Abstract
Background
Hemicelluloses are a diverse group of complex, non-cellulosic polysaccharides, which constitute approximately one-third of the plant cell wall and find use as dietary fibres, food additives and raw materials for biofuels. Genes involved in hemicellulose synthesis have not been extensively studied in small grain cereals.
Results
In efforts to isolate the sequences for the cellulose synthase-like (Csl) gene family from wheat, we identified 108 genes (hereafter referred to as TaCsl). Each gene was represented by two to three homeoalleles, which are named as TaCslXY_ZA, TaCslXY_ZB, or TaCslXY_ZD, where X denotes the Csl subfamily, Y the gene number and Z the wheat chromosome where it is located. A quarter of these genes were predicted to have 2 to 3 splice variants, resulting in a total of 137 putative translated products. Approximately 45% of TaCsl genes were located on chromosomes 2 and 3. Sequences from the subfamilies C and D were interspersed between the dicots and grasses but those from subfamily A clustered within each group of plants. Proximity of the dicot-specific subfamilies B and G, to the grass-specific subfamilies H and J, respectively, points to their common origin. In silico expression analysis in different tissues revealed that most of the genes were expressed ubiquitously and some were tissue-specific. More than half of the genes had introns in phase 0, one-third in phase 2, and a few in phase 1.
Conclusion
Detailed characterization of the wheat Csl genes has enhanced the understanding of their structural, functional, and evolutionary features. This information will be helpful in designing experiments for genetic manipulation of hemicellulose synthesis with the goal of developing improved cultivars for biofuel production and increased tolerance against various stresses.
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1142-z) contains supplementary material, which is available to authorized users.
Keywords: Arabinoxylan, Bioenergy, Biofuels, Cell wall, Cellulose, CesA, Csl, Glucuronoarabinoxylan, Mixed-linked glucan, Wheat
Background
Plant cell wall consists of three main polysaccharide fractions: cellulose, hemicellulose, and pectin, with lignin and proteins being the other two constituents. Grass walls contain mainly two of the three polysaccharide fractions with pectin being a rather minor constituent. Hemicelluloses are plant cell wall matrix polysaccharides that possess diverse linear or branched structures [1, 2]. These mainly encompass 1–4-β-glucan, 1,3;1,4-β-glucan, galactan, and glucomannan in grasses [3]. In addition, glucuronoarabinoxylan is a major grass wall constituent. Because of the presence of heterogeneous substituents or other linkages on their polymer backbones, hemicelluloses are non-crystalline and can be readily hydrolysed in comparison to cellulose. These polysaccharides can interact with cellulose microfibrils through hydrogen bonds [4].
Hemicellulosic polysaccharide backbones in plants are made by the cellulose synthase-like (Csl) enzymes, which are members of a much larger superfamily of genes referred to as glycosyltransferase 2 (GT2) [5]. Several other GTs, i.e., xyloglucan α-1,6-xylosyltransferases (GT34), xyloglucan fucosyltransferases (GT37), and xyloglucan galactosyltransferases (GT47) have been reported to be involved in the biosynthesis of xyloglucans [6]. Genes encoding Csl enzymes share sequence similarity with the cellulose synthase A (CesA) gene family known to form cellulose throughout the plant kingdom [7]. A variable number of Csl genes ranging from 30 to 50 have been reported from different plant species and are classified into nine subfamilies (CslA–CslH and CslJ) [8, 9]. Cereals generally lack CslB and CslG families. Among the remaining families, CslA, CslC, and CslD are conserved in all land plants, whereas CslF, CslH are restricted to grasses [10, 11]. A poorly understood subfamily, CslJ, has been reported in grasses as well as dicots, which contrasts with the previous claims of its occurrence only in grasses [12, 13]. Similarly, the subfamilies CslB and CslG were previously reported to be specific to dicots [14]. However, a recent report established the presence of the CslB subfamily in monocots as well [12]. Several of the Csl subfamilies have been reported to be involved in the biosynthesis of different cell wall polysaccharides. For example, subfamily CslA was shown to form β-1,4-mannan backbone of galactomannan and glucomannan [15, 16]. Similarly, CslF and CslH subfamilies were shown to make 1–3;1–4-β-glucan in grasses [17, 18], whereas CslC genes were associated with the formation of the 1–4-β-glucan backbone of a xyloglucan and some other polysaccharides [19].
Wheat is a major cereal crop grown on the largest area of arable land in the world, is second only to maize in grain production, and feeds approximately 40% of the world population [20]. It has a large genome size (~17 Gb), of which ~80–90% is repetitive [21]. Even after the complete genome sequence became available [22], Csl genes remain unidentified and uncharacterized in bread wheat. In general, homeologous copies of most of the genes are located on each of the three chromosomes belonging to each of the subgenomes (A, B, and D), suggesting that the number of Csl genes is expected to approximately three-times that of a diploid species like rice. We used publicly available resources to retrieve wheat genome sequence. Large-scale data mining was performed using the Pfam domain models for the identification of Csl gene family members, which are reported in this study.
Methods
Data sources and sequence retrieval
Wheat genome data were downloaded from the Ensembl Plants FTP server (ftp://ftp.ensemblgenomes.org/pub/current/plants/fasta/triticum_aestivum/), generated by the International Wheat Genome Sequencing Consortium (IWGSC) and converted into a local BLAST database using the UNIX pipeline. BLAST analyses (BLASTN as well as BLASTP) were performed using the stand-alone command line version of NCBI (National Center for Biotechnology Information) blast 2.2.28+ (ftp://ftp.ncbi.nih.gov/blast/executables/LATEST/), released March 19, 2013. A query file was generated from Pfam domain models; PF00535 (GT2) domain and PF03552 (Cellulose_synt) downloaded from Pfam 30.0 June 2016 release [23]. The sequences of splice variants were also retrieved from Ensembl Plants browser (http://plants.ensembl.org/Triticum_aestivum/Info/Index). Analysis of splice variants was conducted as described by Kim et al. (2007) [24]. Previously known Csl sequences from Arabidopsis thaliana, Oryza sativa, and Zea mays were downloaded from the Cell Wall Navigator database [25]. For Brachypodium, sequences were retrieved from phytomine (https://phytozome.jgi.doe.gov). Amino acid sequences of the aforementioned CSL proteins are given in Additional file 1: Figure S1.
Blast searches for wheat homologs
All query files containing the two Pfam domain models (PF00535 and PF03552) were used to perform the BLASTn searches against the local blast database of bread wheat. All blast hits with E-value >1.0 were removed. Using cut-off E- value <1.0, all previously known CesA genes were retrieved. After the compilation of all the sequences below the cut-off value, CD-hit program was used to obtain non-redundant sequences. Higher cut-off E- value was used to ascertain the identification of all the genes that possessed the Pfam domains PF00535 and PF03552. These genes were further filtered through phylogenetic analysis alongwith previously known CSL proteins from Arabidopsis, Brachypodium, maize, and rice, which reflected some non-targeted genes that were removed from further analysis [26]. Phylogenetic analysis was also implemented to categorize different Csl sub-families. CesA genes were distinguished from the Csl genes with the CXC motif, which is diagnostic of the CesA but absent from the Csl proteins [7, 27]. Presence of the conserved domains Cellulose_synt/GT2 was confirmed using a batch blast search at the CDD (conserved domain database) of NCBI. Homeologous genes from each of the three genomes were named TaCslXY_ZA, TaCslXY_ZB, or TaCslXY_ZD, where X denotes the Csl subfamily, Y the gene number and Z the wheat chromosome where it is located. Alignment of the sequences of all newly identified wheat Csl genes is given in Additional file 2: Figure S2.
Protein structure and motif/domain identification
Protein sequences were downloaded from the Ensembl Plants FTP server (ftp://ftp.ensemblgenomes.org/pub/current/plants/fasta/triticum_aestivum/), developed by the International Wheat Genome Sequencing Consortium (IWGSC) [22]. Multiple protein sequence alignments were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) [28]. The resulting alignments were analysed for the presence of conserved motifs (D, D, DXD, QXXRW) of the GT2 superfamily. Conserved patterns of aligned sequences were highlighted using the sequence manipulation suite: Color align conservation (http://www.bioinformatics.org/sms2/color_align_cons.html) [29]. The conserved domains were predicted using CCD database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) [22, 30, 31]. Wheat Csl genes were named based on their sequence identity, coverage, presence of conserved domains and motifs similar to those of the previously identified rice Csl genes. The number of genes in in a subfamily exceeded that of rice, the additional genes were given new names. Because of the resemblance of CslD genes with CesA genes and their probable role in cellulose synthesis, we specifically focused on the TaCslD subfamily. Gene structures and intron evolution of TaCslD members were predicted using the gene structure display server 2.0 (http://gsds.cbi.pku.edu.cn/) using the genomic and cDNA sequences.
Evolutionary relationships of Csl genes
A total of 215 CSL proteins from Arabidopsis, maize, rice and wheat were aligned using MAAFT (v1.3.6) [32]. Sequences that did not extend over the conserved core region were removed. Positions where more than 40% of the sequences contained a gap were also removed. The phylogeny and 1000 bootstrap replications of these sequences was inferred using Seqboot (v3.696) [33] and FastTree (v2.1.10) implemented on the Guillimin cluster [34].
The phylogeny of the CslD subfamily was also determined separately from Arabidopsis, Brachypodium, maize, rice and wheat. For phylogenetic analysis, the amino acid sequences of CSL proteins were aligned using MUSCLE and their evolutionary history was inferred using Neighbor-Joining methods [35]. The tree was drawn to scale, with branch lengths being equivalent to the evolutionary distances used to infer the phylogenetic tree. Evolutionary distances were computed with a Poisson correction and are given as the number of amino acid substitutions per site. The rate of variation among sites was modeled with a gamma distribution (shape parameter = 1) and all positions containing gaps and missing data were removed. Evolutionary analyses were conducted in MEGA6 [36].
RNA-seq expression analysis
Publicly available RNA-seq data generated from bread wheat (var. Chinese Spring) was used to study the expression of newly identified wheat Csl genes. The data were compiled from five different wheat tissues (spike, leaf, stem, root, and grain) collected at seedling, vegetative and reproductive stages of development [37]. The relative expression of each TaCsl subfamily was presented as a heat map generated from the relative abudnace of transcripts (per 10 million reads) for each gene using wheat expression browser powered by expVIP (http://www.wheat-expression.com).
Results
Identification and classification of Csl gene family members in bread wheat
Database searches for bread wheat using conserved pfam motifs PF00535 and PF03552, which are specific to the GT2 superfamily, resulted in the identification of 108 cellulose synthase-like (TaCsl) genes (Table 1). Two to three homeologous copies of each gene from the A, B and D genomes were common. The identified genes were named following the nomenclature of rice, which shares synteny with wheat. To avoid the complexity of the nomenclature, a suffix corresponding to the chromosome number and the specific wheat genome identifier (A, B, or D) has been used for each gene name [7]. For example, the first gene of subfamily CslA; CslA1 on the long arm of chromosome 1 of genomes A, B, and D is named as TaCslA1_1AL, TaCslA1_1BL, and TaCslA1_1DL, respectively.
Table 1.
Homeologous copies of the bread wheat Csl genes
| No. | Ensembl ID | Gene name | Corresponding gene in rice |
|---|---|---|---|
| 1 | TRIAE_CS42_6BS_TGACv1_513375_AA1639370.1 | TaCslA1_6BS | CslA1 |
| 2 | TRIAE_CS42_6AS_TGACv1_485966_AA1554960.1 | TaCslA1_6AS | CslA1 |
| 3 | TRIAE_CS42_2AL_TGACv1_093375_AA0278800.1 | TaCslA2_2AL | CslOS09G39920 |
| 4 | TRIAE_CS42_2BL_TGACv1_129747_AA0394630.1 | TaCslA2_2BL | CslOS09G39920 |
| 5 | TRIAE_CS42_2DL_TGACv1_160461_AA0550770.1 | TaCslA2_2DL | CslOS09G39920 |
| 6 | TRIAE_CS42_1AS_TGACv1_019142_AA0061550.1 | TaCslA2_1AS | CslOS09G39920 |
| 7 | TRIAE_CS42_7BS_TGACv1_592860_AA1945380.1 | TaCslA3_7BS | CslA3 |
| 8 | TRIAE_CS42_7DS_TGACv1_623146_AA2050070.1 | TaCslA3_7DS | CslA3 |
| 9 | TRIAE_CS42_7AS_TGACv1_569190_AA1809650.1 | TaCslA3_7AS | CslA3 |
| 10 | TRIAE_CS42_6DS_TGACv1_543811_AA1744360.1 | TaCslA4_6DS | CslA10/4/2 |
| 11 | TRIAE_CS42_6AS_TGACv1_487286_AA1569690.1 | TaCslA4_6AS | CslA10/4/2 |
| 12 | TRIAE_CS42_6BS_TGACv1_513376_AA1639390.1 | TaCslA4_6BS | CslA10/4/2 |
| 13 | TRIAE_CS42_2BS_TGACv1_146583_AA0468630.1 | TaCslA5_2BS | CslA5/7 |
| 14 | TRIAE_CS42_2AS_TGACv1_113418_AA0355820.1 | TaCslA5_2AS | CslA5/7 |
| 15 | TRIAE_CS42_2DS_TGACv1_177473_AA0578070.1 | TaCslA5_2DS | CslA5/7 |
| 16 | TRIAE_CS42_3DL_TGACv1_249033_AA0835410.1 | TaCslA6_3DL | CslA11 |
| 17 | TRIAE_CS42_3B_TGACv1_221079_AA0729630.1 | TaCslA6_3B | CslA11 |
| 18 | TRIAE_CS42_3AL_TGACv1_197519_AA0666560.1 | TaCslA6_3AL | CslA11 |
| 19 | TRIAE_CS42_2AS_TGACv1_113300_AA0354190.1 | TaCslA7_2AS | CslA5/7 |
| 20 | TRIAE_CS42_2DS_TGACv1_177798_AA0584795.1 | TaCslA7_2DS | CslA5/7 |
| 21 | TRIAE_CS42_3B_TGACv1_220828_AA0720500.1 | TaCslA8_3B | CslA11 |
| 22 | TRIAE_CS42_3DS_TGACv1_273022_AA0927600.1 | TaCslA8_3DS | CslA11 |
| 23 | TRIAE_CS42_U_TGACv1_642146_AA2112270.1 | TaCslA9 | CslA9 |
| 24 | TRIAE_CS42_7BL_TGACv1_579090_AA1903960.1 | TaCslA9_7BL | CslA9 |
| 25 | TRIAE_CS42_7AL_TGACv1_558725_AA1795700.1 | TaCslA9_7AL | CslA9 |
| 26 | TRIAE_CS42_U_TGACv1_642146_AA2112290.1 | TaCslA10 | CslA9 |
| 27 | TRIAE_CS42_7DL_TGACv1_602617_AA1962870.1 | TaCslA10_7DL | CslA9 |
| 28 | TRIAE_CS42_7AL_TGACv1_557254_AA1778850.1 | TaCslA10_7AL | CslA9 |
| 29 | TRIAE_CS42_7BL_TGACv1_578444_AA1895100.1 | TaCslA10_7BL | CslA9 |
| 30 | TRIAE_CS42_3AS_TGACv1_210508_AA0674280.1 | TaCslA11_3AS | CslA11 |
| 31 | TRIAE_CS42_3DS_TGACv1_272005_AA0912960.1 | TaCslA11_3DS | CslA11 |
| 32 | TRIAE_CS42_3B_TGACv1_223332_AA0780350.1 | TaCslA11_3B | CslA11 |
| 33 | TRIAE_CS42_3DL_TGACv1_251593_AA0882850.1 | TaCslC1_3DL | CslC1 |
| 34 | TRIAE_CS42_3AL_TGACv1_197197_AA0665370.1 | TaCslC1_3AL | CslC1 |
| 35 | TRIAE_CS42_3DS_TGACv1_271926_AA0910940.1 | TaCslC3_3DS | CslC3 |
| 36 | TRIAE_CS42_3B_TGACv1_220758_AA0718310.1 | TaCslC3_3B | CslC3 |
| 37 | TRIAE_CS42_3AS_TGACv1_211225_AA0686890.1 | TaCslC3_3AS | CslC3 |
| 38 | TRIAE_CS42_1DL_TGACv1_061928_AA0205730.1 | TaCslC7_1DL | CslC7 |
| 39 | TRIAE_CS42_1BL_TGACv1_030750_AA0099830.1 | TaCslC7_1BL | CslC7 |
| 40 | TRIAE_CS42_1AL_TGACv1_001272_AA0028090.1 | TaCslC7_1AL | CslC7 |
| 41 | TRIAE_CS42_1DL_TGACv1_062162_AA0209740.1 | TaCslC9_1DL | CslC10/9 |
| 42 | TRIAE_CS42_1BL_TGACv1_030501_AA0092480.1 | TaCslC9_1BL | CslC10/9 |
| 43 | TRIAE_CS42_5BL_TGACv1_404820_AA1311790.1 | TaCslC10_5BL | CslC10/9 |
| 44 | TRIAE_CS42_5DL_TGACv1_435778_AA1454840.1 | TaCslC10_5DL | CslC10/9 |
| 45 | TRIAE_CS42_5AL_TGACv1_374268_AA1195590.1 | TaCslC10_5AL | CslC10/9 |
| 46 | TRIAE_CS42_1BL_TGACv1_030586_AA0094860.1 | TaCslD1_1BL | CslD1 |
| 47 | TRIAE_CS42_1AL_TGACv1_001700_AA0034150.1 | TaCslD1_1AL | CslD1 |
| 48 | TRIAE_CS42_1DL_TGACv1_063091_AA0223780.1 | TaCslD1_1DL | CslD1 |
| 49 | TRIAE_CS42_2BS_TGACv1_148683_AA0494520.1 | TaCslD3_2BS | CslD3 |
| 50 | TRIAE_CS42_2DS_TGACv1_177279_AA0572180.1 | TaCslD3_2DS | CslD3 |
| 51 | TRIAE_CS42_2AS_TGACv1_114244_AA0365360.1 | TaCslD3_2AS | CslD3 |
| 52 | TRIAE_CS42_1BS_TGACv1_049706_AA0160220.1 | TaCslD4_1BS | CslD4 |
| 53 | TRIAE_CS42_5BS_TGACv1_425241_AA1392650.1 | TaCslD4_5BS | CslD4 |
| 54 | TRIAE_CS42_5DS_TGACv1_457675_AA1488780.1 | TaCslD4_5DS | CslD4 |
| 55 | TRIAE_CS42_7BL_TGACv1_577301_AA1871610.1 | TaCslD5_7BL | CslD5 |
| 56 | TRIAE_CS42_7AL_TGACv1_559436_AA1799630.1 | TaCslD5_7AL | CslD5 |
| 57 | TRIAE_CS42_7DL_TGACv1_603510_AA1985050.1 | TaCslD5_7DL | CslD5 |
| 58 | TRIAE_CS42_5DL_TGACv1_433536_AA1415830.1 | TaCslE1_5DL | CslE6/1 |
| 59 | TRIAE_CS42_5BL_TGACv1_406235_AA1342600.1 | TaCslE1_5BL | CslE6/1 |
| 60 | TRIAE_CS42_6DL_TGACv1_526558_AA1687090.1 | TaCslE2_6DL | CslE2 |
| 61 | TRIAE_CS42_6AL_TGACv1_471004_AA1500600.1 | TaCslE2_6AL | CslE2 |
| 62 | TRIAE_CS42_6BL_TGACv1_499967_AA1596110.1 | TaCslE2_6BL | CslE2 |
| 63 | TRIAE_CS42_U_TGACv1_683314_AA2158770.1 | TaCslE3 | CslE6/1 |
| 64 | TRIAE_CS42_6DS_TGACv1_543277_AA1737920.1 | TaCslE4_6DS | CslE6/1 |
| 65 | TRIAE_CS42_5DL_TGACv1_433536_AA1415840.1 | TaCslE6_5DL | CslE6/1 |
| 66 | TRIAE_CS42_5BL_TGACv1_406235_AA1342610.1 | TaCslE6_5BL | CslE6/1 |
| 67 | TRIAE_CS42_5AL_TGACv1_376126_AA1232370.1 | TaCslE6_5AL | CslE6/1 |
| 68 | TRIAE_CS42_2DL_TGACv1_159781_AA0542640.1 | TaCslF1_2DL | CslF1/2/4 |
| 69 | TRIAE_CS42_2AL_TGACv1_094713_AA0301960.1 | TaCslF1_2AL | CslF1/2/4 |
| 70 | TRIAE_CS42_2DL_TGACv1_160109_AA0546890.1 | TaCslF1_2DL | CslF1/2/4 |
| 71 | TRIAE_CS42_2BL_TGACv1_130934_AA0420130.1 | TaCslF1_2BL | CslF1/2/4 |
| 72 | TRIAE_CS42_7BL_TGACv1_580651_AA1914920.1 | TaCslF2_7BL | CslF1/2/4 |
| 73 | TRIAE_CS42_7AL_TGACv1_557532_AA1782680.1 | TaCslF2_7AL | CslF1/2/4 |
| 74 | TRIAE_CS42_7DL_TGACv1_602590_AA1961740.1 | TaCslF2_7DL | CslF1/2/4 |
| 75 | TRIAE_CS42_2AS_TGACv1_113659_AA0359050.1 | TaCslF3_2AS | CslF3 |
| 76 | TRIAE_CS42_2DS_TGACv1_177641_AA0581710.1 | TaCslF3_2DS | CslF3 |
| 77 | TRIAE_CS42_2BS_TGACv1_148608_AA0494060.1 | TaCslF3_2BS | CslF3 |
| 78 | TRIAE_CS42_2BS_TGACv1_146146_AA0456710.1 | TaCslF4_2BS | CslF1/2/4 |
| 79 | TRIAE_CS42_2DS_TGACv1_179076_AA0604160.1 | TaCslF4_2DS | CslF1/2/4 |
| 80 | TRIAE_CS42_2DS_TGACv1_178985_AA0603230.1 | TaCslF5_2DS | CslF3 |
| 81 | TRIAE_CS42_2AS_TGACv1_112790_AA0345230.1 | TaCslF5_2AS | CslF3 |
| 82 | TRIAE_CS42_2BS_TGACv1_148027_AA0489970.1 | TaCslF5_2BS | CslF3 |
| 83 | TRIAE_CS42_7BL_TGACv1_577473_AA1876170.1 | TaCslF6_7BL | CslF6 |
| 84 | TRIAE_CS42_7AL_TGACv1_555973_AA1751470.1 | TaCslF6_7AL | CslF6 |
| 85 | TRIAE_CS42_7DL_TGACv1_607937_AA2011180.1 | TaCslF6_7DL | CslF6 |
| 86 | TRIAE_CS42_5BL_TGACv1_409916_AA1366600.1 | TaCslF7_5BL | CslF7 |
| 87 | TRIAE_CS42_5DL_TGACv1_433902_AA1424880.1 | TaCslF7_5DL | CslF7 |
| 88 | TRIAE_CS42_5AL_TGACv1_374191_AA1193100.1 | TaCslF7_5AL | CslF7 |
| 89 | TRIAE_CS42_2BS_TGACv1_148916_AA0495580.1 | TaCslF8_2BS | CslF8 |
| 90 | TRIAE_CS42_2DS_TGACv1_178471_AA0596060.1 | TaCslF8_2DS | CslF8 |
| 91 | TRIAE_CS42_2AS_TGACv1_112322_AA0335280.1 | TaCslF8_2AS | CslF8 |
| 92 | TRIAE_CS42_2AS_TGACv1_112322_AA0335290.1 | TaCslF9_2AS | CslF9 |
| 93 | TRIAE_CS42_2BS_TGACv1_147667_AA0486240.1 | TaCslF9_2BS | CslF9 |
| 94 | TRIAE_CS42_2DS_TGACv1_177329_AA0573830.1 | TaCslF9_2DS | CslF9 |
| 95 | TRIAE_CS42_U_TGACv1_641498_AA2096480.1 | TaCslF10 | CslF9 |
| 96 | TRIAE_CS42_1BS_TGACv1_049866_AA0163180.1 | TaCslF10_1BS | CslF9 |
| 97 | TRIAE_CS42_2AL_TGACv1_094351_AA0296300.1 | TaCslH1_2AL | Cslh1/2 |
| 98 | TRIAE_CS42_2DL_TGACv1_158387_AA0517170.1 | TaCslH1_2DL | CslH1/2 |
| 99 | TRIAE_CS42_2BL_TGACv1_129372_AA0380770.1 | TaCslH1_2BL | CslH1/2 |
| 100 | TRIAE_CS42_3B_TGACv1_221049_AA0728260.1 | TaCslH2_3B | Csl |
| 101 | TRIAE_CS42_3DS_TGACv1_273502_AA0931770.1 | TaCslH2_3DS | Csl |
| 102 | TRIAE_CS42_3DS_TGACv1_271739_AA0907200.1 | TaCslH3_3DS | Csl |
| 103 | TRIAE_CS42_3AS_TGACv1_212952_AA0704280.1 | TaCslH3_3AS | CslH3 |
| 104 | TRIAE_CS42_3B_TGACv1_222234_AA0760340.1 | TaCslH3_3B | Csl |
| 105 | TRIAE_CS42_3DS_TGACv1_272297_AA0918580.1 | TaCslJ1_3DS | Csl |
| 106 | TRIAE_CS42_3AS_TGACv1_210908_AA0681280.1 | TaCslJ1_3AS | Csl |
| 107 | TRIAE_CS42_3B_TGACv1_221705_AA0747940.1 | TaCslJ2_3B | Csl |
| 108 | TRIAE_CS42_3DS_TGACv1_272756_AA0924850.1 | TaCslJ2_3DS | Csl |
An unrooted neighbor-joining (NJ) tree for the 215 derived Csl proteins from Arabidopsis, maize, rice and wheat is shown in Fig. 1. TaCsl proteins grouped into seven subfamilies: TaCslA (32 proteins), TaCslC (13 proteins), TaCslD (12 proteins), TaCslE (10 proteins), TaCslF (29 proteins), TaCslH (8 proteins), and TaCslJ (4 proteins) (Fig. 2). The TaCslA and TaCslC subfamilies were closely related as shown by their taxonomic distribution and phylogenies. As expected, these subfamilies were conserved across the plant species. Although TaCslD is present in all the plant species whereas TaCslF is specific to grasses, their proximity to each other suggests a common origin [12]. Among the sequences common to both dicots and grasses, subfamily CslA appeared to be the most divergent between these two groups of plants. Whereas the sequences within the subfamilies CslC and CslD were interspersed between Arabidopsis and grasses, all the subfamily CslA sequences of Arabidopsis clustered together, separately from the grass CslA sequences. Proximity of the CslB and CslH subfamilies points to their common origin before the separation of grasses from dicots. Similarly, CslG and CslJ apparently had a common origin.
Fig. 1.
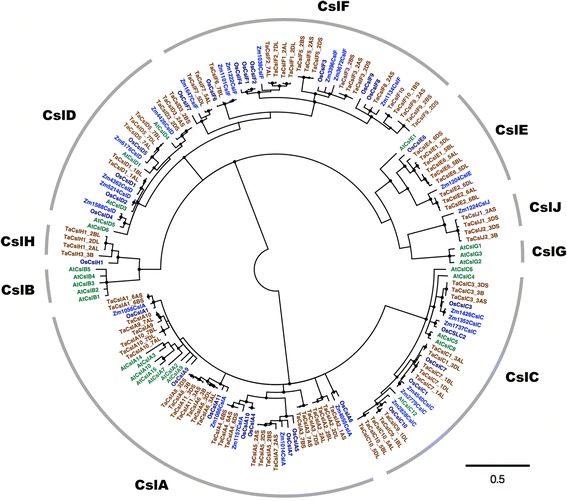
An unrooted maximum likelihood phylogenetic tree of the Cellulose synthase-like (Csl) gene family from Arabidopsis, maize, rice and wheat using FastTree (v2.1.10) according to Price et al. (35). Nodes with more than 70% support from 1000 bootstrap replications were considered significant and indicated by a black circle. Different colors represent CSL proteins from different species. The scale bar indicates a radial distance equal to 0.5 amino acid substitutions per site. To keep the gene family nomenclature uniform, maize gene models from Gramene were renamed as follows: Zm, first four digits of the locus number, Csl, and the class identifier as described in Schwerdt et al. (9)
Fig. 2.
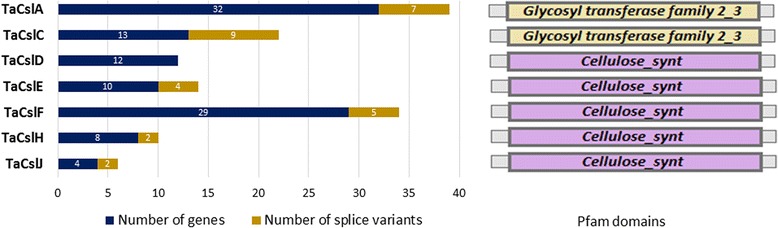
Distribution of the TaCsl genes and their splice variants in seven subfamilies and their corresponding pfam domains used to identify TaCsl gene family members
Splice variants of Csl genes
Twenty two of the 108 genes appeared to encode two or more proteins because of the presence of alternative splicing sites, as predicted by Ensembl database, which would result in 137 probable Csl protein products (Table 2). Splice variants were predicted in all the subfamilies of the TaCsl genes except TaCslD (Table 2). In the subfamily TaCslA, 6 genes alternatively spliced to form 13 putative proteins whereas in the subfamily TaCslC, 5 genes were alternatively spliced resulting in 14 putative proteins. Similarly, for the subfamilies TaCslE and TaCslF, alternative splicing resulted in 7 and 10 splice variants, respectively. Alternative splicing of 1 and 2 genes respectively generated 3 and 4 putative proteins in the CslH and CslJ subfamilies (Fig. 2). More than half (51%) of the splice variants stemmed from exon skipping, ~24% from alternative 5′ and 3′ splice sites, and the rest, ~24%, from intron retention (Table 2).
Table 2.
Splice variants of the bread wheat Csl genes
| Ensembl gene ID | Gene name | Predicted amino acids | Spliced exon/introns | Status |
|---|---|---|---|---|
| TRIAE_CS42_6BS_TGACv1_513375_AA1639370.1 | TaCslA1_6BS | 581 | – | Wild type |
| TRIAE_CS42_6BS_TGACv1_513375_AA1639370.2 | 390 | Exon 1 and 2 | Exon skipping | |
| TRIAE_CS42_6BS_TGACv1_513376_AA1639390.2 | TaCslA4_6BS | 528 | – | Wild type |
| TRIAE_CS42_6BS_TGACv1_513376_AA1639390.1 | 393 | Exon 1 and 2 | Exon skipping | |
| TRIAE_CS42_7AS_TGACv1_569190_AA1809650.1 | TaCslA3_7AS | 551 | – | Wild type |
| TRIAE_CS42_7AS_TGACv1_569190_AA1809650.2 | 380 | Exon 7, 8 and 9 | Exon skipping | |
| TRIAE_CS42_7AS_TGACv1_569190_AA1809650.3 | 503 | Exon 9 | Exon skipping | |
| TRIAE_CS42_7DL_TGACv1_602617_AA1962870.2 | TaCslA10_7DL | 515 | – | Wild type |
| TRIAE_CS42_7DL_TGACv1_602617_AA1962870.1 | 555 | Intron 8 | Intron retention | |
| TRIAE_CS42_3DL_TGACv1_249033_AA0835410.2 | TaCslA6_3DL | 524 | – | Wild type |
| TRIAE_CS42_3DL_TGACv1_249033_AA0835410.1 | 572 | Intron 1 | Intron retention | |
| TRIAE_CS42_3B_TGACv1_221079_AA0729630.1 | TaCslA6_3B | 571 | – | Wild type |
| TRIAE_CS42_3B_TGACv1_221079_AA0729630.2 | 538 | Exon 2 | Exon skipping | |
| TRIAE_CS42_5BL_TGACv1_404820_AA1311790.1 | TaCslC10_5BL | 712 | – | Wild type |
| TRIAE_CS42_5BL_TGACv1_404820_AA1311790.2 | 468 | Exon 5 | Alternative 5′ site | |
| TRIAE_CS42_5BL_TGACv1_404820_AA1311790.3 | 504 | Exon 1 | Exon skipping | |
| TRIAE_CS42_5DL_TGACv1_435778_AA1454840.1 | TaCslC10_5DL | 708 | – | Wild type |
| TRIAE_CS42_5DL_TGACv1_435778_AA1454840.2 | 502 | Exon1 | Exon skipping | |
| TRIAE_CS42_5AL_TGACv1_374268_AA1195590.3 | TaCslC10_5AL | 703 | – | Wild type |
| TRIAE_CS42_5AL_TGACv1_374268_AA1195590.2 | 496 | Exon 5 | Alternative 5′ site | |
| TRIAE_CS42_5AL_TGACv1_374268_AA1195590.1 | 501 | Exon 5 | Exon skipping | |
| TRIAE_CS42_3DL_TGACv1_251593_AA0882850.1 | TaCslC1_3DL | 704 | – | Wild type |
| TRIAE_CS42_3DL_TGACv1_251593_AA0882850.2 | 493 | Exon 5 | Exon skipping | |
| TRIAE_CS42_3DL_TGACv1_251593_AA0882850.3 | 679 | Exon 1 | Alternative 3′ site | |
| TRIAE_CS42_3AL_TGACv1_197197_AA0665370.1 | TaCslC1_3AL | 704 | – | Wild type |
| TRIAE_CS42_3AL_TGACv1_197197_AA0665370.2 | 560 | Exon 5 | Alternative 3′ site | |
| TRIAE_CS42_3AL_TGACv1_197197_AA0665370.3 | 679 | Exon 5 | Alternative 5′ site | |
| TRIAE_CS42_6AL_TGACv1_471004_AA1500600.1 | TaCslE2_6AL | 667 | – | Wild type |
| TRIAE_CS42_6AL_TGACv1_471004_AA1500600.2 | 737 | Intron 8 | Intron retention | |
| TRIAE_CS42_6AL_TGACv1_471004_AA1500600.3 | 635 | Exon 4 | Alternative 5′ site | |
| TRIAE_CS42_5DL_TGACv1_433536_AA1415830.1 | TaCslE1_5DL | 728 | – | Wild type |
| TRIAE_CS42_5DL_TGACv1_433536_AA1415830.2 | 684 | Exon 4 | Exon skipping | |
| TRIAE_CS42_5BL_TGACv1_406235_AA1342600.1 | TaCslE1_5BL | 734 | – | Wild type |
| TRIAE_CS42_5BL_TGACv1_406235_AA1342600.2 | 728 | Exon 1 | Exon skipping | |
| TRIAE_CS42_2DS_TGACv1_177641_AA0581710.1 | TaCslF3_2DS | 847 | – | Wild type |
| TRIAE_CS42_2DS_TGACv1_177641_AA0581710.2 | 735 | Exon 2 | Alternative 3′ site | |
| TRIAE_CS42_2DS_TGACv1_179076_AA0604160.1 | TaCslF4_2DS | 783 | – | Wild type |
| TRIAE_CS42_2DS_TGACv1_179076_AA0604160.2 | 700 | Exon 1 | Exon skipping | |
| TRIAE_CS42_2BS_TGACv1_147667_AA0486240.1 | TaCslF9_2BS | 877 | – | Wild type |
| TRIAE_CS42_2BS_TGACv1_147667_AA0486240.2 | 796 | Exon 1 | Exon skipping | |
| TRIAE_CS42_5BL_TGACv1_409916_AA1366600.1 | TaCslF7_5BL | 745 | – | Wild type |
| TRIAE_CS42_5BL_TGACv1_409916_AA1366600.2 | 815 | Intron 2 | Intron retention | |
| TRIAE_CS42_5AL_TGACv1_374191_AA1193100.1 | TaCslF7_5AL | 792 | – | Wild type |
| TRIAE_CS42_5AL_TGACv1_374191_AA1193100.2 | 807 | Intron 1 | Intron retention | |
| TRIAE_CS42_2AL_TGACv1_094351_AA0296300.1 | TaCslH1_2AL | 737 | – | Wild type |
| TRIAE_CS42_2AL_TGACv1_094351_AA0296300.2 | 660 | Exon 9 | Exon skipping | |
| TRIAE_CS42_2AL_TGACv1_094351_AA0296300.3 | 480 | Exon 6, 7, 8 and 9 | Exon skipping | |
| TRIAE_CS42_3AS_TGACv1_210908_AA0681280.1 | TaCslJ1_3AS | 738 | – | Wild type |
| TRIAE_CS42_3AS_TGACv1_210908_AA0681280.2 | 766 | Intron 4 | Intron retention | |
| TRIAE_CS42_3DS_TGACv1_272756_AA0924850.2 | TaCslJ2_3DS | 609 | – | Wild type |
| TRIAE_CS42_3DS_TGACv1_272756_AA0924850.1 | 734 | Intron 1 | Intron retention |
Conserved motifs and domains
All predicted TaCSL proteins contain either the pfam glycosyltransferase family 2_3 (GT) domain (PF13641) or the cellulose_synt domain (PF03552), considered to be the signature domains of the GT2 superfamily [12, 26]. Subfamilies TaCslA and TaCslC contained GT 2_3, and CslD, CslE, CslF, CslH,and CslJ contained the cellulose_synt domain (Fig. 2). All the TaCsl translanted products contained the motifs D, DXD, D and QXXRW except eight truncated genes that lacked some of these motifs apparently because of the missing sequence (TaCslA7_2DS, TaCslD4_1BS, TaCslD4_5BS, TaCslF2_7BL, TaCslF6_7AL, TaCslF6_7DL, TaCslH3_3AS, TaCslH2_3B). Rice CesA10, 11 and CslH3 also contained only the DXD but lacked the D and QXXRW motifs [38]. The variable amino acids in the conserved motifs DXD and QXXRW were diverse in different subfamilies of Csl genes, for example, for TaCslA (DMD, QQH/FRW); TaCslC (DMD, QQHRW); TaCslD (DCD, QVLRW); TaCslE (DCD, QHKRW); TaCslF (DC/GD, QI/VL/VRW); TaCslH (DCD QF/YKRW); TaCslJ (DCD, QNKRW). These motifs are highlighted in alignment files in the text file S_2a-f.
Phylogenetic analysis of the CslD subfamily
The evolutionary history of the CslD subfamily from Arabidopsis, Brachypodium, rice, maize and wheat was inferred using the Neighbor-Joining method, in MEGA6 [36], after grouping the orthologs from various species into different clades (Fig. 3). Rice Csl genes were used as reference because their complete nomenclature is well documented. All the genes grouped into three clades. The first clade contained CslD2 and CslD1 genes from rice and their orthologs from the remaining species. The three homeologous genes of wheat branched together with OsCslD1; wheat genes under this clade were named TaCslD1_1AL, TaCslD1_1BL, and TaCslD1_1DL. The second clade contained two subgroups with the orthologs of rice genes CslD3 and CslD5 from different species. The genes in the first subgroup were named TaCslD3_2AS, TaCslD3_2BS, and TaCslD3_2DS, and those of the second subgroup TaCslD5_7AL, TaCslD5_7BL, and TaCslD5_7DL. The last clade was composed of the orthologs of the rice CslD4 and wheat genes TaCslD4_5BS, TaCslD4_1BS and TaCslD4_5DS. Here we found only two homeologs of TaCslD4, but a gene from the 1BS genome (TaCslD4_1BS) of wheat grouped together with TaCslD4 genes (bootstrap = 1000), pointing to a translocation from its original A genome (Table 1). This gene shared sequence identity of 85% with TaCslD4_5BS at the amino acid level. OsCslD genes shared 73–86% sequence identity with the corresponding wheat orthologs.
Fig. 3.
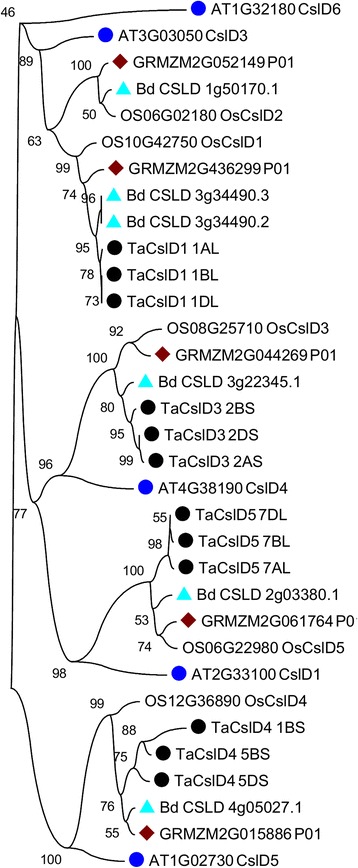
An unrooted phylogenetic tree representing the CslD subfamily from Arabidopsis, Brachypodium, maize, rice and wheat using Neighbour Joining (NJ) method with 1000 replicates to generate bootstrap values that are shown beside the each node forming the Csl clusters. Different colors and shapes represent orthologous Csl genes from different species. Arabidopsis-blue circles, Brachypodium- sky blue triangels, maize-brown rectangles-, rice-no marker, and wheat-black circles
Gene structure and intron evolution of TaCslD subfamily
The 12 TaCslD genes identified from bread wheat ranged in size from 1519 to 5864 bp. The TaCslD4_1BS gene was the shortest and TaCslD1_1AL was the longest. Homeologous copies of all the genes shared sequence identity ranging from 87 to 94% at the nucleotide level. The variation in size among different genes was primarily because of the number and length of introns but also because of a lack of the complete sequences in the database (Fig. 4). The number of introns in all the genes varied from 2 to 4. Two homeologs: TaCslD1_1AL and TaCslD1_1BL each contained three introns whereas, a third homeolog (TaCslD1_1DL) had four. The genes TaCslD3, TaCslD4 and their homeologs contained three introns each, except TaCslD4_1BS with only two introns. TaCslD5 and its homeologs also had two introns each. For the phases of introns, the genes from the TaCslD subfamily exhibited variable patterns of distribution. Introns 1, 2 and 3 of TaCslD1_1AL, TaCslD1_1BL and TaCslD1_1DL were in 2, 0, and 0 phase whereas the 4th intron of TaCslD1_1DL was in 0 phase. Introns 1 and 2 of TaCslD3_2AS, TaCslD3_2BS and TaCslD3_2DS both were in 0 phase. The third intron of these genes was in phase 2, 1 and 2 respectively. The genes TaCslD4_5BS, TaCslD4_5DS, TaCslD5_7AL, TaCslD5_7BL and TaCslD5_7DL had intron 1 and 2 in phases 2 and 0, respectively, and the third intron of TaCslD4_5BS and TaCslD4_5DS was in phase 0 and 2, respectively. TaCslD4_1BS had introns 1 and 2 in phases 1 and 0. The largest proportion of introns (60%) of all the genes was in phase 0, followed by phase 2 (34%) with a few in phase 1 (6%).
Fig. 4.
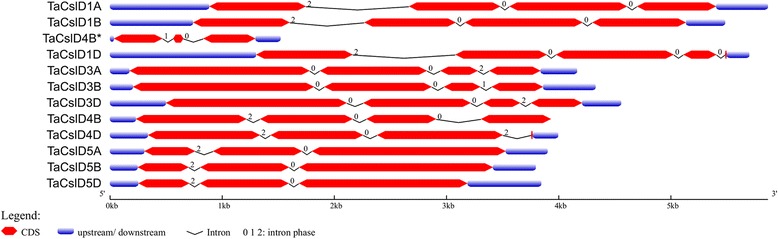
Structural features and phases of intron evolution of the CslD subfamily genes. Drawn to scale, exons are represented by red boxes and introns by back lines. Corresponding phases of intron evolution (0, 1, and 2) for the CslD genes are shown on the top of the black lines
Expression analysis of TaCsl genes from bread wheat
Publicly available RNA-Seq datasets were used to analyse the expression of TaCsl genes over three developmental stages and different tissues of wheat including root, stem, leaf, spike, and grain. Expression data were available for 32 of the TaCslA genes. Two genes (TaCslA1_6AS and TaCslA1_6BS) were expressed in all the tissues except reproductive stem and leaves. Four genes (TaCslA5_2BS, TaCslA5_2DS, TaCslA6_3B, and TaCslA6_3AL) were expressed moderately. TaCslA9 gene was highly expessed in the leaf tissue from the reproductive stage while the transcript abundance of the remaining genes was low (Fig. 5). TaCslC subfamily genes, wtht the exception of TaCslC3, TaCslC9 and two homeologs of TaCslC10, were expressed highly in root and spike tissues. Two genes, TaCslC1 and TaCslC7 and their homeologs displayed moderate to high expression in all the tissues at seeding and vegetative stage. One gene (TaCslC10_5DL) exhibited moderate to high expression in all the tissues studied except reproductive stem and grain (Fig. 6). Expression of most of the genes of the TaCslD subfamily ranged from moderate to a high in the spike and root tissues but was very low in all the other tissues (Fig. 7). Three of the 10 TaCslE subfamily genes (TaCslE2_6AL, TaCslE2_6BL and TaCslE3) were expressed from moderate to high levels in all the tissues.The remaining genes were expressed at a very low level in all the tissues (Fig. 8). A mixed pattern of expression was observed in the large TaCslF subfamily. Three genes (TaCslF6_7AL, TaCslF6_7BL and TaCslF6_7DL) were highly expressed in all the tissues except the leaves at the reproductive stage. Two genes (TaCslF4_2BS and TaCslF4_2DS) were highly expressed in the stem tissue, but only at a low or moderate level in all other tissues. All other genes expressed at low or moderate levels in one or more tissues (Fig. 9). In the TaCslH subfamily, one of the eight genes, TaCslH1_2BL, was expressed from moderate to high levels in the leaf, stem and spike tissues. The remaining genes were expressed from low to moderate levels in all the tissues (Fig. 10). Three out of four members of the subfamily TaCslJ were expressed from low to moderate levels in the leaf and root tissues while one gene (TaCslJ1_3DS) was poorly expressed in all the tissues studied (Fig. 10).
Fig. 5.
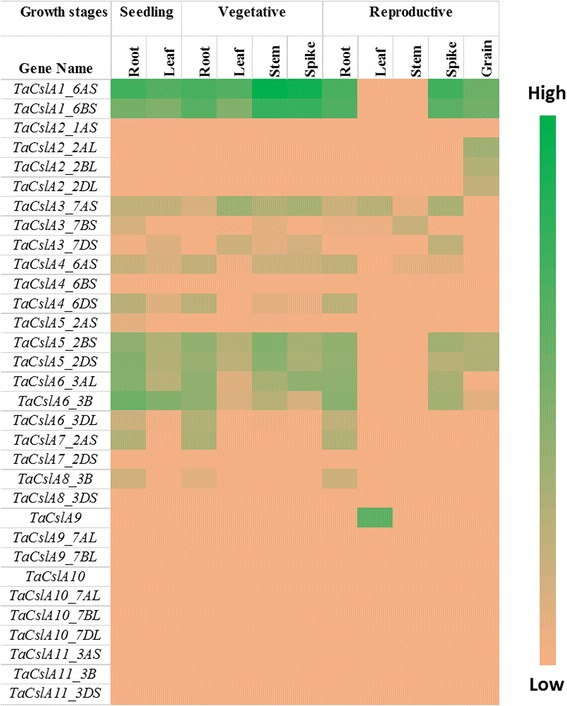
Heat map showing the expression profiling of wheat TaCslA genes at seedling, vegetative and reproductive stages. RNA-seq data were obtained from root, leaf, stem, spike and grain of the Chinese spring cultivar. The respective transcripts per 10 million values were used to construct heat map with the scale bar showing expression of the genes
Fig. 6.
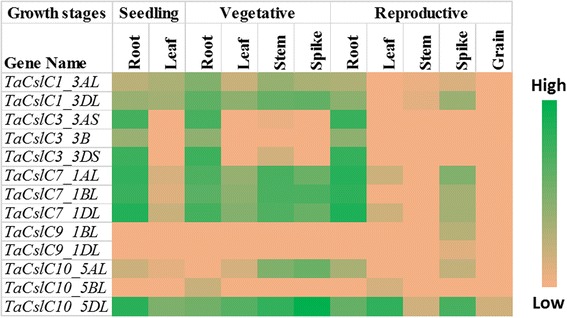
Heat map of the expression profiling of wheat TaCslC genes at seedling, vegetative and reproductive stages. RNA-seq data were obtained from root, leaf, stem, spike and grain of Chinese spring cultivar. The respective transcripts per 10 million values were used to construct heat map with scale bar showing expression of the genes
Fig. 7.
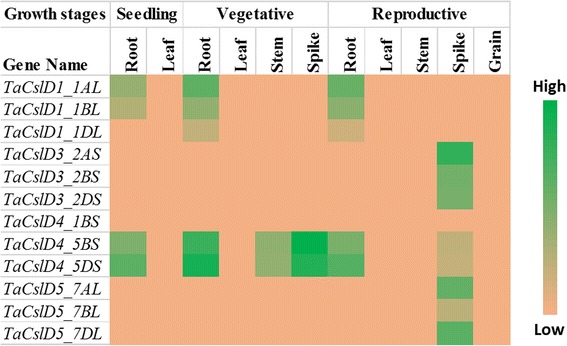
Heat map of the expression profiling of wheat TaCslD genes at seedling, vegetative and reproductive stages. RNA-seq data were obtained from root, leaf, stem, spike and grain of Chinese spring cultivar. The respective transcripts per 10 million values were used to construct heat map with scale bar showing expression of the genes
Fig. 8.
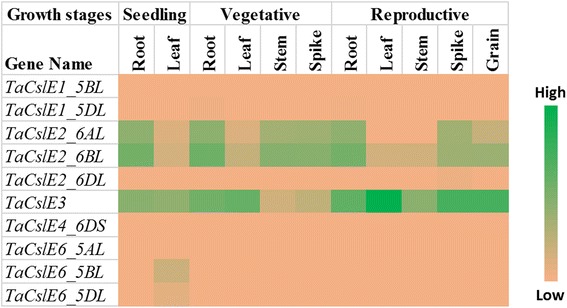
Heat map of the expression profiling of wheat TaCslE genes at seedling, vegetative and reproductive stages. RNA-seq data were obtained from root, leaf, stem, spike and grain of Chinese spring cultivar. The respective transcripts per 10 million values were used to construct heat map with scale bar showing expression of the genes
Fig. 9.
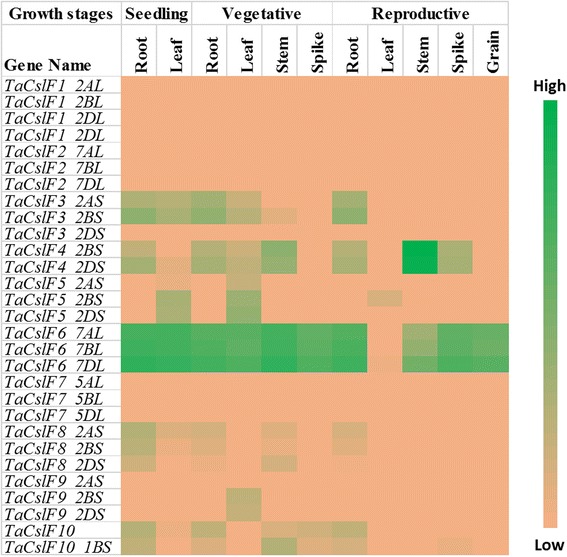
Heat map of the expression profiling of wheat TaCslF genes at seedling, vegetative and reproductive stages. RNA-seq data were obtained from root, leaf, stem, spike and grain of Chinese spring cultivar. The respective transcripts per 10 million values were used to construct heat map with scale bar showing expression of the genes
Fig. 10.
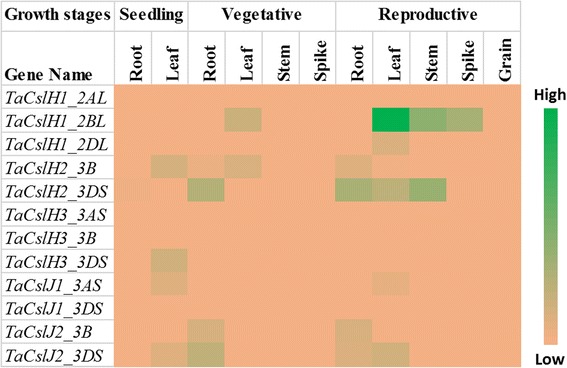
Heat map of the expression profiling of wheat TaCslH and TaCslJ genes at seedling, vegetative and reproductive stages. RNA-seq data were obtained from root, leaf, stem, spike and grain of Chinese spring cultivar. The respective transcripts per 10 million values were used to construct heat map with scale bar showing expression of the genes
Discussion
Grass cell walls contain 20–40% non-cellulosic polysaccharides. The proportion and composition of these polysaccharides varies in different plant species [39]. After the first report demonstrating the β-glucan synthase activity in a Csl-encoded protein was published [15], several members of the Csl gene family have been reported to be involved in the formation of the backbone of the hemicellulosic polysaccharides [16, 18, 19, 26, 38, 40, 41]. As information on the identify of the Csl genes in wheat was lacking, we undertook this study to fill this gap.
We retrieved 108 TaCsl genes from wheat using two conserved domains, PF00535, and PF03552, which were previously shown to be present in the derived proteins of all the Csl genes [12]. These genes include homeologs from A, B and D genome of bread wheat. Similar patterns of homeologous genes were found for FLOWERING LOCUS T (FT), Pairing homeologous 1 (Ph1) and ADP-glucose pyrophosphorylase (AGPase) gene families of hexaploid wheat. Approximately, a quarter of the identified Csl genes were predicted to be alternatively spliced, possibly contributing to the diversity of encoded enzymes. A recent study suggested that alternative splicing was common in plants and accounted for about 20% of the loci transcribed in the leaf and spike tissues of Aegilops tauschii. In the case of germinating barley embryos, 14–20% of intron-containing genes were alternatively spliced [42]. This phenomenon, apparently meant to increase the fitness of an organism, has not thus far been reported for the Csl genes from other species [43].
The TaCsl genes were distributed across all the wheat chromosomes except one, chromosome 4 (Fig. 11). A similar trend of Csl gene distribution was observed in barley [9, 44, 45]. More than half the TaCsl genes were located on only two chromosomes: 2 (32%) and 3 (22%). This suggests hyper-multiplication of the Csl genes on these chromosomes although the reasons for this phenomenon are unknown. It appears, though, that cis duplication of the Csl genes was favored over trans duplication in wheat. Five of the nine CslF genes in barley were located on chromosome 2H [40]. In fact, the barley CslF gene was assigned its role in mixed-linked glucan (MLG) formation via syntenic orthology with rice long before the barely genome sequence became available [40] A detailed analysis of the rice syntenic region corresponding to a known QTL for MLG from barley, which had been published previously, initially led to the breakthrough of the role of CslF in the formation of this polysaccharide [40]). A similar cluster of CslF genes was also detected in the conserved syntenic regions of Brachypodium and sorghum on chromosomes 1and 2, respectively [9].
Fig. 11.
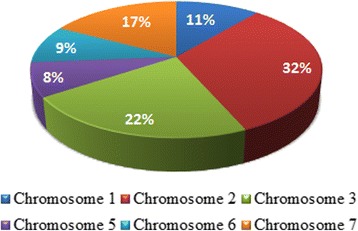
Pie chart showing the percentage of TaCsl genes on wheat chromosomes
The observation that only half of genes from the subfamily CslA were expressed at varying levels in the studied tissues suggests that the apparently silent genes may provide a backup under stressful conditions. Alternatively, they may express only transiently in specialized cells or cell parts at levels too low to be detected by the method used to study expression. The first biochemical evidence for the relationship of CslA genes with mannan synthase activity came from the expression of a guar CSLA cDNA in soybean somatic embryos [15]. Subsequent studies in insect cells demonstrated the role of CslA family members in the glucomannan synthases [16, 46]. Reverse genetic and biochemical approaches in Arabidopsis and Dendrobium officinale have also allowed association of certain CslA genes with glucomannan biosynthesis [41, 47]. A recent study in wheat suggested the involvement of a gene from the CslA subfamily in the development of tillers, cell wall composition and stem strength. This study further suggested the probable role of CslA gene transcript levels in carbon partitioning throughout the plant [48].
For the subfamilies TaCslC and TaCslD, most of the genes were relatively highly expressed in the root and spike tissues during the vegetative as well as reproductive phases. Heterologous expression in Pichia revealed that the CslC-encoded enzymes made β-1,4-glucan, the backbone of xyloglucan [19]. The CslD subfamily is conserved in all land plants and is most closely related to the CesA gene family with 40–50% sequence similarity at the amino acid level [49]. Similar to CesAs, the CslD subfamily is ubiquitous in all plant genomes examined to date, unlike other, taxa-specific Csl subfamilies [50]. Previous reports also showed the involvement of certain members of the CslD subfamily in tip growth, for example development of root hairs and pollen tube elongation [51, 52], normal plant growth [50, 53], and meristem morphology [53, 54]. More recently, their role in resistance against biotic stresses has been described [55]. Adding to this discussion, our in silico expression analysis suggests the involvement of certain TaCslD genes in spike development. This suggestion is supported by the observation that a mutant, slender leaf 1 (sle1), which encodes the CSLD4 protein in rice, reduces the number and width of spikelets in the panicle [56].
Two groups of Csl genes, CslF and CslH, have evolved independently in grasses [57]. A third group CslJ, originally believed to be specific to grasses, was recently identified in some dicots [11, 13]. Although TaCslF6 gene showed higher expression in all the studied tissues except the leaf tissue from reproductive stage, it was the only member of the TaCslF subfamily which expressed highly in the grain tissue. Several studies have demonstrated the functional role of CslF6 and CslH in the synthesis of MLG [18, 44, 58, 59]. Only one member of all the genes in these families, CslF6, was expressed in the grain, suggesting that it was responsible for MLG formation. MLG is a desirable polysaccharide as a dietary fiber but undesirable for the brewery industry because it causes haze in beer. It should be possible to select natural variants for the expression of the CslF6 gene to select for an increased or reduced MLG content depending upon the target market for the grain.
Differential expression patterns were observed among homeologous copies from three different genomes of bread wheat, which agree with the previous studies reporting unequal contributions of the three genomes toward gene expression. Interestingly, the homeologous copies of TaCslD genes also differed from each other in terms of intron phase evolution, indicating structural and functional divergence of homeologous gene copies (Fig. 4). Most introns were present in phase 0, which is in accordance with previous findings showing an intron bias in favour of phase 0 [7, 60, 61]. The three homeologs of each gene were not observed for all the genes reported in this study. This could be because of the incomplete sequencing information or because of the elimination of the genes during the allopolyploidization of wheat.
Conclusions
We have identified 108 TaCsl genes in bread wheat and classified them into seven subfamilies (CslA, CslC, CslD, CslE, CslF, CslH, and CslJ). Two or three homeoalleles were identified for most of the Csl genes. Although located on all the wheat chromosomes except chromosome 4, the Csl genes were especially concentrated on chromosomes 2 and 3, suggesting selective, localized duplication in cis phase. Only one of the 29 CslF genes, CslF6, was expressed in the grain, suggesting its role in mixed-linked glucan formation. Neither CslJ nor CslH was expressed in the grain. Information in this report will be helpful in designing experiments to alter wall composition in wheat for improving grain quality, culm strength, or culm composition for biofuels.
Additional files
FASTA sequences of CSL proteins used for the phylogenetic analysis. (PDF 453 kb)
List of Csl subfamily genes, their protein sizes (number of amino acids), and multiple protein sequence alignments for different subfamilies. The conserved motifs (D, D, DXD, QXXRW) diagnostic of CSL proteins are highlighted with red boxes for each of the subfamilies. (PDF 465 kb)
Acknowledgements
This work was supported by the CGIAR’s Consortium Research Program WHEAT (KSD), Canada Foundation for Innovation (CFI), the ministère de l’Économie, de la science et de l’innovation du Québec (MESI) and the Fonds de recherche du Québec - Nature et technologies (FRQ-NT) (RB), and Natural Sciences and Engineering Research Council of Canada through discovery program (JS).
Computations were made on the supercomputer Guillimin from McGill University, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation (CFI), the ministère de l’Économie, de la science et de l’innovation du Québec (MESI) and the Fonds de recherche du Québec - Nature et technologies (FRQ-NT).
Funding
Natural sciences and engineering research council of Canada.
Availability of data and materials
Yes, all the data are included in the supplement already.
Abbreviations
- CesA
Cellulose synthase
- Csl
Cellulose synthase-like
- GT
Glycosyltransferase
- MLG
Mixed-linked glucan
Authors’ contributions
SK extracted the sequences, analyzed them, and wrote the paper; KSD conceived the project along with JS, analyzed the data, wrote parts of the paper, and edited the manuscript; RB carried out the phylogenetic analysis and constructed the phylogenetic tree; JS conceived and supervised the project, and helped write the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
N/A
Consent for publication
N/A
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1142-z) contains supplementary material, which is available to authorized users.
Contributor Information
Simerjeet Kaur, Email: Simerjeet@mcgill.ca.
Kanwarpal S. Dhugga, Email: k.dhugga@cgiar.org
Robin Beech, Email: robin.beech@mcgill.ca.
Jaswinder Singh, Email: Jaswinder.singh@mcgill.ca.
References
- 1.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54(4):559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- 2.Sandhu APS, Randhawa GS, Dhugga KS. Plant cell wall matrix polysaccharide biosynthesis. Mol Plant. 2009;2(5):840–850. doi: 10.1093/mp/ssp056. [DOI] [PubMed] [Google Scholar]
- 3.Sorek N, Yeats TH, Szemenyei H, Youngs H, Somerville CR. The implications of Lignocellulosic biomass chemical composition for the production of advanced biofuels. Bioscience. 2014;64(3):192–201. doi: 10.1093/biosci/bit037. [DOI] [Google Scholar]
- 4.Pauly M, Gille S, Liu L, Mansoori N, de Souza A, Schultink A, Xiong G. Hemicellulose biosynthesis. Planta. 2013;238(4):627–642. doi: 10.1007/s00425-013-1921-1. [DOI] [PubMed] [Google Scholar]
- 5.Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124(2):495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rai KM, Thu SW, Balasubramanian VK, Cobos CJ, Disasa T, Mendu V. Identification, characterization, and expression analysis of Cell Wall related genes in Sorghum Bicolor (L.) Moench, a food, fodder, and biofuel crop. Front Plant Sci. 2016;1287. [DOI] [PMC free article] [PubMed]
- 7.Kaur S, Dhugga KS, Gill K, Singh J. Novel structural and functional motifs in cellulose synthase (CesA) genes of bread wheat (Triticum Aestivum, L.). PLoS One. 2016;11(1):e0147046. [DOI] [PMC free article] [PubMed]
- 8.Hazen SP, Scott-Craig JS, Walton JD. Cellulose synthase-like genes of rice. Plant Physiol. 2002;128(2):336–340. doi: 10.1104/pp.010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwerdt JG, MacKenzie K, Wright F, Oehme D, Wagner JM, Harvey AJ, Shirley NJ, Burton RA, Schreiber M, Halpin C. Evolutionary dynamics of the cellulose synthase gene superfamily in grasses. Plant Physiol. 2015;168(3):968–983. doi: 10.1104/pp.15.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton RA, Collins HM, Kibble NA, Smith JA, Shirley NJ, Jobling SA, Henderson M, Singh RR, Pettolino F, Wilson SM, et al. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-beta-d-glucans and alters their fine structure. Plant Biotechnol J. 2011;9(2):117–135. doi: 10.1111/j.1467-7652.2010.00532.x. [DOI] [PubMed] [Google Scholar]
- 11.Farrokhi N, Burton RA, Brownfield L, Hrmova M, Wilson SM, Bacic A, Fincher GB. Plant cell wall biosynthesis: genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol J. 2006;4(2):145–167. doi: 10.1111/j.1467-7652.2005.00169.x. [DOI] [PubMed] [Google Scholar]
- 12.Yin Y, Johns MA, Cao H, Rupani M. A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genomics. 2014;15(1):1. doi: 10.1186/1471-2164-15-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel JP, Garvin DF, Mockler TC, Schmutz J, Rokhsar D, Bevan MW, Barry K, Lucas S, Harmon-Smith M, Lail K. Genome sequencing and analysis of the model grass Brachypodium Distachyon. Nature. 2010;463(7282):763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 14.Dhugga KS. Biosynthesis of non-cellulosic polysaccharides of plant cell walls. Phytochemistry. 2012;74:8–19. doi: 10.1016/j.phytochem.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Dhugga KS, Barreiro R, Whitten B, Stecca K, Hazebroek J, Randhawa GS, Dolan M, Kinney AJ, Tomes D, Nichols S. Guar seed ß-mannan synthase is a member of the cellulose synthase super gene family. Science. 2004;303(5656):363–366. doi: 10.1126/science.1090908. [DOI] [PubMed] [Google Scholar]
- 16.Liepman AH, Wilkerson CG, Keegstra K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc Natl Acad Sci U S A. 2005;102(6):2221–2226. doi: 10.1073/pnas.0409179102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1, 3; 1, 4)-ß-D-glucans. Science. 2006;311(5769):1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- 18.Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin E, Bacic A. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc Natl Acad Sci U S A. 2009;106(14):5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cocuron JC, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc Natl Acad Sci U S A. 2007;104(20):8550–8555. doi: 10.1073/pnas.0703133104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta PK, Mir RR, Mohan A, Kumar J. Wheat genomics: present status and future prospects. Int J Plant Genomics. 2008;2008:896451. doi: 10.1155/2008/896451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer KF, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Gill B, Friebe B, Lukaszewski AJ, Sourdille P. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum Aestivum) genome. Science. 2014;345(6194):1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 22.Consortium IWGS. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum Aestivum) genome. Science. 2014;345(6194):1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35(1):125–131. doi: 10.1093/nar/gkl924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girke T, Lauricha J, Tran H, Keegstra K, Raikhel N. The cell wall navigator database. A systems-based approach to organism-unrestricted mining of protein families involved in cell wall metabolism. Plant Physiol. 2004;136(2):3003–3008. doi: 10.1104/pp.104.049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Y, Huang J, Xu Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009;9:99. doi: 10.1186/1471-2229-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richmond T. Higher plant cellulose synthases. Genome Biol. 2000;1(4):REVIEWS3001. doi: 10.1186/gb-2000-1-4-reviews3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol. 2011;7(1):539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28(6):1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2014:gku1221. [DOI] [PMC free article] [PubMed]
- 31.Kaur R, Singh K, Singh J. A root-specific wall-associated kinase gene, HvWAK1, regulates root growth and is highly divergent in barley and other cereals. Funct Integr Genomics. 2013;13(2):167–177. doi: 10.1007/s10142-013-0310-y. [DOI] [PubMed] [Google Scholar]
- 32.Katoh K, Misawa K, Ki K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felsenstein J. Phylogeny inference package (version 3.2) Cladistics. 1996;5:164–166. [Google Scholar]
- 34.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, Pingault L, Sourdille P, Couloux A, Paux E. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345(6194):1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- 38.Wang L, Guo K, Li Y, Tu Y, Hu H, Wang B, Cui X, Peng L. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010;10 [DOI] [PMC free article] [PubMed]
- 39.Saxena IM, Brown R. Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J Bacteriol. 1995;177(18):5276–5283. doi: 10.1128/jb.177.18.5276-5283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-beta-D-glucans. Science. 2006;311(5769):1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- 41.Goubet F, Barton CJ, Mortimer JC, Yu X, Zhang Z, Miles GP, Richens J, Liepman AH, Seffen K, Dupree P. Cell wall glucomannan in Arabidopsis is synthesised by CSLA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 2009;60(3):527–538. doi: 10.1111/j.1365-313X.2009.03977.x. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Zhang X, Wang S, Tan C, Zhou G, Li C. Involvement of alternative splicing in barley seed germination. PLoS One. 2016;11(3):e0152824. doi: 10.1371/journal.pone.0152824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Zhou C, Ye L, Dong J, Xu H, Cai L, Zhang L, Wei L. Database and analyses of known alternatively spliced genes in plants. Genomics. 2003;82(6):584–595. doi: 10.1016/S0888-7543(03)00204-0. [DOI] [PubMed] [Google Scholar]
- 44.Schreiber M, Wright F, MacKenzie K, Hedley PE, Schwerdt JG, Little A, Burton RA, Fincher GB, Marshall D, Waugh R. The barley genome sequence assembly reveals three additional members of the CslF (1, 3; 1, 4)-β-glucan synthase gene family. PLoS One. 2014;9(3):e90888. doi: 10.1371/journal.pone.0090888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB. The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiol. 2008;146(4):1821–1833. doi: 10.1104/pp.107.114694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki S, Li L, Sun Y-H, Chiang VL. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus Trichocarpa. Plant Physiol. 2006;142(3):1233–1245. doi: 10.1104/pp.106.086678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He C, Wu K, Zhang J, Liu X, Zeng S, Yu Z, Zhang X, da Silva JAT, Deng R, Tan J. Cytochemical localization of polysaccharides in Dendrobium Officinale and the involvement of DoCSLA6 in the synthesis of Mannan polysaccharides. Front Plant Sci. 2017;8:173. [DOI] [PMC free article] [PubMed]
- 48.Hyles J, Vautrin S, Pettolino F, MacMillan C, Stachurski Z, Breen J, Berges H, Wicker T, Spielmeyer W. Repeat-length variation in a wheat cellulose synthase-like gene is associated with altered tiller number and stem cell wall composition. J Exp Bot. 2017;68(7):1519–1529. doi: 10.1093/jxb/erx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doblin MS, De Melis L, Newbigin E, Bacic A, Read SM. Pollen tubes of Nicotiana Alata express two genes from different β-glucan synthase families. Plant Physiol. 2001;125(4):2040–2052. doi: 10.1104/pp.125.4.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunter CT, Kirienko DH, Sylvester AW, Peter GF, McCarty DR, Koch KE. Cellulose Synthase-like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiol. 2012;158(2):708–724. doi: 10.1104/pp.111.188466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim CM, Park SH, Je BI, Park SH, Park SJ, Piao HL, Eun MY, Dolan L, Han CD. OsCSLD1, a cellulose synthase-like D1 gene, is required for root hair morphogenesis in rice. Plant Physiol. 2007;143(3):1220–1230. doi: 10.1104/pp.106.091546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuo T, Shiotani K, Shitsukawa N, Miyao A, Hirochika H, Ichii M, Taketa S. Root hairless 2 (rth2) mutant represents a loss-of-function allele of the cellulose synthase-like gene OsCSLD1 in rice (Oryza Sativa L.) Breed Sci. 2011;61(3):225–233. doi: 10.1270/jsbbs.61.225. [DOI] [Google Scholar]
- 53.Li M, Xiong G, Li R, Cui J, Tang D, Zhang B, Pauly M, Cheng Z, Zhou Y. Rice cellulose synthase-like D4 is essential for normal cell-wall biosynthesis and plant growth. Plant J. 2009;60(6):1055–1069. doi: 10.1111/j.1365-313X.2009.04022.x. [DOI] [PubMed] [Google Scholar]
- 54.Bernal AJ, Jensen JK, Harholt J, Sørensen S, Moller I, Blaukopf C, Johansen B, De Lotto R, Pauly M, Scheller HV. Disruption of ATCSLD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007;52(5):791–802. doi: 10.1111/j.1365-313X.2007.03281.x. [DOI] [PubMed] [Google Scholar]
- 55.Douchkov D, Lueck S, Hensel G, Kumlehn J, Rajaraman J, Johrde A, Doblin MS, Beahan CT, Kopischke M, Fuchs R. The barley (Hordeum Vulgare) cellulose synthase-like D2 gene (HvCslD2) mediates penetration resistance to host-adapted and nonhost isolates of the powdery mildew fungus. New Phytol. 2016;212:421–33. [DOI] [PubMed]
- 56.Yoshikawa T, Eiguchi M, Hibara K-I, Ito J-I, Nagato Y. Rice SLENDER LEAF 1 gene encodes cellulose synthase-like D4 and is specifically expressed in M-phase cells to regulate cell proliferation. J Exp Bot. 2013;64(7):2049–2061. doi: 10.1093/jxb/ert060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burton RA, Collins HM, Kibble NA, Smith JA, Shirley NJ, Jobling SA, Henderson M, Singh RR, Pettolino F, Wilson SM. Over-expression of specific HVCSLF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1, 3; 1, 4)-β-D-glucans and alters their fine structure. Plant Biotechnol J. 2011;9(2):117–135. doi: 10.1111/j.1467-7652.2010.00532.x. [DOI] [PubMed] [Google Scholar]
- 58.Taketa S, Yuo T, Tonooka T, Tsumuraya Y, Inagaki Y, Haruyama N, Larroque O, Jobling SA. Functional characterization of barley betaglucanless mutants demonstrates a unique role for CslF6 in (1,3;1,4)-beta-D-glucan biosynthesis. J Exp Bot. 2012;63(1):381–392. doi: 10.1093/jxb/err285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nemeth C, Freeman J, Jones HD, Sparks C, Pellny TK, Wilkinson MD, Dunwell J, Andersson AAM, Aman P, Guillon F, et al. Down-regulation of the CSLF6 gene results in decreased (1,3;1,4)-beta-D-Glucan in endosperm of wheat. Plant Physiol. 2010;152(3):1209–1218. doi: 10.1104/pp.109.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lynch M. Intron evolution as a population-genetic process. Proc Natl Acad Sci U S A. 2002;99(9):6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhattachan P, Dong B. Origin and evolutionary implications of introns from analysis of cellulose synthase gene. J Syst Evol. 2017;55(2):142–148. doi: 10.1111/jse.12235. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FASTA sequences of CSL proteins used for the phylogenetic analysis. (PDF 453 kb)
List of Csl subfamily genes, their protein sizes (number of amino acids), and multiple protein sequence alignments for different subfamilies. The conserved motifs (D, D, DXD, QXXRW) diagnostic of CSL proteins are highlighted with red boxes for each of the subfamilies. (PDF 465 kb)
Data Availability Statement
Yes, all the data are included in the supplement already.


