Abstract
One major challenge encountered with interpreting human genetic variants is the limited understanding of the functional impact of genetic alterations on biological processes. Furthermore, there remains an unmet demand for an efficient survey of the wealth of information on human homologs in model organisms across numerous databases. To efficiently assess the large volume of publically available information, it is important to provide a concise summary of the most relevant information in a rapid user-friendly format. To this end, we created MARRVEL (model organism aggregated resources for rare variant exploration). MARRVEL is a publicly available website that integrates information from six human genetic databases and seven model organism databases. For any given variant or gene, MARRVEL displays information from OMIM, ExAC, ClinVar, Geno2MP, DGV, and DECIPHER. Importantly, it curates model organism-specific databases to concurrently display a concise summary regarding the human gene homologs in budding and fission yeast, worm, fly, fish, mouse, and rat on a single webpage. Experiment-based information on tissue expression, protein subcellular localization, biological process, and molecular function for the human gene and homologs in the seven model organisms are arranged into a concise output. Hence, rather than visiting multiple separate databases for variant and gene analysis, users can obtain important information by searching once through MARRVEL. Altogether, MARRVEL dramatically improves efficiency and accessibility to data collection and facilitates analysis of human genes and variants by cross-disciplinary integration of 18 million records available in public databases to facilitate clinical diagnosis and basic research.
Keywords: rare diseases, genetic diseases, variants of unknown significance, FlyBase, Zfin, MGI, Diopt, ExAC, ClinVar, Geno2MP
Introduction
One major challenge encountered with interpreting human genetic variants is the limited understanding of the functional impact of genetic alterations on biological processes. Traditional variant interpretation methodology relies on restricting clinical interpretation to known Mendelian diseases and employing in silico prediction algorithms. For most genes, few variants have reliable and validated clinical significance designation, resulting in difficulties in differentiating between benign and pathogenic variants or determining whether variants in a candidate gene are causative.1 The wealth of available biological information across multiple model organisms could aid in the interpretation of variants such as known molecular functions of the candidate gene. However, there are major barriers to search for biological data in specific model organism databases due to the intricacies of evaluating orthologs and navigating seven different websites’ different organization, different approaches, and different use of gene or protein identifiers (Figure S1). This limits the efficiency of incorporating known model organism data into analysis of candidate genes.
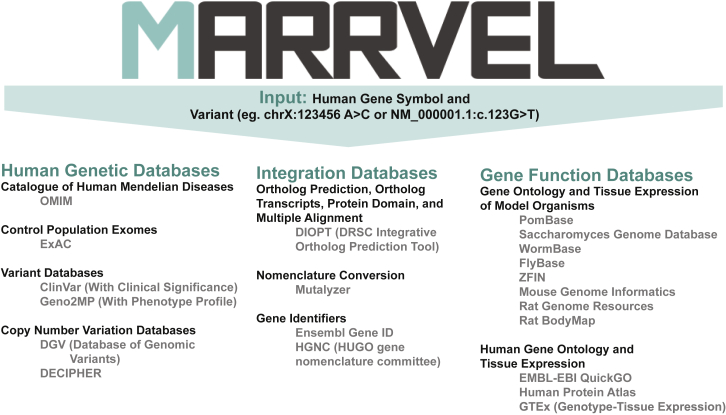
Therefore, there is an unmet demand for resources to facilitate rapid curation of available human gene and variant information, to determine conservation, and to gather relevant information on homologous genes in model organisms. Furthermore, such data compilation is relevant to evaluating the consequences of human genetic variation in model organisms.2 To provide a concise and user-friendly curation of pertinent and publicly available knowledge, we created MARRVEL (model organism aggregated resources for rare variant exploration). MARRVEL is an open-access resource that synthesizes genetic and model organism information from several public databases into a single user-friendly website (Figure 1).
Figure 1.
Overall Structure of MARRVEL
MARRVEL integrates 21 different databases to facilitate human gene and variant analysis for further study in model organisms. Human genetic databases are selected to provide data on disease association, statistics on variants found in a gene of interest, and exact matches with a variant of interest. Integration databases are important to the overall structure of MARRVEL due to the complicated structures and connections between each database that require homology prediction, specific gene identifiers, and nomenclature. Gene function databases are selected to provide a concise summary of what is known about a gene of interest across organisms.
The major impetus for developing MARRVEL arose from growing efforts to analyze the potential pathogenicity of genetic alterations in genes that are either not previously associated with human genetic disease or associated with different clinical features. A wide range of efforts for the discovery of disease-causing variants include the research consortiums for rare (e.g., Center for Mendelian Genomics3 and Undiagnosed Diseases Network4) or common (CHARGE consortium3) diseases, clinical genetics laboratories, large-scale sequencing projects,5, 6 and collaborations between human geneticists and model organism researchers.7 Together, these research efforts generate growing numbers of large human genomic datasets that require the development of resources and tools to facilitate efficient data analysis.
For example, the Undiagnosed Diseases Network4 combines the expertise of clinicians, sequencing centers (e.g., whole-exome, whole-genome, RNA-seq), metabolomics laboratories, and model organism scientists (fruit fly, zebrafish, and mouse) to diagnose individuals with rare disorders that eluded traditional diagnostic modalities. Many of these cases are predicted to have a primary genetic cause but the suspected causative variant may not be in disease-associated genes. When candidate pathogenic gene variants are identified, model organism data available for predicted orthologs of the human gene are an invaluable resource for interpreting the biological significance of the genetic alterations. However, this model organism-based resource is underutilized due to limited accessibility by non-model organism researchers. Currently, researchers need to visit and navigate separate model organism-specific databases (e.g., FlyBase,8 MGI,9 ZFIN10) that utilize distinct genotype and phenotype nomenclature as well as data organization. Moreover, in the study of genes or variants linked to human diseases, model organisms provide powerful platforms for mechanistic studies. Hence, a user-friendly open-access web-based resource to curate and synthesize current knowledge and resources from model organisms and human genomics databases is invaluable.11, 12, 13
Material and Methods
Human Genetics Databases
Human genetics data are extracted from Online Mendelian Inheritance in Man (OMIM),14 Exome Aggregation Consortium (ExAC),15 Genotype to Mendelian Phenotype (Geno2MP), ClinVar,16 Database of Genomic Variants (DGV),17 and DECIPHER (database of genomic variation and phenotype in humans using Ensembl resources).18
We display the human gene description, gene-phenotype relationships, and reported alleles from OMIM. Next, control population gene summary from the ExAC15 database is displayed. ExAC is a public collection of more than 60,000 exomes that have been selected against individuals with severe early-onset Mendelian phenotypes.15 When MARRVEL is primarily applied to early-onset pediatric phenotypes and used to evaluate candidate genes for Mendelian disease, the ExAC data can be considered as a “control” dataset. We will refer to this data as “control” throughout the paper though it should be noted these samples should not be considered similarly for adult neurodegenerative phenotypes, for example. Within the control population gene summary, we include the pLI (the probability of being loss-of-function [LoF] intolerant) score of a gene, which assesses the probability that a gene is extremely intolerant to loss of function variants (nonsense, splice acceptor, and splice donor variants) caused by single-nucleotide changes.15
We next display data from the Geno2MP database. Geno2MP is a database sponsored by the University of Washington Center for Mendelian Genetics displaying variants from Mendelian gene discovery projects and provide phenotype information for individuals with specific genotypes, including affected and unaffected family members.
Next, we extract data from ClinVar16 containing more than 255,000 unique variants annotated with clinical significance and review status (i.e., level of evidence). When a user searches for a gene and variant, MARRVEL displays all ClinVar variants reported in the gene of interest, summarizes the number of variants in each category of clinical significance, and highlights any variant(s) that match the location of the variant of interest. We provide both a high-level summary of the variants in terms of its reported clinical significance as well as a table with details for each reported variant. In addition, any alleles that overlap with the location of the variant of interest is highlighted in blue.
We then display data from the Database of Genomic Variants (DGV)17 database, which contains a large collection of structural variants from more than 54,000 individuals. The database includes samples of reportedly healthy individuals, at the time of ascertainment, from up to 72 different studies. Using the DGV database, we report all copy-number variants (CNVs) that overlap the input gene. If a CNV containing the gene of interest exists, we display the frequency, type of CNV, and publications associated with the CNV.
Finally, we display additional CNV information from the DECIPHER18 database based on the variant coordinate that includes common variants from the control population. Due to data display restrictions, we are able to provide the users only with control population data from DECIPHER.
Gene Function Databases
Biological and genetic features of human genes and their putative orthologous genes, including tissue expression pattern and Gene Ontology (GO) terms, are extracted from the following model organism databases: Saccharomyces Genome Database (SGD)19 for the budding yeast Saccharomyces cerevisiae, PomBase20 for the fission yeast Schizosaccharomyces pombe, WormBase21 for the nematode worm Caenorhabditis elegans, FlyBase8 for the fruit fly Drosophila melanogaster, ZFIN10 for the zebrafish Danio rerio, Mouse Genome Informatics (MGI)9 for mouse Mus musculus, and Rat Genome Database22 and Bodymap23 for rat Rattus norvegicus. For humans, we extract GO terms from QuickGO24 and tissue expression data from GTEx25 and Protein Atlas.26 To identify the putative orthologs of the human gene, we incorporate information from DIOPT (Drosophila RNAi Screening Center [DRSC] Integrative Ortholog Prediction Tool),27 an online tool integrating 14 ortholog prediction tools to provide a homology score for each predicted ortholog pair. Additionally, DIOPT is used to display a multiple protein alignment that is generated with MAFFT and human gene functional domains.27
Data Processing
MARRVEL search allows three types of inputs: a single HUGO gene symbol,28 a single human variant, or a combination of both. The human variant input can be in the format conforming to HGVS nomenclature29 or in the genomic variant format [chromosome number]:[genomic coordinate] [Reference nucleotide]>[Alternate nucleotide]), for example 6:99365567T>C. If the variant is input in HGVS nomenclature format, then the Mutalyzer Position Converter tool30 is used to transform the variant input into genomic coordinate, as variants stored in our database follow the genomic variant format.
If the input to MARRVEL includes both variant and gene symbol, data from OMIM14 are retrieved using the OMIM API and gene summary table is extracted from the ExAC website in real time. Variant data from the ExAC15 and Geno2MP databases are retrieved from our MySQL31 database as explained in the following section. Regarding ClinVar16 alleles, MARRVEL searches by the gene symbol and reports all alleles that overlap with the input gene. MARRVEL also provides a summary on clinical significance from these alleles. MARRVEL displays DGV17 copy-number variants based on the genes that are encompassed by the copy-number variants. Variant data from DECIPHER18 are retrieved from our MySQL database based on the chromosomal location.
If the input includes only a gene symbol, MARRVEL retrieves the gene summary table from the ExAC website. For Geno2MP, it shows all variants overlapping the gene in the database and its heterozygote count, homozygote count, and their sum. For DGV, it shows all CNV regions overlapping the gene. DECIPHER data are not retrieved since it does not provide report data associated with genes.
When the input includes only a variant, MARRVEL first searches the ExAC database to retrieve the variant information. It then shows gene-related information such as OMIM, orthologs and their functions, and protein alignment of the first gene the ExAC database matches.
For any combination of gene and variant input, the gene function table includes the following columns: the orthologous genes column, the DIOPT27 score column, the tissue expression column, and the associated GO terms’ columns. The orthologous genes column displays the putative orthologs predicted using DIOPT with a link to each organism database as well as a PubMed link. The PubMed link is generated from the NCBI32 gene page’s sub-link “Related articles in PubMed - See all citations in PubMed” under the “Bibliography” section of the NCBI gene page. By default, the gene function table shows only the putative orthologs with the best DIOPT score. All predicted orthologs can also be displayed by deselecting the check-box for this option at the top of the gene function table. The tissue expression column displays the tissue expression data for human and six model organisms. Expression data shown in the table list the names of tissues that highly express the gene of interest. For humans, there is an option to show all tissues with high protein expression levels from Protein Atlas26 and a bar graph of mRNA expression data from GTEx,25 including tissue names and its median value of RPKM. The mouse and zebrafish expression show only tissues expressed in wild-type. For fly, the tissues with high expression levels are displayed.
MARRVEL also provides human gene protein domain information and protein alignments for the gene and its homolog genes, which is extracted from DIOPT.
Server and Data Storing
MARRVEL is hosted on Amazon Web Service (AWS) EC2. We extracted data from the databases either by the database’s API or by downloading and storing files publicly available into a MySQL database. Multiple protein alignment and domain information from DIOPT are stored using AWS S3. Human variant data from the ExAC and Geno2MP databases were extracted by downloading and processing their respective VCF files and storing them in our database while ExAC gene summary data is pulled on demand from the ExAC website. Human copy-number variation data were extracted from the DGV17 and DECIPHER18 databases by downloading the databases’ tab delimited files and storing them in our database as well. MARRVEL’s usage of DECIPHER data adheres to the DECIPHER Data Access Agreement. ClinVar16 data were pulled from the ClinVar website and stored in the MySQL database. MARRVEL retrieves updated data from ClinVar bi-weekly.
Additional gene function data were obtained by accessing and extracting data from the DIOPT website.27 MARRVEL uses DIOPT’s ortholog prediction, protein alignment, and domain information. Human tissue expression data were obtained from Protein Atlas26 website API and median values from GTEx25 downloadable files. Human GO terms are directly downloaded from the QuickGO24 web pages. Rat expression data and GO terms are from RGD downloadable files.22 Mouse expression data and GO terms are from MGI website.9 Zebrafish data are from ZFIN downloadable files.10 Fly expression data and GO terms are pulled from the FlyBase website.8 S. cerevisiae data and S. pombe data are from SGD19 and PomBase.20
MARRVEL’s interface is implemented using the Twitter bootstrap framework v.4.0.0, jQuery v.2.2.0, and Angular JS v.1.6.1. The server backend was implemented using the Node.js framework v.6.7.0.
For the exact database versions, please see Tables S1 and S2.
Results
MARRVEL Integrates Data from Human and Model Organism Databases
MARRVEL builds upon and complements existing tools by integrating population genetics, model organism functional data, multiple protein alignments, and other information into one web- and mobile device-friendly site (Figure 1). The simple interface at MARRVEL allows entry of a human gene or variant to begin the survey with the results falling into two main categories. First, MARRVEL aggregates information from widely used human genetic databases (ExAC, Geno2MP, ClinVar, DGV, DECIPHER-control population), including sources of control and disease population data, to facilitate gene variant analysis. Second, MARRVEL displays a concise summary of available information for putative orthologs across yeast, worm, fly, fish, mouse, and rat (see Material and Methods). For genes that are not previously associated with human disease, there is often limited in vivo human functional data. However, there is often a wealth of model organisms data that can be used to infer the human gene function. By integrating biological and biochemical data across multiple model organisms, we provide links between human disease and gene function through a comprehensive overview of publicly available data. In total, MARRVEL integrates variants from 115,000 control individuals, 12.3 million variants, 6.95 million genotype-phenotype relationships, and 20,683 GO terms used to describe 235,928 model organism homolog-human gene pairs.
MARRVEL Facilitates Human Genomic Analysis
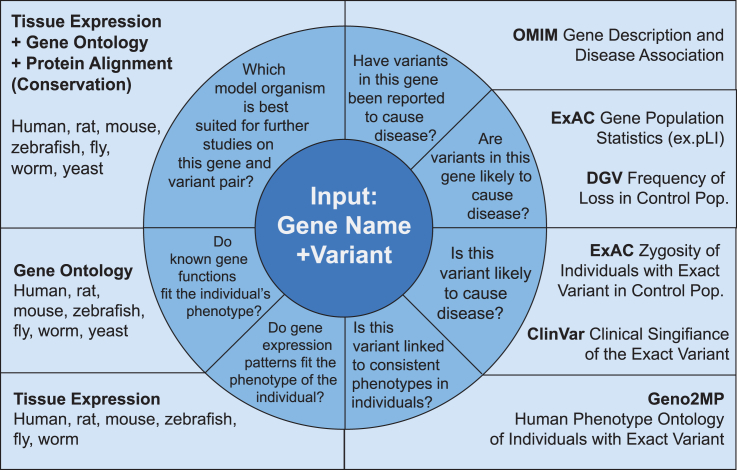
MARRVEL collects a wide range of data that can be used for multiple purposes for users from all fields. Here, we present just one of many ways that MARRVEL assists in human gene and variant analysis (Figure 2). In our approach, MARRVEL is used downstream of initial Whole Exome/Genome Sequencing bioinformatics analysis that results in a short list of candidate variants for a given individual’s phenotype. MARRVEL first extracts key data from public human databases for gene-based analysis. We first display results from OMIM (Online Mendelian Inheritance in Man).14 If the gene is documented at OMIM to be associated with a disease and the individual’s phenotype is consistent, then the variant is likely causative. However, there is the caveat that in some cases the variant may be benign and this does not exclude the possibility that genetic alterations in other genes may also result in similar clinical phenotype. If a unique variant is in a disease-associated gene but the phenotypes are inconsistent with previously reported phenotypes, then this suggests a possible phenotypic expansion. If there are no known diseases or phenotypes associated with the gene in OMIM, then this may represent a potential disease-association for the gene.
Figure 2.
Example of an Approach for Variant Analysis using MARRVEL
An example of how MARRVEL output can be used to analyze human genes and variants is illustrated by a question asked by the user in the inner ring and the answer that can be found in MARRVEL’s output in the outer boxes. We start at the noon position and advance clockwise.
The next set of data is used to assess whether variants in a specific gene is potentially pathogenic. The pLI score from ExAC expresses the probability that a gene is intolerant to loss-of-function alterations. For CNVs, the data that we collect are the deletion/duplications in the control population that contains the gene of interest. We obtain datasets from DGV and DECIPHER. The data obtained by DECIPHER is restricted to CNVs found in control population. DGV (Database of Genomic Variants) contains copy-number variations from a large number of non-disease individuals (control populations from many published cohorts). A high frequency of deletions in the gene of interest in this population suggests that the gene tolerates haploinsufficiency. Similarly, a high frequency of duplications in the gene of interest in DGV suggests that gain of one copy is likely tolerated, depending on the specific location of duplications. DECIPHER similarly provides copy-number variations for a control population.
Next, MARRVEL displays the presence or absence of the variant of interest in ExAC, displayed as an estimate of allele frequency in a large cohort of individuals without early-onset disease. For candidate gene variants in individuals with early-onset disease and a proposed dominant mode of inheritance, the presence of the same variant in ExAC decreases the likelihood that the variant is pathogenic especially if the disease is early onset. However, ExAC does include data from populations known to be affected by adult-onset diseases, including schizophrenia and cardiovascular diseases. In contrast, if the variant is absent in ExAC, then the variant may be a potential candidate for further analysis. For candidate gene variants with a proposed recessive mode of inheritance, the presence of individuals homozygous for the variant of interest in ExAC suggests that different gene variants may need to be considered in evaluating disease pathogenesis.
ClinVar16 is a valuable resource for researchers and clinicians to deposit gene variants and associated phenotypes. It contains more than 255,000 unique variants that are annotated with clinical significance and review status (i.e., level of evidence). When a user searches for a gene and variant, MARRVEL displays all ClinVar variants reported in the gene of interest, summarizes the number of variants in each category of clinical significance, and highlights the variant(s) that match the location of the variant of interest. If the variant of interest is documented in ClinVar as “benign” or “likely benign” with review status of “criteria provided, multiple submitters, no conflicts,” then the variant is unlikely pathogenic. However, if the variant is designated as “risk factor,” “likely pathogenic,” or “uncertain significance” and with review status such as “no assertion criteria provided” or “single submitter,” then the variant should remain a pathogenic candidate.
Geno2MP (Genotype to Mendelian Phenotype Browser) provides gene variant and phenotype correlation. Geno2MP provides cursory phenotypes for each sequenced individual in an affected population, as well as their unaffected relatives (if available), with human phenotype ontology (HPO) profiles. If a variant of interest is also present in an affected individual in Geno2MP, then the HPO terms would allow determination if similar biological systems are affected (i.e., potential phenotypic similarity). When both the genotype and phenotype of an affected individual in Geno2MP are consistent with the variant or gene of interest and the variant is not identified in an unaffected relative, then the variant is a pathogenic candidate.
MARRVEL Curates Gene Function Data in Humans and Model Organisms
MARRVEL summarizes human and model organism data relevant to gene function in three main steps. The first step compares expression patterns in specific organs or tissues across human and model organisms (except for yeasts). For human expression data, the source of the data in MARRVEL is protein levels from the Protein Atlas.26 In addition, GTEx provides quantitative expression data, RPKM (reads per kilobase per million mapped reads), of each gene in 53 human tissues. Model organism tissue expression data are obtained from individual model organism databases. Detailed information about the data sources can be found in Table S2. The tissue expression data serve at least three purposes. First, genes for which the pattern of expression is similar in humans and model organisms (e.g., expressed in comparable tissues) might be more likely to be informative in the context of human variant analysis. Second, display of the human tissue data allows for quick assessment of gene expression in the tissues affected in the individual under study. Third, expression patterns in human and model organism tissues can be used to design tissue-specific studies in model systems. One caveat to note is that the expression of a gene does not indicate necessity of the gene product in a specific tissue. In addition, reported developmental expression patterns often cover only specific stages and therefore may not provide valuable information. Moreover, many genes are only transiently expressed or their expression is induced only under specific environmental or physiological conditions. Finally, expression of many genes is below detection levels of current techniques.
The second step compares GO terms across human and model organisms for biological process, molecular function, and cellular component.33 GO terms are useful to quickly compare biological and molecular functions of the gene across species. In many cases, a gene may be well studied in one or more model systems but not in others. Data from the model organisms can be compared to provide insight into the degree of conservation, reveal possible disease mechanisms, and assist in the selection of one or more specific model organism for further mechanistic study.
The third step examines the conservation of specific amino acids and protein domains among orthologs based on multiple alignments of the human protein sequence and putative orthologs in model organisms (Figure S2). The alignment provides information on conservation of the amino acid or functional domain affected by the missense variant. MARRVEL also lists functional domains present in the human protein, highlighted in the multiple alignment. These steps further assist in determining whether there is evolutionary selection against variation at the residues analogous to the human variant of interest and in the selection of model organisms for pursuing further study of disease mechanism.
Case Example of How MARRVEL Facilitates Gene and Variant Analysis
Here, we provide an example of how MARRVEL displays information useful for variant prioritization in an output to facilitate analysis of genes and variants. We describe a specific case for which MARRVEL can be used to facilitate manual analysis of putative human disease-causing variants in an individual (Figure 3).
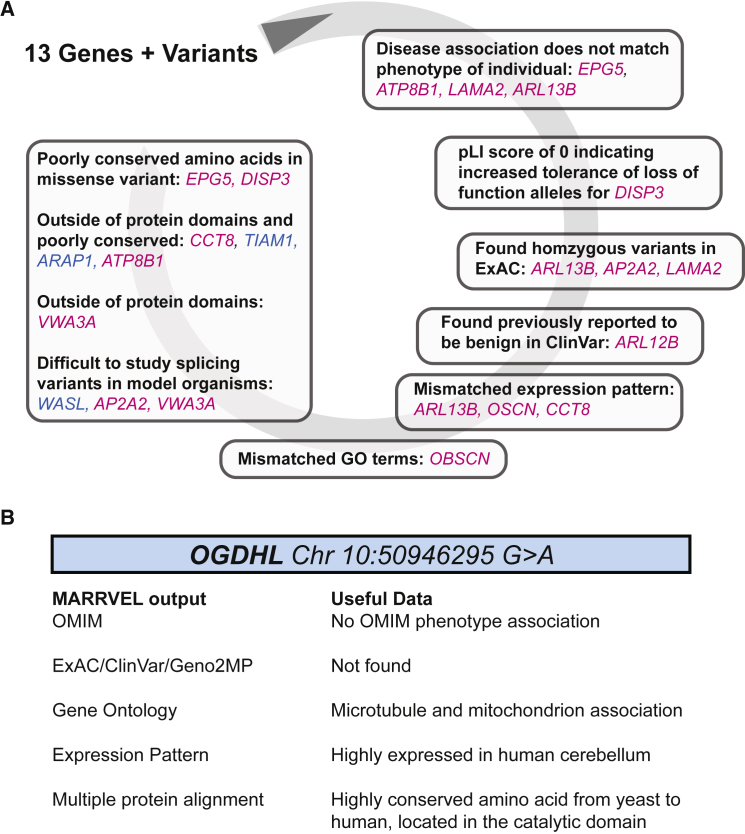
Figure 3.
Example of Variant Analysis using MARRVEL
We re-analyzed a previously published case by Yoon et al.13 by following our strategy outlined in Figure 2.
(A) Magenta genes and variants are eliminated based on multiple criteria. Blue genes and variants are eliminated with only one criteria and users may consider further analysis. The arrangement of the chart is reflective of Figure 2 which explains our strategy of analysis.
(B) OGDHL is the most likely candidate out of the 13 genes and variants to cause the individual’s phenotypes based on MARRVEL data. For more details on the genes and variants, see Table S3.
Yoon et al.13 recently successfully performed functional studies in Drosophila to demonstrate the pathogenicity of a gene variant found in a proband with a neurodegenerative phenotype. Previous analysis performed by an expert identified a homozygous variant in OGDHL (HGNC:25590) as the likely cause of the proband’s phenotype.34 Yoon et al.13 subsequently showed that loss of Ogdh, the Drosophila homolog of OGDHL, exhibits a neurodegenerative phenotype consistent with the proband’s neurological disorder. As an example of how MARRVEL can be used to conduct variant analysis, we obtained a list of 13 candidate variants for this case13 and conducted a variant analysis using output from MARRVEL to determine whether we were able to come to a similar conclusion.
The example presented is a 15-year-old girl with developmental delay, microcephaly, ataxia, motor impairment, hypotonia, language impairments, brain abnormalities, and hypoplasia of the corpus callosum. She was identified in a consanguineous family in a Turkish brain malformation cohort.34 The proband, her unaffected parents, and an unaffected sibling received whole-exome sequencing. After filtering for variants that were both unique to the proband and rare in the population (at least <0.01 minor allele frequency), variants in 13 different genes remained. Subsequent steps illustrate how MARRVEL can be incorporated downstream of whole-exome or -genome analysis pipelines.
We manually filtered and analyze Yoon et al.’s list of 13 candidates13 through MARRVEL’s synopsis of publicly available databases including OMIM and ExAC; tissue expression patterns; and the location of the amino acid change relative to known functional domains. Table S3 shows the manual analysis of the MARRVEL output of these 13 genes in comparison to the original analysis by an expert (see Table S2 in Yoon et al.13). The first step is to examine any existing phenotypic associations with the gene. Of the 13 genes, 4—EPG5 (MIM: 615068, HGNC:29331), ATP8B1 (MIM: 602397, HGNC:3706), ARL13B (MIM: 608922, HGNC:25419), and LAMA2 (MIM: 156225, HGNC:6482)—have a disease association that is partially or completely inconsistent with the individual’s phenotype. Although phenotypic expansion may still be possible, our current strategy defers that possibility until all other candidate genes are ruled out.
Based on the ExAC data, a gene suspected to have a de novo variant, DISP3 (MIM: 611251, HGNC:29251), has a pLI score of 0, indicating a high tolerance of loss-of-function variants. In addition, there are three candidate genes—ARL13B, AP2A2 (MIM: 607242, HGNC:562), and LAMA2—in the individual that are either homozygous or compound heterozygous variants. For these three variants the same homozygous mutations are listed in ExAC, suggesting that these variants are unlikely to result in early-onset developmental disorders. The ARL13B variant was also reported in ClinVar as benign and likely benign by multiple submitters. Through curation of human genomics information, the list of 13 candidate genes was narrowed to 6 remaining genes.
Model organism gene expression data and biological function GO terms were analyzed next. For three genes—ARL13B, OBSCN (MIM: 608616, HGNC:15719), and CCT8 (HGNC:1623)—the tissue expression pattern did not match the nervous system involvement in the individual of interest. Additionally, for OBSCN the GO terms across model organisms exclusively focuses on muscle development, structure, and function, making it less likely to be involved in nervous system-related pathology.
Further analysis of the missense variants revealed that the affected amino acid residues in EPG5 and DISP3 are poorly conserved amino acids across model organisms. In addition, the variant in ARL13B affects a site outside of the coding regions or protein domains, and variants in CCT8, TIAM1 (MIM: 600687, HGNC:11805), ARAP1, and ATP8B1 are poorly conserved and encode residues located outside of protein domains. These variants are therefore less likely to disrupt protein function. Splicing variants such as those found in WASL (MIM: 605056, HGNC: 12735) are difficult to study in model organisms and should be pursued in alternative approaches such as quantitative measure of mRNA in human samples.
Altogether, the homozygous variant in OGDHL emerged as the best pathogenic candidate for further study based on the human and model organism output from MARRVEL. In the model organism output from MARRVEL, three lines of information suggested that OGDHL is a promising candidate for further study in model organisms (Figure 3B). (1) Although the gene had not been functionally studied in vertebrate or Drosophila, the gene has been linked to mitochondria function in C. elegans and yeast, consistent with some of the neurodegenerative phenotypes. (2) Expression data in human and model organisms suggest that the gene is highly expressed in the affected tissue (brain). (3) The amino acid is highly conserved throughout evolution and is located in a highly conserved stretch of the protein. Indeed, Yoon et al.13 showed that flies with a null allele of Ogdh exhibit neurodegenerative phenotypes consistent with a neurological disorder. Importantly, the variant found in the individual corresponds to a severe loss-of-function allele based on gene humanization and rescue experiments in Drosophila,8 indicating that OGDHL is the likely candidate responsible for the neurological phenotype. In summary, MARRVEL displays information that provides input for the prioritization of potentially disease-causing variants for functional validation (Figure 3).
We recognize that there are multiple approaches to the analysis of possible disease-causing variants. Above, we provided one example of using reanalysis of published data for how MARRVEL can be applied to the downstream analysis of sequencing data for determination of candidate disease genes. If inheritance pattern is unclear, then multiple parallel analyses should be performed assuming that the variant could result in either dominant or recessive phenotypes. Furthermore, the variant interpretation can be evaluated for possible functional consequences including gain of function, haploinsufficiency, and dominant negative.
Discussion
In summary, MARRVEL affords an efficient aggregation of information from multiple human genomics and model organism databases to allow for rapid view and assessment of candidate genes and variants. OMIM provides fundamental information about disease association for the gene of interest. ExAC provides a powerful resource for examining the allele frequency of rare variants and can be used to prioritize the frequency of a coding variant and potential pathogenicity.15 Geno2MP and ClinVar provide unique sources of phenotypic and interpretation data for a variant of interest. DGV and DECIPHER control population provide publically available data, copy-number variants in apparently healthy individuals, which complements the data from ExAC, Geno2MP, and ClinVar. MARRVEL displays all of this information in a concise format providing highly integrated, convenient, and fast access (Figure S1). For potential genes in which disruption may cause disease, there is often limited in vivo human gene functional data; however, there is a wealth of information in model organisms that can be used to develop meaningful hypotheses regarding human gene function and to inform the likelihood that a variant causes or contributes to a disease phenotype. For example, in the case of OGDHL, integrating human and model organism data in MARRVEL allows us to aggregate all the information needed to prioritize this gene and variant to be tested experimentally. One key benefit of MARRVEL is allowing the data from model organism databases to be reviewed in a concise format. In MARRVEL, key biological and genetic features of putative orthologous genes, including tissue expression pattern and Gene Ontology (GO) terms, are extracted from model organism databases. MARRVEL displays all the relevant information normally assessed in a manual analysis pipeline described in Figure 2.
Several bioinformatics tools exist for aggregating available data to increase efficiency of variant analysis. For example, GeneCards is an aggregation of human gene-centric data. MARRVEL and GeneCards have a number of overlapping datasets. However, MARRVEL places more emphasis on human variant data (ExAC, ClinVar, etc.) and has a much broader range of data from model organisms. Combined Annotation Dependent Depletion (CADD)35 and PolyPhen36 focus on predicting the pathogenicity of an amino acid change. These two tools incorporate a combination of homology, structural, and machine learning analysis to predict whether or not a single amino acid change is likely to disrupt protein function.37, 38 However, there are cases where additional population frequency data and model organism phenotypic data are needed to improve variant interpretation.35, 39
The Monarch Initiative40 addresses the challenge of annotating the human genome by gathering data on known phenotypes in other organisms (phenotype-centric) to assist in variant analysis whereas MARRVEL provides a gene-centric toolkit including non-vertebrate model organisms and protein alignments. Although most bioinformatics tools and strategies are useful guides, combining multiple resources often provides a better view of the variant and higher predictive value when analyzing variants and genes.
Clinical genetics labs and research sequencing centers have access to well-established variant analysis and annotation pipelines such as Exomiser/PHenotypic Interpretation of Variants in Exomes (PHIVE),41 ANNOVAR,42 and Codified Genomics (see Web Resources) that utilize existing tools to analyze entire sets of sequencing data. These require familiarity with bioinformatics data processing and access to these resources. By contrast, clinicians and model organism researchers often have access only to variants reported in clinical sequencing reports and in the literature. Furthermore, the majority of clinicians and model organism researchers lack training in bioinformatics data analysis. In the absence of an integrated pipeline, it is difficult for clinicians and basic scientists to efficiently obtain information on candidate disease variants, as the information needed is spread across various databases and tools for variant analysis (Figure S1).
Despite an increasing interest to utilize model organism data in human genetic analysis pipelines such as in the Monarch Initiative and Exomiser/PHIVE, the current major focus is on matching phenotypic information.41, 43 Although the similarities between human and model organism mutant phenotype can be informative, this approach may miss numerous opportunities in which the protein functions are part of conserved pathways among organisms when the orthologous phenotypes are not obviously analogous.44 For example, a yeast model for angiogenesis44 and a worm model for breast cancer44 revealed molecular pathways that contribute to these disorders based on the “phenology” concept. Therefore, we adapted a gene-centric rather than phenotypic-centric approach to study gene function by integrating model organism and human data in a single aggregated web-based resource.
Many model organism databases, such as FlyBase,8 WormBase,21 and ZFIN,10 are comprehensive and contain a monumental amount of data accumulated over numerous decades.45 However, the extremely valuable information in these databases is not easily accessible by those outside the field. Importantly, there is a barrier to search specific model organism databases due to the intricacies of evaluating orthologs and navigating different websites and the different use of gene or protein identifiers (Figure S1). MARRVEL organizes this information across multiple species in a clear and concise way and also provides the best predicted orthologs.
In recent years, whole-exome or -genome sequencing has increasingly been used to assist in the diagnosis of human diseases.46 Meaningful analysis and interpretation of the sequencing results require a team of dedicated experts. Current bioinformatics pipelines are efficient at identifying previously reported pathogenic variants in known human genes in which disruption causes disease. By filtering out previously identified benign variants, as well as those appearing at a high frequency in control populations, the number of potentially disease-causing variants can be narrowed down significantly. Further analysis to identify variants to functionally test in model organisms will benefit from a survey of currently available model organism data. In conclusion, MARRVEL is a flexible web resource that provides a useful and accessible tool for efficiently matching an input against 18 million records of human variants and genes as well as model organism homologs. MARRVEL provides a step toward the overarching goal of integrating model organism databases with human gene-centric user interfaces41 to improve the accessibility and evaluation of data typically used by experts fluent in specific data formats and software. Our future goals for MARRVEL include continuing to integrate additional human genomics and model organism resources as they become publicly available to ensure that MARRVEL remains a valuable and up-to-date analytical resource.
Consortia
Members of the Undiagnosed Diseases Network are Christopher J. Adams, David R. Adams, Mercedes E. Alejandro, Patrick Allard, Euan A. Ashley, Mashid S. Azamian, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Alan H. Beggs, Hugo J. Bellen, Jonathan A. Bernstein, Anna Bican, David P. Bick, Camille L. Birch, Braden E. Boone, Lauren C. Briere, Donna M. Brown, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Katherine R. Chao, Gary D. Clark, Joy D. Cogan, Cynthia M. Cooper, William J. Craigen, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Daniel C. Dorset, David D. Draper, Annika M. Dries, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Paul G. Fisher, Trevor S. Frisby, Kate Frost, William A. Gahl, Valerie Gartner, Rena A. Godfrey, Mitchell Goheen, Gretchen A. Golas, David B. Goldstein, Mary G. Gordon, Sarah E. Gould, Jean-Philippe F. Gourdine, Brett H. Graham, Catherine A. Groden, Andrea L. Gropman, Mary E. Hackbarth, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Isabel Hardee, Matthew R. Herzog, Ingrid A. Holm, Ellen M. Howerton, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Alanna E. Koehler, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, Lea Latham, Yvonne L. Latour, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Paul R. Lee, Shawn E. Levy, Denise J. Levy, Richard A. Lewis, Adam P. Liebendorfer, Sharyn A. Lincoln, Carson R. Loomis, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Paul Mazur, Alexandra J. McCarty, Allyn McConkie-Rosell, Alexa T. McCray, Thomas O. Metz, Matthew Might, Paolo M. Moretti, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Loren D.M. Pena, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Rachel B. Ramoni, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Sarah Sadozai, Katherine E. Schaffer, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Edwin K. Silverman, Janet S. Sinsheimer, Ariane G. Soldatos, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Kimberly A. Strong, Jennifer A. Sullivan, David A. Sweetser, Sara P. Thomas, Cynthia J. Tifft, Nathanial J. Tolman, Camilo Toro, Alyssa A. Tran, Zaheer M. Valivullah, Eric Vilain, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Michael F. Wangler, Mike Warburton, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Alec A. Weech, Monte Westerfield, Matthew T. Wheeler, Anastasia L. Wise, Lynne A. Wolfe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, and Patricia A. Zornio.
Acknowledgments
We thank the Undiagnosed Diseases Network Model Organism Working Group and Coordinating Center, Jim Lupski, Zeynep Akdemir, Ender Karaca, John Seavitt, George Eisenhoffer, Swathi Arur, Grzegorz Ira, and Karen Schulze for providing input in the design of MARRVEL. We thank Wan Hee Yoon for providing input in the manuscript. This work was supported by the NINDS (1U54NS093793-01) to the Model Organisms Screening Center of the UDN and NIH/ORIP (1R24 OD022005-01). J.W. is supported by The Robert and Janice McNair Foundation McNair MD/PhD Student Scholar Program and Baylor College of Medicine Medical Scientist Training Program. H.J.B. and N.P. are Investigators of the Howard Hughes Medical Institute. S.-Y.K., S.Y., M.F.W., and H.J.B. are supported by NIH (3U54NS093793-02S1). H.J.B. is supported by NIH (R01 GM067858). Z.L., R.A.-O., and W.-W.W. are supported by NSF (DMS 1263932), CPRIT (RP170387), NIH (R01 GM120033), Houston Endowment, Huffington Foundation, Belfer Foundation, and T T Chao Family Foundation. N.P., Y.H., A.C., and S.E.M. are supported by NIH NIGMS (R01 GM067761, NIGMS R01 GM084947), NIH (R24 RR032668, R24 OD021997 to N.P., P.I.), and Dana Farber/Harvard Cancer Center (NCI Cancer Center Support Grant # NIH 5 P30 CA06516 to S.E.M.). H.-T.C. is supported by the Pediatric Neurology Basic Neuroscience Research Track residency training program at Baylor College of Medicine. S.Y. is supported by the Texas Children’s Hospital (NRI Fellowship) and Alzheimer’s Association (New Investigator Research Grant NIRG-15-364099). M.F.W. is supported by NIH (U01HG007709). S.Y. and M.F.W. are supported by the Simons Foundation (#368479 SFARI Functional Screen of Autism-Associated Variants Award).
Published: May 11, 2017
Footnotes
Supplemental Data include three figures, three tables, and Supplemental Acknowledgments and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2017.04.010.
Contributor Information
Zhandong Liu, Email: zhandonl@bcm.edu.
Hugo J. Bellen, Email: hbellen@bcm.edu.
UDN:
Christopher J. Adams, David R. Adams, Mercedes E. Alejandro, Patrick Allard, Euan A. Ashley, Mashid S. Azamian, Carlos A. Bacino, Ashok Balasubramanyam, Hayk Barseghyan, Alan H. Beggs, Hugo J. Bellen, Jonathan A. Bernstein, Anna Bican, David P. Bick, Camille L. Birch, Braden E. Boone, Lauren C. Briere, Donna M. Brown, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Katherine R. Chao, Gary D. Clark, Joy D. Cogan, Cynthia M. Cooper, William J. Craigen, Mariska Davids, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Katrina M. Dipple, Laurel A. Donnell-Fink, Naghmeh Dorrani, Daniel C. Dorset, David D. Draper, Annika M. Dries, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Cecilia Esteves, Tyra Estwick, Paul G. Fisher, Trevor S. Frisby, Kate Frost, William A. Gahl, Valerie Gartner, Rena A. Godfrey, Mitchell Goheen, Gretchen A. Golas, David B. Goldstein, Mary G. Gordon, Sarah E. Gould, Jean-Philippe F. Gourdine, Brett H. Graham, Catherine A. Groden, Andrea L. Gropman, Mary E. Hackbarth, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Lori H. Handley, Isabel Hardee, Matthew R. Herzog, Ingrid A. Holm, Ellen M. Howerton, Howard J. Jacob, Mahim Jain, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Alanna E. Koehler, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Donna M. Krasnewich, Elizabeth L. Krieg, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, Lea Latham, Yvonne L. Latour, C. Christopher Lau, Jozef Lazar, Brendan H. Lee, Hane Lee, Paul R. Lee, Shawn E. Levy, Denise J. Levy, Richard A. Lewis, Adam P. Liebendorfer, Sharyn A. Lincoln, Carson R. Loomis, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Paul Mazur, Alexandra J. McCarty, Allyn McConkie-Rosell, Alexa T. McCray, Thomas O. Metz, Matthew Might, Paolo M. Moretti, John J. Mulvihill, Jennifer L. Murphy, Donna M. Muzny, Michele E. Nehrebecky, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Jordan S. Orange, J. Carl Pallais, Christina G.S. Palmer, Jeanette C. Papp, Loren D.M. Pena, John A. Phillips, III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Rachel B. Ramoni, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Sarah Sadozai, Katherine E. Schaffer, Kelly Schoch, Molly C. Schroeder, Daryl A. Scott, Prashant Sharma, Vandana Shashi, Edwin K. Silverman, Janet S. Sinsheimer, Ariane G. Soldatos, Rebecca C. Spillmann, Kimberly Splinter, Joan M. Stoler, Nicholas Stong, Kimberly A. Strong, Jennifer A. Sullivan, David A. Sweetser, Sara P. Thomas, Cynthia J. Tifft, Nathanial J. Tolman, Camilo Toro, Alyssa A. Tran, Zaheer M. Valivullah, Eric Vilain, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Michael F. Wangler, Mike Warburton, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Alec A. Weech, Monte Westerfield, Matthew T. Wheeler, Anastasia L. Wise, Lynne A. Wolfe, Elizabeth A. Worthey, Shinya Yamamoto, Yaping Yang, Guoyun Yu, and Patricia A. Zornio
Web Resources
Angular JS v.1.6.1, https://angularjs.org/
Bootstrap v.4.0.0, v4-alpha.getbootstrap.com
Codified Genomics, http://codifiedgenomics.com/
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
DECIPHER, http://decipher.sanger.ac.uk/
DIOPT, http://www.flyrnai.org/diopt
Ensembl GRCh37 Rest API, http://grch37.rest.ensembl.org
ExAC Browser, http://exac.broadinstitute.org/
FlyBase, http://flybase.org/
Geno2MP (March 2017 accessed), http://geno2mp.gs.washington.edu/Geno2MP/#/
HUGO Gene Nomenclature Committee, http://www.genenames.org/
jQuery v.2.2.0, https://jquery.com/
MARRVEL, http://marrvel.org/
Mouse Genome Informatics, http://www.informatics.jax.org/
Mutalyzer, https://mutalyzer.nl/index
Node.js framework v.6.7.0, https://nodejs.org/en/
OMIM, http://www.omim.org/
PomBase, https://www.pombase.org/
QuickGO, https://www.ebi.ac.uk/QuickGO/
Saccharomyces Genome Database, http://www.yeastgenome.org/
The Human Protein Atlas, http://www.proteinatlas.org/
Undiagnosed Diseases Network, https://undiagnosed.hms.harvard.edu/
WormBase, http://www.wormbase.org/
ZFIN, http://zfin.org
Supplemental Data
References
- 1.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellen H.J., Yamamoto S. Morgan’s legacy: fruit flies and the functional annotation of conserved genes. Cell. 2015;163:12–14. doi: 10.1016/j.cell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gahl W.A., Mulvihill J.J., Toro C., Markello T.C., Wise A.L., Ramoni R.B., Adams D.R., Tifft C.J., UDN The NIH Undiagnosed Diseases Program and Network: applications to modern medicine. Mol. Genet. Metab. 2016;117:393–400. doi: 10.1016/j.ymgme.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Roak B.J., Deriziotis P., Lee C., Vives L., Schwartz J.J., Girirajan S., Karakoc E., Mackenzie A.P., Ng S.B., Baker C. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright C.F., Fitzgerald T.W., Jones W.D., Clayton S., McRae J.F., van Kogelenberg M., King D.A., Ambridge K., Barrett D.M., Bayzetinova T., DDD study Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385:1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramocki M.B., Zoghbi H.Y. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marygold S.J., Crosby M.A., Goodman J.L., FlyBase Consortium Using FlyBase, a database of Drosophila genes and genomes. Methods Mol. Biol. 2016;1478:1–31. doi: 10.1007/978-1-4939-6371-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eppig J.T., Smith C.L., Blake J.A., Ringwald M., Kadin J.A., Richardson J.E., Bult C.J. Mouse genome informatics (MGI): resources for mining mouse genetic, genomic, and biological data in support of primary and translational research. Methods Mol. Biol. 2017;1488:47–73. doi: 10.1007/978-1-4939-6427-7_3. [DOI] [PubMed] [Google Scholar]
- 10.Ruzicka L., Bradford Y.M., Frazer K., Howe D.G., Paddock H., Ramachandran S., Singer A., Toro S., Van Slyke C.E., Eagle A.E. ZFIN, the zebrafish model organism database: updates and new directions. Genesis. 2015;53:498–509. doi: 10.1002/dvg.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao H.-T., Davids M., Burke E., Pappas J.G., Rosenfeld J.A., McCarty A.J., Davis T., Wolfe L., Toro C., Tifft C. A syndromic neurodevelopmental disorder caused by de novo variants in EBF3. Am. J. Hum. Genet. 2017;100:128–137. doi: 10.1016/j.ajhg.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen K., Ho T.S.-Y., Lin G., Tan K.L., Rasband M.N., Bellen H.J., Ackermann E., Guo S., Booten S., Alvarado L. Loss of Frataxin activates the iron/sphingolipid/PDK1/Mef2 pathway in mammals. eLife. 2016;5:43–44. doi: 10.7554/eLife.20732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon W.H., Sandoval H., Nagarkar-Jaiswal S., Jaiswal M., Yamamoto S., Haelterman N.A., Putluri N., Putluri V., Sreekumar A., Tos T. Loss of Nardilysin, a mitochondrial co-chaperone for α-Ketoglutarate Dehydrogenase, promotes mTORC1 activation and neurodegeneration. Neuron. 2017;93:115–131. doi: 10.1016/j.neuron.2016.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacDonald J.R., Ziman R., Yuen R.K.C., Feuk L., Scherer S.W. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry J.M., Hong E.L., Amundsen C., Balakrishnan R., Binkley G., Chan E.T., Christie K.R., Costanzo M.C., Dwight S.S., Engel S.R. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood V., Harris M.A., McDowall M.D., Rutherford K., Vaughan B.W., Staines D.M., Aslett M., Lock A., Bähler J., Kersey P.J., Oliver S.G. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–D699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44(D1):D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimoyama M., De Pons J., Hayman G.T., Laulederkind S.J.F., Liu W., Nigam R., Petri V., Smith J.R., Tutaj M., Wang S.-J. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–D750. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y., Fuscoe J.C., Zhao C., Guo C., Jia M., Qing T., Bannon D.I., Lancashire L., Bao W., Du T. A rat RNA-seq transcriptomic BodyMap across 11 organs and 4 developmental stages. Nat. Commun. 2014;5:3230. doi: 10.1038/ncomms4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huntley R.P., Sawford T., Mutowo-Meullenet P., Shypitsyna A., Bonilla C., Martin M.J., O’Donovan C. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 2015;43:D1057–D1063. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carithers L.J., Ardlie K., Barcus M., Branton P.A., Britton A., Buia S.A., Compton C.C., DeLuca D.S., Peter-Demchok J., Gelfand E.T., GTEx Consortium A novel approach to high-quality postmortem tissue procurement: the GTEx Project. Biopreserv. Biobank. 2015;13:311–319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 27.Hu Y., Flockhart I., Vinayagam A., Bergwitz C., Berger B., Perrimon N., Mohr S.E., McKusick V., Hamosh A., Scott A. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yates B., Braschi B., Gray K.A., Seal R.L., Tweedie S., Bruford E.A. Genenames.org: the HGNC and VGNC resources in 2017. Nucleic Acids Res. 2017;45(D1):D619–D625. doi: 10.1093/nar/gkw1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.den Dunnen J.T. Describing sequence variants using HGVS nomenclature. Methods Mol. Biol. 2017;1492:243–251. doi: 10.1007/978-1-4939-6442-0_17. [DOI] [PubMed] [Google Scholar]
- 30.Wildeman M., van Ophuizen E., den Dunnen J.T., Taschner P.E.M. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum. Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 31.Axmark, D., and Widenius, M. (2002). MySQL Reference Manual, P. DuBois, ed. (O’Reilly & Assoc.).
- 32.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44(D1):D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gene Ontology Consortium Gene Ontology Consortium: going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karaca E., Harel T., Pehlivan D., Jhangiani S.N., Gambin T., Coban Akdemir Z., Gonzaga-Jauregui C., Erdin S., Bayram Y., Campbell I.M. Genes that affect brain structure and function identified by rare variant analyses of Mendelian neurologic disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miosge L.A., Field M.A., Sontani Y., Cho V., Johnson S., Palkova A., Balakishnan B., Liang R., Zhang Y., Lyon S. Comparison of predicted and actual consequences of missense mutations. Proc. Natl. Acad. Sci. USA. 2015;112:E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katsonis P., Koire A., Wilson S.J., Hsu T.-K., Lua R.C., Wilkins A.D., Lichtarge O. Single nucleotide variations: biological impact and theoretical interpretation. Protein Sci. 2014;23:1650–1666. doi: 10.1002/pro.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katsonis P., Lichtarge O. A formal perturbation equation between genotype and phenotype determines the evolutionary action of protein-coding variations on fitness. Genome Res. 2014;24:2050–2058. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun S., Yang F., Tan G., Costanzo M., Oughtred R., Hirschman J., Theesfeld C.L., Bansal P., Sahni N., Yi S. An extended set of yeast-based functional assays accurately identifies human disease mutations. Genome Res. 2016;26:670–680. doi: 10.1101/gr.192526.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mungall C.J., McMurry J.A., Köhler S., Balhoff J.P., Borromeo C., Brush M., Carbon S., Conlin T., Dunn N., Engelstad M. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2017;45(D1):D712–D722. doi: 10.1093/nar/gkw1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson P.N., Köhler S., Oellrich A., Wang K., Mungall C.J., Lewis S.E., Washington N., Bauer S., Seelow D., Krawitz P., Sanger Mouse Genetics Project Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014;24:340–348. doi: 10.1101/gr.160325.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mungall C., McMurry J., Koehler S., Balhoff J., Borromeo C., Brush M., Carbon S., Conlin T., Dunn N., Engelstad M. The Monarch Initiative: Insights across species reveal human disease mechanisms. bioRxiv. 2016 [Google Scholar]
- 44.McGary K.L., Park T.J., Woods J.O., Cha H.J., Wallingford J.B., Marcotte E.M. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl. Acad. Sci. USA. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaiser J., Lock A., Harris M.A., Nurse P., Wood V., Kaiser J., Bond M., Holthaus S.-M., Tammen I., Tear G. BIOMEDICAL RESOURCES. Funding for key data resources in jeopardy. Science. 2016;351 doi: 10.1126/science.351.6268.14. 14–14. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.