Abstract
Symbiodinium is best-known as the photosynthetic symbiont of corals, but some clades are symbiotic in other organisms or include free-living forms. Identifying similarities and differences among these clades can help us understand their relationship with corals, and thereby inform on measures to manage coral reefs in a changing environment. Here, using sequences from 24 publicly available transcriptomes and genomes of Symbiodinium, we assessed 78,389 gene families in Symbiodinium clades and the immediate outgroup Polarella glacialis, and identified putative overrepresented functions in gene families that (1) distinguish Symbiodinium from other members of Order Suessiales, (2) are shared by all of the Symbiodinium clades for which we have data, and (3) based on available information, are specific to each clade. Our findings indicate that transmembrane transport, mechanisms of response to reactive oxygen species, and protection against UV radiation are functions enriched in all Symbiodinium clades but not in P. glacialis. Enrichment of these functions indicates the capability of Symbiodinium to establish and maintain symbiosis, and to respond and adapt to its environment. The observed differences in lineage-specific gene families imply extensive genetic divergence among clades. Our results provide a platform for future investigation of lineage- or clade-specific adaptation of Symbiodinium to their environment.
Introduction
Dinoflagellates of genus Symbiodinium are known for their mutualistic relationships with corals and other marine organisms. Association with Symbiodinium enables corals to inhabit nutrient-poor tropical waters, grow and build up coral reefs; breakdown of the relationship leads to coral bleaching and, unless the relationship is re-established, death. Reef ecosystems in turn provide diverse benefits and services both to the environment, and to the economy of nearby communities1. A clear understanding of the relationship between Symbiodinium and corals is thus indispensable if we are to take a knowledge-driven approach to protect and manage these valuable ecosystems in the face of global environmental change.
Symbiodinium has been classified into nine distinct groups, clades A through I2,3. Studies based on genome and transcriptome data (generated so far only for Symbiodinium clades A through F) have contributed substantially to understanding the biology of each of these clades. One of those studies, based on transcriptome data, revealed that clades A and B use a smaller number of transcription factors than do other eukaryotes, implying particular gene regulation mechanisms4. Other studies report the genetic basis of thermal tolerance in clades C and D5, and gene homologs and pathways shared among clades A, B, C and D6. Another transcriptome-based study revealed that divergence of Symbiodinium within the same clade (clade B in this case) can be mirrored as extensive differences in gene expression7. To date, three draft genomes of Symbiodinium are available, revealing the presence of unique splice sites and a unidirectional gene arrangement in S. minutum (clade B)8, and that retrotransposition and gene duplication are the main drivers of gene family expansion in S. kawagutii (clade F)9. Comparative analysis of the S. microadriaticum (clade A) genome with the two others, together with additional sequence data from other dinoflagellates, supports the hypothesis that the symbiotic lifestyle of Symbiodinium was predisposed by an abundance of membrane transporters in all dinoflagellates, rather than being an adaptive novelty10.
Although some genome and transcriptome data are available from representatives of Symbiodinium, little is known of how gene content or biological function may differ within and between clades. Key questions about Symbiodinium biology remain largely unexplored, including what features distinguish them from other dinoflagellates, and what attributes are shared by all Symbiodinium or are exclusive to one or a few clades. To link genomic information to functions of cells, organisms and ecosystems, a comparative approach using gene families can be adopted11.
Here, using available genome8–10 and transcriptome4,5,7,12–15 data from dinoflagellates within Order Suessiales (Symbiodinium and Polarella glacialis) we systematically assess the gene families and inferred biological functions that are represented in one or more of Symbiodinium clades A through F, and investigate whether these functions are overrepresented in each analysed group. This represents the first comprehensive analysis of shared intra- and inter-cladal gene families in Symbiodinium.
Results and Discussion
Genome and transcriptome data
For this study we assembled 24 datasets (Table 1) of Symbiodinium genomes (3) and transcriptomes (19), and P. glacialis transcriptomes (2), with a total of 1,300,300 sequences (total length 1,333.87 Mbp; Supplementary Table S1). The N50 of each set of predicted coding sequences (CDS) ranges between 219 and 3987 bp (average 1480 bp). Fewer than 3% of the sequences from each dataset have significant BLASTn matches (E ≤ 10−10) against bacterial genomes, implying that there is little bacterial contamination. The completeness of each dataset was examined by comparison against core eukaryote genes in CEGMA16 and BUSCO17 (see Methods). On average, 72% of the 234 alveolate-stramenopile BUSCO genes17 and 89% of the 458 CEGMA genes16 were recovered by BLASTx from the datasets (Supplementary Table S1).
Table 1.
Summary of the selected datasets for analysis in the present study.
| Species (Isolates) | Clade | Number of sequences | N50 (bp) | Total length (Mbp) | Data type |
|---|---|---|---|---|---|
| S. microadriaticum (CCMP2467)10 | A | 49,109 | 3,987 | 166.722 | Genome |
| Symbiodinium sp. (CassKB8)4 | A | 72,152 | 1,087 | 61.921 | Transcriptome |
| Symbiodinium sp. (CCMP2430)15 | A | 44,733 | 1,356 | 42.483 | Transcriptome |
| Symbiodinium aenigmaticum (mac04-487)7 | B | 45,343 | 1,355 | 44.628 | Transcriptome |
| Symbiodinium sp. (SSB01)12 | B | 59,669 | 1,752 | 71.172 | Transcriptome |
| S. pseudominutum (rt146)7 | B | 47,411 | 1,508 | 51.270 | Transcriptome |
| S. psygmophilum (HIAp, Mf10.14b.02, PurPFlex, rt141)7 | B | 50,745 | 1,618 | 51.37 | Transcriptome |
| S. minutum (Mac703, Mf1.05b, rt002, rt351)7 | B1 | 51,199 | 1,597 | 57.248 | Transcriptome |
| S. minutum (Mf1.05b)8 | B1 | 47,014 | 2,675 | 97.202 | Genome |
| S. minutum (Mf1.05b)4 | B1 | 76,284 | 741 | 45.335 | Transcriptome |
| Symbiodinium sp.5 | C | 26,986 | 534 | 12.546 | Transcriptome |
| Symbiodinium sp.54 | C | 55,588 | 687 | 30.570 | Transcriptome |
| Symbiodinium sp.13 | C | 65,838 | 1,746 | 97.581 | Transcriptome |
| Symbiodinium sp.15 | C1 | 45,782 | 1,443 | 45.706 | Transcriptome |
| Symbiodinium sp. (MI-SCF055)14 | C1 | 116,479 | 1,323 | 106.160 | Transcriptome |
| Symbiodinium sp. (WSY)14 | C1 | 131,066 | 1,239 | 113.375 | Transcriptome |
| Symbiodinium sp.15 | C15 | 37,277 | 1,299 | 33.008 | Transcriptome |
| Symbiodinium sp.5 | D | 23,777 | 920 | 16.609 | Transcriptome |
| Symbiodinium sp.15 | D1a | 43,662 | 804 | 25.956 | Transcriptome |
| Symbiodinium voratum (CCMP421)15 | E2 | 71,624 | 1,701 | 86.612 | Transcriptome |
| S. kawagutii (CCMP2468)9 | F | 36,850 | 1,467 | 38.379 | Genome |
| S. kawagutii (CCMP2468)15 | F | 11,679 | 219 | 2.666 | Transcriptome |
| Polarella glacialis (CCMP1383)15 | — | 57,865 | 1,581 | 57.733 | Transcriptome |
| Polarella glacialis (CCMP2088)15 | — | 32,168 | 1,161 | 21.755 | Transcriptome |
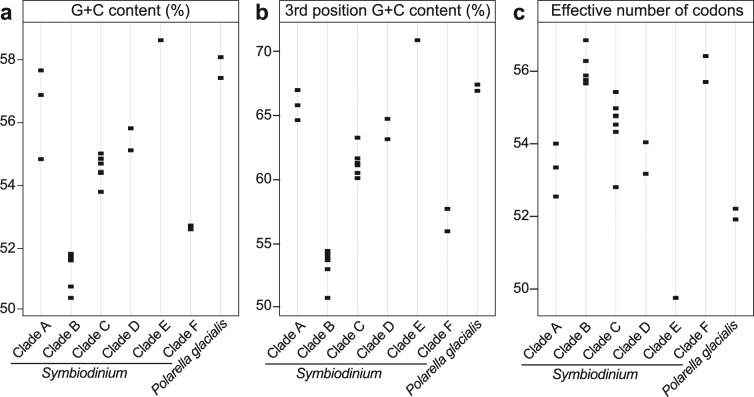
Where available, CDS predictions from the original source were used; otherwise CDS were predicted with TransDecoder v2.0.1 (see Methods). The overall sequence data yielded a total of 1,131,289 CDS with a total length of 1.21 Gbp; these correspond to the same number of predicted protein sequences. Completeness analyses returned results similar to those of the overall data (Supplementary Table S1). Overall G + C content ranges from 50.43% to 58.62% over all lineages (where lineage is defined as any clade of Symbiodinium or P. glacialis: Fig. 1a), G + C content in the third codon position between 50.81% and 70.86% (Fig. 1b), and the effective number of codons between 49.76 and 56.85 (Fig. 1c). These values differ between clades but fall within a relatively narrow range within each clade (Fig. 1).
Figure 1.
Overall G + C content (a), G + C content in third codon positions (b) and effective number of codons (c) are shown for the complete CDS of each dataset.
Due to the heterogeneity and incomplete (and fragmented) nature of the transcriptome data, we carefully scope our analysis at the clade (instead of species or isolate) level. However, we note that most genes in dinoflagellates including Symbiodinium have been found to be constitutively expressed irrespective of growth conditions18,19. After pooling datasets by clade and removing redundant sequences (see Methods), our final datasets consist of 584,272 predicted proteins (Supplementary Tables S2 and S3). Details on the contribution of each individual dataset to the clade pools are shown in Supplementary Fig. S1.
Delineation of gene families
Functions of proteins from each clade pool were assessed based on similarity search against the UniProt database, following Aranda et al.10 (see Methods). Of the 584,272 inferred proteins, 228,391 have significant (E ≤ 10−10) matches against sequences in UniProt (Swiss-Prot + TrEMBL); of these, 139,188 find a top match against an entry in Swiss-Prot, and a further 89,203 have a top match against an entry in TrEMBL. The matched UniProt identifiers were used to retrieve their associated KEGG Orthology (KO)20 and Gene Ontology (GO)21 terms. We define a UniProt Homolog Group (UP-HoG) as a set of proteins that share a common UniProt top match that has not been assigned a KO term, and a KEGG Homolog Group (KO-HoG) as those proteins for which the UniProt top match(es) have the same assigned KO term. We clustered proteins that show no significant (E ≤ 10−10) match against any UniProt entry using orthAgogue22 and MCL23, and define each of the resulting groups as an orthAgogue-MCL Homolog Group (OM-HoG).
The 228,391 proteins with UniProt matches were grouped into 40,688 UP-HoGs (mean size 3.85, sd 15.87) and 5679 KO-HoGs (mean size 15.39, sd 32.18), while those with no matches in the database were clustered into 37,483 OM-HoGs (mean size 3.47, sd 2.99); see Supplementary Table S4 for further details on size of the gene sets in each category. Because some dinoflagellate genes are similar in sequence to bacterial genes24,25, we carefully filtered these groups to minimize bacterial contamination (Supplementary Figure S2; see Methods) while attempting to retain true dinoflagellate genes. Most (83%) of the clusters with functional annotation (KO-HoGs and UP-HoGs) show no significant match against any bacterial sequence. Of those that do match a bacterial sequence, nearly one-third are eukaryote-like, with evidence of a multi-exonic CDS. We identified 4296 protein sets as having evidence of putative bacterial contamination, and excluded them from subsequent analysis. Of the 37,483 OM-HoGs, 36,318 (96.9%) have more than one representative in each lineage, and were retained for subsequent analyses. These steps yielded 78,389 protein sets (5331 KO-HoGs, 36,740 UP-HoGs and 36,318 OM-HoGs) for subsequent analysis. We provisionally refer to these sets as gene families.
Functional annotation of gene families
For KO-HoGs and UP-HoGs, function was annotated at the protein level based on 62,339 distinct UniProt matches and their associated Gene Ontology terms (see Methods). For all gene families including OM-HoGs, we also searched for Pfam protein domains as additional support. In total, 33,766 of 78,389 (43%) families were annotated with 48,669 Pfam domains.
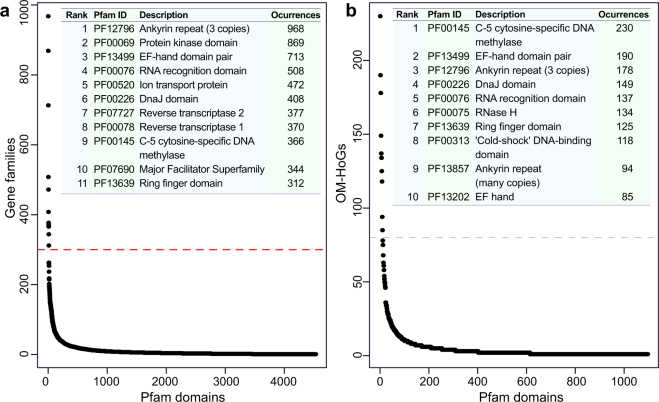
Figure 2a shows the ranked distribution of 4532 distinct Pfam domains found across all gene families, and the identity of those found in >300. Ankyrin repeat (3 copies) (PF12796), protein kinase (PF00069) and EF-hand domain pair (PF13499) domains were found in 968, 869 and 713 gene families. These functions are known to be prevalent in Symbiodinium 8,12. Ankyrins are important for protein-protein interaction (and potentially host-symbiont recognition), and the EF-hand domains are involved in calcium-binding and metabolism8,26. Membrane transport also appears prevalent in these gene families, i.e. ion transport protein (PF00520) and major facilitator superfamily (PF07690) in 472 and 344 respectively. The DnaJ domain found in 408 families are known to be involved in the response of Symbiodinium to photo- and thermal stress14,27,28. Similarly, reverse transcriptase (PF07727 and PF00078) domains in 370 and 377 families respectively may be involved in stress-response mechanisms29–31. The presence of C-5 cytosine-specific DNA methylase (PF00145) domains in 344 families agrees with the hypermethylated state of DNA in Symbiodinium 32 that might be also related to the regulation of gene expression in dinoflagellates33.
Figure 2.
Number of gene families (y-axis) in which each Pfam domain (x-axis) was found in (a) all gene families, and (b) only OM-HoGs. The dashed red line separates the most-prevalent domains, >300 for all gene families and >80 in OM-HoGs, in each case. Identities of these domains are given in the top-right inset of each plot.
We used Pfam domains as a proxy of putative function of OM-HoGs. We recovered 1097 distinct domains distributed among 6283 OM-HoGs. Of these domains, ten were found in >80 families (Fig. 2b). These prevalent domains are largely similar to what we observed in the overall gene families (Fig. 2a). RNase H (PF00075) and RNA recognition (PF00076) domains, found in 134 and 137 families respectively, have been shown to regulate reverse transcription34,35 and splicing36. Two types of ankyrin repeat domains, PF12796 and PF13857, were found in 178 and 94 OM-HoGs respectively. As the functions implicated by these domains are critical to growth and survival, we included OM-HoGs in further analyses.
Dynamics of gene families among Symbiodinium clades
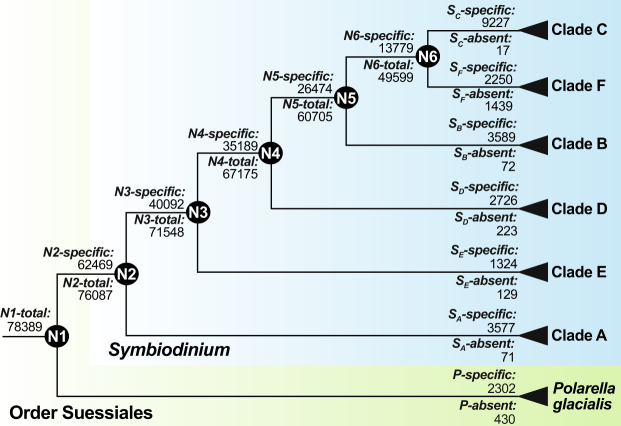
To explore the dynamics of gene families among Symbiodinium clades, we numbered each node (N1 through N6: Fig. 3) on the accepted phylogeny37 and counted the families inferred to be represented at each node (Table 2). We infer a family to be part of Node-total at a node if a member of that family is identified in any lineage descendant from that node, regardless of whether or not the family is represented elsewhere in our dataset. Node-specific families are a subset of these, represented in one or more descendant clades but otherwise not observed in our dataset. The N1-total and N1-specific gene family sets are by this definition identical (Fig. 3); for simplicity we refer to these as N1-total. The membership of all gene sets is given in Supplementary Table S5.
Figure 3.
Changes in gene family numbers in Suessiales shown for Symbiodinium phylogeny (simplified cladogram based on Pochon et al.37), with Polarella glacialis as outgroup. The notation in this diagram is used throughout the text. Numbers of total and specific gene families at each node are shown to the left of the node in question. Numbers of specific and absent gene families for each lineage are correspondingly shown at the tips (right).
Table 2.
Number of gene families in which each lineage is found, shown for annotated (KO-HoGs or UP-HoGs) and non-annotated (OM-HoGs) gene families.
| Lineage | Total | KO-HoGs/UP-HoGs | OM-HoGs |
|---|---|---|---|
| Symbiodinium clade A | 30,409 | 15,399 | 15,010 |
| Symbiodinium clade B | 35,152 | 15,506 | 10,646 |
| Symbiodinium clade C | 43,412 | 23,121 | 20,291 |
| Symbiodinium clade D | 20,833 | 12,434 | 8,399 |
| Symbiodinium clade E | 17,481 | 10,910 | 6,571 |
| Symbiodinium clade F | 14,967 | 7,658 | 7,309 |
| Polarella glacialis | 15,920 | 9,195 | 6,725 |
The count of gene families differs substantially among clades (Table 2). These results may reflect actual genome dynamics (e.g. changes in genome size, gene content and/or sequence divergence) in the various lineages. However, for transcriptomes that lack genome data support, biases arising from the amount or quality of data (Supplementary Figure S3), taxon sampling, or details of data generation or processing (Supplementary Table S1) cannot be dismissed.
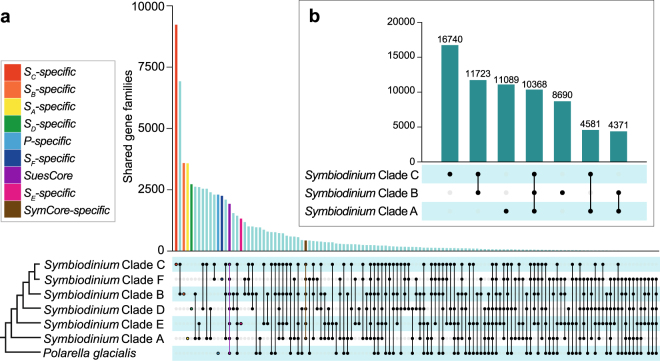
To further explore the differences in gene family number among clades, we define lineage-specific gene families (L-specific, where L is an identified lineage, e.g. L = S A denotes Symbiodinium clade A) as those represented in only that lineage, and L-absent families as those represented in all these lineages except L. The latter have either been lost from L, or are present but were not recovered in these data. Notation of gene families in individual lineages is given in Supplementary Table S6, and the number of shared gene families among all lineages is shown in Fig. 4a. The number of gene families specific to each lineage does not necessarily resemble the changes in gene family number displayed by the nodes. We assessed the effects of unbalanced taxon sampling and of differences in amount of data on the number of gene families inferred as specific to each clade (Supplementary Note and Supplementary Figure S4). Although we observed that taxon sampling could bias our results, the natural diversity of Symbiodinium could also contribute to the observed patterns. The amount of data, on the other hand, seems to impact our results less.
Figure 4.
Gene families shared by (a) all individual lineages within Suessiales, and (b) clades A, B and C when compared among each other. The bars represent the number of gene families shared exclusively by the lineages marked below in the box with dots and connected by lines. In (a), lineage-specific gene families, those shared by all lineages in Suessiales (SuesCore) and SymCore-specific gene families are highlighted according to the colour code at the top right. The simplified topology shown at the bottom left depicts phylogenetic relationships among lineages.
As transcriptome data are inherently incomplete, inferring the gain or loss of genes based on these potentially biased data is not straightforward. Here we discuss our results focusing on clades A, B (for which genome data are available) and C (the most data-rich lineage, with seven transcriptome datasets). S c -specific gene families (9227) are the most abundant overall (Fig. 3), compared to 3577 S A -specific and 3589 S B -specific families. In contrast, S c -absent (17) gene families are the least overall, compared to S A -absent (71) and S B -absent (72) families. The number of gene families specific to and absent from clades A and B are similar despite higher number of gene families in B (Table 2), suggesting no drastic gain of gene families between them.
In an independent analysis of shared gene families among clades A, B and C (Fig. 4b), the more-recently diverged C shares more gene families (11,723) with B than with the basal A (4581). Interestingly, S A -specific families are more abundant than those shared by clade A with B, with C or with both, suggesting either a substantial gain of gene families in A, or an extensive loss of gene families between nodes N2 and N5 (Fig. 3). The latter alternative is supported by fewer families shared between clades A and B than between B and C. Under this scenario, our results suggest that clade C has retained more gene families and undergone further functional diversification than has clade B.
Similar gene-family dynamics are also observed in the numbers of L-specific and L-absent families in all other lineages (Fig. 3), although at this broad scale we cannot dismiss the impact of systematic and data biases such as poor taxa sampling in clades D, E and F (Supplementary Table S1 and Fig. S3), which could also contribute to this observation.
We categorized the gene families according to the lineages in which they are represented, defining those common to all Symbiodinium clades and P. glacialis as SuesCore, and those shared by all Symbiodinium clades (regardless of their presence or absence in P. glacialis) as SymCore. Families shared by all Symbiodinium clades but not P. glacialis, i.e. those exclusive to Symbiodinium, were annotated as SymCore-specific, and in this dataset are equivalent to P-absent. Given the possible combinations of gene-family sharing among these lineages (Fig. 4a), it is remarkable that families exclusive to one lineage are always amongst the major fractions, and more abundant than SymCore-specific. This could be explained by extensive divergence among lineages caused by differential recruitment (or preservation coupled with loss in other lineages), or alternatively by an extent of sequence variation so great as to prevent family members from clustering together with the strategies we employed. The low level of variation (sd ≤ 5 × 10−11) among E values, relative to the top Swiss-Prot match within each UP-HoG, renders the latter alternative less likely. Interestingly, the number of gene families shared by two lineages does not necessary correlate to their phylogenetic proximity. For instance, Symbiodinium clades C and F are closely related (Fig. 3, N6) but exclusively share fewer gene families (2302) than do clades C and B (6923), despite clade B diverging from the C-F lineage at a more-ancestral node (N5). However, clade F is the lineage with the fewest high-quality predicted proteins and the least-complete dataset (Supplementary Figure S1, Supplementary Table S2).
What makes SymbiodiniumSymbiodinium?
For a functional overview at ordinal rank, we tested first for generality versus specificity by comparing SuesCore against all gene families in Suessiales (see Methods). GO terms enriched in SuesCore correspond to a wide variety of biological processes related to cytoplasmic translation, response to environmental factors (salt, temperature, nutrients, bacteria), and regulation of transcription and life cycle (Supplementary Table S7). Among the most-significantly overrepresented Pfam domains in SuesCore we found pentatricopeptide domains (PPRs), ankyrins, domains of AAA chaperone-like ATPases, several dynein domains and a kinesin motor domain (Supplementary Table S8); proteins carrying the latter three types of domain are necessary for movement and assembly of eukaryotic flagella38,39.
To determine what functions characterize Symbiodinium, and distinguish Symbiodinium from Polarella within Order Suessiales, we tested for enrichment of Pfam domains and GO terms in the SymCore and SymCore-specific gene families. GO terms and Pfam domains in SymCore were very similar to those enriched in SuesCore; this was expected, given that P. glacialis contributes few sequences to the latter gene family set (Supplementary Tables S9 and S10).
Amongst SymCore-specific gene families, enriched GO terms describe biological processes that are required for the maintenance of Symbiodinium symbiosis, including transmembrane transport of ions, amino acids and proteins10, mechanisms of response to reactive oxygen species (ROS), and protection against ultraviolet radiation (Supplementary Table S11). Reef habitats of Symbiodinium are typically characterised by high photon flux, and large amounts of ROS are generated during photosynthesis40. Mechanisms involved in nucleotide-excision DNA repair are overrepresented (and in SuesCore and SymCore), suggesting the critical involvement of this process in counteracting the mutagenic effects of UV radiation and free radicals in Symbiodinium. Enriched protein domains included those associated with transmembrane transport, protein-protein interaction potentially involved in host recognition (ankyrins and leucine-rich repeats)41–43, DNA repair, and protection from free radicals (Supplementary Table S12).
Since the enrichment tests compare general versus specific attributes, we expect lineage-specific functions to be underrepresented. For instance, multiple copies of genes encoding components of reverse transcription pathways have so far been reported only in S. kawagutii (clade F)9, and several domains annotated with that function are underrepresented in our SuesCore, SymCore and SymCore-specific gene families. Our results further suggest that certain protein domains considered as abundant in Symbiodinium may be dominant in specific genomes or clades; for instance, the domains involved in DNA methylation and transmembrane amino acid transport were underrepresented in SuesCore and SymCore, as is an EF-hand domain in SymCore-specific gene families.
Lineage-specific enrichment of function
To assess lineage-specific attributes, we systematically identified gene families that are exclusive to, or absent in, each lineage (Supplementary Table S6). The G + C content distribution of the CDS in lineage-specific gene families resembles that of all gene families for that lineage (Supplementary Figure S5), suggesting non-exogenous origins (i.e. there is no evidence for systematic lateral gene transfer). GO terms and Pfam domains enriched in gene families exclusive to each lineage are not necessarily lineage-specific. For example, retrotransposition facilitated by reverse transcription has been reported in Symbiodinium clade F, a conclusion supported by our results (Supplementary Tables S13 and S14). However, gene families exclusive to clades A and B also display enriched GO terms and protein domains related to reverse transcription and retrotransposition (Supplementary Tables S15–18). Although viruses have been found in tight relationship with some Symbiodinium isolates in culture44, our results are not obviously the result of recent viral contamination since the G + C content of CDS associated with retrotransposition and reverse transcription does not differ from that of all CDS in Suessiales (Supplementary Figure S6). In addition, some retrotransposons are known to be activated under stress conditions in other eukaryotes including diatoms29 and plants30,31; our findings may reflect functions relevant to stress-response mechanisms in Symbiodinium. Other examples of GO terms and protein domains enriched in lineage-specific gene families that are not exclusive to a certain lineage include DNA methylation in Symbiodinium clades A and F (Supplementary Tables S15 and S19), and amino acid transmembrane transport in clades B and E (Supplementary Tables S18 and S20).
Among the biological processes annotated in clade A-specific gene families (S A -specific), mechanisms related to adaptation to light conditions and avoidance of photodamage were enriched, including the GO terms Chloroplast avoidance movement, Chloroplast localization, Establishment of plastid localization, Plastid localization, Chloroplast relocation and Phototropism. Free-living isolates have been described in clade A45,46. These capabilities could be beneficial for free-living as well as symbiotic lifestyles, or for the ability to switch between the two. On the other hand, gene families absent only from clade A (S A -absent) are rich in ribosomal protein domains and translational functions (Supplementary Tables S21 and S22).
Free-living isolates have been reported in Symbiodinium clade E as well, including the only isolate in this study. However, adaptive thermal regulation is the only biological process enriched in gene families exclusive to this clade that is obviously associated with the free-living habit (Supplementary Table S20). Many of the enriched functions are related to transmembrane transport, and the most-enriched protein domain in the exclusive gene families was the major facilitator superfamily (Supplementary Tables S20 and S23), a diverse family of membrane transporters implicated in the transport of metabolites and nutrients, including nitrate and nitrite47. Although membrane transport is a characteristic process of Symbiodinium symbioses and members of the major facilitator superfamily have been already reported for other Symbiodinium isolates27, this superfamily seems to have functions of particular relevance in this isolate from clade E.
Several of the most-enriched biological processes and protein domains in S C -specific are linked to GTPase activity or its regulation, more specifically to the Rho GTPase family (Supplementary Tables S24 and S25). Rho GTPases function as molecular switches that activate responses to a wide variety of stimuli including changes in the cytoskeleton, regulation of gene expression, control of the cell cycle and transmembrane trafficking48. Rho-GTPase has been attributed to the rapid evolution of the Atlantic killifish Fundulus heteroclitus by facilitating adaptation to the presence of toxic compounds in the environment49. We therefore hypothesize that the overrepresentation of proteins with Rho GTPase-related functions, and the subsequent capability to respond effectively to different stimuli, could have contributed to the great genetic diversity observed in Symbiodinium clade C and its dominance in the Indo-Pacific ocean50,51.
Symbiodinium clade D are known for their high tolerance to thermal stress52,53. The molecular basis of this resilience has been linked to high proportions of unsaturated fatty acids in the cell membranes, protein folding, and chloroplast proteins involved in photosynthesis or constituents of the thylakoid membrane5. In this study we did not find any overrepresentation in S D -specific of GO terms or Pfam domains annotated with plastid-related functions. However, the GO term Unsaturated fatty acid elongation is overrepresented in S D -specific gene families. Among the overrepresented protein domains are a transcription factor DNA binding domain that regulates expression of heat shock proteins, and a heat shock protein (HSP20), both involved in protein folding in response to thermal stress (Supplementary Tables S26 and S27).
Conclusions
The study of Symbiodinium from a genomic perspective, using both transcriptome and genome data, has broadened our understanding of its evolution, its capability to establish symbiosis and its response to a wide variety of conditions. Here we examined the gene families of six Symbiodinium clades (A-F) to identify functional attributes either shared among, or exclusive to, each of them. We also used data from the closely related species Polarella glacialis to determine which features are characteristic of Symbiodinium within the order Suessiales. Gene families shared among all these Symbiodinium are enriched in functions essential to the establishment and maintenance of symbiosis, and survival in a high-energy environment. At the same time, clade-specific differences in the presence or absence of gene families, and in the enrichment of functions, offer potential for members of distinct clades to specialize in diverse environments. Our results provide a foundation for future investigation of lineage- or clade-specific adaptation of Symbiodinium to their environment, and emphasize the need for more high-quality genomic data from understudied Symbiodinium clades and closely related species (such as Polarella glacialis).
Methods
Data collection and preparation
We collected a total of 30 datasets (Supplementary Figure S1), from which 24 were selected for this study (Table 1) based on quality of assembled sequences and certainty of taxonomic assignment (Supplementary Figure S7), including the published genomes of S. minutum (clade B)8, S. kawagutii (clade F)9 and S. microadriaticum (clade A)10, and 21 transcriptomes (19 from Symbiodinium spp. and two from Polarella glacialis) from previous studies4,5,7,12–14,54 and from the Marine Microbial Eukaryote Transcriptome Sequencing Projects database (MMETSP)15. Characteristics of the datasets are summarised in Supplementary Table S1. Because different methods yield different estimates of completeness55, we compared each dataset with the 458 CEGMA genes16 (BLASTx, E ≤ 10−10) and the BUSCO17 datasets for eukaryotes, alveolates-stramenopiles, and protists (using BUSCO v3.0.2b and by BLASTx, E ≤ 10−10). Sequences in each dataset were additionally searched (BLASTn, E ≤ 10−10) against all bacterial genomes in RefSeq release 76 to assess the proportion of sequences from bacterial sources (putative contaminants).
Where available (i.e. for the three genomes and the transcriptomes from MMETSP), the predicted CDS and proteins were used for the analyses. For the other transcriptome data, we used TransDecoder v2.0.1 (transdecoder.github.io) to predict CDS and proteins at default settings. Completeness of the protein datasets was assessed with CEGMA16 and BUSCO17 genes, as for the original data but using BLASTp instead of BLASTx. Detail for each CDS/protein dataset is shown in Supplementary Table S1. Codon usage of full-length CDS (i.e. CDS that begin with a start codon and end with a stop codon) in each dataset was assessed using chips and cusp from the EMBOSS software suite (emboss.sourceforge.net). Proteins from Symbiodinium isolates within the same clade were pooled together: three datasets in clade A, seven in B, seven in C, two in D, one in E and two in clade F. Similarly, all proteins from the two P. glacialis isolates were pooled as one. Redundant sequences from each clade pool were removed using CD-HIT56 to cluster similar sequences at default settings (sequence identity threshold = 0.90); the longest sequence in each group was kept as representative.
Homolog clusters
To assess protein functions we followed Aranda et al.10 using BLASTp search (E ≤ 10−10) against the UniProt database (release 2016_01). Briefly, protein sequences were first searched against Swiss-Prot, and those with no matches were subsequently searched against the TrEMBL database. The UniProt identifier of the best match for each protein was used to retrieve its associated KEGG Orthology (KO)20 term using UniProtKB ID mapping release 2015_03 (ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping) and the Gene Ontology (GO)21 terms in UniProt-GOA release 163 (ftp.ebi.ac.uk/pub/databases/GO/goa). Proteins without functional annotation were clustered using orthAgogue22 v1.0.3 (e-value cut-off = 10−10) and MCL23 (I = 1.2, scheme 7), as recommended for extensive genetic divergence (expected among Symbiodinium clades1); here we define a group of two or more such proteins as an OM-HoG.
To minimize the inclusion of sequences from potential bacterial sources (i.e. contaminants) in our analysis, we carefully selected a high-confidence set of putative homolog groups of Symbiodinium and Polarella glacialis for subsequent analysis based on the schema detailed in Supplementary Fig. S2. All KO-HoGs and UP-HoGs in which no member matched a bacterial sequence were included in subsequent analysis. For groups in which one or more members matched a bacterial sequence, we referred to the genome data of the corresponding isolate where available. We took the presence of multiple exons in these CDS as evidence of eukaryote origin; homolog groups containing any protein with such evidence were retained for subsequent analyses. For homolog groups in which one or more members matched a bacterial sequence but no genome data were available, and those for which some members had bacterial hits but no multi-exon evidence, we kept any groups in which the bacterial hits are present in two or more lineages; these are potential real dinoflagellate proteins that may have arisen through lateral genetic transfer from a bacterial source24,25. Among the non-annotated OM-HoGs, we considered only those containing proteins from two or more of the original 24 datasets.
Functional analysis of gene families
For our purposes here, we refer to the selected homolog clusters as gene families. We based our functional annotation on the Gene Ontology (GO) terms. Pfam domains57 were annotated for the proteins corresponding to each gene family using PfamScan58 (E ≤ 0.001). For each category in Supplementary Table S6, GO and Pfam-domain enrichment analyses were performed against N1-total as the reference background; here we consider a gene family as the unit of analysis. GO enrichment analysis was performed using the topGO Bioconductor package59 implemented in R v3.2.1, applying Fisher’s Exact test with the ‘elimination’ method to correct for the dependence structure among GO terms. A one-tailed Fisher’s Exact test was used to assess over- and under-representation of Pfam protein domains independently, with adjustment of p-values for multiple tests following Benjamini and Hochberg60.
Data availability
Datasets analysed during the current study are identified and cited in this published article (and its Supplementary Information files).
Electronic supplementary material
Acknowledgements
RAGP is supported by an International Postgraduate Research Scholarship and a University of Queensland Centenary Scholarship. This project is supported by an Australian Research Council Discovery Project grant (DP150101875) awarded to MAR and CXC.
Author Contributions
R.A.G.P. conducted all experiments, and prepared all figures, tables, and the first draft of this manuscript. R.A.G.P., M.A.R. and C.X.C. conceived the study, designed the experiments, analysed and interpreted the results. All authors prepared, wrote, reviewed, commented on and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15029-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baker AC. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 2003;34:661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417. [DOI] [Google Scholar]
- 2.Pochon X, LaJeunesse T, Pawlowski J. Biogeographic partitioning and host specialization among foraminiferan dinoflagellate symbionts (Symbiodinium; Dinophyta) Mar. Biol. 2004;146:17–27. doi: 10.1007/s00227-004-1427-2. [DOI] [Google Scholar]
- 3.Pochon X, Gates RD. A new Symbiodinium clade (Dinophyceae) from soritid foraminifera in Hawai’i. Mol. Phylogenet. Evol. 2010;56:492–497. doi: 10.1016/j.ympev.2010.03.040. [DOI] [PubMed] [Google Scholar]
- 4.Bayer T, et al. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One. 2012;7:e35269. doi: 10.1371/journal.pone.0035269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ladner JT, Barshis DJ, Palumbi SR. Protein evolution in two co-occurring types of Symbiodinium: an exploration into the genetic basis of thermal tolerance in Symbiodinium clade D. BMC Evol. Biol. 2012;12:217. doi: 10.1186/1471-2148-12-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosic N, et al. Unfolding the secrets of coral–algal symbiosis. ISME J. 2015;9:844–856. doi: 10.1038/ismej.2014.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkinson JE, et al. Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus. Symbiodinium. Genome Biol. Evol. 2016;8:665–680. doi: 10.1093/gbe/evw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shoguchi E, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 9.Lin S, et al. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350:691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- 10.Aranda M, et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016;6:39734. doi: 10.1038/srep39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiang T, Nelson W, Rodriguez J, Tolleter D, Grossman AR. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 2015;82:67–80. doi: 10.1111/tpj.12789. [DOI] [PubMed] [Google Scholar]
- 13.Davies SW, Marchetti A, Ries JB, Castillo KD. Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 2016;3:112. doi: 10.3389/fmars.2016.00112. [DOI] [Google Scholar]
- 14.Levin RA, et al. Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 2016;33:2201–2215. doi: 10.1093/molbev/msw119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keeling PJ, et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 17.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 18.Moustafa A, et al. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS One. 2010;5:e9688. doi: 10.1371/journal.pone.0009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liew YJ, Li Y, Baumgarten S, Voolstra CR, Aranda M. Condition-specific RNA editing in the coral symbiont Symbiodinium microadriaticum. PLoS Genet. 2017;13:e1006619. doi: 10.1371/journal.pgen.1006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekseth OK, Kuiper M, Mironov V. OrthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics. 2014;30:734–734. doi: 10.1093/bioinformatics/btt582. [DOI] [PubMed] [Google Scholar]
- 23.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beauchemin M, et al. Dinoflagellate tandem array gene transcripts are highly conserved and not polycistronic. Proc. Natl. Acad. Sci. USA. 2012;109:15793–15798. doi: 10.1073/pnas.1206683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan CX, et al. Analysis of Alexandrium tamarense (Dinophyceae) genes reveals the complex evolutionary history of a microbial eukaryote. J. Phycol. 2012;48:1130–1142. doi: 10.1111/j.1529-8817.2012.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourne, D. G. & Webster, N. S. Coral reef bacterial communities in The Prokaryotes (eds Rosenber, E., DeLong, E. F., Lory, S., Stackebrandt, E. & Thompson, F.) 163–187 (Springer, 2013).
- 27.Leggat W, Hoegh‐Guldberg O, Dove S, Yellowlees D. Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef‐building corals. J. Phycol. 2007;43:1010–1021. doi: 10.1111/j.1529-8817.2007.00387.x. [DOI] [Google Scholar]
- 28.Baumgarten S, et al. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics. 2013;14:704. doi: 10.1186/1471-2164-14-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maumus F, et al. Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics. 2009;10:624. doi: 10.1186/1471-2164-10-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramallo E, Kalendar R, Schulman AH, Martínez-Izquierdo JA. Reme1, a Copia retrotransposon in melon, is transcriptionally induced by UV light. Plant Mol. Biol. 2008;66:137. doi: 10.1007/s11103-007-9258-4. [DOI] [PubMed] [Google Scholar]
- 31.Ito H, et al. A stress-activated transposon in Arabidopsis induces transgenerational abscisic acid insensitivity. Sci. Rep. 2016;6:23181. doi: 10.1038/srep23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ten Lohuis MR, Miller DJ. Hypermethylation at CpG‐motifs in the dinoflagellates Amphidinium carterae (Dinophyceae) and Symbiodinium microadriaticum (Dinophyceae): evidence from restriction analyses, 5‐azacytidine and ethionine treatment. J. Phycol. 1998;34:152–159. doi: 10.1046/j.1529-8817.1998.340152.x. [DOI] [Google Scholar]
- 33.ten Lohuis MR, Miller DJ. Light-regulated transcription of genes encoding peridinin chlorophyll a proteins and the major intrinsic light-harvesting complex proteins in the dinoflagellate Amphidinium carterae Hulburt (Dinophycae). Changes in cytosine methylation accompany photoadaptation. Plant Physiol. 1998;117:189–196. doi: 10.1104/pp.117.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goedken ER, Marqusee S. Folding the ribonuclease H domain of Moloney murine leukemia virus reverse transcriptase requires metal binding or a short N‐terminal extension. Proteins. 1998;33:135–143. doi: 10.1002/(SICI)1097-0134(19981001)33:1<135::AID-PROT12>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Lemay J, et al. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology. 2008;5:47. doi: 10.1186/1742-4690-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pochon X, Putnam HM, Gates RD. Multi-gene analysis of Symbiodinium dinoflagellates: a perspective on rarity, symbiosis, and evolution. PeerJ. 2014;2:e394. doi: 10.7717/peerj.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindemann CB, Lesich KA. Flagellar and ciliary beating: the proven and the possible. J. Cell Sci. 2010;123:519–528. doi: 10.1242/jcs.051326. [DOI] [PubMed] [Google Scholar]
- 39.Blaineau C, et al. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr. Biol. 2007;17:778–782. doi: 10.1016/j.cub.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 40.Dykens JA, Shick JM, Benoit C, Buettner GR, Winston GW. Oxygen radical production in the sea anemone Anthopleura elegantissima and its endosymbiotic algae. J. Exp. Biol. 1992;168:219–241. [Google Scholar]
- 41.Schwarz JA, et al. Coral life history and symbiosis: functional genomic resources for two reef building Caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genomics. 2008;9:97. doi: 10.1186/1471-2164-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jernigan KK, Bordenstein SR. Ankyrin domains across the Tree of Life. PeerJ. 2014;2:e264. doi: 10.7717/peerj.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen MT, Liu M, Thomas T. Ankyrin‐repeat proteins from sponge symbionts modulate amoebal phagocytosis. Mol. Ecol. 2014;23:1635–1645. doi: 10.1111/mec.12384. [DOI] [PubMed] [Google Scholar]
- 44.Lawrence SA, Wilson WH, Davy JE, Davy SK. Latent virus‐like infections are present in a diverse range of Symbiodinium spp. (Dinophyta) J. Phycol. 2014;50:984–997. doi: 10.1111/jpy.12242. [DOI] [PubMed] [Google Scholar]
- 45.Hirose M, Reimer JD, Hidaka M, Suda S. Phylogenetic analyses of potentially free-living Symbiodinium spp. isolated from coral reef sand in Okinawa, Japan. Mar. Biol. 2008;155:105–112. doi: 10.1007/s00227-008-1011-2. [DOI] [Google Scholar]
- 46.Yamashita H, Koike K. Genetic identity of free-living Symbiodinium obtained over a broad latitudinal range in the Japanese coast. Phycol. Res. 2013;61:68–80. doi: 10.1111/pre.12004. [DOI] [Google Scholar]
- 47.Quistgaard EM, Löw C, Guettou F, Nordlund P. Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat. Rev. Mol. Cell Biol. 2016;17:123–132. doi: 10.1038/nrm.2015.25. [DOI] [PubMed] [Google Scholar]
- 48.Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/S0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 49.Nacci D, Proestou D, Champlin D, Martinson J, Waits ER. Genetic basis for rapidly evolved tolerance in the wild: adaptation to toxic pollutants by an estuarine fish species. Mol. Ecol. 2016;25:5467–5482. doi: 10.1111/mec.13848. [DOI] [PubMed] [Google Scholar]
- 50.Stat M, Carter D, Hoegh-Guldberg O. The evolutionary history of Symbiodinium and scleractinian hosts—symbiosis, diversity, and the effect of climate change. Perspect. Plant Ecol. Evol. Syst. 2006;8:23–43. doi: 10.1016/j.ppees.2006.04.001. [DOI] [Google Scholar]
- 51.Pochon X, Pawlowski J. Evolution of the soritids-Symbiodinium symbiosis. Symbiosis. 2006;42:77–88. [Google Scholar]
- 52.Rowan R. Coral bleaching: thermal adaptation in reef coral symbionts. Nature. 2004;430:742–742. doi: 10.1038/430742a. [DOI] [PubMed] [Google Scholar]
- 53.Berkelmans R, Van Oppen MJ. The role of zooxanthellae in the thermal tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. Proc. R. Soc. B Biol. Sci. 2006;273:2305–2312. doi: 10.1098/rspb.2006.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.González-Pech, R. A., Vargas, S., Francis, W. & Wörheide, G. Transcriptomic resilience of a coral holobiont to low pH. bioRxiv, 157008, 10.1101/157008 (2017).
- 55.Veeckman E, Ruttink T, Vandepoele K. Are we there yet? Reliably estimating the completeness of plant genome sequences. Plant Cell. 2016;28:1759–1768. doi: 10.1105/tpc.16.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 57.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li W, et al. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 2015;43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alexa, A. & Rahnenfuhrer, J. TopGO: enrichment analysis for gene ontology. R Package Version 2 (2010).
- 60.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B Met. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets analysed during the current study are identified and cited in this published article (and its Supplementary Information files).