Abstract
Cumulative evidence from biological experiments has confirmed that microRNAs (miRNAs) are related to many types of human diseases through different biological processes. It is anticipated that precise miRNA-disease association prediction could not only help infer potential disease-related miRNA but also boost human diagnosis and disease prevention. Considering the limitations of previous computational models, a more effective computational model needs to be implemented to predict miRNA-disease associations. In this work, we first constructed a human miRNA-miRNA similarity network utilizing miRNA-miRNA functional similarity data and heterogeneous miRNA Gaussian interaction profile kernel similarities based on the assumption that similar miRNAs with similar functions tend to be associated with similar diseases, and vice versa. Then, we constructed disease-disease similarity using disease semantic information and heterogeneous disease-related interaction data. We proposed a deep ensemble model called DeepMDA that extracts high-level features from similarity information using stacked autoencoders and then predicts miRNA-disease associations by adopting a 3-layer neural network. In addition to five-fold cross-validation, we also proposed another cross-validation method to evaluate the performance of the model. The results show that the proposed model is superior to previous methods with high robustness.
Introduction
MiRNAs are a special type of short endogenous non-coding RNA with a length of ~22 nt. MiRNAs are usually regarded as gene repressors at the post-transcriptional level through binding to the 3′-UTRs of the target mRNAs1–4. Nevertheless, miRNAs show a positive influence in regulating genes according to some studies5. Enormous numbers of miRNAs have previously been discovered, including 2588 human genome miRNAs reported in miRBase6. Substantial evidence indicates that miRNAs play a vital role in regulating biological processes such as cell development7, proliferation8, differentiation9, etc. In particular, miRNA dysregulation is related to many human diseases through many factors, including, for example, miRNA-mRNA interactions10, miRNA-lncRNA (long non-coding RNA) interactions11,12, miRNA-protein interactions13, miRNA-environmental factors interactions14, and so on. Among these miRNAs, miR-15 and miR-16 are the first two miRNAs reported to interact with cancers. Calin et al. clarified that miRNAs are deleted in over half of B-cell chronic lymphocytic leukaemia (B-CLL)15 instances. MiR-129, miR-142-5p and miR-25 showed differentially expressed phenomenon in all paediatric brain tumour types16. Accumulated evidence also revealed that the regulation of Ad6 by miR-122 could be significantly helpful in improving the anticancer efficacy of prostate cancer17. Therefore, the inferring associations between miRNAs and diseases could effectively boost the detection of disease biomarkers for cancer treatment, diagnosis, and furthermore, prevention.
By collecting data from biological experiments and raising evidence of connections between miRNAs and diseases, two databases called HMDD18 and miR2Disease19 were constructed. These databases may provide a comprehensive resource for miRNA deregulation in various human diseases. Due to the fast development of computational intelligence, various types of powerful computational methods have emerged with the goal of predicting miRNA-disease associations14,20. Among these methods, many require similarity computation for miRNAs and diseases to accomplish association prediction. Zou et al.21 reviewed the main similarity computational methods. By integrating different source datasets, many approaches have been developed to compute the similarity matrices and give a final prediction regarding the miRNA-disease association. Both machine learning based methods and network-based methods are frequently used in these approaches.
The machine-learning-based methods mainly focus on improving classification accuracy and prediction performance using features extracted from raw data22,23. Many studies have proposed approaches that involve machine-learning algorithms. A typical support vector machine based method was used to predict each miRNA-disease relation24. The lasso regression method was proposed using protein related diseases and miRNAs connections to infer miRNA-disease associations25. Furthermore, Jiang et al. adopted a Naive Bayes model to prioritize disease-related miRNAs using genomic data26. However, these methods require both positive and negative samples to train the models; therefore, a semi-supervised method using regularized least squares (RLS) was proposed to make predictions without using negative samples27. A similar method called Laplacian Regularized Least Squares (LRLS) was adopted to predict long non-coding RNAs and disease association prediction28. This method achieved good success. Another machine-learning based method was RBMMMDA, which utilized a restricted Boltzmann machine (RBM) to predict miRNA-disease association types; however, this method required lots of parameters29.
Network approaches to predict diseases have been successfully applied to explore the relationship between diseases and genes30. Observing the influence and causes of diseases such as cancers via networks has become popular. Network-based methods are typically based on the common assumption that functional related miRNAs tend to be associated with phenotypically similar diseases, and vice versa31,32. Jiang et al. proposed a computational model based on hyper-geometric distribution to detect human miRNA-disease associations33. However, this method relied heavily on predicted association, which results in high false-positive and false-negative rates. Considering that many miRNAs can regulate target genes that may finally cause different diseases if not properly expressed, those gene-related diseases could also have potential associations with gene-related miRNAs. Based on that, Shi et al. presented a random walk algorithm by integrating a protein-protein interaction (PPI) network and utilizing functional links between disease genes and miRNA targets genes34. Furthermore, protein related diseases and miRNAs could also contribute to the inference of novel miRNA-disease associations35. However, the methods mentioned above were restricted by the incomplete disease-related gene/protein network and result in low true-positive rates. Thus, using more data is necessary. Consequently, Wang et al. proposed a method to calculate miRNA functional similarity (MISIM) using miRNA-disease associations and a directed acyclic graph (DAG) of disease annotation. Chen et al. discussed a model called WBSMDA that combined miRNA functional similarity, disease semantic similarity, and Gaussian interaction profile kernel similarity to obtain a more reliable prediction result36. Chen et al. also presented a network similarity-based model using a random walk with restart algorithm (RWRMDA) to predict miRNA-disease associations37. Similarly, Chen further proposed an improved random walk with restart algorithm called HGIMDA to boost the performance of the traditional RWRMDA38. Recently, two methods that used k-nearest-neighbour (KNN) methods, SDMMDA39 and RKNNMDA40, were proposed and gained satisfactory results. You et al. proposed a path-based method and adopted depth-first algorithms to infer potential miRNA-disease associations41 and further improve the performance. Furthermore, Li et al. applied a low-rank matrix recovery method to uncover missing miRNA-disease associations42, and Chen et al. proposed a model called DRMDA43 that utilized a sparse auto-encoder to obtain the representation of miRNA and disease and then chose a SVM classifier to predict miRNA-disease associations.
Previous computational models were limited to low true-positive rates because of the lack of non-linear relationship capture. Deep learning is a recently developed approach that provides applicable solutions, especially for large datasets and non-linear pattern analyses44,45. Deep learning models often play a role of hybrid multiple-layer abstractors by mapping the data to a high-level feature space where the prediction model can be constructed. These approaches have been successfully applied in mutiple bioinformatics scenarios, such as gene expression46, DNA-protein binding prediction47, etc. Deep learning models have been shown to greatly enhance the performance and they show satisfactory results. Due to their powerful ability to capture hidden, complicated, non-linear connections from original data, deep learning models can even act as feature extractors to supplement the original feature input48. The number of miRNA/disease samples is quite small compared to other biological problems; however, the total number of miRNA-disease pair-wise connections is quite large. Therefore, to implement the proposed model using deep learning, every pair-wise miRNA-disease connection is used as one sample to train the network, which helps overcome the lack of training samples.
Integrating more heterogeneous data is also helpful in enhancing the prediction performance49. Therefore, in this study, we integrated multiple datasets related to miRNAs or diseases and then developed a novel deep ensemble framework, DeepMDA, to predict miRNA-disease associations using deep learning. DeepMDA first utilizes a deep learning model to extract high-level features from miRNA and disease similarity matrices; then, a three-layer neural network classifier was implemented to make predictions between miRNA-disease pairs. DRMDA was also observed using an auto-encoder to extract features, but its structure, computational cost and robustness are different from DeepMDA; our proposed model gained better results compared to DRMDA. Overall, our model showed superior performance than did the other five classical miRNA-disease association prediction models in both 5-fold cross-validation and leave-one-disease-out cross-validation.
Results
Five-fold cross validation
Cross-validation is a frequently used method in machine learning and can greatly reduce the bias caused by sample selection. In this case, evaluating the performance of different models is crucial and practical when some of the positive miRNA-disease associations are missing or a new miRNA-disease association is added. To evaluate the prediction performance of DeepMDA, we adopted 5-fold cross-validation compared with other five state-of-the-art computational models (i.e., RLSMDA, HGIMDA, NCPMDA, PBMDA, RKNNMDA). RWRMDA was a representative approach in the domain and was often considered as a standard method to validate performance. HGIMDA was an improved version of RWRMDA that incorporated changes in the data pre-processing procedure. RLSMDA was a semi-supervised method often listed as a compared method in miRNA-disease studies. NCPMDA was a network consistency projection method that showed superior performance compared to HDMP50 and NetCBI51. PBMDA is a path-based method that adopted a depth-first search algorithm to infer potential miRNA-disease associations41. RKNNMDA is a KNN-based model that was combined with the SVM rank method to predict miRNA-disease associations40. Other recently developed methods such as MCMDA52 and ILRMR42, whose datasets used in their studies differ from ours; therefore, we did not choose them for comparison. In the 5-fold cross-validation, all the known interactions were randomly split into 5 subsets with equal size. In each fold, one subset was left out as testing samples, and the remaining four subsets were treated as training sets. The entire procedure was repeated until the entire subset was used for training. The average performance was adopted for evaluation.
The receiver-operating characteristics (ROC) curve was chosen by plotting the true positive rate (TPR, sensitivity) curve against the false positive rate (FPR, specificity) at different thresholds. Specificity is the proportion of samples below the given threshold and sensitivity represents the percentage of samples higher than the threshold. The area under the ROC curve (AUC) was also calculated to evaluate the ability of the prediction model. An AUC value of 1 denotes that the performance is perfect, and an AUC value of 0.5 indicates random prediction performance. Furthermore, we also adopted another type of quality measure used in these types of studies called AUPR (Area Under the Precision vs. Recall Curve). Due to the unbalanced phenomenon of the dataset, the positive data were smaller compared to the negative data. Therefore, AUPR was proposed to reduce the impact caused by a high proportion of false positive data. Similar to the AUC score, AUPR values closer to 1 indicate that the performance is better.
The 152 miRNAs in SM T and 255 miRNAs in SM F are separately utilized with 383 disease similarities to evaluate the performance in small datasets of miRNAs. The experiments implemented 5-fold cross-validation, and the results are shown in Tables 1 and 2. The results show that DeepMDA achieved the highest AUC and AUPR scores in both datasets compared to other algorithms. Furthermore, the three deep models that will be mentioned later had advantages compared to other network based algorithms.
Table 1.
Results on the SM T miRNA datasets.
| Method | AUC | AUPR |
|---|---|---|
| RLSMDA | 0.7715 ± 0.027 | 0.0321 ± 0.004 |
| HGIMDA | 0.7040 ± 0.025 | 0.0402 ± 0.006 |
| NCPMDA | 0.7760 ± 0.012 | 0.0578 ± 0.006 |
| PBMDA | 0.8194 ± 0.032 | 0.2106 ± 0.017 |
| RKNNMDA | 0.5678 ± 0.016 | 0.1581 ± 0.038 |
| DeepMDA | 0.9126* ± 0.004 | 0.2297* ± 0.040 |
| SAE + ADA | 0.8996 ± 0.010 | 0.1859 ± 0.026 |
| RAW + DNN | 0.9102 ± 0.007 | 0.1991 ± 0.028 |
The AUC and AUPR scores are listed above. The * indicates the highest AUC/AUPR score. Generally, the three deep learning models performed better than other five models.
Table 2.
Results on the SM F miRNA datasets.
| Method | AUC | AUPR |
|---|---|---|
| RLSMDA | 0.8325 ± 0.007 | 0.2457 ± 0.008 |
| HGIMDA | 0.7169 ± 0.005 | 0.1182 ± 0.004 |
| NCPMDA | 0.8849 ± 0.006 | 0.3473 ± 0.010 |
| PBMDA | 0.8925 ± 0.005 | 0.4867 ± 0.014 |
| RKNNMDA | 0.7044 ± 0.004 | 0.3365 ± 0.013 |
| DeepMDA | 0.9270* ± 0.005 | 0.5853* ± 0.031 |
| SAE + ADA | 0.8982 ± 0.005 | 0.4376 ± 0.024 |
| RAW + DNN | 0.9153 ± 0.004 | 0.5293 ± 0.026 |
The AUC and AUPR scores are listed above. The * indicates the highest AUC/AUPR score. Generally, the three deep learning models performed better than the other five models.
Next, we integrated multiple datasets and used all the 495 miRNAs in SM with the 383 disease similarities to evaluate the performances. The results of the five different approaches together with DeepMDA are shown in Table 3. DeepMDA, RLSMDA, HGIMDA, NCPMDA, PBMDA, RKNNMDA obtained average AUCs of 0.9486 ± 0.002, 0.8475 ± 0.005, 0.7689 ± 0.011, 0.8731 ± 0.007, 0.9086 ± 0.004, 0.7076 ± 0.005, respectively, in 5-fold cross-validation. DeepMDA also showed the highest AUPR score compared to the five previous methods in all three datasets.
Table 3.
Results on the full miRNA-disease datasets.
| Method | AUC | AUPR |
|---|---|---|
| RLSMDA | 0.8475 ± 0.005 | 0.1157 ± 0.004 |
| HGIMDA | 0.7689 ± 0.011 | 0.1120 ± 0.007 |
| NCPMDA | 0.8731 ± 0.007 | 0.2801 ± 0.011 |
| PBMDA | 0.9086 ± 0.004 | 0.4378 ± 0.016 |
| RKNNMDA | 0.7076 ± 0.005 | 0.3534 ± 0.011 |
| DeepMDA | 0.9486* ± 0.002 | 0.5917* ± 0.014 |
| SAE + ADA | 0.9211 ± 0.002 | 0.4075 ± 0.011 |
| RAW + DNN | 0.9368 ± 0.001 | 0.4933 ± 0.014 |
The AUC and AUPR scores are listed above. The * indicates the highest AUC/AUPR score. Generally, the three deep learning models performed better than the other five models.
To validate the reasonability of proposed model, another two models were implemented based on the deep learning framework. In the first model, we constructed Stacked AutoEncoder(SAE) with Adaboost, called SAE-ADA, as one alternative classifier. Adaboost is an ensemble method often used in machine learning that obtains a more satisfactory result compared to other methods during experiments. In the second model, we used the latter part of DeepMDA, that is, the raw similarity data was used as input, removing the two stacked autoencoders and directly feeding the feature vector to a three-layer fully connected network to construct the classifier (RAW-DNN). We measured these two deep models (SAE + ADA, RAW + DNN) and calculated their AUC values for comparison with DeepMDA. As shown in Table 3, SAE + ADA achieved an average AUC score of 0.9211 ± 0.002 and RAW + DNN obtained an average AUC score of 0.9386 ± 0.001. Therefore, the two deep network models show promising results. We also compared the DRMDA model (i.e., single auto-encoder with SVM) and single auto-encoder with DNN model to validate their performances. As shown in Table 4, DeepMDA still achieved the highest AUC and AUPR scores.
Table 4.
Comparison between DRMDA and DeepMDA on the full miRNA-disease datasets in five-fold cross validation.
| Method | AUC | AUPR |
|---|---|---|
| Single SAE + SVM(DRMDA) | 0.8812 ± 0.006 | 0.4614 ± 0.004 |
| Single SAE + DNN | 0.9394 ± 0.002 | 0.5003 ± 0.010 |
| DeepMDA | 0.9486* ± 0.002 | 0.5917* ± 0.014 |
The AUC and AUPR scores are listed above. Generally, DeepMDA performed better than the other two models.
From these results, it is clear that adopting the SAE as a high-level feature extractor is an essential aspect for improving performance when comparing DeepMDA with RAW + DNN. On the other hand, DNN still inevitably played the role of final classifier when we compared the result of the proposed model with SAE + ADA. Overall, DeepMDA showed a better result in ensemble deep network frameworks compared with two other deep models, and it achieved the best performance of all the compared methods. The standard error of each AUC was small in the five CVs’; therefore, we randomly chose one of the ROC results during 5-fold cross-validation, as depicted in Fig. 1.
Figure 1.
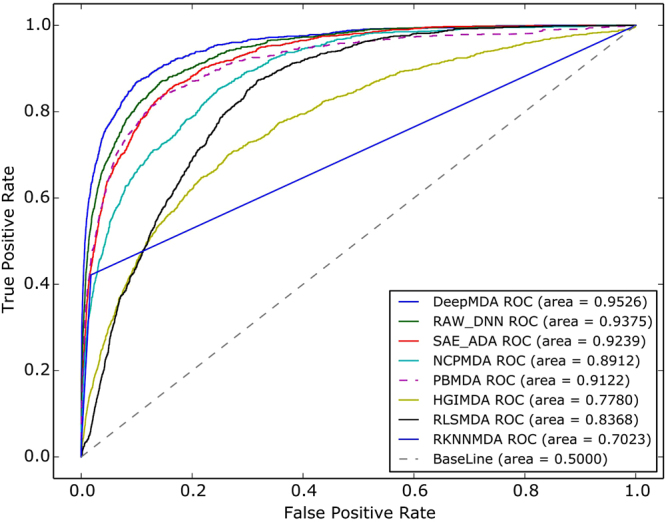
ROC curves and AUC values of eight different methods based on 5-fold cross-validation. The three deep learning models performed better than other 4 network-based methods in general. DeepMDA had the best performance in all the eight models.
Leave-one-disease-out cross-validation
The traditional leave-one-out method of cross-validation (LOOCV) leaves one known miRNA-disease association out in each turn and uses other known associations for model training, and the method then uses that test sample ranking with all the other associations in every iteration. However, training samples were separated from test samples during every recursion in the proposed model, because it could induce bias if we used only one miRNA-disease association as the test sample. Thus, instead of leaving each known miRNA-disease association out and predicting it among all the unknown miRNA-disease association w.r.t. of the investigated disease in each turn, we left every column samples about one disease each time. This method was called Leave-One-Disease-Out Cross-Validation (LODOCV). In every iteration, we tried to predict all the chosen disease-associated miRNAs using the information of other disease-related miRNAs. To our knowledge, LODOCV is considerably more difficult to use than traditional LOOCV because we tried to uncover every miRNA-disease association w.r.t to each disease without any known miRNA-disease information. To be specific, we left all of the diseases as test samples in one iteration. Using other disease-related miRNA information to predict all the unknown disease-associated miRNA associations is a challenging and meaningful problem for researchers and medical diagnoses. Network-based models and three deep learning models were selected to evaluate the overall performance. The results are shown in Table 5. DeepMDA achieved an average AUC score of 0.8729, SAE + ADA obtained an average AUC score of 0.8552, and RAW + DNN reached an AUC score of 0.8633. Five network-based models (RLSMDA, NCPMDA, HGIMDA, PBMDA, RKNNMDA) achieved average AUC scores of 0.8530, 0.6374, 0.7616, 0.6902 and 0.5680, respectively. Regarding the AUPR scores, DeepMDA still achieved the highest AUPR score compared with the other methods. Likewise, we randomly picked one ROC result and drew the ROC curve as shown in Fig. 2. Overall, DeepMDA obtained the best performance in LODOCV compared with the other 7 methods.
Table 5.
Results on the full miRNA-disease datasets in LODOCV.
| Method | AUC | AUPR |
|---|---|---|
| RLSMDA | 0.8530 ± 0.133 | 0.2066 ± 0.240 |
| HGIMDA | 0.7616 ± 0.164 | 0.1025 ± 0.142 |
| NCPMDA | 0.6374 ± 0.220 | 0.0596 ± 0.104 |
| PBMDA | 0.6902 ± 0.223 | 0.2918* ± 0.289 |
| RKNNMDA | 0.5680 ± 0.131 | 0.2085 ± 0.273 |
| DeepMDA | 0.8729* ± 0.118 | 0.2556 ± 0.271 |
| SAE + ADA | 0.8552 ± 0.124 | 0.1914 ± 0.220 |
| RAW + DNN | 0.8633 ± 0.121 | 0.2180 ± 0.248 |
The AUC and AUPR scores are listed above. Generally, DeepMDA performed better than the other seven models in LODOCV.
Figure 2.
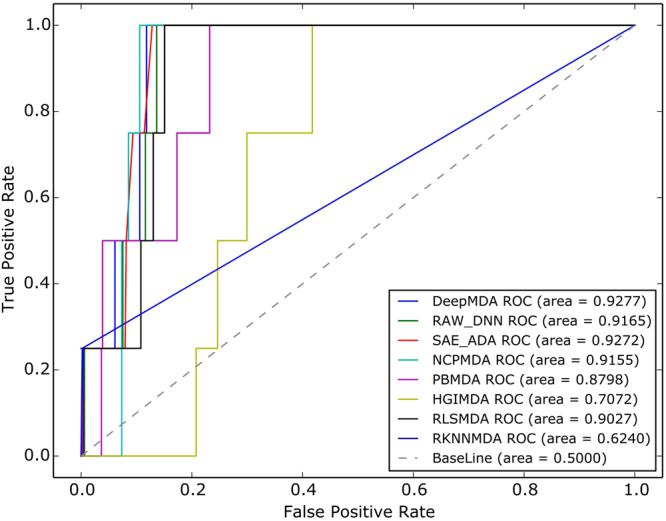
ROC curves and AUC values of eight different methods based on LODOCV. The three deep learning models performed better than other five network-based methods in general. DeepMDA had the best performance in all the eight models.
Robustness in DeepMDA
The deep learning models showed powerful abilities in high-level feature extraction, especially in complex relationship analysis. To measure their abilities to capture the data structure and interaction relationship, we further implemented five-fold cross-validation using noisy data. We added some white noise data to the trained data obtained from the autoencoders, and then implemented a deep neural network classifier, an AdaBoost classifier and a random forest classifier separately to compare their performances. The latter two classifiers were chosen because they are both ensemble classifiers and they achieved more satisfactory prediction results during the experiments. The results in Table 6 show that the AUC score dropped from 0.9486 to 0.9334 using DeepMDA, but the AUC score dropped from 0.9211 to 0.8235 using SAE + ADA and from 0.9249 to 0.8122 using SAE + RF. The AUC score of DRMDA dropped from 0.8812 to 0.7757. This result illustrated that DeepMDA could capture the complex relationships and be robust when noise data were introduced.
Table 6.
Results on the noisy miRNA-disease datasets.
| Method | AUC | AUPR |
|---|---|---|
| Noised DeepMDA | 0.9334 ± 0.005 | 0.4558 ± 0.019 |
| Noised SAE + ADA | 0.8235 ± 0.012 | 0.1613 ± 0.015 |
| Noised SAE + RF | 0.8122 ± 0.015 | 0.1320 ± 0.014 |
| Noised DRMDA | 0.7757 ± 0.044 | 0.1290 ± 0.034 |
| DeepMDA | 0.9486* ± 0.002 | 0.5917* ± 0.014 |
| SAE + ADA | 0.9211 ± 0.002 | 0.4075 ± 0.011 |
| SAE + RF | 0.9249 ± 0.003 | 0.5674 ± 0.012 |
| DRMDA | 0.8812 ± 0.006 | 0.4614 ± 0.004 |
The AUC and AUPR scores are listed above. Generally, DeepMDA performed better than other three models when adding noise.
Case studies
We further investigated some complex human diseases to determine the disease-related miRNAs using the proposed model for measuring model prediction ability. The results showed that human digestive and urinary systems are occasionally deregulated through miRNA functional expression. The oesophagus and colon belong to the digestive system, while the kidneys belong to the urinary system. Therefore, we investigated the potential association between miRNAs and three different diseases, i.e., oesophageal neoplasms, kidney neoplasms and colon neoplasms. The prediction results were validated by checking the experimental results presented in two databases, miR2Disease19 and dbDEMC53, which record many experimentally verified miRNA-disease associations. We implemented LODOCV to predict candidate disease-related miRNAs for these three disease-related cases, and many miRNAs could be precisely predicted using DeepMDA. In total, 47, 42 and 44 out of the top 50 validated miRNAs were predicted w.r.t. colon neoplasms, oesophageal neoplasms, and kidney neoplasms, respectively (see details in Supplementary Table S1, Supplementary Table S2 and Supplementary Table S3).
Colon neoplasms are one of the most severe diseases worldwide54. It was reported that almost half of the patients with colon neoplasms die of metastatic disease within 5 years from diagnosis55,56. Increased evidence has indicated that miRNAs have potential associations with colon neoplasms. For instance, miR-145 may inhibit cell growth in colon neoplasms by targeting the insulin receptor substrate-1 57. Furthermore, tumour specimens showed highly significant and large-fold change differential expression of the levels of several miRNAs, including miR-135b, miR-133a, miR-1, miR-31, and others58. MiR-20a and miR-155 were confirmed to be up-regulated in Colon Neoplasms59. By using DeepMDA, the potential colon neoplasm-related miRNAs were identified, and the results are listed in Supplementary Table S1, which shows that 10 out of the top 10 and 46 out of the top 50 predicted miRNAs were confirmed based on miR2Disease and dbDEMC. For example, an inverse correlation of miR-21 was found in 10 colorectal cell lines suggesting that it might play a role as a useful diagnostic biomarker for colon neoplasms prognosis60,61. To further validate the relationship between predicted miRNAs and cancers, various cancer hallmarks were verified, such as genes that are associated with miRNAs. For instance, some genes such as BRAF, APC, and TP53 can be regarded as colon cancer hallmarks62, and these gene-related miRNAs associations could be validated by miRTarBase63, showing that these miRNAs could possibly regulate these genes. We also found that many disease-related miRNAs are likely to be enriched together; this pattern is similar to disease-associated genes that play roles in some cancer hallmarks30, suggesting that these miRNAs may co-regulate some diseases such as cancers.
Oesophageal neoplasms are one of the most common malignant tumours worldwide and are ranked as the sixth main cause of cancer related deaths64. It has been reported that the overall 5-year survival rate is approximately 20% despite advanced treatments56,65. Improving the understanding of the biological mechanism underlying oesophageal cancer is crucial for diagnosis and disease prevention66. Experimental evidence has revealed that several human miRNAs are located at genomic regions related to the expression of tumour genes such as oesophageal neoplasms67. For instance, miR-155 and miR-103 are highly expressed in tumour tissues and could be correlated with different clinic pathologic classifications68. miR-98 may suppress migration and invasion in human oesophageal squamous cell carcinomas69. Using DeepMDA to predict potential oesophageal neoplasm-related miRNAs could help validate the prediction ability of our model. As a result, 8 out of the top 10 candidates and 42 out of the top 50 predicted miRNAs were selected as having potential relationships with oesophageal neoplasms, according to miR2Disease and dbDEMC (see Supplementary Table S2).
Kidney neoplasms are a type of cancer with an incidence increase of 43% since 1973 in the US70. The risk of the disorder increases with age and differs between men and women. The diagnosed number of kidney neoplasms every year has exceeded 250,000 cases71, among which over 80% are found to have renal-cell-carcinoma (RCC). Recent studies have found that miR-34a can be over-expressed in patients with RCC who suffer from kidney neoplasms72. It also showed that a combination of miR-141 with miR-155 resulted in a 97% correct classification rate, which implied reliable evidence of potential associations between miR-141/miR-155 and kidney neoplasms. To discover the potential associations between miRNAs and kidney neoplasms, we implemented DeepMDA to accomplish the prediction. The results, shown in Supplementary Table S3, were that 8 out of the top 10 and 44 out of the top 50 candidates were chosen as the kidney neoplasms related miRNAs. For example, miR-155, miR-126 and miR-20a were found over-expressed in malignant samples such as clear-cell type human renal cell carcinoma73. miR-145 was reported to down-regulate its target mRNA and the corresponding protein in kidney tissues74.
Overall, the results from LODOV and the separate case studies on three typical diseases showed satisfactory performances using DeepMDA. Unlike the traditional method, which uses prior knowledge to perform the prediction, the proposed method was capable of capturing one specific potential disease related to the relationship miRNAs without relying on any known information. Therefore, DeepMDA can be applied to a wide range of applications. To make the model more useful for the community and biologists, we also developed a web server to search for each potential disease-related miRNA that our model predicted (https://laiyifu.shinyapps.io/DeepMDA/).
Discussions
Increasing evidence shows that miRNA genes are located at genomic regions involved in cancer, indicating that miRNAs play significant roles in the development of various diseases. Due to the limitations of previous computational models, a more effective and less costly way to predict miRNA-disease associations is required. In this study, a deep ensemble miRNA-disease association prediction (DeepMDA) framework was proposed by synthesizing heterogeneous biological networks. First, miRNA functional similarity and heterogeneous Gaussian interaction profile kernel similarities were integrated to form the miRNA similarity data and the disease semantic information. In addition, heterogeneous disease-related data were utilized to construct disease similarity data. Second, two similarity data matrices were segmented by lines separately and fed into two stacked autoencoders to learn complex high-level features. Then, the two output feature vectors from two SAEs were concatenated to form an independent feature vector, whose corresponding label was picked from a known miRNA-disease association matrix. The latter part of DeepMDA used a three-layer fully connected neural network to make the final predictions of the potential miRNA-disease associations with the feature vectors gained from two autoencoders. Both LODOCV and 5-fold cross-validation were implemented to validate DeepMDA performance. Compared with five state-of-the-art computational models and two other deep models, DeepMDA showed the best performance and good robustness compared to the other deep models. Furthermore, case studies were also implemented using several complex human diseases (colon neoplasms, oesophageal neoplasms, and kidney neoplasms), in which 47, 42 and 44 out of the top 50 predicted miRNAs, respectively, had experimentally supported evidence based on previous literature. DeepMDA can also be used to predict the miRNAs associated with isolated diseases, which could benefit human disease diagnoses and prevention.
There are several reasons that account for the reliable performance of DeepMDA. First, multiple dataset sources (more knowledge) were adopted to enlarge the miRNA and disease similarity matrices, and more data could provide more evidence when trying to predict the associations of disease-related miRNAs. Second, a deep ensemble framework was proposed to extract high-level features from traditional feature vectors and predict the potential associations using these non-linear high-level features, which improved the model’s performance compared to other state-of-the-art models.
Furthermore, the proposed model can be regarded as a more general model that may play a potential role in predicting other kinds of associations, such as lncRNA-disease, drug-targets, and so on.
Methods
Datasets
Biological experiments have collected many miRNA-disease associations, and multiple databases were constructed for researchers to verify the research. The human miRNA-disease dataset used in this study was downloaded from the HMDD database (June 2013)18. It consists of 5430 validated distinct experimental human miRNA-disease associations of approximately 495 miRNAs and 383 diseases. We used adjacency matrix A md to represent miRNA-disease associations. For instance, if miRNA m(i) is reported to be associated with disease d(j) in the database HMDD, the value of a md (i,j) is 1; otherwise, it is 0. The number of miRNAs and diseases in the database are denoted as nm and nd, respectively.
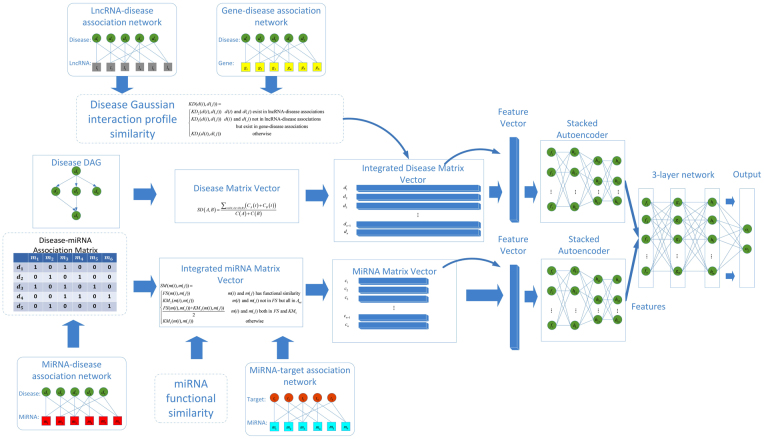
We also adopted disease related long noncoding RNAs (lncRNAs) data from LncRNADisease75. The LncRNADisease database has integrated more than 1000 lncRNA-disease entries including 321 lncRNAs and 221 diseases from ~500 publications. Furthermore, disease-related gene data was retrieved from the DisGeNET (Version 4.0) database76. We chose curated gene-disease association containing 14412 genes and 10757 unique diseases from DisGeNET. Using these two data sets, we constructed two adjacency matrixes A ld and A gd to denote the lncRNA-disease associations and the gene-disease associations, respectively. Furthermore, due to the close relationship between miRNAs and their corresponding targets, we utilized the experimentally validated miRNA-target interaction data from miRTarBase63. MiRTarBase has collected more than 41000 human miRNA-target interactions, including 2649 miRNAs and 14894 targets that are validated through various studies. The adjacency matrix A mt was constructed to represent miRNA-target associations. The overall design of the dataset integration is shown in Fig. 3.
Figure 3.
The flowchart of proposed DeepMDA. The miRNA similarity was integrated using miRNA-functional similarity and miRNA-disease association. As for disease similarity, we adopted DAG information and Gaussian interaction profile similarity information. The two input data was fed into two stacked autoencoders to learn high-level features, then merged and finally utilized a 3-layer network to infer the association between miRNAs and diseases.
Gaussian interaction profile kernel similarity for miRNAs
Based on the assumption that similar miRNAs with similar functions tend to be associated with similar diseases, and vice versa, the interaction profile of miRNA m(i) is denoted by a binary vector IP(m(i)) representing whether miRNA m(i) is interacted with each disease or not. Then, the kernel for the two miRNAs m(i) and m(j) are defined to calculate the Gaussian kernel similarity based on their interaction profiles, which are defined as follows:
| 1 |
| 2 |
where γ m used to control the kernel bandwidth and is obtained by normalizing a new bandwidth parameter by the average number of associated diseases for all the miRNAs. Here, is set to 1 according to previous research38.
Likewise, similar miRNAs with similar functions tend to be related to similar target genes, and vice versa. Thus, by using miRNA-target interaction matrix A mt, we could also obtain 2649 miRNAs Gaussian interaction profile kernel similarity matrix. The calculation is the same as before.
| 3 |
| 4 |
where γ m is calculated by normalizing , which divided the average number of associated targets for all miRNAs. is set to 1 again.
Integrate similarity for miRNAs
Because of the lack of data concerning the 495 miRNAs similarity scores, we chose to integrate miRNA functional similarity and two Gaussian interaction profile kernel similarity matrices for a new mixed similarity for each pair of miRNAs. Specifically, for a miRNA pair m(i) and m(j) that exist only in the functional similarity matrix, the miRNA functional similarity is chosen as their integrated similarity score: if m(i) and m(j) do not exist in the miRNA functional similarity matrix but both exist in the KM 2 matrix, we chose their Gaussian profile kernel similarity score from KM 2 as their integrated similarity score, and if m(i) and m(j) both exist in the miRNA functional similarity matrixes FS and KM 2, the average score was calculated. If the two miRNAs do not exist in the matrixes FS or KM 2, we adopted their Gaussian profile kernel similarity score from KM 1 as the integrated similarity. The overall integrated similarity score between miRNA m(i) and m(j) is as follows:
| 5 |
Other similarity for small dataset of miRNAs
In addition, we also obtained two miRNA similarity matrices SM F and SM T separately from the miRNA functional similarity matrix FS and the miRNA Gaussian profile similarity matrix KM 2 gained from the miRNA-target associations. Specifically, 255 miRNAs in SM F both appeared in HMDD and the miRNA functional similarity matrix FS. Similarly, there are 152 miRNAs in SM T that exist in both HMDD and miRNA-target associations. These two similarity matrices SM F and SM T were also used to train and test our model, and their performance was evaluates using the same procedure as was used for SM.
Disease semantic similarity
Many diseases’ MeSH descriptors are collected in the MeSH database, which can be downloaded from the National Library of Medicine (http://www.nlm.nih.gov)77. Each disease can be described as an entry item in a Directed Acyclic Graph (DAG), such as DAG(D) = (D, T(D), E(D)), where T(D) stands for the node set that includes node D itself and its ancestor nodes,E(D) represents the corresponding edge set that directly links the parent nodes to the child nodes. Here, we chose the MeSH descriptor starting with the capital letter “C” to acquire the diseases to construct the disease DAGs. Each tree number corresponds to a specific position in the DAG collected from each MeSH descriptor. In the traditional disease semantic similarity calculation78, disease terms in the same layer would contribute the same to the disease semantic value of disease A as an example. However, if two disease terms (disease A and B) occur in the same layer of disease DAGs but their frequency varies in all the DAGs, this causes an inaccurate measurement of the contributions of the two disease terms. Consequently, we adopted an alternative way to calculate the semantic value based on the assumption that a more frequent disease term should have a greater contribution to the semantic value of disease A, which is shown as follows:
| 6 |
The semantic value of disease A was calculated by summing the contribution from all the disease terms in DAG(A).
| 7 |
Finally, the semantic similarity between diseases A and B can be obtained by summing the contributions of disease terms shared by the following two DAGs:
| 8 |
Gaussian interaction profile kernel similarity for diseases
Similar to the miRNA Gaussian similarity matrix construction, the disease Gaussian interaction profile kernel similarity matrices were also computed using three association matrices, the miRNA-disease association matrix, the lncRNA-disease association matrix, and the gene-disease association matrix. Three Gaussian interaction profile kernel matrices, KD 1, KD 2 and KD 3, were obtained and integrated to determine the overall 383 diseases in the Gaussian profile kernel similarity matrix, defined as follows:
| 9 |
The integrated disease Gaussian interaction profile kernel similarity matrix can be used to adjust our model and improve its performance, which will be discussed in the next section.
DeepMDA
In this study, we proposed a deep ensemble framework for miRNA-disease association prediction (DeepMDA). DeepMDA is a neural network structure composed of two parts. First, for every miRNA-disease pair a md (i, j), we assigned the labels of all the known miRNA-disease association pair (positive samples) to 1; otherwise, to 0. Second, the ith of the miRNA similarity matrix (i.e., the similarity data between miRNA i and all the other miRNAs) was fed into a stacked autoencoder to learn another representation, the jth row of disease similarity matrix and the jth row of integrated disease Gaussian interaction profile similarity matrix (i.e., the similarity data between disease j and all the other diseases) were concatenated into a feature vector and regarded as an independent training sample, which was fed to another stacked autoencoders. The autoencoder was a stacked deep neural network that can be trained to learn high-level biological patterns and was already implemented in some bioinformatics field such as yeast microarrays analysis79 and DNA Methylation state prediction48. Third, two separate features were then merged and integrated into a three-layer fully connected neural network to predict the label of each pair sample, which indicated whether it has a connection or not. The flowchart of DeepMDA is shown in Fig. 3.
Stacked autoencoder
The nm miRNA samples correspond to nm rows of miRNA similarity data and the nd disease samples correspond to nd rows of the disease similarity data. These were fully connected to form a large dataset of nm × nd samples. These pair-wise samples were separately fed into two autoencoders consisting of multiple layers. Autoencoders have been widely used in capturing complex biological patterns80.
Assume an input data x has d dimensions. Its mapping formation is constructed as follows:
| 10 |
where f is a non-linear function that maps the linear result of x to a non-linear space.
The output y is then projected back to form the reconstruction output z, which has the same shape as original input x. The equation is as follows:
| 11 |
where is the reconstruction-weighting matrix, is the reconstruction bias, and g is another non-linear function same as f. The entire reconstruction procedure needs to calculate the error; therefore, we chose the mean squared error between x and z, which can be optimized using stochastic gradient descent (SGD)81. All the parameters used in the network implemented greedy layer-wise learning, which learns the parameters of one layer while freezing the parameters of the other layers.
In this study we did not utilize a commonly used deep network module, such as a convolutional neural network (CNN)47 or a recurrent neural network (RNN)82 because our input was purely a similarity item corresponding to a miRNA or a disease without sequence or positional information. Therefore, CNN and RNN do not show great improvements and introduce large computational cost. As an alternative, we used fully connected layers with an activation function and dropout layers83 to construct an autoencoder. Dropout layers mainly help to avoid the possibility of over-fitting by randomly dropping some neuron units. The dropout rates were all set to 0.5 during model training. Finally, for a pairwise sample, we obtained two pairs of extracted high-level features using the autoencoders; these were input to the classifier to make the final predictions.
Deep neural network
After the two autoencoders extracted two parts of high-level features, they were concatenated to form an integrated sample feature vector. Altogether, there were nm × nd samples, and the label of each sample was 1 if the miRNA-disease pair has connection according to the known relationship in the miRNA-disease association matrix otherwise 0. The combined feature vector was then fed into a feed-forward neural network consisting of three fully connected layers. We set the output dimension of each autoencoder to 64; therefore, a 128-dimensional feature vector was fed to the network. In the fully connected layer, a three-layer neural network was implemented to obtain the final prediction of the association between each miRNA and disease. The number of layers we chose here was dependent on the experiments, and the best results were obtained when the three-layer network was utilized. The predicted association possibility of each pairwise miRNA-disease sample exceeding the threshold was considered as a potential disease-related miRNA, and vice versa.
In the fully connected layer, a three-layer neuron network was implemented to get the final prediction of the association between each miRNA and disease. The predicted association possibility of each pairwise miRNA-disease sample exceeded the threshold was considered as a potential disease-related miRNA and vice versa. All the neuron units in the layer i was connected to the previous layer (i − 1) and generated outputs using non-linear transformation function f.
| 12 |
where H is the number of hidden neurons, are the weights and bias of neuron j which sums up all the hidden units. After one fully-connected layer, the network executed a rectified linear activation function(ReLU).
| 13 |
Activation function ReLU is a non-linear function that can capture hidden patterns within the data46 and can reduce gradient vanishing in the meantime. Dropout was also used behind every fully connected layer to avoid overfitting. The final output utilized sigmoid function to make prediction of each sample, which is shown as follows:
| 14 |
To train the model, we need to minimize the objective function in order to minimize the loss. We chose one function frequently used called cross-entropy cost function C 84.
| 15 |
where C is the loss function output called cross-entropy cost function. And x is the index of the training examples and t indicates the index of different labels, y represents the true label for sample x, 0 or 1 respectively and a indicates the predicted output of the model for 0 or 1 label given input sample x. The more the predicted outputs approaches the true values, the less value C gets. As the cross-entropy function is non-negative, our goal is to minimize the function to get the best prediction.
Neural network models were trained using the Keras 1.0.1 library (https://github.com/fchollet/keras) with Tensorflow as the backend. The ADADELTA algorithm85 with a mini batch size of 200 was used to minimize the loss on the training set. The batch number was set to 200 because the model achieved the best performance using the 200-batch size. All the weights were initialized using a Gaussian distribution with a standard deviation of 0.05, and its corresponding bias was initialized ranging from unif (−1.0,0.0) as is typical. A computer with an NVIDIA Tesla K80 GPU was used to train the model. The Python code and the datasets are all available at https://github.com/sperfu/DeepMDA.
Electronic supplementary material
Acknowledgements
This work was jointly supported by the National Natural Science Foundation of China (Grant Numbers 61173111).
Author Contributions
L.F. conceived the project, developed the prediction model, designed and implemented the experiments, analyzed the result, and wrote the paper. Q.P. designed the prediction model and provided scientific advice. All authors reviewed the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15235-6.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hammond SM. An overview of microRNAs. Advanced Drug Delivery Reviews. 2015;87:3. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Subbiah R, Pattarayan D, Rajaguru P, Sudhakar Gandhi PS, Thimmulappa RK. MicroRNA Regulation of Acute Lung Injury and Acute Respiratory Distress Syndrome. Journal of Cellular Physiology. 2016;231:2097. doi: 10.1002/jcp.25316. [DOI] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffithsjones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Research. 2010;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp X, Ambros V. Encountering MicroRNAs in Cell Fate Signaling. Science. 2005;310:1288–1289. doi: 10.1126/science.1121566. [DOI] [PubMed] [Google Scholar]
- 8.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Research. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miska EA. How microRNAs control cell division, differentiation and death. Current Opinion in Genetics & Development. 2005;15:563. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Liang C, Wong KC, Luo J, Zhang Z. Mirsynergy: detecting synergistic miRNA regulatory modules by overlapping neighbourhood expansion. Bioinformatics. 2014;30:2627–2635. doi: 10.1093/bioinformatics/btu373. [DOI] [PubMed] [Google Scholar]
- 11.Chen, X., Yan, C. C., Xu, Z. & You, Z. H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Briefings in Bioinformatics (2016). [DOI] [PMC free article] [PubMed]
- 12.Chen X, Huang YA, Wang XS, You ZH, Chan KC. FMLNCSIM: fuzzy measure-based lncRNA functional similarity calculation model. Oncotarget. 2016;7:45948–45958. doi: 10.18632/oncotarget.10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H, et al. Integration of Multiple Genomic and Phenotype Data to Infer Novel miRNA-Disease Associations. Plos One. 2015;11:e0148521. doi: 10.1371/journal.pone.0148521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X, Liu MX, Cui QH, Yan GY. Prediction of disease-related interactions between microRNAs and environmental factors based on a semi-supervised classifier. Plos One. 2012;7:e43425. doi: 10.1371/journal.pone.0043425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GA C, et al. Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sredni ST, Huang CC, Bonaldo MF, Tomita T. MicroRNA expression profiling for molecular classification of pediatric brain tumors. Pediatric Blood & Cancer. 2011;57:183–184. doi: 10.1002/pbc.23105. [DOI] [PubMed] [Google Scholar]
- 17.Zhang, Z., Zhang, X., Newman, K. & Liu, X. MicroRNA Regulation of Oncolytic Adenovirus 6 for Selective Treatment of Castration-Resistant Prostate Cancer. in 8th Congress of the European Association for Palliative Care (EAPC). 34–34 (2012). [DOI] [PubMed]
- 18.Li Y, et al. HMDDv2.0: a database for experimentally supported human microRNA and disease associations. Nucleic Acids Research. 2013;42:1070–1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Q, et al. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Research. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng, X., Zhang, X. & Zou, Q. Integrative approaches for predicting microRNA function and prioritizing disease-related microRNA using biological interaction networks. Briefings in Bioinformatics17 (2016). [DOI] [PubMed]
- 21.Zou Q, Li J, Song L, Zeng X, Wang G. Similarity computation strategies in the microRNA-disease network: a survey. Briefings in Functional Genomics. 2016;15:55. doi: 10.1093/bfgp/elv024. [DOI] [PubMed] [Google Scholar]
- 22.Gu B, et al. Incremental learning for v-Support Vector Regression. Neural Networks the Official Journal of the International Neural Network Society. 2015;67:140–150. doi: 10.1016/j.neunet.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Wen X, Shao L, Xue Y, Fang W. A rapid learning algorithm for vehicle classification. Information Sciences. 2015;295:395–406. doi: 10.1016/j.ins.2014.10.040. [DOI] [Google Scholar]
- 24.Jiang, Q., Wang, G., Zhang, T. & Wang, Y. Predicting human microRNA-disease associations based on support vector machine. in IEEE International Conference on Bioinformatics and Biomedicine. 467–472 (2010). [DOI] [PubMed]
- 25.Qabaja A, Alshalalfa M, Bismar TA, Alhajj R. Protein network-based Lasso regression model for the construction of disease-miRNA functional interactions. EURASIP J Bioinform Syst Biol. 2013;2013:3. doi: 10.1186/1687-4153-2013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, Q. H. W., G.H., Wang, Y.D. An approach for prioritizing disease-related microRNAs based on genomic data integration. In 3rd International Conference on Biomedical Engineering and Informatics (2010).
- 27.Chen X, Yan GY. Semi-supervised learning for potential human microRNA-disease associations inference. Scientific Reports. 2014;4:5501. doi: 10.1038/srep05501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen X. Novel human lncRNA–disease association inference based on lncRNA expression profiles. Bioinformatics. 2013;29:2617–2624. doi: 10.1093/bioinformatics/btt426. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, et al. RBMMMDA: predicting multiple types of disease-microRNA associations. Scientific Reports. 2015;5:13877. doi: 10.1038/srep13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang E, et al. Predictive genomics: a cancer hallmark network framework for predicting tumor clinical phenotypes using genome sequencing data. Seminars in cancer biology. 2015;30:4–12. doi: 10.1016/j.semcancer.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 31.Lu M, et al. An Analysis of Human MicroRNA and Disease Associations. Plos One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics. 2010;26:1644–1650. doi: 10.1093/bioinformatics/btq241. [DOI] [PubMed] [Google Scholar]
- 33.Jiang Q, et al. Prioritization of disease microRNAs through a human phenome-microRNAome network. BMC Systems Biology. 2010;4(Suppl 1):S2. doi: 10.1186/1752-0509-4-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi H, et al. Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Systems Biology. 2013;7:1–12. doi: 10.1186/1752-0509-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mørk S, Pletscherfrankild S, Caro AP, Gorodkin J, Jensen LJ. Protein-driven inference of miRNA–disease associations. Bioinformatics. 2014;30:392. doi: 10.1093/bioinformatics/btt677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, et al. WBSMDA: Within and Between Score for MiRNA-Disease Association prediction. Scientific Reports. 2016;6:21106. doi: 10.1038/srep21106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X. RWRMDA: predicting novel human microRNA-disease associations. Molecular Biosystems. 2012;8:2792–2798. doi: 10.1039/c2mb25180a. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, et al. HGIMDA: Heterogeneous graph inference for miRNA-disease association prediction. Oncotarget. 2016;7:65257–65269. doi: 10.18632/oncotarget.11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, et al. A novel computational model based on super-disease and miRNA for potential miRNA-disease association prediction. Molecular Biosystems. 2017;13:1202. doi: 10.1039/C6MB00853D. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Wu QF, Yan GY. RKNNMDA: Ranking-based KNN for MiRNA-Disease Association prediction. RNA biology. 2017;14:952–962. doi: 10.1080/15476286.2017.1312226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You ZH, et al. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. Plos Computational Biology. 2017;13:e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng L, et al. Improved low-rank matrix recovery method for predicting miRNA-disease association. Scientific Reports. 2017;7:6007. doi: 10.1038/s41598-017-06201-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen, X., Gong, Y., Zhang, D. H., You, Z. H. & Li, Z. W. DRMDA: deep representations-based miRNA-disease association prediction. Journal of Cellular & Molecular Medicine (2017). [DOI] [PMC free article] [PubMed]
- 44.Hinton GE, Salakhutdinov RR. Reducing the Dimensionality of Data with Neural Networks. Science. 2006;313:504. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 45.Lecun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 46.Chen Y, Li Y, Narayan R, Subramanian A, Xie X. Gene expression inference with deep learning. Bioinformatics. 2016;32:1832. doi: 10.1093/bioinformatics/btw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng H, Edwards MD, Liu G, Gifford DK. Convolutional neural network architectures for predicting DNA–protein binding. Bioinformatics. 2016;32:i121. doi: 10.1093/bioinformatics/btw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, et al. Predicting DNA Methylation State of CpG Dinucleotide Using Genome Topological Features and DeepNetworks. Scientific Reports. 2016;6:19598. doi: 10.1038/srep19598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Zeng X, He Z, Zou Q. Inferring microRNA-disease associations by random walk on a heterogeneous network with multiple data sources. IEEE/ACM transactions on computational biology and bioinformatics. 2016 doi: 10.1109/TCBB.2016.2550432. [DOI] [PubMed] [Google Scholar]
- 50.Xuan P, et al. Prediction of microRNAs associated with human diseases based on weighted k most similar neighbors. PLoS One. 2013;8:e70204. doi: 10.1371/journal.pone.0070204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H, Zhang Z. Similarity-based methods for potential human microRNA-disease association prediction. BMC Medical Genomics. 2013;6:1–9. doi: 10.1186/1755-8794-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li, J.-Q., Rong, Z.-H., Chen, X., Yan, G.-Y. & You, Z.-H. MCMDA: Matrix Completion for MiRNA-Disease Association prediction.pdf. Oncotarget (2017). [DOI] [PMC free article] [PubMed]
- 53.Yang Z, et al. dbDEMC: a database of differentially expressed miRNAs in human cancers. BMC genomics. 2010;11(Suppl 4):S5. doi: 10.1186/1471-2164-11-S4-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogatakawata H, et al. Circulating Exosomal microRNAs as Biomarkers of Colon Cancer. Plos One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drusco A, et al. MicroRNA Profiles Discriminate among Colon Cancer Metastasis. Plos One. 2014;9:e96670. doi: 10.1371/journal.pone.0096670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. Ca A Cancer Journal for Clinicians. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 57.Shi B, et al. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. Journal of Biological Chemistry. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 58.Sarver AL, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:1–15. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Asangani IA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 61.Schetter AJ, et al. MicroRNA Expression Profiles Associated With Prognosis and Therapeutic Outcome in Colon Adenocarcinoma. Jama. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao S, et al. Identification and Construction of Combinatory Cancer Hallmark-Based Gene Signature Sets to Predict Recurrence and Chemotherapy Benefit in Stage II Colorectal Cancer. JAMA oncology. 2016;2:37–45. doi: 10.1001/jamaoncol.2015.3413. [DOI] [PubMed] [Google Scholar]
- 63.Chou CH, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Research. 2015;44:gkv1258. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He B, et al. MicroRNAs in esophageal cancer (review) Molecular medicine reports. 2012;6:459–465. doi: 10.3892/mmr.2012.975. [DOI] [PubMed] [Google Scholar]
- 65.Enzinger PC, Mayer RJ. Esophageal cancer. New England Journal of Medicine. 2004;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 66.Xie Z, et al. Salivary MicroRNAs as Promising Biomarkers for Detection of Esophageal Cancer. Plos One. 2013;8:e57502. doi: 10.1371/journal.pone.0057502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wan J, Wu W, Che Y, Kang N, Zhang R. Insights into the potential use of microRNAs as a novel class of biomarkers in esophageal cancer. Diseases of the Esophagus. 2015;29:412–420. doi: 10.1111/dote.12338. [DOI] [PubMed] [Google Scholar]
- 68.Guo Y, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Research. 2008;68:26. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 69.Huang SD, et al. MicroRNA-98 and microRNA-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Molecular Cancer. 2012;11:51. doi: 10.1186/1476-4598-11-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogelzang NJ, Stadler WM. Kidney cancer. Lancet. 1998;352:1691. doi: 10.1016/S0140-6736(98)01041-1. [DOI] [PubMed] [Google Scholar]
- 71.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. Ca A Cancer Journal for Clinicians. 2012;60:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 72.Jung M, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. Journal of Cellular & Molecular Medicine. 2009;13:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chow TFF, et al. Differential expression profiling of microRNAs and their potential involvement in renal cell carcinoma pathogenesis. Clinical Biochemistry. 2010;43:150. doi: 10.1016/j.clinbiochem.2009.07.020. [DOI] [PubMed] [Google Scholar]
- 74.Lu MC, et al. Decreased microRNA(miR)-145 and increased miR-224 expression in T cells from patients with systemic lupus erythematosus involved in lupus immunopathogenesis. Clinical & Experimental Immunology. 2013;171:91–99. doi: 10.1111/j.1365-2249.2012.04676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen G, et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Research. 2013;41:983–986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Piñero J, et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Research. 2017;45:D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lipscomb CE. Medical Subject Headings (MeSH) Bulletin of the Medical Library Association. 2000;88:265–266. [PMC free article] [PubMed] [Google Scholar]
- 78.Chen, X. et al. Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. 5, 11338 (2015). [DOI] [PMC free article] [PubMed]
- 79.Chen L, Cai C, Chen V, Lu X. Learning a hierarchical representation of the yeast transcriptomic machinery using an autoencoder model. BMC bioinformatics. 2016;17:S9. doi: 10.1186/s12859-015-0852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan, J., Hammond, J. H., Hogan, D. A. & Greene, C. S. ADAGE-Based Integration of Publicly Available Pseudomonas aeruginosa Gene Expression Data with Denoising Autoencoders Illuminates Microbe-Host Interactions. mSystems1, 10.1128/mSystems.00025-15 (2016). [DOI] [PMC free article] [PubMed]
- 81.Le, Q. V. et al. Building high-level features using large scale unsupervised learning. 8595–8598 (2011).
- 82.Quang D, Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Research. 2016;44:e107. doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. Journal of Machine Learning Research. 2014;15:1929–1958. [Google Scholar]
- 84.Boer PTD, Kroese DP, Mannor S, Rubinstein RY. A Tutorial on the Cross-Entropy Method. Annals of Operations Research. 2014;134:19–67. doi: 10.1007/s10479-005-5724-z. [DOI] [Google Scholar]
- 85.Zeiler, M. D. ADADELTA: An Adaptive Learning Rate Method. Computer Science (2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.