Abstract
Background
Serological tests for gambiense human African trypanosomiasis (gHAT) detect antibodies to antigens on the cell surface of bloodstream trypanosomes. As trypanosomes that cause animal African trypanosomiasis (AAT) also express related antigens, we have evaluated two rapid diagnostic tests (RDTs) on cattle in trypanosomiasis endemic and non-endemic regions, to determine whether gHAT serological tests could also be used to screen for AAT.
Methods
Two RDTs, 1G RDT, made with native antigens, and p2G RDT, made with recombinant antigens, were tested on 121 cattle in a trypanosomiasis-free region, and on 312 cattle from a rhodesiense HAT and AAT endemic region. A subset of samples from the endemic region were also tested with two immune trypanolysis (TL) tests. The sensitivity of the tests was estimated by evaluating the result of the RDT on samples that were positive by both microscopy and internal transcribed spacer (ITS) PCR, whilst specificity was the result of the RDT on samples that were negative by ITS PCR and microscopy, and others from the non-endemic region.
Results
The specificity of the p2G RDT on cattle from the non-endemic region was 97.5% (95% CI: 93.0–99.2%), compared to only 57.9% (95% CI: 48.9–66.3%) for 1G RDT. The specificities of 1G RDT, p2G RDT and TL on endemic control cattle were 14.6% (95% CI: 9.7–21.5%), 22.6% (95% CI: 16.4–30.3%) and 68.3% (95% CI: 59.6–75.9%), respectively. The sensitivities of the tests on trypanosome positive samples were 85.1% (95% CI: 79.1–89.7%), 89.1% (95% CI: 83.7–93.0%) and 59.3% (95% CI: 51.8–66.4%), respectively. Among the same samples, 51.7% were positive by both TL and the 1G RDT.
Conclusions
These serological tests detect cross-reacting antibodies in cattle. The p2G RDT based on recombinant antigens had a high specificity in a non-endemic region, while the 1G RDT had a lower specificity, suggesting cross-reactivity with other pathogens.
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2487-8) contains supplementary material, which is available to authorized users.
Keywords: Serological tests, Trypanosomes, Specificity, Variant surface glycoproteins, Invariant surface glycoproteins
Background
Human African trypanosomiasis (HAT) and animal African trypanosomiasis (AAT) are endemic in resource-limited regions of at least 36 African countries. Both diseases are caused by protozoan parasites of the genus Trypanosoma, which is transmitted mainly by tsetse fly vectors of the genus Glossina. The cell surface of bloodstream trypanosomes is covered by a dense monolayer of a single protein coat that consists of millions of copies of a variant surface glycoprotein (VSG) that frequently switches to a new one to enable the parasite to evade the host immune response [1–7]. VSGs are highly immunogenic and are associated with a strong antibody response that is easily detected in circulation [8]. Trypanosomes switch VSGs through differential expression of an archive of hundreds of silent VSG genes and pseudogenes that reconstitute functional genes upon conversion and recombination events. The rate that trypanosomes belonging to one variant antigenic type (VAT) switch expression from one VSG to another is less than 1 in 100 trypanosomes per generation, and as a result, most trypanosomes in a wave of parasitaemia that fail to switch VSGs are destroyed by the host immune system [9–15]. The rate is, however, fast enough for some parasites to switch VSG and become a different VAT, thus evading the host immunological response, to establish the next wave of parasitaemia. Expression of VSGs occurs hierarchically, such that some VATs tend to appear early in infection, others in late disease, and others somewhere in between [11, 14, 16–18]. As a result, in an infected host, there are simultaneously several sub-populations of trypanosomes belonging to different VATs, with estimates that at any point during infection, up to 100 VSGs could be expressed, and as many as 83 have been definitively measured [8, 19, 20]. The variant-specific antibody response tends to persist for long periods during infection, and therefore antibodies to different VATs are found in serum at the same time. One consequence of switching VATs is that there is a constant regeneration of previously expressed variants and their rapid elimination by existing antibodies [14].
Invariant surface glycoproteins (ISGs) are another class of glycoproteins, also attached to the plasma membrane of the trypanosome, but unlike VSGs, they remain unchanged from one cell cycle to another, shielded from host antibodies by the VSG. ISGs also undergo rapid turnover and recycling, with half-lives that are shorter than the generation time of the parasite [21, 22]. The number of copies of individual ISGs is estimated at between 50,000 and 70,000 [23–25], and if this level of expression is extended to the entire ISG family, it is possible that one trypanosome has approximately 200,000 ISGs in total, equivalent to one ISG for every 50 VSG molecules on one trypanosome [26]. In live trypanosomes, most ISGs are shielded from the host immune response by the VSG coat. However, when trypanosomes are killed by a complement-mediated antibody response, the ISGs become exposed, presenting an opportunity for the body to mount an anti-ISG antibody response, making them good candidates for test development [27].
The serological tests used to screen for gambiense HAT (gHAT) detect antibodies to VSGs and ISGs [28–30]. These include the card agglutination test for trypanosomiasis (CATT), developed by the Institute of Tropical Medicine (Belgium) [31], first and second generation (1G and p2G) rapid diagnostic tests (RDTs) developed by Alere/Standard Diagnostics (SD; South Korea) [32, 33], and the HAT Sero-K-SeT RDT developed by Coris BioConcept (Belgium) [34]. The CATT test detects antibodies against freeze-dried, fixed trypanosomes expressing VSG LiTat 1.3, while both the 1G SD and Sero-K-SeT RDTs detect antibodies against native VSG LiTat 1.3 and native VSG LiTat 1.5 antigens [35]. The p2G RDT is a prototype developed using recombinant antigens, which detects antibodies to VSG LiTat 1.5 and ISG65 antigens [32]. The immune trypanolysis test (TL), which until now has been used in research settings, is highly specific and has been proposed for use in epidemiological studies, and for surveillance in gHAT elimination programmes [36–38].
This study was carried out to determine whether serological tests for gHAT could also be used in the diagnosis of AAT. We tested cattle in trypanosomiasis endemic and non-endemic regions, to determine whether the tests would exhibit different degrees of cross-reactivity between trypanosome-exposed and non-exposed cattle. We also assessed whether animal-infective trypanosomes express VATs against which an antibody response would be detected by the TL test.
Methods
Test samples
Two categories of samples were used in this study. The first category was blood and sera collected from communally grazed indigenous cattle in a region of mid-northern Uganda that is endemic for rhodesiense HAT, and highly endemic for AAT (Fig. 1). The samples were collected in previous surveys of whole herds, comprising animals that were 6 months and older. No data on ages were collected from these animals, but since the animals were grazed communally and whole herds were tested, the majority were more than 12 months old. Venous blood was taken from each animal and immediately examined using the haematocrit centrifugation technique (HCT) to determine the infection status [39]. The blood was stored in liquid nitrogen for subsequent analysis by PCR to determine the parasite species, and to confirm that the animals found negative by HCT were indeed negative (endemic controls). The second category of samples comprised fresh blood obtained by an ear prick from 121 cattle owned by small-scale farmers in a region of central Kenya that is free from tsetse and trypanosomiasis (non-endemic controls). The animals were mainly exotic and cross-bred, aged from 1 week to 10 years, and kept at different levels of husbandry, some in well managed zero-grazing systems in which calves were separated from cows, and others in open paddocks. Of these, 39 (32.0%) were less than 1 year old, 41 (33.6%) between one and 4 years, and 42 (34.4%) were 4 years or older.
Fig. 1.
Map of Kenya and Uganda showing the counties of origin of samples analysed (Ugandan samples are shown in red, Kenyan Samples in green). The geographical layers were obtained from CC-BY License compatible sources: GADM (http://www.gadm.org) and the RCMRD geoportal (http://geoportal.rcmrd.org/)
Test kits
Two gHAT RDTs developed by SD were used in the study. The 1G RDT, named SD BIOLINE HAT, was developed using native VSGs (Nat-LiTat 1.3 and Nat-LiTat 1.5) obtained from the Institute of Tropical Medicine (ITM) in Antwerp, Belgium, and detects anti-VSG LiTat 1.3 (band 1) and anti-VSG LiTat 1.5 (band 2) antibodies [32, 33, 35]. The p2G RDT is a prototype developed using the recombinant antigens rISG65 (band 1) and rLiTat 1.5 (band 2) and detected the corresponding antibodies to ISG65 and VSG LiTat 1.5 [32]. The formats of both tests allow reading of results of each antigen individually (see the example of 1G RDT in Fig. 2). The intensity of positive test bands is estimated using a colour chart provided by the manufacturer. When the RDTs are used to screen patients, however, a test is considered positive if the result of any of the two antigens is positive.
Fig. 2.
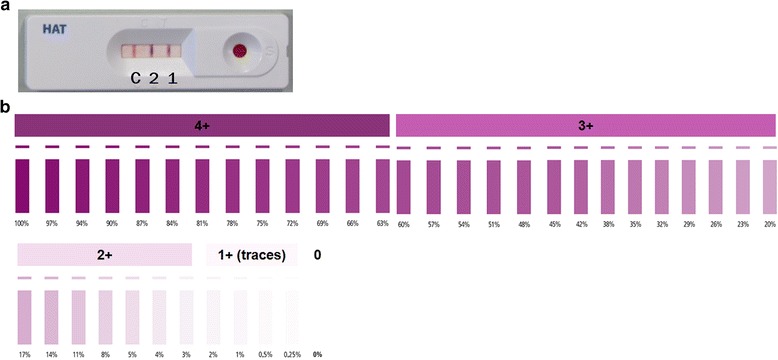
A 1G RDT for gHAT (a) and colour chart (b) for estimating the intensity of positive results. The RDT shows positive results on both antigen bands (1 and 2) after testing fresh blood. C, control band
Test procedures
Detection of parasite DNA
Whole blood samples collected in Uganda were thawed and DNA extracted using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), then tested by PCR targeting the internal transcribed spacer (ITS1) as described by Njiru et al. [40]. For this study, cases were defined as cattle that were positive by both microscopy and ITS PCR; endemic controls were animals from the same herds that were negative by both microscopy and PCR. Samples whose parasitological and ITS PCR results did not agree were excluded in the subsequent analysis. A subset of samples whose ITS PCR indicated the presence of the T. brucei group (Trypanozoon) were tested further using the serum resistance associated PCR (SRA-PCR) and the T. gambiense-specific glycoprotein PCR (TgsGP-PCR) [41] to determine the presence of T. b. rhodesiense or T. b. gambiense, respectively. These tests were not performed on the non-endemic control samples from Kenya because the study region is free from tsetse and trypanosomiasis.
Rapid diagnostic tests
The rapid diagnostic tests were performed according to the manufacturers’ instructions. Briefly, in Kenya, 20 μl of fresh blood samples from 121 cattle were obtained from an ear-prick and immediately put in the sample wells of both RDTs in parallel. Four drops of buffer were added, and the results were read after 15 to 20 min. In Uganda, the serum was thawed and 10 μl used to perform the test. The technician who performed the tests on samples from Uganda was blinded to their identity. In both countries, the result for each antigen was recorded and when positive, the intensity of the bands was estimated using a colour scale, from +1 (lowest) to +4 (highest) (Fig. 2b). The results were also recorded digitally by taking a photograph.
Immune trypanolysis tests
The immune trypanolysis tests (TL) were performed only on samples from the endemic region, as they only detect antibodies to trypanosomes. A total of 188 frozen blood and 163 serum samples were shipped to the Institute of Tropical Medicine (ITM) in Antwerp, Belgium. The samples were tested for antibodies against VSG LiTat 1.3 and VSG LiTat 1.5 using two TL tests as described by Van Meirvenne et al. [9]. Briefly, 25 μl of the undiluted test sample was pipetted into separate wells of microplates. 25 μl of complement-rich guinea pig serum (GPS) was added and plates pre-incubated at room temperature for 30 min. Next, 50 μl of a 107/ml suspension of trypanosomes in GPS were added and plates incubated at room temperature for another 90 min. A drop of the reaction mixture was then examined under a phase contrast microscope (250×). The highest antiserum dilution, defined as the final dilution in the reaction mixture still causing 50% lysis of trypanosomes was arbitrarily considered as the end-point titre, and samples with a higher figure were recorded as positive. A sample was recorded as positive if either of the TL tests was positive. Only samples with sufficient volumes for at least one of the TL tests were included in this assay. The testing laboratory was blinded to the origin of the samples.
Case/control definitions and statistical methods
For this study, ‘cases’ were defined as cattle that were positive by both microscopy and ITS PCR; ‘endemic controls’ were animals from the same herds that were negative by both microscopy and PCR. Any samples from the endemic region for which the results of microscopy and ITS PCR were discordant were excluded. ‘Non-endemic controls’ were also included in the analyses.
All statistical analyses were performed in the R statistical environment [42]. 95% confidence intervals (CI) were calculated using the Wilson interval. Statistical differences between the percentage of tests that were positive and those negative on unpaired tests were evaluated using Fisher’s exact test and using McNemar’s chi-square test on paired 2 × 2 table data. The intensity of the RDT bands was analysed by converting the categorical scale to a numeric scale with 0 = negative, 1 = 1+ to 4 = 4 + based on the manufacturers’ chart (Fig. 2b).
Results
Microscopy and PCR on endemic samples
The samples were from two batches. In the first batch, 194 samples were pre-selected from a larger set of samples and comprised 99 that were positive by both ITS1 PCR for T. brucei, as well as microscopy and so, were included as cases, while 95 were negative by both tests and so were therefore included as endemic controls.
The second batch of 159 samples included samples that were positive with other animal infective trypanosome species (T. congolense and T. vivax) that co-exist with T. brucei. For this, we tested the 159 samples with no pre-selection, including 76 that met the case definition criteria; 42 controls, and 41 that gave discordant results and were therefore excluded. Of the 76 cases in this batch, 47 (61.8%) were positive for only one species (T. brucei, T. congolense or T. vivax), 16 (21.1%) were positive for any two, while 13 cattle (17.1%) were positive for all the three species. Overall in the second batch, 31 animals (40.8%) were infected with T. brucei, either as a single infection or as a co-infection with other species (Additional file 1: Table S1). In total, there were 175 cases and 137 controls from the endemic region.
Performance of serological tests
RDT positivity
When 121 samples from the non-endemic region were tested with the 1G RDT, 51 (42.1%; 95% CI: 33.7–51.1%) were positive on at least one band (Nat-LiTat1.3 and/or Nat-LiTat 1.5). This was significantly lower (Fisher’s exact test, P < 0.001) than the 266 out of 312 (85.3%; 95% CI: 80.9–88.8%) samples from the endemic region that were positive by the 1G RDT. Furthermore, the band intensities (a qualitative indication of the concentration of detectable antibodies) of the positive samples from the non-endemic region were fainter (in the range 1.07–1.45), compared to those from the endemic sera (range 2.07–2.44) (Table 1).
Table 1.
Percentage positivity and intensity of positive bands of the 1G HAT RDT on cattle samples from AAT endemic and non-endemic regions
| Cases (n = 175) | Endemic controls (n = 137) | Non-endemic controls (n = 121) | ||||
|---|---|---|---|---|---|---|
| Device | % positive (95% CI) | Mean band intensity ± SD (+ve’s) | % positive (95% CI) | Mean band intensity ± SD (+ve’s) | % positive (95% CI) | Mean band intensity ± SD (+ve’s) |
| 1G RDT (either band +ve) | 85.1 (79.1–89.7) (n = 149) | 2.15 ± 1.11 | 85.4 (78.5–90.3) (n = 117) | 2.07 ± 1.09 | 42.1 (33.7–51.1) (n = 51) | 1.07 ± 0.46 |
| Band 1 (Nat-LiTat 1.3) | 81.1 (74.7–86.2) (n = 142) | 2.44 ± 1.11 | 76.6 (68.9–82.9) (n = 105) | 2.35 ± 1.13 | 38.8 (30.6–47.7) (n = 47) | 1.45 ± 0.50 |
| Band 2 (Nat-LiTat 1.5) | 72.6 (65.5–78.6) (n = 127) | 2.31 ± 1.15 | 71.5 (63.5–78.4) (n = 98) | 2.42 ± 1.14 | 24.0 (17.2–32.3) (n = 29) | 1.41 ± 0.50 |
Abbreviation: SD standard deviation
There was a significant difference between the results of bands 1 and 2 of the 1G RDT in the non-endemic samples (Tables 2 and 3; McNemar’s χ 2 1 = 11.1, P < 0.001) with greater numbers positive on band 1 (Nat-LiTat 1.3), and only four positives on band two alone. Similar patterns were seen in the samples from the endemic region.
Table 2.
Agreement between bands 1 and 2 in the 1G RDT on samples from Kenya and Uganda
| 1G RDT | Band 2 LiTat 1.5 | |||
|---|---|---|---|---|
| -ve | +ve | |||
| Band 1 LiTat 1.3 | Non-endemic samples (n = 121) | -ve | 70 | 4 |
| +ve | 22 | 25 | ||
| Endemic samples (n = 312) | -ve | 46 | 19 | |
| +ve | 41 | 206 | ||
Table 3.
Agreement between bands 1 and 2 in the p2G RDT on samples from Kenya and Uganda
| p2G RDT | Band 2 rLiTat 1.5 | |||
|---|---|---|---|---|
| -ve | +ve | |||
| Band 1 rlSG65 | Non-endemic samples (n = 121) | -ve | 118 | 1 |
| +ve | 2 | 0 | ||
| Endemic samples (n = 312) | -ve | 50 | 15 | |
| +ve | 60 | 187 | ||
Only 3 out of 121 cattle (2.5%; 95% CI: 0.8–7.0%) from the non-endemic region were positive with the p2G RDT (Table 4). Two of the three animals, aged 15 months and 4 years, were positive in band 1 (rlSG65) only. The third, aged 10 years, was positive on band 2 (rLiTat 1.5). Significantly fewer were positive on band 2 (rLiTat 1.5) in the p2G RDT (0.8%; 95% CI: 0–4.5%) than band 2 (Nat-LiTat 1.5) in the 1G RDT (24.0%; 95% CI: 17.2–32.3%) yet both bands detect antibodies to the same antigen (McNemar’s χ 2 1 = 5.31, P = 0.021). These results were significantly different from the results on sera from the endemic region (Fisher’s exact test, P < 0.001), in which 89.1% of cases and 77.4% of controls were positive by the p2G RDT, mainly due to band 1 (rISG65) (Table 4). A significantly larger proportion of the sera were positive in band one compared to band 2 of the p2G RDT in samples from the endemic region (McNemar’s χ 2 1 = 25.8, P < 0.001).
Table 4.
Percentage positivity of the p2G HAT RDT on cattle sera from the endemic and non-endemic regions (S2)
| Cases (n = 175) | Endemic controls (n = 137) | Non-endemic controls (n = 121) | ||||
|---|---|---|---|---|---|---|
| % positive (95% CI) | Mean band intensity ± SD (+ve’s) | % positive (95% CI) | Mean band intensity ± SD (+ve’s) | % positive (95% CI) | Mean band intensity (+ve’s) | |
| p2G RDT | 89.1 (83.7–92.9) (n = 156) | 2.16 ± 0.94 | 77.4 (69.7–83.6) (n = 106) | 1.69 ± 0.86 | 2.5 (0.8–7.0) (n = 3) | 0.67 |
| Band 1 (ISG65) | 86.3 (80.4–90.6) (n = 151) | 2.83 ± 1.08 | 70.1 (61.9–77.1) (n = 96) | 2.36 ± 1.16 | 1.7 (0.5–5.8) (n = 2) | 1 |
| Band 2 (rLiTat 1.5) | 66.9 (59.6–73.4) (n = 117) | 2.10 ± 1.05 | 62.0 (53.7–69.7) (n = 85) | 1.54 ± 0.80 | 0.8 (0–4.5) (n = 1) | 2 |
Abbreviation: SD standard deviation
Standard deviation is not shown for the endemic control region owing to the small number of positives
The results of testing sera from cattle in an endemic region with the 1G and p2G RDTs are shown in Table 5. There was no significant difference in the percentage positivity between cases (85.1%) and endemic controls (85.4%) (Fisher’s exact test, P = 1). Among the cases, 168 (96%) were positive by one or both RDTs, while among the endemic controls, 121 (89.1%) were positive with one or both RDTs, altogether, 290 (92.9%) animals in the endemic area were positive with one or both RDTs.
Table 5.
Some endemic sera that were positive by individual antigens and RDTs
| RDT | Infection status | No. of samples | RDT | By RDT band | ||
|---|---|---|---|---|---|---|
| Either band | Band 1 only | Band 2 only | Bands 1 & 2 | |||
| 1G | Cases | 175 | 149 (85.1%) | 22 (12.6%) (Nat-LiTat 1.3) | 7 (4.0%) (Nat-LiTat 1.5) | 120 (68.6%) |
| Endemic controls | 137 | 117 (85.4%) | 19 (13.9%) (Nat-LiTat 1.3) | 12 (8.8%) (Nat-LiTat 1.5) | 86 (62.8%) | |
| Total | 312 | 266 (85.3%) | 41(13.1%) | 19 (6.1%) | 206 (66.0%) | |
| p2G | Cases | 175 | 156 (89.1%) | 39 (22.3%) (rISG65) | 5 (2.9%) (rLiTat 1.5) | 112 (64.0%) |
| Endemic controls | 137 | 106 (77.4%) | 21 (15.3%) (rISG65) | 10 (7.3%) (rLiTat 1.5) | 75 (54.7%) | |
| Total | 312 | 262 (84.0%) | 60 (19.2%) | 15 (4.8%) | 187 (59.9%) | |
When the results of native and recombinant VSG LiTat 1.5 were compared, Nat-LiTat 1.5 (band 2 of the 1G RDT) was positive in a larger number of cases (127, comprising 7 on band 2 only and 120 on bands 1 and 2, i.e. 72.6%) than rLiTat 1.5 (band 2 on the p2G RDT) (117, comprising 5 on band 2 only and 112 on bands 1 and 2, i.e. 66.9%). The difference was however not significant (McNemar’s χ 2 1 = 1.45, P = 0.229). Similarly, among endemic controls, 98 (comprising 12 on band 2 only and 86 on bands 1 and 2, i.e. 71.5%) sera were positive for Nat-LiTat 1.5 (band 2 on the 1G RDT) compared to 85 (comprising 10 on band 2 only and 75 on bands 1 and 2, i.e. 62.0%) for rLiTat 1.5 (band 2 of the p2G RDT), a difference that was significant (McNemar’s χ 2 1 = 4.11, P = 0.045) (Table 5).
A comparison of the results of individual antigens on both RDTs with PCR and microscopy are shown in Table 6. The results for both RDTs agreed in 56% (n = 7+ 4 + 87 = 98) of cases and 62.0% (n = 15 + 5 + 4 + 61 = 85) of endemic controls. 148 (87 + 61) (47.4%) cattle were positive with all four antigens; 87 (49.7%) of these were cases and 61 (44.5%) endemic controls.
Table 6.
Comparison of results of antigens on the 1G and p2G RDTs on samples from the endemic area
| p2G RDT | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases (n = 175) | Endemic controls (n = 137) | ||||||||
| −/− | +/− | −/+ | +/+ | −/− | +/− | −/+ | +/+ | ||
| 1G RDT | −/− | 7 | 10 | 0 | 9 | 15 | 3 | 0 | 2 |
| +/− | 4 | 4 | 0 | 14 | 5 | 5 | 3 | 6 | |
| −/+ | 0 | 5 | 0 | 2 | 0 | 2 | 4 | 6 | |
| +/+ | 8 | 20 | 5 | 87 | 11 | 11 | 3 | 61 | |
Abbreviations: −/−, negative both bands; +/−, positive band 1 only; −/+, positive band 2 only; +/+, positive both bands
The number and intensity scores of positive bands in the p2G RDT were higher on band 1 (rISG65) than band 2 (rLiTat 1.5), particularly among cattle that were trypanosome positive (Wilcoxon rank sum test, P < 0.001). This difference in intensity scores was not observed with the 1G RDT (P = 0.618) (Tables 1 and 4).
Five of the T. brucei (Trypanozoon) positive samples from Uganda (n = 130) that were tested with SRA-PCR were positive, indicating the presence of T. b. rhodesiense, while the other 125 were T. b. brucei. These five were also positive by both bands of the p2G RDT. Four of these samples were positive by both bands of the 1G RDT and one by band one only. None of these samples was positive with TgsGP-PCR (T. b. gambiense).
Effect of age on the positivity of RDTs
The number of cattle in the non-endemic region that were positive with the 1G RDT increased with age (Fig. 3). There was a significant difference on both antigens separately, and on the overall RDT result (Fisher’s exact P-value < 0.001) between animals aged less than 12 months and those that were 12 months or older. In the older age groups (12 months and older), the percentage positivity was similar to that seen in cattle in the endemic region, but the band intensity among positive animals in this age group remained low (1.41 and 1.40 in bands 1 and 2). The effect of age on the results of the p2G RDT could not be assessed because only three animals were positive with this RDT.
Fig. 3.
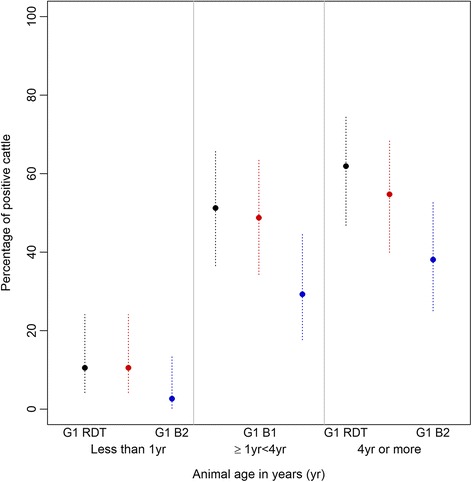
Percentage and 95% confidence interval of samples found positive with 1G RDT in cattle in a non-endemic region of Kenya. The results are shown by age group, overall RDT result and individual bands
Immune trypanolysis tests on sera from the endemic region
The immune trypanolysis (TL) tests were performed on 295 sera from Uganda, including 172 that were cases, and 123 endemic controls (Table 7). Of the 295, 141 (47.8%) were positive by TL (comprising 102 cases and 39 endemic controls). Among the sera from cases, TL Nat-LiTat 1.5 was positive in 52.3%, while 82.9% of endemic controls tested negative, the difference in numbers testing positive (52.3% of cases vs 100–82.9% = 17.1% endemic control sera) was statistically significant (Fisher’s exact P-value <0.001). However, TL Nat-LiTat 1.3 was positive in only 23.3% of the case sera, but the specificity among the endemic control sera was 79.7%, the difference in numbers testing positive (23.3% of cases vs 100–79.7% = 20.3% endemic control sera) was not significant (Fisher’s exact P-value = 0.572). The high sensitivity among trypanosome positive sera was mainly driven by VSG LiTat 1.5 (Tables 7, 8) (McNemar’s χ 2 1 = 32.4, P < 0.001) with 23.3% positive by VSG LiTat 1.3 and 52.3% by VSG LiTat 1.5, but there was no difference among the negative sera (McNemar’s χ 2 1 = 0.281, P = 0.60), corresponding specificities were 79.7 and 82.9%.
Table 7.
Results of the TL tests on cattle sera from the endemic region
| Cases | Endemic controls | |||||
|---|---|---|---|---|---|---|
| TL result | TLa | TL Nat-LiTat 1.3 | TL Nat-LiTat 1.5 | TLa | TL Nat-LiTat 1.3 | TL Nat-LiTat 1.5 |
| Tested | 172 | 172 | 172 | 123 | 123 | 123 |
| Negative | 70 | 132 | 82 | 84 | 98 | 102 |
| Positive | 102 | 40 | 90 | 39 | 25 | 21 |
| Sensitivity (%) (95% CI) | 59.3 (51.8–66.4) | 23.3 (17.6–30.1) | 52.3 (44.9–59.7) | – | – | – |
| Specificity (%) (95% CI) | – | – | – | 68.3 (59.6–75.9) | 79.7 (71.7–85.8) | 82.9 (75.3–88.6) |
aResults of both TL tests combined
Table 8.
Cross-tabulation of TL results by antigen
| TL Nat-LiTat 1.5 | ||||
|---|---|---|---|---|
| Negative | Positive | Total | ||
| Cases | ||||
| TL Nat-LiTat 1.3 | Negative | 70 | 62 | 132 |
| Positive | 12 | 28 | 40 | |
| Total | 82 | 90 | 172 | |
| Endemic controls | ||||
| TL Nat-LiTat 1.3 | Negative | 84 | 14 | 98 |
| Positive | 18 | 7 | 25 | |
| Total | 102 | 21 | 123 | |
As both TL tests detect antibodies against the antigens that are included in the 1G RDT, results of TL were compared to those obtained using the 1G RDT. Among cases, 51.7% were also positive by both TL and the 1G RDT, with the highest agreement being due to the VSG Nat-LiTat 1.5 (Table 9). Among endemic controls, a smaller number (30.9%) were positive by both 1G RDT and by TL.
Table 9.
Results of TL tests and 1G RDT, and the level of agreement between tests
| Testa | Positive TL (%) | Positive RDT (%) | Positive agreement (%) | Negative agreement (%) |
|---|---|---|---|---|
| Cases (n = 172) | ||||
| Test | 102 (59.3) | 148 (86.0) | 89 (51.7) | 11 (6.4) |
| Nat-LiTat 1.3 | 40 (23.3) | 141 (82.0) | 36 (20.9) | 27 (15.7) |
| Nat-LiTat 1.5 | 90 (52.3) | 127 (73.8) | 69 (40.1) | 24 (14.0) |
| Endemic controls (n = 123) | ||||
| Test | 39 (31.7) | 105 (85.3) | 38 (30.9) | 17 (13.8) |
| Nat-LiTat 1.3 | 25 (20.3) | 94 (76.4) | 22 (17.9) | 26 (21.1) |
| Nat-LiTat 1.5 | 21 (17.1) | 89 (72.4) | 20 (16.2) | 33 (26.8) |
a“Test” refers to the test being positive, in TL and the RDT, the test is interpreted as positive if either band is positive. Nat-LiTat 1.3 and 1.5 refer to the results of each antigen in TL and the RDT. Positive and negative agreement are the numbers where the test results agreed
Discussion
This study has demonstrated a variability of the specificity of RDTs for gHAT when tested on cattle, depending on the antigens used, and on the presence or absence of AAT in a region. The specificity of the p2G RDT, which is made using recombinant antigens, on cattle in a non-endemic region was very good. In contrast, the specificity of the 1G RDT, made using native antigens, decreased with the age of the animals. However, when the three serological tests used in this study were performed on samples from cattle in an AAT endemic region, they all had a low specificity. Antibodies to ISG65 were detected in the largest number of cattle in the endemic region. This could be because, unlike VSGs, ISG65 is constantly expressed by all trypanosome populations in the host, unlike VSGs, which switch with each wave of parasitaemia; the ISG family was previously investigated as T. congolense diagnostics with promising results [43]. In live trypanosomes, ISGs are largely protected from the host immune response by the VSG coat. However, as parasites are continuously killed by the host anti-VSG antibody response, this process exposes ISGs, eliciting a persistent antibody response, which could explain the larger number of positive cattle.
The results of this study also suggest that all the antigens tested are likely to have cross-reacting epitopes from other trypanosome species, but perhaps not predominantly. This would not be surprising, considering that recently, a high similarity between different trypanosome species has been demonstrated at the genomic level [44, 45]. In other studies, the CATT test, which has the VSG LiTat 1.3 antigen, was also positive in animals infected with either T. congolense, T. b. brucei or T. evansi [46–48]. In another study, a large number of trypanosome antigens, including VSG LiTat 1.3 and VSG LiTat 1.5, reacted to sera from both T. b. gambiense and T. b. rhodesiense patients [49]. Another candidate, rISG65-2 had a sensitivity of 92% and a specificity of 85% for T. b. rhodesiense [50, 51], indicating that, while the antigens in these studies had good sensitivity for both T. b. gambiense and T. b. rhodesiense, their sensitivity was not restricted to the two sub-species of trypanosomes.
The specificity of the recombinant antigens on non-endemic cattle was very good, while that of native antigens was low. As a result, the p2G RDT had a very good specificity when compared to the 1G RDT. This difference may be attributed to the manner in which the antigens used to develop the tests are produced. The VSGs used in the 1G RDT are produced from pathogenic T. b. gambiense in a process that involves infecting rodents and harvesting the parasites during the first peak of parasitaemia. The parasites are purified by gel chromatography, and the VSG is separated through a process of freeze-thawing and fractionation [52]. While the final sample would comprise mainly VSGs of the particular VAT, proteins that could cross-react with antigens expressed by related organisms would likely be present. The possibility of such cross-reactivity would explain the lower specificity of the 1G RDT on samples from the non-endemic region. In that husbandry system, cattle were under different levels of exposure to infections such as anaplasmosis, theileriosis and babesiosis; and besides, the presence of the tabanid-transmitted T. theileri cannot be ruled out. This hypothesis is supported by our observation that the likelihood of an animal in the non-endemic region being positive with the 1G RDT increased with age. In the non-endemic region, young calves are usually kept in sheds where the risk of being bitten by ticks that transmit such diseases is low. As the calves grow older, they are kept in zero-grazing systems or in open paddocks in close contact with older animals, where the risk of tick infestation and transmission of disease is comparatively higher. Indeed, previous reports have suggested a possible cross-reactivity of the CATT reagents with antibodies against other infections such as malaria [53, 54]. There could, however, be other reasons for the low specificity of the 1G RDT in these animals, such as the likely presence of anti-VSG native antibodies in animals that had never been exposed to trypanosomes, a phenomenon that has been observed in mice, bovines and humans [53]. Noteworthy is that although LiTat1.3 is considered specific to T. b. gambiense it is identical to many other Trypanozoon VSGs. A BLAST search with the respective protein sequence (accession no. AHW98113.1) indicates considerable identity with several T. brucei VSGs, peaking at 79–81% with four different VSGs (accession nos. APD74596.1, APD75145.1, AGH60298.1, APD73903.1, AGH59165.1). The search also picks out putative VSGs from T. equiperdum albeit at lower identity (40–45%). Thus cross-reactivity should be expected even with other trypanosomes expressing different VSGs. Besides, all VSGs share the so-called cross-reactive determinant (CRD) epitope on the GPI anchor that can also contribute to cross-reactivity.
Unlike the 1G RDT, the p2G RDT is made using highly purified recombinant antigens, which could explain the high specificity observed in cattle from the non-endemic region. Indeed when the results of the native VSG LiTat 1.5 antigens are compared with those of the recombinant variant, the native antigen was positive in more samples than the recombinant protein in both the endemic and non-endemic regions, regardless of the infection status, although the difference was not significant. The additional positive samples with native antigens in the endemic region could be due to cross-reactivity with antibodies against other pathogens, otherwise the native and recombinant forms of the same antigen detect the same antibodies.
Among the trypanosome positive samples, more than half were also positive by TL, a clear indication that TL positive results are associated with trypanosome infection. A proportion of endemic controls were also positive by TL. As with RDTs, it is likely that some of these controls were true infections that were missed by microscopy and PCR, attributed to the higher sensitivity of tests that detect host antibodies, which would be abundant in blood.
The number of trypanosome positive and negative samples from the endemic region that were positive with one or both RDTs was not significantly different. This could be confusing, considering that animals were identified as negative when the results of microscopy and PCR were negative. However, RDTs detect antibody responses to exposure to parasites, and not necessarily active infection. This, coupled with a lower sensitivity of parasitological and molecular tests when compared to detection of antibodies, means that some of the animals that were RDT positive but trypanosome negative could have been missed infections. That some animals had an antibody response to each of the test antigens may be related to the duration of infection. Waves of parasitaemia are associated with different VATs, and the longer an animal remains infected, the more VATs appear in the blood, each eliciting an antibody response [30, 33, 36]. Sometimes farmers also treat their animals with trypanocides, and in such cases, recently treated animals would be positive for antibodies but trypanosome negative.
The p2G RDT detected 89.1% of the sera from the endemic region that were trypanosome positive, which was significantly higher than those positive among the endemic controls. This is a striking correlation, considering that at present, no RDT for diagnosis of AAT is commercially available. The possibility that this test could be used for passive screening of cattle that have clinical signs in AAT endemic regions needs further investigation. The test would not be used for herd screening since not all animals with an antibody response would develop the disease. In the context of elimination of both gHAT and AAT however, such a test would be useful for confirming total clearance of trypanosomes in a region, and in detecting likely re-emergence when used on sentinel herds.
The sensitivity of the 1G RDT in trypanosome positive samples was higher than TL, which uses live parasites expressing the same VSGs as those used to develop the RDT. Indeed when the results of each of the TL tests were considered separately, the sensitivity decreased further. This would be expected, since TL detects antibodies that are specific to the VATs in the test [11], in this case only anti-VSG LiTat 1.3 and anti-VSG LiTat 1.5 antibodies. A similar difference between TL tests and other screening tests has been reported in other studies [47]. The difference in sensitivity between the 1G RDT and TL could be because the VSGs used to develop the 1G RDT are detached from trypanosomes, exposing epitopes that in the live parasites used in the TL test, would not be available to detect antibodies against them [52]. In the animal, however, such epitopes would have been exposed when trypanosomes are killed by the host immune system, eliciting an antibody response that would be detected by the RDT but not by the TL test.
The sensitivity of TL was largely driven by TL Nat-LiTat 1.5. The comparatively low sensitivity of TL Nat-LiTat 1.3 could be an indication that this antigen is expressed at a lower frequency by trypanosomes that cause AAT than the other antigens used in this study. T. b. gambiense parasites expressing LiTat 1.3 have in the past been among the most suitable for developing a gHAT screening test as opposed to those expressing LiTat 1.5 or LiTat 1.6 [31, 37, 52, 55]. The TL tests were the least sensitive among the serological tests used in the present study, which is understandable, because the tests are VAT-specific, only detecting antibodies to the specific parasite VATs. A similar low sensitivity of TL has been reported in other studies [17].
The low specificity of the 1G RDT observed in a non-endemic region could have implications on the interpretation of positive screening test results in animals living in gHAT endemic regions. In previous studies in gHAT endemic regions, animals found positive with the CATT test were considered to be infected with T. b. gambiense, and therefore potential reservoirs of the disease [48]. The results of this study, however, show that besides the possible existence of T. b. gambiense in the animal reservoir, cross-reactivity with other infections or with host proteins could influence the performance of the test. Conversely, the rLiTat 1.5 antigen on the p2G RDT was positive in only one animal (99.2% specificity) in the non-endemic region, indicating that in a region that is endemic for trypanosomiasis, a positive result with this antigen could most likely be due to the presence of anti-trypanosome antibodies. Other molecular methods such as sub-species-specific PCR would, however, be required to confirm the presence of human-infective trypanosomes in such samples.
Five of the T. brucei positive samples from the endemic region were also positive with SRA-PCR, indicating the presence of T. b. rhodesiense, further evidence that livestock is an important reservoir of T. b. rhodesiense HAT [39, 56]. None of the samples was positive with TgsGP-PCR, indicating that T. b. gambiense was not circulating in the region, and further indirect evidence that the results obtained with the RDTs were due to the presence of cross-reacting antibodies to parasites other than T. b. gambiense.
This study has demonstrated that serological tests for gHAT when used on cattle, cross-react with antibodies to other pathogens. These results, if reproduced in humans, could have important implications on the selection of samples suitable for development of tests for gHAT, interpretation of screening test results, control of the disease, and surveillance to sustain elimination. Plasma and serum samples that are used during development of RDTs for gHAT are taken from people living in regions that are endemic for both gHAT and AAT. The distinction between the samples has been that gHAT cases are those that are confirmed by microscopy, in most cases following a positive screening test, while controls are individuals found negative by screening tests that are in use at that time. Current screening tests detect antibodies to only one or two antigens, while there are likely to be many more anti-trypanosome antibodies in the samples, depending on the number of VATs that had been expressed in different waves of parasitaemia, and to exposure to various trypanosome species. Such antibodies could be detected by prototype tests during development, with the risk that the prototypes would be discarded on account of low specificity. Indeed in a recent study in which newly developed RDTs were tested using stored samples, their specificity was found to be low, while the sensitivity was very good [35]. The control samples in that study had been classified as controls because they were negative with CATT, which mainly detects antibodies to VSG LiTat 1.3. To minimize such challenges, we recommend that control samples for development of serological tests be obtained from regions that are geographically similar to gHAT endemic ones, but free from tsetse flies, and both HAT and AAT.
Clinical testing of new tests for gHAT is carried out at multiple sites, to correct for the potential geographic variability of trypanosome strains. Due to the continued decline in the number of gHAT cases reported annually, data from multiple sites is pooled to meet the required number of study participants. The specificity of such tests has often varied from region to region [33, 35, 55]. The results of this study show that it is possible to have antibody responses to trypanosomes that do not cause gHAT, and to non-target organisms, which should be taken into account when designing clinical trials on new tests.
This study had some limitations. The serological tests are designed for use on human samples. Additional studies are needed, to determine whether their performance is affected by factors that could be present in cattle serum. The communally grazed cattle in the AAT endemic region are occasionally treated with trypanocides, and due to the persistence of antibodies, such animals would have been positive with the serological test but trypanosome negative, creating a bias on the number of endemic controls that were positive by serology. The TL tests were included to determine whether they can detect VAT-specific antibodies to trypanosomes that are not infective to humans, and because they are highly specific to trypanosomes, they were not included in the samples from the non-endemic region. This omission could have biased the TL results and was a missed opportunity to confirm the specificity of the tests.
Conclusions
While the specificity of an RDT for gHAT made using recombinant antigens was very good when used on cattle in a trypanosomiasis-free region, this decreased dramatically in an AAT endemic region, limiting its potential as a diagnostic test. The specificity of an RDT made using native parasite antigens decreases with the age of the cattle, probably due to cross-reacting antibodies arising from exposure to other pathogens. Trypanosomes that are not infective to humans are capable of switching to VATs that are detectable by the current serological tests for gHAT, rendering the tests of less utility for identification of animal reservoirs for gHAT.
Acknowledgements
The authors are grateful to the COVAB team that tested cattle in the field and laboratory identification of infecting trypanosomes. We acknowledge the free access given by cattle owners, and their assistance in handling the animals during sampling and testing. We thank Professor Mike Barrett, Glasgow University, UK, for his comments and suggestions on the manuscript.
Funding
This study was funded by UBS Optimus Foundation through COVAB, and through FIND, the Swiss Government and UK aid from the UK Government.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 1G
First generation
- AAT
Animal African trypanosomiasis
- CATT
Card agglutination test for trypanosomiasis
- CI
Confidence interval
- ELISA
Enzyme-linked immunosorbent assay
- gHAT
gambiense human African trypanosomiasis
- GPS
Guinea pig serum
- HAT
Human African trypanosomiasis
- HCT
Haematocrit centrifugation technique
- ISG65
Invariant surface glycoprotein 65
- ITM
Institute of Tropical Medicine
- ITS
Internal transcribed spacer
- Nat
Native
- p2G
Prototype second generation
- RDT
Rapid diagnostic test
- rHAT
rhodesiense human African trypanosomiasis
- SD
Alere/standard diagnostics
- SRA
Serum resistance associated
- TgsGP
T. gambiense-specific glycoprotein
- TL
Immune trypanolysis tests
- VAT
Variant antigenic type
- VSG
Variant surface glycoprotein
Additional file
Results of ITS1-PCR on the 76 PCR and parasitology positive samples from the second set of sera. (DOCX 15 kb)
Authors’ contributions
JMN conceived the hypothesis. JMN, EM and AK designed the study. AK, EM and JMN conducted the experiments. PB, SB and JMN analysed the data. EM, JMN, AK, PB, AP and SB wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Consent to take samples from cattle was given verbally by the owners.
Consent for publication
Not applicable.
Competing interests
AP, JMN and SB are employees of FIND, which co-developed the RDTs used in the present study. PB is a consultant for FIND. The authors declare no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13071-017-2487-8) contains supplementary material, which is available to authorized users.
Contributor Information
Enock Matovu, Email: matovue@covab.mak.ac.ug.
Annah Kitibwa, Email: kitty.ann34@yahoo.com.
Albert Picado, Email: albert.picado@finddx.org.
Sylvain Biéler, Email: sylvain.bieler@finddx.org.
Paul R. Bessell, Email: paul.bessel@finddx.org
Joseph Mathu Ndung’u, Email: joseph.ndungu@finddx.org.
References
- 1.Carrington M, Miller N, Blum M, Roditi I, Wiley D, Turner M. Variant specific glycoprotein of Trypanosoma brucei consists of two domains each having an independently conserved pattern of cysteine residues. J Mol Biol. 1991;221:823–835. doi: 10.1016/0022-2836(91)80178-W. [DOI] [PubMed] [Google Scholar]
- 2.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 3.Marcello L, Barry D. Analysis of the VSG gene silent archive in Trypanosoma brucei reveals that mosaic gene expression is prominent in antigenic variation and is favored by archive substructure. Gen Res. 2007;17:1344–1352. doi: 10.1101/gr.6421207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones NG, Nietlispach D, Sharma R, Burke DF, Eyres I, Mues M, et al. Structure of a glycosylphosphatidylinositol-anchored domain from a trypanosome variant surface glycoprotein. J Biol Chem. 2008;283:3584–3593. doi: 10.1074/jbc.M706207200. [DOI] [PubMed] [Google Scholar]
- 5.Rudenko G. African trypanosomes: the genome and adaptations for immune evasion. Essays Bioch. 2011;51:47–62. doi: 10.1042/bse0510047. [DOI] [PubMed] [Google Scholar]
- 6.Jackson AP, Berry A, Aslett M, Allison HC, Burton P, Vavrova-Anderson J, et al. Antigenic diversity is generated by distinct evolutionary mechanisms in African trypanosome species. Proc Natl Acad Sci USA. 2012;109:3416–21. doi: 10.1073/pnas.1117313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Günzl A, Kirkham JK, Nguyen TN, Badjatia N, Park SH. Mono-allelic VSG expression by RNA polymerase I in Trypanosoma brucei: expression site control from both ends? Gene. 2015;556:68–73. [DOI] [PMC free article] [PubMed]
- 8.Mugnier MR, Stebbins CE, Papavasiliou FN. Masters of disguise: antigenic variation and the VSG coat in Trypanosoma brucei. PLoS Pathog. 2016;12:e1005784. doi: 10.1371/journal.ppat.1005784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner CM, Barry JD. High frequency of antigenic variation in Trypanosoma brucei rhodesiense infections. Parasitology. 1989;99:67–75. doi: 10.1017/S0031182000061035. [DOI] [PubMed] [Google Scholar]
- 10.Turner CM. The rate of antigenic variation in fly-transmitted and syringe-passaged infections of Trypanosoma brucei. FEMS Microbiol Lett. 1997;153:227–231. doi: 10.1111/j.1574-6968.1997.tb10486.x. [DOI] [PubMed] [Google Scholar]
- 11.Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol Cell Biol. 1999;19:5839–5846. doi: 10.1128/MCB.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry JD, McCulloch R. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv Parasitol. 2001;49:1–70. doi: 10.1016/S0065-308X(01)49037-3. [DOI] [PubMed] [Google Scholar]
- 13.Hutchinson OC, Picozzi K, Jones NG, Mott H, Sharma R, Welburn SC, et al. Variant surface glycoprotein gene repertoires in Trypanosoma brucei have diverged to become strain-specific. BMC Gen. 2007;8:234. doi: 10.1186/1471-2164-8-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lythgoe KA, Morrison LJ, Read AF, Barry JD. Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc Natl Acad Sci USA. 2007;104:8095–100. doi: 10.1073/pnas.0606206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aresta-Branco F, Pimenta S, Figueiredo LM. A transcription-independent epigenetic mechanism is associated with antigenic switching in Trypanosoma brucei. Nucleic Acids Res. 2015;44:3131–3146. doi: 10.1093/nar/gkv1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadjuk SL. Antigenic variation during the developmental cycle of Trypanosoma brucei. J Protozool. 1984;31:41–47. doi: 10.1111/j.1550-7408.1984.tb04287.x. [DOI] [PubMed] [Google Scholar]
- 17.Morrison LJ, Majiwa P, Read AF, Barry JD. Probabilistic order in antigenic variation of Trypanosoma brucei. Int J Parasitol. 2005;35:961–972. doi: 10.1016/j.ijpara.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Hovel-Miner G, Mugnier MR, Goldwater B, Cross GAM, Papavasiliou FN. A conserved DNA repeat promotes selection of a diverse repertoire of Trypanosoma brucei surface antigens from the genomic archive. PLoS Genet. 2016;12(5):e1005994. doi: 10.1371/journal.pgen.1005994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall JPJ, Wang H, Barry JD. Mosaic VSGs and the scale of Trypanosoma brucei antigenic variation. PLoS Pathog. 2013;9(7):e1003502. [DOI] [PMC free article] [PubMed]
- 20.McCulloch R, Field MC. Quantitative sequencing confirms VSG diversity as central to immune evasion by Trypanosoma brucei. Trends Parasitol. 2015;31(8):346–349. doi: 10.1016/j.pt.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koumandou VL, Boehm C, Horder KA, Field MC. Evidence for recycling of invariant surface transmembrane domain proteins in African trypanosomes. Euk Cell. 2013;12:330–342. doi: 10.1128/EC.00273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zoltner M, Leung KF, Alsford S, Horn D, Field MC. Modulation of the surface proteome through multiple ubiquitylation pathways in African trypanosomes. PLoS Pathog. 2015;11(10):e1005236. doi: 10.1371/journal.ppat.1005236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegelbauer K, Overath P. Organization of two invariant surface glycoproteins in the surface coat of Trypanosoma brucei. Infect Immun. 1992;61:4540–4545. doi: 10.1128/iai.61.11.4540-4545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziegelbauer K, Multhaup G, Overath P. Molecular characterization of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J Biol Chem. 1992;267:10797–10803. [PubMed] [Google Scholar]
- 25.Jackson DG, Windle HJ, Voorheis HP. The identification, purification, and characterization of two invariant surface glycoproteins located beneath the surface coat barrier of bloodstream forms of Trypanosoma brucei. J Biol Chem. 1993;268:8085–8095. [PubMed] [Google Scholar]
- 26.Schwede A, Jones N, Engstler M, Carrington M. The VSG C-terminal domain is inaccessible to antibodies on live trypanosomes. Mol Biochem Parasitol. 2011;175:201–204. doi: 10.1016/j.molbiopara.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwede A, Carrington M. Bloodstream form trypanosome plasma membrane proteins: antigenic variation and invariant antigens. Parasitology. 2010;137:2029–2039. doi: 10.1017/S0031182009992034. [DOI] [PubMed] [Google Scholar]
- 28.Chappuis F, Loutan L, Simarro P, Lejon V, Buscher P. Options for field diagnosis of human African trypanosomiasis. Clin Microbiol Rev. 2005;18:133–146. doi: 10.1128/CMR.18.1.133-146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndung’u JM, Biéler S, Roscigno G. “Piggy-backing” on diagnostic platforms brings hope to neglected diseases: the case of sleeping sickness. PLoS Negl Trop Dis. 2010;4:e715. doi: 10.1371/journal.pntd.0000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitashi P, Lutumba P, Lumbala C, Bessell P, Biéler S, Ndung’u JM. Improved detection of sleeping sickness cases by LED fluorescence microscopy: evidence from a prospective multicentric study in the Democratic Republic of the Congo. Microscopy Res. 2015;3:17–25. doi: 10.4236/mr.2015.32003. [DOI] [Google Scholar]
- 31.Magnus E, Vervoort T, van Meirvenne N. A card agglutination test with stained trypanosomes (C.A.T.T.) for the serological diagnosis of T. b. gambiense trypanosomiasis. Ann Soc Belge Med Trop. 1978;58:169–176. [PubMed] [Google Scholar]
- 32.Sternberg JM, Gierlinski M, Bieler S, Ferguson MAJ, Ndung’u JM. Evaluation of the diagnostic accuracy of prototype rapid tests for human African trypanosomiasis. PLoS Negl Trop Dis. 2014;8:e3373. doi: 10.1371/journal.pntd.0003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bisser S, Lumbala C, Nguertoum E, Kande V, Flevaud L, Vatunga G, et al. Sensitivity and specificity of a prototype rapid diagnostic test for the detection of Trypanosoma brucei gambiense infection: a multi-centric prospective study. PLoS Negl Trop Dis. 2016;10:e0004608. doi: 10.1371/journal.pntd.0004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buscher P, Mertens P, Leclipteux T, Gilleman Q, Jacquet D, Mumba-Ngoyi D, et al. Sensitivity and specificity of HAT Sero-K-SeT, a rapid diagnostic test for serodiagnosis of sleeping sickness caused by Trypanosoma brucei gambiense: a case-control study. Lancet Glob Health. 2014;2(6):e359–e363. doi: 10.1016/S2214-109X(14)70203-7. [DOI] [PubMed] [Google Scholar]
- 35.Jamonneau V, Camara O, Ilboudo H, Peylhard M, Koffi M, Sakande H, et al. Accuracy of individual rapid tests for serodiagnosis of gambiense sleeping sickness in West Africa. PLoS Negl Trop Dis. 2015;9:e0003480. doi: 10.1371/journal.pntd.0003480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Meirvenne N, Magnus E, Buscher P. Evaluation of variant-specific immune trypanolysis tests for serodiagnosis of human infections with Trypanosoma brucei gambiense. Acta Trop. 1995;60:189–199. doi: 10.1016/0001-706X(95)00127-Z. [DOI] [PubMed] [Google Scholar]
- 37.Jamonneau V, Truc P, Garcia A, Magnus E, Buscher P. Preliminary evaluation of LATEX/T. b. gambiense and alternative versions of CATT/T. b. gambiense for the serodiagnosis of human African trypanosomiasis in a population at risk in Cote d'Ivoire: considerations for mass-screening. Acta Trop. 2000;76:175–183. doi: 10.1016/S0001-706X(00)00095-4. [DOI] [PubMed] [Google Scholar]
- 38.Jamonneau V, Bucheton B, Kabore’ J, Ilboudo H, Camara O, Courtin F, et al. Revisiting the immune trypanolysis test to optimise epidemiological surveillance and control of sleeping sickness in West Africa. PLoS Negl Trop Dis. 2010;4:e917. doi: 10.1371/journal.pntd.0000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simarro PP, Franco JR, Diarra A, Ruiz Postigo JA, Jannin J. Diversity of human African trypanosomiasis epidemiological settings requires fine-tuning control strategies to facilitate disease elimination. Res Rep Trop Med. 2013;4:1–6. doi: 10.2147/RRTM.S40157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RCA, Dávila AMR. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res. 2005;95(3):186–192. doi: 10.1007/s00436-004-1267-5. [DOI] [PubMed] [Google Scholar]
- 41.Maina NWN, Oberle M, Otieno C, Kunz C, Maeser P, Ndung’u JM, Brun R. Isolation and propagation of Trypanosoma brucei gambiense from sleeping sickness patients in South Sudan. Trans R Soc Trop Med Hyg. 2007;101:540–546. doi: 10.1016/j.trstmh.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Core R Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2015; URL https://www.R-project.org/.
- 43.Fleming JR, Sastry L, Crozoier TWM, Napier GB, Sullivan L, Ferguson MA. Proteomic selection of immunodiagnostic antigens from Trypanosoma congolense. PLoS Negl Trop Dis. 8(6):e2936. [DOI] [PMC free article] [PubMed]
- 44.Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, Quail MA, et al. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human African trypanosomiasis. PLoS Negl Trop Dis. 2010;4:e658. doi: 10.1371/journal.pntd.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sistrom M, Evans B, Benoit J, Balmer O, Aksoy S, Caccone A. De novo genome assembly shows genome wide similarity between Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS One. 2016;11:e0147660. doi: 10.1371/journal.pone.0147660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zweygarth E, Sabwa C, Rottcher D. Serodiagnosis of trypanosomiasis in dromedary camels using a card agglutination test set (Testryp® CATT) Ann Soc BeIge Med Trop. 1984;64:309–313. [PubMed] [Google Scholar]
- 47.Holland WG, Thanh NG, Do TT, Sangmeneeddet S, Goddeeris B, Vercruysse J. Evaluation of diagnostic tests for Trypanosoma evansi in experimentally infected pigs and subsequent use in field surveys in North Vietnam and Thailand. Trop Anim Hlth Prod. 2005;37:457–467. doi: 10.1007/s11250-005-1217-y. [DOI] [PubMed] [Google Scholar]
- 48.Noireau F, Gouteux JP, Frezil JL. Sensibilite du test d’agglutination sur carte (TESTRYP CATT) dans les infections porcines a Trypanosoma (nannomonas) congolense en Republique Populaire du Congo. Ann Soc Belge Med Trop. 1986;66:63–68. [PubMed] [Google Scholar]
- 49.Biéler S, Waltenberger H, Barrett MP, McCulloch R, Mottram JC, Carrington M, et al. Evaluation of antigens for development of a serological test for human African trypanosomiasis. PLoS One. 2016;11:e0168074. doi: 10.1371/journal.pone.0168074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sullivan L, Wall SJ, Carrington M, Ferguson MA. Proteomic selection of immunodiagnostic antigens for human African trypanosomiasis and generation of a prototype lateral flow immunodiagnostic device. PLoS Negl Trop Dis. 2013;7:e2087. doi: 10.1371/journal.pntd.0002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan L, Fleming J, Sastry L, Mehlert A, Wall SJ, Ferguson MAJ. Identification of VSG117 as an immunodiagnostic antigen and evaluation of a dual-antigen lateral flow test for the diagnosis of human African trypanosomiasis. PLoS Negl Trop Dis. 2014;8:e2976. doi: 10.1371/journal.pntd.0002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buscher P, Lejon V, Magnus E, Van Meirvenne N. Improved latex agglutination test for detection of antibodies in serum and cerebrospinal fluid of Trypanosoma brucei gambiense infected patients. Acta Trop. 1999;73:11–20. doi: 10.1016/S0001-706X(99)00009-1. [DOI] [PubMed] [Google Scholar]
- 53.Gillet P, Mumba Ngoyi D, Lukuka A, Kande V, Atua B, van Griensven J, et al. False positivity of non-targeted infections in malaria rapid diagnostic tests: the case of human African trypanosomiasis. PLoS Negl Trop Dis. 2013;7:e2180. doi: 10.1371/journal.pntd.0002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller N, Mansfield JM, Seebeck T. Trypanosome variant surface glycoproteins are recognized by self-reactive antibodies in uninfected hosts. Infect Immun. 1996;64:4593–4597. doi: 10.1128/iai.64.11.4593-4597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Truc P, Lejon V, Magnus E, Jamonneau V, Nangouma A, Verloo D, et al. Evaluation of the micro-CATT, CATTITrypanosoma brucei gambiense, and LATEX/T. b. gambiense methods for serodiagnosis and surveillance of human African trypanosomiasis in West and Central Africa. Bull World Health Organization. 2002;80:882–886. [PMC free article] [PubMed] [Google Scholar]
- 56.von Wissmann B, Fyfe J, Picozzi K, Hamill L, Waiswa C, Welburn SC. Quantifying the association between bovine and human trypanosomiasis in newly affected sleeping sickness areas of Uganda. PLoS Negl Trop Dis. 2014;8:e2931. doi: 10.1371/journal.pntd.0002931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.


