000 times and reporting the SD of the bootstrapped median estimation. The error bars provided for all log-binned histograms of the probability density distribution of the dwell times (e.g., Figure 1D) represent 1-SD confidence intervals extracted from 1,000 bootstraps of each indicated dwell-time distribution.
We calculate the errors in our fitted parameter estimates by bootstrapping (Press et al., 1992) each dataset 200 times, applying an MLE to these bootstrapped datasets, and determining the asymmetric one-sigma confidence intervals among the bootstrapped datasets. The fitted parameters are presented in the main text as either , where indicates the range of the confidence interval, or .
PV-RdRp-Catalyzed Single-Nucleotide Incorporation Assays
Reactions were performed as described in detail previously (Arnold and Cameron, 2000). Briefly, elongation complexes were assembled by incubating 2 μM WT PV PVdRp with a 0.5 μM sym/sub RNA primer-template (1 μM duplex) for 3 min, at which time reactions were initiated with a 16 μM heparin trap and 100 μM NTP substrate. Reactions were quenched at various times by addition of 50 mM EDTA. All reactions were performed at 30°C in 50 mM HEPES (pH 7.5), 10 mM 2-mercaptoethanol and 5 mM MgCl2. Products were analyzed by denaturing PAGE. Gels were visualized by using a Phosphor Imager and quantified by using ImageQuant TL software (GE Healthcare).
Inhibition of PV Replication by Ribavirin and T-1106
HeLa cell monolayers in 6-well plates were pretreated for 1 hr with various concentrations of ribavirin and T-1106 and then infected with 50 plaque-forming units (PFUs) WT, H273R, or G64S PV, incubated for 20 min to allow for virus adsorption, and then washed and overlaid with 0.5% agarose media containing the same concentration of ribavirin or T-1106. Plates were incubated for 3–4 days at 37°C before overlays were removed and monolayers were stained with crystal violet. Plaques were counted and compared to the untreated control dish. To determine the IC50 value, the percentage of plaques was plotted as a function of ribavirin concentration and fit to a sigmoidal dose response equation:
where is the percentage of plaques relative to untreated cells, is the minimum percentage of plaques, and is the Hill slope.
Cytotoxicity Assays
HeLa cell monolayers in 24-well plates were treated with various concentrations of ribavirin and T-1106 for 7 hr. Cells were then washed, media without drug was added, and cells allowed to grow for 24 hr. Cells were detached using trypsin, counted, and compared to untreated control cells to determine the CC50 value.
Author Contributions
D.D., J.J.A., C.E.C., and N.H.D. designed the research. D.D. designed and performed the single-molecule experiments. D.D. and T.v.L. designed the RNA construct. J.J.A., H.-S.O., C.L., and C.E.C. provided the purified poliovirus polymerases and antiviral nucleotide analogs and performed the bulk biochemical experiments and/or biological experiments with PV. A.L.P. and D.A.H. performed nucleoside syntheses. D.D. and M.D. analyzed and interpreted the single-molecule data. D.D., J.J.A., C.E.C., M.D., and N.H.D. discussed the results and wrote the manuscript.
Acknowledgments
We acknowledge Eric Snijder, Clara Posthuma, Bojk Berghuis, and Behrouz Eslami Mossalam for fruitful discussions. We thank Bojk Berghuis for experimental assistance, Richard Janissen for figure revisions, and Leo Beigelman and Jerome Deval from Alios BioPharma for providing T-705-TP. D.D. was supported by the Interdisciplinary Center for Clinical Research (IZKF) at the University Hospital of the University of Erlangen-Nuremberg. J.J.A. and C.E.C. were supported in part by the NIH/NIAID (grant AI045818). M.D. acknowledges support from a TU Delft startup grant. C.E.C. was the recipient of a Burroughs Wellcome Fund 2015 Collaborative Research Travel Grant. Funding to N.H.D. was provided by the Netherlands Organisation for Scientific Research (NWO) via its TOP-GO program and by the European Union via an ERC consolidator grant (DynGenome, 312221).
Published: October 24, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.10.005.
Contributor Information
Martin Depken, Email: s.m.depken@tudelft.nl.
Craig E. Cameron, Email: cec9@psu.edu.
Nynke H. Dekker, Email: n.h.dekker@tudelft.nl.
Supplemental Information
References
- Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol) is sufficient for template switching in vitro. J. Biol. Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub) J. Biol. Chem. 2000;275:5329–5336. doi: 10.1074/jbc.275.8.5329. [DOI] [PubMed] [Google Scholar]
- Arnold J.J., Cameron C.E. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+ Biochemistry. 2004;43:5126–5137. doi: 10.1021/bi035212y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J.J., Vignuzzi M., Stone J.K., Andino R., Cameron C.E. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 2005;280:25706–25716. doi: 10.1074/jbc.M503444200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold J.J., Sharma S.D., Feng J.Y., Ray A.S., Smidansky E.D., Kireeva M.L., Cho A., Perry J., Vela J.E., Park Y. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012;8:e1003030. doi: 10.1371/journal.ppat.1003030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batabyal D., McKenzie J.L., Johnson K.A. Role of histidine 932 of the human mitochondrial DNA polymerase in nucleotide discrimination and inherited disease. J. Biol. Chem. 2010;285:34191–34201. doi: 10.1074/jbc.M110.156182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., Einav S. Infectious disease. Combating emerging viral threats. Science. 2015;348:282–283. doi: 10.1126/science.aaa3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghuis B.A., Dulin D., Xu Z.Q., van Laar T., Cross B., Janissen R., Jergic S., Dixon N.E., Depken M., Dekker N.H. Strand separation establishes a sustained lock at the Tus-Ter replication fork barrier. Nat. Chem. Biol. 2015;11:579–585. doi: 10.1038/nchembio.1857. [DOI] [PubMed] [Google Scholar]
- Butcher S.J., Grimes J.M., Makeyev E.V., Bamford D.H., Stuart D.I. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- Cameron C.E., Moustafa I.M., Arnold J.J. Fidelity of Nucleotide Incorporation by the RNA-Dependent RNA Polymerase from Poliovirus. Enzymes. 2016;39:293–323. doi: 10.1016/bs.enz.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.S., Olsen D.B. Nucleoside analog inhibitors of hepatitis C virus replication. Infect. Disord. Drug Targets. 2006;6:17–29. doi: 10.2174/187152606776056698. [DOI] [PubMed] [Google Scholar]
- Carroll S.S., Tomassini J.E., Bosserman M., Getty K., Stahlhut M.W., Eldrup A.B., Bhat B., Hall D., Simcoe A.L., LaFemina R. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 2003;278:11979–11984. doi: 10.1074/jbc.M210914200. [DOI] [PubMed] [Google Scholar]
- Cnossen J.P., Dulin D., Dekker N.H. An optimized software framework for real-time, high-throughput tracking of spherical beads. Rev. Sci. Instrum. 2014;85:103712. doi: 10.1063/1.4898178. [DOI] [PubMed] [Google Scholar]
- Crotty S., Maag D., Arnold J.J., Zhong W., Lau J.Y., Hong Z., Andino R., Cameron C.E. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000;6:1375–1379. doi: 10.1038/82191. [DOI] [PubMed] [Google Scholar]
- Crotty S., Cameron C.E., Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA. 2001;98:6895–6900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Abramo C.M., Cellai L., Götte M. Excision of incorporated nucleotide analogue chain-terminators can diminish their inhibitory effects on viral RNA-dependent RNA polymerases. J. Mol. Biol. 2004;337:1–14. doi: 10.1016/j.jmb.2004.01.030. [DOI] [PubMed] [Google Scholar]
- de Ávila A.I., Gallego I., Soria M.E., Gregori J., Quer J., Esteban J.I., Rice C.M., Domingo E., Perales C. Lethal mutagenesis of hepatitis C virus induced by favipiravir. PLoS ONE. 2016;11:e0164691. doi: 10.1371/journal.pone.0164691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Avila A.I., Moreno E., Perales C., Domingo E. Favipiravir can evoke lethal mutagenesis and extinction of foot-and-mouth disease virus. Virus Res. 2017;233:105–112. doi: 10.1016/j.virusres.2017.03.014. [DOI] [PubMed] [Google Scholar]
- De Vlaminck I., Henighan T., van Loenhout M.T., Burnham D.R., Dekker C. Magnetic forces and DNA mechanics in multiplexed magnetic tweezers. PLoS ONE. 2012;7:e41432. doi: 10.1371/journal.pone.0041432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debing Y., Neyts J., Delang L. The future of antivirals: broad-spectrum inhibitors. Curr. Opin. Infect. Dis. 2015;28:596–602. doi: 10.1097/QCO.0000000000000212. [DOI] [PubMed] [Google Scholar]
- Depken M., Galburt E.A., Grill S.W. The origin of short transcriptional pauses. Biophys. J. 2009;96:2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J., Powdrill M.H., D’Abramo C.M., Cellai L., Götte M. Pyrophosphorolytic excision of nonobligate chain terminators by hepatitis C virus NS5B polymerase. Antimicrob. Agents Chemother. 2007;51:2920–2928. doi: 10.1128/AAC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval J., Symons J.A., Beigelman L. Inhibition of viral RNA polymerases by nucleoside and nucleotide analogs: therapeutic applications against positive-strand RNA viruses beyond hepatitis C virus. Curr. Opin. Virol. 2014;9:1–7. doi: 10.1016/j.coviro.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D., Berghuis B.A., Depken M., Dekker N.H. Untangling reaction pathways through modern approaches to high-throughput single-molecule force-spectroscopy experiments. Curr. Opin. Struct. Biol. 2015;34:116–122. doi: 10.1016/j.sbi.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Dulin D., Cui T.J., Cnossen J., Docter M.W., Lipfert J., Dekker N.H. High spatiotemporal-resolution magnetic tweezers: calibration and applications for DNA dynamics. Biophys. J. 2015;109:2113–2125. doi: 10.1016/j.bpj.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulin D., Vilfan I.D., Berghuis B.A., Hage S., Bamford D.H., Poranen M.M., Depken M., Dekker N.H. Elongation-competent pauses govern the fidelity of a viral RNA-dependent RNA polymerase. Cell Rep. 2015;10:983–992. doi: 10.1016/j.celrep.2015.01.031. [DOI] [PubMed] [Google Scholar]
- Dulin D., Vilfan I.D., Berghuis B.A., Poranen M.M., Depken M., Dekker N.H. Backtracking behavior in viral RNA-dependent RNA polymerase provides the basis for a second initiation site. Nucleic Acids Res. 2015;43:10421–10429. doi: 10.1093/nar/gkv1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep P.A., Johnson K.A. Effect of the Y955C mutation on mitochondrial DNA polymerase nucleotide incorporation efficiency and fidelity. Biochemistry. 2011;50:6376–6386. doi: 10.1021/bi200280r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y., Takahashi K., Shiraki K., Sakamoto K., Smee D.F., Barnard D.L., Gowen B.B., Julander J.G., Morrey J.D. T-705 (favipiravir) and related compounds: novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galburt E.A., Grill S.W., Wiedmann A., Lubkowska L., Choy J., Nogales E., Kashlev M., Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- Garriga D., Ferrer-Orta C., Querol-Audí J., Oliva B., Verdaguer N. Role of motif B loop in allosteric regulation of RNA-dependent RNA polymerization activity. J. Mol. Biol. 2013;425:2279–2287. doi: 10.1016/j.jmb.2013.03.034. [DOI] [PubMed] [Google Scholar]
- Jin Z., Leveque V., Ma H., Johnson K.A., Klumpp K. NTP-mediated nucleotide excision activity of hepatitis C virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA. 2013;110:E348–E357. doi: 10.1073/pnas.1214924110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Smith L.K., Rajwanshi V.K., Kim B., Deval J. The ambiguous base-pairing and high substrate efficiency of T-705 (Favipiravir) Ribofuranosyl 5′-triphosphate towards influenza A virus polymerase. PLoS ONE. 2013;8:e68347. doi: 10.1371/journal.pone.0068347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.A. Rapid quench kinetic analysis of polymerases, adenosinetriphosphatases, and enzyme intermediates. Methods Enzymol. 1995;249:38–61. doi: 10.1016/0076-6879(95)49030-2. [DOI] [PubMed] [Google Scholar]
- Korboukh V.K., Lee C.A., Acevedo A., Vignuzzi M., Xiao Y., Arnold J.J., Hemperly S., Graci J.D., August A., Andino R., Cameron C.E. RNA virus population diversity, an optimum for maximal fitness and virulence. J. Biol. Chem. 2014;289:29531–29544. doi: 10.1074/jbc.M114.592303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta R.D., Mizrahi V., Benkovic P.A., Johnson K.A., Benkovic S.J. Kinetic mechanism of DNA polymerase I (Klenow) Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- Kuchta R.D., Benkovic P., Benkovic S.J. Kinetic mechanism whereby DNA polymerase I (Klenow) replicates DNA with high fidelity. Biochemistry. 1988;27:6716–6725. doi: 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- Lee H., Hanes J., Johnson K.A. Toxicity of nucleoside analogues used to treat AIDS and the selectivity of the mitochondrial DNA polymerase. Biochemistry. 2003;42:14711–14719. doi: 10.1021/bi035596s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maale G., Stein G., Mans R. Effects of cordycepin and cordycepintriphosphate on polyadenylic and ribonucleic acid-synthesising enzymes from eukaryotes. Nature. 1975;255:80–82. doi: 10.1038/255080a0. [DOI] [PubMed] [Google Scholar]
- Makeyev E.V., Bamford D.H. The polymerase subunit of a dsRNA virus plays a central role in the regulation of viral RNA metabolism. EMBO J. 2000;19:6275–6284. doi: 10.1093/emboj/19.22.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa I.M., Korboukh V.K., Arnold J.J., Smidansky E.D., Marcotte L.L., Gohara D.W., Yang X., Sánchez-Farrán M.A., Filman D., Maranas J.K. Structural dynamics as a contributor to error-prone replication by an RNA-dependent RNA polymerase. J. Biol. Chem. 2014;289:36229–36248. doi: 10.1074/jbc.M114.616193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S.D. Designing defenses against deadly viruses. Cell. 2014;157:279. [Google Scholar]
- Ng K.K., Arnold J.J., Cameron C.E. Structure-function relationships among RNA-dependent RNA polymerases. Curr. Top. Microbiol. Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press W.H., Flannery B.P., Teukolsky S.A., Vetterling W.T. Cambridge University Press; 1992. Numerical Recipes in C: the Art of Scientific Computing. [Google Scholar]
- Sangawa H., Komeno T., Nishikawa H., Yoshida A., Takahashi K., Nomura N., Furuta Y. Mechanism of action of T-705 ribosyl triphosphate against influenza virus RNA polymerase. Antimicrob. Agents Chemother. 2013;57:5202–5208. doi: 10.1128/AAC.00649-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin L.P., Poranen M.M., Lehti N.M., Ravantti J.J., Koivunen M.R., Aalto A.P., van Dijk A.A., Stuart D.I., Grimes J.M., Bamford D.H. Insights into the pre-initiation events of bacteriophage phi 6 RNA-dependent RNA polymerase: towards the assembly of a productive binary complex. Nucleic Acids Res. 2009;37:1182–1192. doi: 10.1093/nar/gkn1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaevitz J.W., Abbondanzieri E.A., Landick R., Block S.M. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sholders A.J., Peersen O.B. Distinct conformations of a putative translocation element in poliovirus polymerase. J. Mol. Biol. 2014;426:1407–1419. doi: 10.1016/j.jmb.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia M.J., Bao D., Chang W., Du J., Nagarathnam D., Rachakonda S., Reddy P.G., Ross B.S., Wang P., Zhang H.R. Discovery of a β-d-2′-deoxy-2′-α-fluoro-2′-β-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J. Med. Chem. 2010;53:7202–7218. doi: 10.1021/jm100863x. [DOI] [PubMed] [Google Scholar]
- Steitz T.A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- Streeter D.G., Witkowski J.T., Khare G.P., Sidwell R.W., Bauer R.J., Robins R.K., Simon L.N. Mechanism of action of 1- -D-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc. Natl. Acad. Sci. USA. 1973;70:1174–1178. doi: 10.1073/pnas.70.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Wells J., Panchal R.G., Stuthman K.S., Garza N.L., Van Tongeren S.A., Dong L., Retterer C.J., Eaton B.P., Pegoraro G. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature. 2014;508:402–405. doi: 10.1038/nature13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Poranen M.M., Bamford D.H., Stuart D.I., Grimes J.M. Noncatalytic ions direct the RNA-dependent RNA polymerase of bacterial double-stranded RNA virus ϕ6 from de novo initiation to elongation. J. Virol. 2012;86:2837–2849. doi: 10.1128/JVI.05168-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Dulin D., Cnossen J., Köber M., van Oene M.M., Ordu O., Berghuis B.A., Hensgens T., Lipfert J., Dekker N.H. A force calibration standard for magnetic tweezers. Rev. Sci. Instrum. 2014;85:123114. doi: 10.1063/1.4904148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.