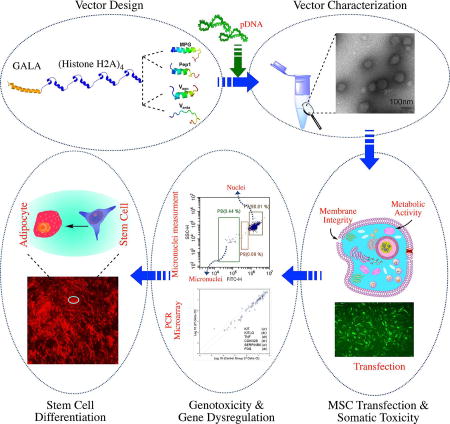
Abstract
Vectors used for stem cell transfection must be non-genotoxic, in addition to possessing high efficiency, because they could potentially transform normal stem cells into cancer-initiating cells. The objective of this research was to bioengineer an efficient vector that can be used for genetic modification of stem cells without any negative somatic or genetic impact. Two types of multifunctional vectors, namely targeted and non-targeted were genetically engineered and purified from E. coli. The targeted vectors were designed to enter stem cells via overexpressed receptors. The non-targeted vectors were equipped with MPG and Pep1 cell penetrating peptides. A series of commercial synthetic non-viral vectors and an adenoviral vector were used as controls. All vectors were evaluated for their efficiency and impact on metabolic activity, cell membrane integrity, chromosomal aberrations (micronuclei formation), gene dysregulation, and differentiation ability of stem cells. The results of this study showed that the bioengineered vector utilizing VEGFR-1 receptors for cellular entry could transfect mesenchymal stem cells with high efficiency without inducing genotoxicity, negative impact on gene function, or ability to differentiate. Overall, the vectors that utilized receptors as ports for cellular entry (viral and non-viral) showed considerably better somato- and genosafety profiles in comparison to those that entered through electrostatic interaction with cellular membrane. The genetically engineered vector in this study demonstrated that it can be safely and efficiently used to genetically modify stem cells with potential applications in tissue engineering and cancer therapy.
Keywords: stem cells, cell transfection, genotoxicity, vector engineering, non-viral, nanoparticles
Graphical abstract
Introduction
Stem cells have the ability to self-renew and transform into various cell types in an organism. Owing to this unique characteristic, they have been used as a source of donor cells to replace damaged organs. In addition, current evidence indicates that systemically administered mesenchymal stem cells (MSCs) can migrate to primary and metastatic solid tumors and deliver therapeutic molecules to tumor foci [1–4]. It is envisioned that stem cell-mediated gene delivery could emerge as a strategy to improve the efficacy and minimize the toxicity of various cancer gene therapy approaches [4, 5]. For such purposes, MSCs are first harvested from donors and then genetically modified ex-vivo to express a variety of bioactive agents. For example, MSCs can be engineered to express prodrug converting enzymes or antiproliferative, pro-apoptotic, anti-angiogenic agents [6, 7]. Vectors used for stem cell transfection need to be highly efficient because the methods to rapidly produce unlimited quantities of undifferentiated stem cells have not yet been perfected. Moreover, stem cells in cell culture change/mutate over time (usually after eight to ten passages), thereby providing a limited window of opportunity for processing.
Vectors that are currently used for stem cell engineering can be categorized into viral (adenovirus, lentivirus, and AAV) and non-viral (polymer and lipid based) vectors. Adenoviral (Ad) vectors can be used to mediate transient and high-level transgene expression. However, for adenoviral vectors to achieve a transduction efficiency beyond 50% in MSCs, the multiplicity of infection (MOI) needs to be increased to a value greater than 5000. Unfortunately, the presence of such large amounts of viral proteins in transfected cells could elicit immune response after implantation into the human body, resulting in rapid clearance of transfected MSCs [8]. Integrating vectors such as lentivirus and AAVs can transfect stem cells efficiently but this method is marred by the potential for insertional mutagenesis [9]. Consequently, the transfected stem cells require an extensive screening process to ensure safety, which in turn, raises the concern of cost-effectiveness in clinical trials. Electroporation is another method for MSC transfection, but it leads to excessive cell death [10].
Commercially available non-viral vectors based on polymers and lipids bear a positive surface charge and have the ability to condense plasmid DNA (pDNA) into nanosized particles suitable for cellular uptake. While such nanoparticles may not show significant toxicity in terms of impact on metabolic activity, as evaluated by MTT or similar assays, recent studies show that nanoparticles may cause genotoxicity [11, 12]. This could become notably problematic when dealing with stem cells because such vectors could theoretically turn a normal MSC into a cancer-initiating cell. Therefore, high levels of safety are expected from vectors that are used in stem cell engineering. Unfortunately, for demonstration of safety, non-viral vectors have been mostly evaluated by using simple assays such as MTT and/or dye inclusion/exclusion (e.g., eosin and trypan blue) while there has been no comprehensive study that has closely examined vectors’ potential for genotoxicity.
The objective of this research was to develop an efficient non-viral vector that can be used for genetic modification of stem cells without any negative somatic or genetic impact. To achieve the objective, two types of designer biomimetic vectors (DBVs) were engineered: targeted and non-targeted. As described previously, DBVs are genetically engineered biomimetic non-viral vectors that are composed of motifs from diverse biological and synthetic origins [13–15]. The targeted vectors were composed of four repeating units of histone H2A to condense DNA (H4), a pH-dependent endosomolytic fusogenic peptide GALA (G), and either a vascular endothelial growth factor receptor 1 (VEGFR-1) agonist targeting peptide (Vago) or antagonist peptide (Vanta). The rationale for targeting VEGFR-1 is that this receptor is overexpressed on the surface of stem cells and internalizes via receptor mediated endocytosis. The non-targeted vectors are composed of the same motifs as mentioned above, but instead of the VEGFR-1 targeting peptide, they have non-cationic cell penetrating peptides such as Pep1 (tryptophan-rich cluster with high affinity for membranes) and MPG (derived from the fusion sequence of the HIV glycoprotein 41). While many other cell-penetrating peptides are reported in literature (e.g., Tat), the rationale behind choosing these two peptides are as follows: 1) non-cationic nature, 2) high efficiency in membrane fusion and cellular entry, and 3) negligible cytotoxicity [16–20]. The role of the cell penetrating peptides is to facilitate internalization of the vector through the stem cell membrane. To evaluate the efficiency and safety of the vectors, adipose-derived MSCs (ADSCs) were selected for this study because in the clinical setting, they can be obtained from patients in large amounts using minimally painful procedures (in contrast to bone marrowderived). The following widely used commercially available non-viral vectors were selected as controls: GeneIn™, Lipofectamine® LTX with Plus, Attractene, FuGENE® HD and jetPRIME®. A commercially available adenoviral vector (Ad-GFP) was used as a viral vector control. This research addresses two significant deficiencies that currently exist. The first is the low efficiency of non-viral vectors in MSC transfection, and the second is a lack of comprehensive toxicity data related to the cell proliferation rate, membrane integrity, micronuclei formation, gene dysregulation, and cell differentiation.
Materials and Methods
Genetic engineering and production of recombinant vectors
We used standard genetic engineering techniques similar to our previous reports in order to clone, express, and purify the DBVs [15, 21, 22]. In brief, the genes encoding untargeted vectors H4G, MPG-H4G, Pep1-H4G and targeted Vago-H4G, and Vanta-H4G with 6x-histidine tag at the c-terminus, were designed and then chemically synthesized by Integrated DNA Technologies (Coralville, IA, US). The corresponding amino acid sequences of the vectors are shown in Table 1. The genes were restriction digested by NdeI and XhoI enzymes and cloned into a pET21b bacterial expression vector (Novagen®, EMD Millipore, MA, US). The fidelity of each gene sequence to the original design was verified by DNA sequencing.
Table 1.
The amino acid sequences of the designed recombinant vectors for stem cell transfection.
| Name | Cell Targeting Peptide |
Cell Penetrating Peptide |
DNA Condensing Motif | Fusogenic Peptide |
|---|---|---|---|---|
| H4G | N/A | N/A | (SGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRK)4 | WEAALAEALAEALAEHLAEALAEALEALAA |
| MPG-H4G | N/A | GALFLGFLGAAGSTMGAWSQPKKKRKV | (SGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRK)4 | WEAALAEALAEALAEHLAEALAEALEALAA |
| Pep1-H4G | N/A | KETWWETWWTEWSQPKKKRKV | (SGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRK)4 | WEAALAEALAEALAEHLAEALAEALEALAA |
| Vago-H4G | KLTWQELYQLKYKGI | N/A | (SGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRK)4 | WEAALAEALAEALAEHLAEALAEALEALAA |
| Vanta-H4G | NGYEIEWYSWVTHGMY | N/A | (SGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRK)4 | WEAALAEALAEALAEHLAEALAEALEALAA |
To express the vectors, the expression plasmids were transformed into the LOBSTR BL21(DE3) E. coli expression strain (Kerafast Inc., MA, US). The protein expression protocol is optimized for the production of highly cationic vectors in E. coli as described previously by our group [23]. In brief, one colony was picked from the LB agar plate and inoculated overnight in a 5 mL Miller’s LB media supplemented with 100 µg/mL carbenicillin (Sigma-Aldrich, MO, US). The next day, the starter culture was transferred into 500 mL terrific broth (TB) supplemented with 100 µg/mL carbenicillin. The culture was incubated at 37 °C under vigorous shaking until the OD600 reached 0.4–0.6. To induce protein expression, isopropyl β-D-1-thiogalactopyranoside (IPTG, Teknova, CA, US) was added to the culture at the final concentration of 1 mM. After 2.5–4 hours of induction, the E. coli pellet was collected by centrifugation at 5000 g (10 min, 4 °C) weighed and stored in −80 °C freezer.
To purify the peptides, a method based on Ni-NTA immobilized metal affinity chromatography (QIAGEN, MD, US) was developed. A lysis buffer was formulated beforehand, containing 8 M urea, 2 M NaCl, 100 mM NaH2PO4,10 mM Tris, 1% (v/v) Triton X-100, and 10 mM imidazole. The bacterial pellet was lysed by the lysis buffer (5 mL buffer per 1 gram pellet) for one hour at room temperature under vigorous stirring. Then, the supernatant was collected by centrifuging the slurry for one hour, at 20,000 rpm, 4°C. Meanwhile, the Ni-NTA resin was washed with 10 mL distilled/deionized water and preconditioned with 2 mL of lysis buffer. Afterwards, the supernatant was mixed with the preconditioned Ni-NTA resin and incubated on ice with gentle shaking. After one hour of incubation, the mixture was diluted with 3 times lysis buffer and passed through a 10 mL polypropylene filter column (Bio-Rad Inc., US) by vacuum driven filtration. The column was washed by 100 mL of lysis buffer followed by 50 mL wash buffer (5 M Urea, 1.5 M NaCl, 100 mM NaH2PO4, 10 mM Tris and 40 mM imidazole). Finally, the purified vector was eluted by 5 mL of elution buffer (3 M Urea, 0.5 M NaCl, 100 mM NaH2PO4, 10 mM Tris and 300 mM imidazole) and collected in 500 µL fractions. The concentration of the peptide within each fraction was measured by the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, US). The purity of each peptide was determined by SDS-PAGE analysis.
Peptide desalting and preparation of stock solution
To desalt, a disposable PD-10 desalting column with Sephadex G-25 resin (GE Healthcare’s Life Sciences, MA, US) was preconditioned with 25 mL of 10mM L-Glu/ L-Arg buffer (pH 5.8–6.0). Then, each purified peptide fraction was loaded onto the column and eluted with additional 5 mL of buffer driven by gravity. The concentration of each peptide was measured by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, US) using the molecular weight and corresponding extinction coefficient as calculated by the ProtParam tool from the ExPASy Bioinformatics Resource Portal (http://web.expasy.org/protparam/). The conductivity of the peptide solution was determined by Laser Doppler Velocimetry using Malvern Nano-ZS Zetasizer (Malvern Instruments, UK).
Nanoparticle formation and particle size, charge and shape analysis
The DNA/peptides nanoparticles were formed by the Flash Mixing method [22]. In brief, the required amount of each peptide to condense 1µg of pEGFP plasmid DNA (pDNA) at various N:P ratios was calculated beforehand. For example, to prepare a N:P ratio of 1, the required amounts of H4G, MPG-H4G, Pep1-H4G, Vago-H4G and Vanta-H4G were 1.17 µg, 1.22 µg, 1.29 µg, 1.27 µg, and 1.35 µg, respectively. Then, pEGFP was diluted to a volume of 50µL using distilled/deionized water. Concurrently, predetermined amount of each peptide was diluted to 50µL volume using distilled/deionized water and placed in another microfuge tube. The peptide solution was added to the pDNA solution rapidly and flash mixed. After 5–10 minutes of incubation, the nanoparticle size was measured by Dynamic Light Scattering and surface charge by Laser Doppler Velocimetry using Malvern Nano-ZS Zetasizer (Malvern Instruments, UK). To make nanoparticles with the commercial transfection reagents including GeneIn™ (MTI-GlobalStem, MD, US), Lipofectamine® LTX with Plus (Thermo Fisher Scientific, MA, US), Attractene (QIAGEN, MD, US), FuGENE® HD (Promega Corporation, WI, US) and jetPRIME® (Polyplus-transfection, France), we followed the corresponding manufacturers’ protocols. Once nanoparticles were formed, the surface charges were measured in 5mM NaCl solution. The data are presented as mean±s.d. (n=3). Each mean is the average of 15 measurements while n represents the number of independent batches prepared for the measurements.
To study the morphology of the nanoparticles, transmission electron microscopy (TEM) was utilized [22]. First, nanoparticles were formed and then one drop of the mixture was loaded onto a carbon type B coated copper grid. As soon as the sample dried on the surface, the solution of 1% sodium phosphotungstate was added to stain the nanoparticles. The detailed images were recorded by 1200EX electron microscope (JEOL, US).
ADSC Characterization
The ADSCs (Lonza, NJ, US) were cultured in ADSC™ Growth Medium Bullet kit (Lonza, NJ, US) which contains the basal media and the necessary supplements for proliferation of human adipose derived mesenchymal stem cells. ADSCs were characterized for cell cycle and VEGFR-1 expression by flow cytometry. The cell cycle study was performed using propidium iodide (PI) DNA staining protocol. In brief, cells were seeded in 96-well plates at the density of 6000 cells per well. After 16, 20, 24, 26, 28 hours incubation with ADSC™ Growth Medium Bulletkit at 37 °C and 5% CO2, cells were detached through trypsinization. Cells were then fixed by 70% cold ethanol. After 1-hour, cells were collected by centrifugation, re-suspended in PBS and treated with 0.5 mg/mL RNase A. Finally, cells were stained by PI (10 µg/mL) for 1 hour. The cell cycle distribution was determined by flow cytometry (Beckman Coulter GALLIOS Cytometer, CA, US).
To determine the level of VEGFR-1 expression, ADSCs were detached by Accutase® Cell Detachment Solution (Innovative Cell Technologies, CA, US). Cells were fixed by 4% formaldehyde solution in PBS and then permeabilized by 0.1% Tween 20/PBS solution. Cells were washed and re-suspended in the staining buffer (0.3M glycine and 10% normal goat serum in PBS solution). 2 µL of Anti-VEGFR-1 rabbit monoclonal antibody conjugated with Alexa Fluor® 488 (abcam, MA, US) was added to each sample. Rabbit monoclonal IgG conjugated with Alexa Fluor® 488 (abcam, MA, US) was used as isotype control. Samples were incubated overnight at 4°C and then washed extensively with PBS. The expression level of VEGFR-1 was determined by flow cytometry (Beckman Coulter GALLIOS Cytometer, CA, US). The unstained sample was also included as a negative control.
Evaluation of cell transfection efficiency
The day before transfection, ADSCs were seeded in 96-well tissue culture plates at the density of 6000 cells per well and incubated for 24 hours. In a microfuge tube, nanoparticles were prepared at various N:P ratios as described above in a total volume of 50 µL and incubated for 5–10 minutes at room temperature. Each tube was further supplemented with 200 µL of ADSC basal media, 1µM dexamethasone (Sigma-Aldrich, MO, US) and 1X ITS Liquid Media. A 100X ITS solution includes 1.0 mg/mL recombinant human insulin, 0.55 mg/mL human transferrin and 0.5 µg/mL sodium selenite (Sigma-Aldrich, MO, US). Next, the old media in each well was removed and replaced with the 250 µL nanoparticle mixture. Twenty four hours post transfection, the media in each well was replaced with 200 µL full growth media and the cells were allowed to grow for another twenty four hours. The green fluorescent protein (GFP) expression was visualized and qualitatively evaluated by a fluorescent microscope (Olympus, FL, US). To quantify GFP expression and percent transfection, cells were trypsinized and analyzed by flow cytometry (Beckman Coulter CytoFLEX Cytometer, CA, US). The ratio of GFP positive cells to untransfected cells was calculated by Kaluza flow analysis software (Beckman Coulter, CA, US).
To measure the transfection efficiency of commercially available transfection reagents including GeneIn™ , Lipofectamine® LTX with Plus, Attractene, FuGENE® HD and jetPRIME®, cells were seeded in 96-well plates at the density of 6,000 cells/well. Twenty four hours later, cells were transfected following each manufacturer’s cell transfection protocol.
To measure transduction efficiency of adenoviruses, cells were seeded as above. Adenovirus particles encoding GFP (Ad-GFP) were purchased from Baylor College of Medicine (TX, US), and the transduction process was performed according to the manufacturer’s protocol. In brief, the multiplicity of infection (MOI) was calculated based on viral titer (plaque-forming units, PFU/mL). The Ad-GFP particles were mixed thoroughly with 300 µL of ADSC basal media. Next, the old media in each well was replaced by the transduction mixture. Four hours post transduction, the media in each well was replaced by the full growth media and the GFP expression was quantified after forty eight hours by flow cytometry as described above. The data are presented as mean±s.d. (n=3).
Evaluation of vectors’ impact on cell proliferation rate, membrane integrity and morphology
The impact of each vector on ADSC proliferation rate was evaluated by the WST-1 cell proliferation assay. Cells were seeded in the 96-well plates at the density of 6,000 cells per well. After twenty four hours of incubation, ADSCs were transfected with vectors as described above. Forty eight hours post-transfection, the old media was replaced with 100 µL of fresh media containing 10 µL WST-1 reagent (1:10 dilution). After one hour of incubation at 37 °C / 5% CO2, the absorbance of each well was measured by Infinite® M200 PRO NanoQuant microplate reader (Tecan, Switzerland) at 440nm/600nm. The absorbance of each treatment was normalized to the negative control (untreated cells) to measure the percentage of cell viability.
To evaluate the impact of each vector on ADSC membrane integrity, a lactate dehydrogenase (LDH) release assay (Roche, IN, US) was performed using manufacturer’s kit and protocol. In brief, cells were seeded and transfected as described above. Cells were incubated in ADSC basal media for 48 hours post transfection since the LDH reagent is not compatible with serum. Media in each well was collected and centrifuged at 250g for 5 minutes to pellet the debris. The supernatants were transferred into a 96-well plate with 100 µL per well. Next, 100 µL LDH reagent was added into each well and incubated for 30 minutes at room temperature. The absorbance at wavelengths of 490nm and 600nm was measured using Infinite® M200 PRO NanoQuant (Tecan, Switzerland) microplate reader. The media, without contacting any cells, served as the background control. The media from the untransfected cells was used as the negative control (spontaneous LDH release). The media from the cells incubated with the 2% Triton X-100 was served as the positive control (maximum LDH release). After subtracting the background control, the percentage of impact on membrane integrity was calculated as follows: %membrane integrity= (Positive-Treatment)/ (Positive-Negative) × 100. The data are presented as mean±s.d. (n=3).
The morphology of ADSCs before and after transfection was studied by using phase-contrast microscopy (Olympus, FL, US).
Evaluation of vectors’ impact on micronuclei formation (genotoxicity)
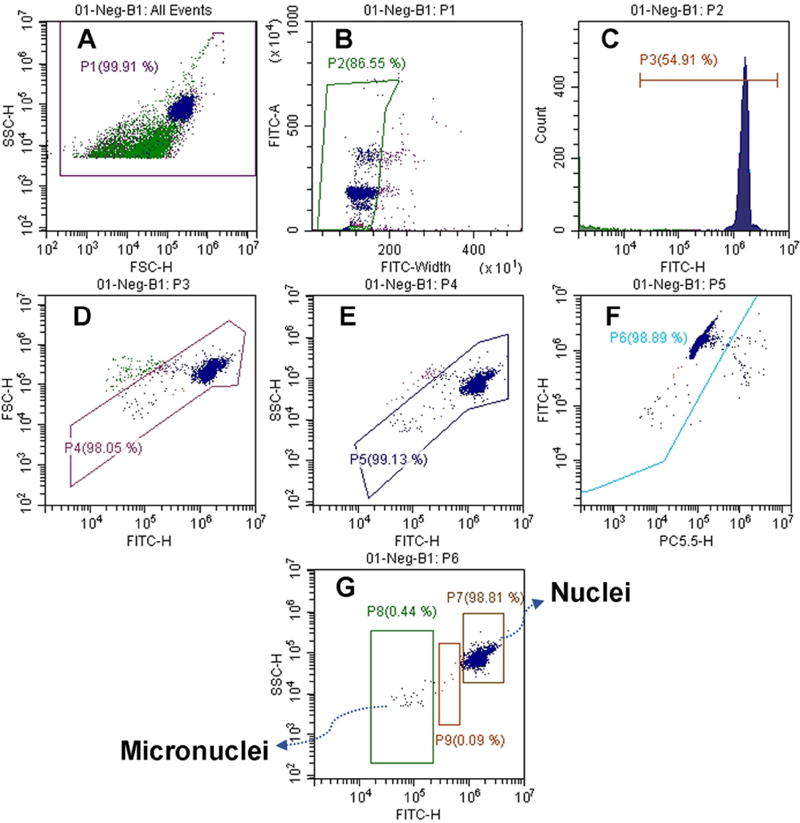
To quantify the percentage of micronuclei formation, cells were seeded and transfected as described above. Twenty four hours post-transfection (equivalent to 1–1.5 doubling time), cells were harvested and stained using an In Vitro MicroFlow® Kit (Litron Lab., NY). The staining was performed according to the manufacturer’s protocol with several modifications. Briefly, cells were detached, transferred into a microfuge tube, and centrifuged for 6 min at 300g. The supernatant was removed and the pellet was placed on ice for 20 min. Next, ADSCs were resuspended in 50 µL of ethidium monoazide (EMA) solvent (Dye A) and incubated while exposed to fluorescent light. EMA is a DNA staining fluorescent dye that cannot pass through the cell membrane of live cells. As a result, it can only stain the late apoptotic or dead cells helping to distinguish them from live cells. After 30 min of incubation with EMA, cells were washed by the Kit’s wash buffer, lysed by lysis buffer, and treated with RNase enzyme. Cells were then exposed to SYTOX green fluorescent dye that stains all nuclei and micronuclei. The lysis and SYTOX green staining process were performed at 37 °C while samples were protected from light. After staining, samples were analyzed by CytoFlex Flow Cytometer (Beckman Coulter, Brea, CA) using an optimized acquisition protocol according to the guideline of In Vitro Microflow® Kit (Figure 1). The detailed information about the gating protocol can be found elsewhere [24]. Briefly, the process started by gating the majority of events from side scatter vs. forward scatter plots (Figure 1A) and continued with the second plot in which the doublet nuclei were discriminated and excluded by FITC width vs. FITC area plot (Figure 1B). Next, the SYTOX Green positive events were selected (Figure 1C) and the two different dot plots represented in Figures 1D and 1E illustrate nuclei and micronuclei populations with the correct size and pattern. This excludes other interfering events, such as smaller fluorescent particles, green fluorescent protein aggregates, and stained plasmids or nanoparticles. Figure 1F shows exclusion of the EMA-positive events which originated from dead or late apoptotic cells. At this point, the number and percentage of micronuclei and nuclei shown in Figure 1G can be quantified. In general, micronuclei are defined as events showing 1/10 to 1/100 of the mean intensity of SYTOX Green fluorescence found in nuclei of viable (i.e. EMA-negative) cells. The gating protocols were kept unchanged during the analysis and for each sample, at least 1000 EMA negative nuclei events were counted. Accordingly, %MN= Number of MN/ Number of viable nuclei × 100. The data are presented as mean±s.d. (n=4).
Figure 1.
The gating protocol that was designed for quantification of micronuclei formation in transfected stem cells.
Determination of vectors’ impact on gene regulation (microarray analysis)
The effects of vectors on the expression of 84 genes associated with cell growth regulation were analyzed by using the Human Genes RT2 Profiler™ PCR Array (Qiagen, MD, US). The names of the tested genes are as follows: SERPINB5, MYCN, ABL1, AKT1, APC, ATM, BAX, BCL2, BCL2L1, BCR, BRCA1, BRCA2, CASP8, CCND1, CDH1, CDK4, CDKN1A, CDKN2A, CDKN2B, CDKN3, CTNNB1, E2F1, EGF, ELK1, ERBB2, ESR1, ETS1, FHIT, FOS, FOXD3, HGF, HIC1, HRAS, IGF2R, JAK2, JUN, JUNB, JUND, KIT, KITLG, KRAS, MCL1, MDM2, MEN1, MET, MGMT, MLH1, MOS, MYB, MYC, NF1, NF2, NFKB1, NFKBIA, NRAS, PIK3C2A, PIK3CA, PML, PRKCA, RAF1, RARA, RASSF1, RB1, REL, RET, ROS1, RUNX1, RUNX3, S100A4, SH3PXD2A, SMAD4, SRC, STAT3, STK11, TGFB1, TNF, TP53, TP73, TSC1, VHL, WT1, WWOX, XRCC1, ZHX2. ADSCs were seeded in the 96-well plates at the density of 6,000 cells per well and then transfected with selected DBVs. For adenovirus, ADSCs were seeded in a 6-well plate at the density of 100,000 cells per well and incubated for twenty four hours. Cells were transduced by Ad-GFP at MOI of 5,000 and 50,000 in a serum free media (ADSC basal media). Four hours post transduction, the media was removed and replaced with full growth media. Forty eight hours after, ADSCs were collected and the GFP positive cells were separated from the general population by the Moflo XDP Cell Sorter (Beckman Coulter, CA, US). The GFP positive cells were reseeded in a 6-well plate at the density of 45,000 cells per well and allowed to fully recover from the process until they reached 80% confluency (4 to 8 days). The mRNAs of transfected and untransfected cells were extracted by RNeasy Mini Kit (Qiagen, MD, US). The genome DNA was eliminated by the RNase-Free DNase Set (Qiagen, MD, US) during the RNA isolation process. The concentration and purity of mRNA were evaluated by measuring the absorbance at wavelength 260 nm and 280 nm. Concurrently, an agarose gel (1%) electrophoresis was performed to examine the mRNA integrity. Then, 0.5 µg of mRNA was reverse transcribed into complementary DNA (cDNA) by RT2 First Strand Kit (Qiagen, MD, US). The cDNA of each sample with RT2 SYBR Green ROX PCR Master mix (Qiagen, MD, US) was loaded onto the PCR array. The real-time PCR reactions were performed using StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific, MA, US). The program settings on temperature cycling were followed as instructed by the manufacturer. The raw data and gene profile expression was analyzed by “Double Delta Ct Method” using the manufacturer’s online software tool (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php). Here, five housekeeping genes (ACTB, B2M, GAPDH, HPRT1 and RPLP0) were used as controls. All experiments were performed in triplicates while a two-fold change in RNA levels served as the cut-off point (*, p<0.05).
Evaluation of vectors’ impact on ADSC differentiation
To examine whether the transfection process had a negative impact on ADSC differentiation, the cells were induced to differentiate into adipocytes and osteocytes. ADSCs were transfected with the developed vectors and after 48 h post-transfection, were harvested and sorted by flow cytometry according to their respective GFP expression. The sorted GFP-positive cells were then reseeded in 96-well plates at the density of 10,000 cells per well and incubated at 37 °C with ADSC full media. The media was changed every other day until cells reached maximum confluency.
For adipogenesis study, the ADSC full growth media was removed and replaced with adipogenesis differentiation media cocktail (Lonza Inc., NJ) containing 1 µM dexamethasone, 0.5 mM isobutyl-methylxanthine (IBMX), 1 µg/mL insulin, and 100 µM indomethacin. The differentiation media was gently replaced every 3 days for 12 days. Next, ADSCs were washed by PBS and stained with AdipoRed™ fluorescent staining reagent (Lonza Inc., NJ). The production of intracellular oil vesicles was visualized by fluorescent microscopy (Olympus Co., USA) and the percentage of highly differentiated ADSCs was quantified by flow cytometry.
For osteogenesis differentiation study, the media was removed and replaced with osteogenesis hMSC differentiation BulletKit™ media cocktail (Lonza Inc., NJ). The differentiation media was gently replaced every 3 days for two weeks. Next, the ADSCs were gently washed with PBS and stained with Alizarin Red S (Sigma). The intracellular calcium deposits were first visualized by light microscopy. Then, the stained cells were washed twice by PBS and incubated with 500 µL of 100 mM cetylpyridinium chloride (Sigma) for 1 h to dissolve and release the Alizarin Red S/calcium complexes. The percentage of differentiated ADSCs was determined by measuring the absorbance at 570 nm wavelength. Untransfected ADSCs were subjected to the same differentiation protocol and used as the positive control. The data are presented as mean ± s.d. (n=3).
Evaluation of vectors’ impact on surface biomarker expression
ADSCs were seeded in 96-well plates and transfected with vectors as described above. The transfected cells were transferred into a 6-well plate and incubated for 48 hours. Cells were detached by Accutase® and washed twice with cell staining buffer (BioLegend, CA, US). Cells were resuspended in 100µL of cell staining buffer and incubated with 5µL of Human TruStain FcX™ (BioLegend, CA, US) for 5 minutes at room temperature to block the Fc Receptor. Afterwards, cells were washed once and resuspended in another 100µL cell staining buffer. Then, 5 µL isotype control or antibodies conjugated with fluorophore phycoerythrin (PE) including anti-human CD13, anti-human CD29, anti-human CD105, and anti-human CD271 were added into the mixture and incubated on ice for 30 minutes. Cells were washed extensively and the expression level of each surface marker was determined by flow cytometry (Beckman Coulter GALLIOS Cytometer, CA, US). The untreated ADSCs went through the same process and used as controls.
Results and Discussion
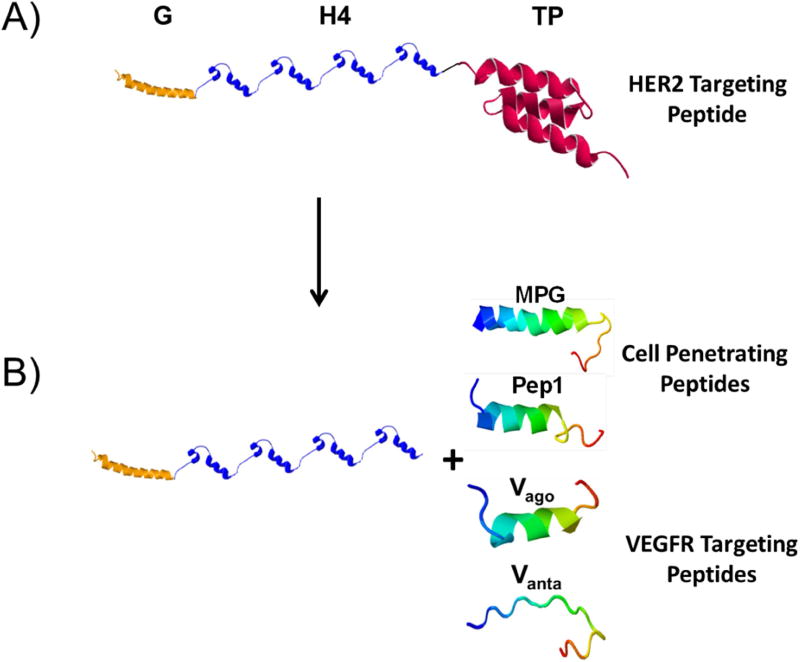
The concept of engineering recombinant fusion vectors for gene delivery dates back to the late 1990s [25]. However, owing to significant technical difficulties related to recombinant production of highly cationic vectors and formulation of stable and efficient nanoparticles, recombinant fusion vectors remained ineffective for more than a decade (reviewed in reference [26]). Since 2006, we have worked to overcome these challenges and have successfully created highly efficient targeted fusion vectors for various gene delivery applications including the targeting of different cancer cell types or compartments within the cell [13, 14, 27–29]. We have previously reported the structure of a DBV composed of four repeating units of histone H2A (H4) for efficient condensation of DNA into nanosized particles and a pH-dependent fusogenic peptide (GALA) for disruption of endosome membranes facilitating the escape of cargo into the cytoplasm. Due to the presence of an inherent nuclear localization signal in the structure of histone H2A [30], the vector also uses microtubules to actively transport the nanoparticles toward the cell nuclear membrane [15]. To make the above mentioned vector (i.e., H4G) suitable for targeted gene transfer to HER2 positive mammalian cells (e.g., SKOV-3), a HER2 targeting affibody was fused with the vector sequence (Figure 2A) [29]. We have demonstrated that this vector can target and transfect SKOV-3 cancer cells at an efficiency greater than 95% [21]. To make this vector suitable for transfection of stem cells which is a primary cell line without HER2 expression, we replaced the HER2 targeting peptide in the vector structure with the VEGFR-1 targeting peptides and cell penetrating peptides (Figure 2B). The sequences of the VEGFR targeting peptides (agonist and antagonist) are previously reported and also shown in the method section (Table 1) [31, 32]. To achieve the objective, we first genetically engineered the DBVs as described below.
Figure 2.
A) Schematics of the fusion vector composed of a fusogenic peptide GALA (G) to disrupt endosomal membranes, a DNA condensing motif with inherent nuclear localization signal (H4) and a HER2 targeting peptide (TP). B) By removing the HER2 targeting peptide and replacing it with VEGFR targeting or cell penetrating peptides, the vector is tailor-made for carrying genes into MSCs. The 3-D structure of each motif was simulated independently by I-TASSER server for protein structure and function prediction [33].
Genetic engineering and production of fusion vectors
Considering that the above mentioned DBVs are highly cationic, their production in E. coli expression systems is marred by low expression yield, which complicates the possibility of obtaining pure products. For example, SlyD and ArnA endogenous E. coli proteins are considered the major culprits that co-purify with low-expressing DBVs during metal affinity chromatography [34]. The inability to produce highly pure vectors and in sufficient quantities are among the major obstacles that significantly hampered the progress of this field of research. To overcome this obstacle, we developed and previously reported an optimized protocol for the recombinant production of cationic fusion vectors [23]. Using this protocol, all constructs in this study were expressed in an E. coli expression system, purified by Ni-NTA affinity columns and analyzed for purity by SDS-PAGE. The results of this study showed that by using E. coli BL21(DE3) LOBSTR strain in combination with the developed stringent expression and Ni-NTA purification methods, highly pure products in one purification step (>95% purity) could be obtained (Supporting Figure 1). In the next step, we examined the ability of the vectors to condense pDNA into nanosized particles.
Nanoparticle formation and particle size, charge and shape analysis
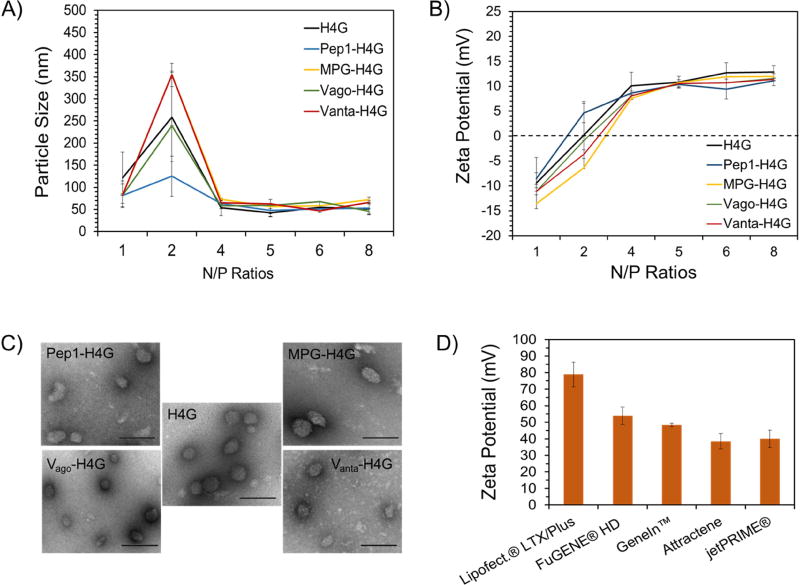
We performed a peptide desalting step before forming nanoparticles. The desalting step is crucial as it helps remove the excess ions from the system. This procedure stabilizes the nanoparticles’ diameters by minimizing the possibility of inter-particle salt bridge formation and ensuing aggregation. In addition, the presence of excess ions in the media interferes with the electrostatic interactions between cationic residues in the vector sequence and anionic residues in the pDNA resulting in the formation of pseudo-condensed DNA. Therefore, we performed a desalting step to significantly reduce the ionic strength of the DBV solution, which brought down the solution conductivity from 33.7±0.6 mS/cm to 0.45±0.01 mS/cm without compromising solubility. We have previously shown that this level of conductivity is equivalent to that of a 5 mM NaCl solution [35]. The low conductivity value allows for efficient condensation of pDNA by DBVs and production of stable nanoparticles. The purified/desalted DBVs were then complexed with pDNA (i.e., pEGFP) at various N:P ratios and characterized in terms of size, surface charge and morphology. The results of this study showed that all DBVs were able to condense pEGFP into floccus, spherical particles with sizes of less than 100nm and surface charges below +15 mV (Figure 3A-C). The analysis of data showed that all nanoparticles beyond the N:P ratio of 4 were statistically the same in terms of size and charge (p>0.05). Maintaining the nanoparticle surface charge below +20mV is critically important as it has been shown that the potential for genetic aberrations (genotoxicity) increases when the surface charge goes beyond +20mV [36]. This goal could be reached due to the unique structure of histone H2A in the DBV sequence. Histone H2A is a basic peptide with an amino sequence of SGRGKQGGKARAKAKTRSSRAGLQFPVGRVHRLLRKG. Even though only 33% of amino acid residues in the histone H2A sequence are cationic, it can efficiently condense pDNA into nanosized particles. This efficiency in DNA condensation is attributed to the alpha-helix secondary structure at the H2A N-terminal domain [30]. As a result, less amount of vector is required to efficiently condense pDNA into compact nanoparticles.
Figure 3.
Characterization of nanoparticles in terms of size, charge, and shape. A) Size of DBV/pEGFP nanocomplexes as determined by dynamic light scattering. B) Surface charge of DBV/pEGFP nanocomplexes as determined by laser Doppler velocimetry. C) Shape of DBV/pEGFP nanocomplexes captured by TEM. The scale bar is 100nm (magnification: 75000×). D) Surface charge analysis of commercial vectors in complex with pEGFP.
The commercial vectors used in this study generated nanoparticles with surface charges ranging from +30 mV to +80 mV (Figure 3D). While this high surface charge guarantees production of stable nanoparticles even in the presence of serum, there remains significant potential for toxicity in primary mammalian cell lines such as stem cells.
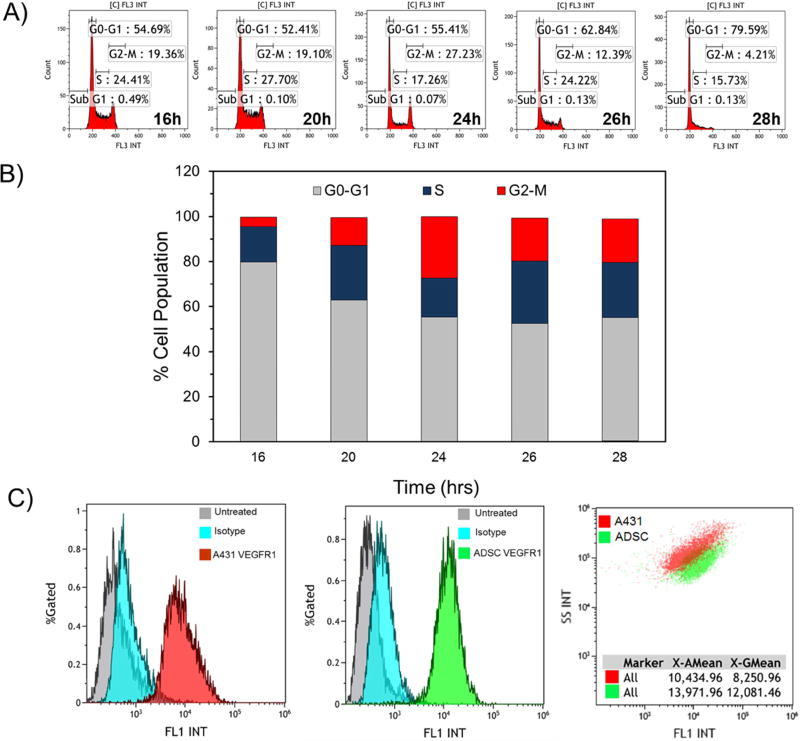
Characterization of ADSCs in terms of cell cycle and VEGFR-1 expression
Before cell transfection, we performed a cell cycle analysis to determine the optimum time for transfection of ADSCs because non-viral vectors can mainly transfect dividing cells that are in the mitotic state. For this purpose, we analyzed the cell cycle status of the ADSCs from 16 to 28 h post-seeding. This study revealed that the optimum time for transfecting ADSCs is 24 h post-seeding because at this point, significant numbers of ADSCs are in G2-M phase where the nuclear membrane starts dissolving (Figure 4A-B). Furthermore, we characterized the ADSCs in terms of VEGFR-1 expression to confirm that this receptor is expressed on the surface of ADSCs in abundance. This is important because our targeted DBVs are expected to rely on these receptors for entry into the cells. The results of this study showed a very high expression of VEGFR-1 on the surface of the ADSCs (Figure 4C). The VEGFR-1 expression level in ADSCs appeared to be even higher than A431 (human squamous carcinoma) cancer cells, which are known to have high expression levels of VEGFR-1 [37].
Figure 4.
Characterization of ADSCs in terms of cell cycle and expression of VEGFR-1. A) Flow cytometry histograms showing the percentage of cells in each phase at different time points (i.e., 16–28 h). B) Bar chart summarizing the percentage of cell population in each cell cycle phase at different time points. As the percentages of cells in Sub G1 phase are very low, they are not observable in the bar chart. C) Flow cytometry histogram/dotplot showing the overexpression of VEGFR-1 on the surface of ADSCs (left panel), A431 cells (middle panel) and in comparison (right panel).
Evaluation of transfection efficiency
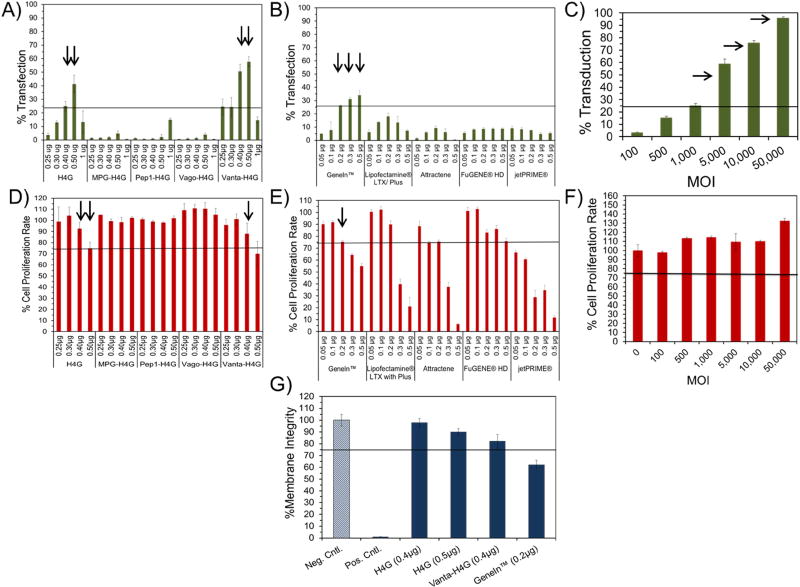
Learning from the studies mentioned above, we initiated the ADSC transfection studies. We used the DBVs (N:P 5) to transfect ADSCs with pEGFP 24 h post-cell seeding. As controls, we also transfected the ADSCs with commercial non-viral and viral vectors to help us better understand the efficiencies of currently available vector technologies. Using fluorescent microscopy, we first qualitatively evaluated the transfection rates of the different vectors and observed that there were noticeable differences among the vectors’ efficiencies (Supporting Figures 2 and 3). This prompted us to use flow cytometry in order to quantify the percentage of transfected cells in each group. For practical purposes and to assist in identifying the most efficient vector, we drew a line at 25% efficiency. This means that the constructs that could transfect ADSCs at rates higher than 25% were considered efficient. It is noteworthy that ADSCs are primary cells and considered as difficult to transfect; in contrast to cells that are easy to transfect such as HEK293 or HeLa (Supporting Figure 4). The results of this study demonstrated that the H4G and Vanta-H4G vectors carrying 0.4 and 0.5µg pEGFP were among the most efficient DBVs with Vanta-H4G surpassing 50% transfection efficiency (Figure 5A). A complementary cell transfection study using U87 glioblastoma, which does not express the VEGFR-1 receptor [38], confirmed the ability of Vanta-H4G to transfect VEGFR-1 positive ADSCs but not U87 cells (Supporting Figure 5). Among the non-viral commercial vectors, GeneIn™ carrying 0.2, 0.3 and 0.5µg of pEGFP was the most efficient (Figure 5B).
Figure 5.
Evaluation of transfection efficiency and impact on cell proliferation rate of DBVs and commercial vectors. A-C) Bar charts that quantitatively demonstrate the percentage of transfected cells using DBVs and commercial non-viral and viral vectors. The arrows point at the most efficient vectors. D-F) Bar charts that demonstrate the impact of DBVs and commercial vectors on the proliferation rate of ADSCs. The arrows highlight the vectors that had high efficiencies (>25%) with acceptable impacts on cell proliferation rate. G) LDH release assay demonstrating the impact of vectors on cell membrane integrity.
One curious observation was that we did not observe significant cell transfection rates with Pep1-H4G and MPG-H4G. Muller et al. (2012), have previously emphasized that not only does the chemical nature of the peptides’ C-terminus determine the cell penetration efficacy of the Pep1 and MPG peptides, but also the type of cell line [19]. Therefore, the data in Figure 5A suggest that either the ADSC is not a suitable cell model for transfection by Pep1 and MPG, or the Pep1 and MPG should have been positioned at the H4G C-terminus (i.e., H4G-Pep1 and H4G-MPG). If the former is true and the cell type has played a role, then the MPG-H4G and Pep1-H4G vectors should be able to effectively transfect other mammalian cell lines. To examine this hypothesis, we selected Pep1-H4G carrying 0.5µg pEGFP as an example along with a fast growing cancer cell line model such as SKOV-3 (ovarian cancer). Interestingly, the results showed that Pep1-H4G could easily transfect 35% of SKOV-3 cells (Supporting Figure 6). This rate of transfection efficiency is far higher than what was observed in ADSCs (i.e., <5%) (Figure 5A). This shows that the cell type played a significant role in limiting the efficiency of Pep1-H4G. To examine whether the positioning of Pep1 and MPG at the C-terminus would make a difference, we genetically engineered H4G-Pep1 and H4G-MPG. Unfortunately, due to the co-expression and co-purification of prematurely terminated H4G-Pep1 and H4G-MPG peptide sequences, we could not obtain pure products to test the latter hypothesis. As a side note and theoretically speaking, we believe that the positioning of MPG and Pep1 at the H4G C-terminus is not an appropriate design for gene delivery as both cell penetrating peptides (CPPs) have their cationic residues clustered at their C-terminus (i.e., KKKRKV). As a result, the KKKRKV cluster will interact with the pDNA and participate in DNA condensation; therefore, it is unavailable for interaction with negatively charged phospholipids in the cell membrane. Nonetheless, our data show that ADSCs may not be easily transfected with vectors that are decorated with Pep1 and MPG and perhaps other types of CPPs could produce better results.
Another interesting observation was the inability of Vago-H4G to efficiently transfect ADSCs. We believe that this could be due to the presence of hree lysine residues in the Vago sequence (20% cationic residue content), particularly the presence of one lysine at the N-terminus and one at the C-terminus. Cationic-charged lysine residues could electrostatically interact with pDNA inhibiting the protrusion of the VEGFR-1 agonist peptide from the surface of the nanoparticles rendering them unavailable for receptor binding. Considering that a non-cationic high affinity VEGFR-1 agonist has not been developed yet, this would be an interesting venue to pursue in order to design the next generation of VEGFR-1 targeted DBVs for stem cell transfection.
With regard to the adenoviral vector, we used Ad-GFP at extremely high MOIs (>5K) in order to transfect ADSCs beyond 50% (Figure 5C). Adenoviral vectors are known to be very efficient in transfecting mammalian cells and can render beyond 50% efficiency at MOIs as low as 50 [39]. The fact that such high numbers of adenoviral particles are required to achieve high transfection efficiency indicates that the coxsackie adenovirus receptor (CAR) is not expressed in abundance on the surface of ADSCs. Consequently, the downside of using adenoviral vectors at such high MOIs is not only the elevated costs, but also the presence of large amounts of viral proteins inside the stem cells which could induce immune response after reintroduction into a patient’s body.
Assessment of cell proliferation rate, morphology and membrane integrity
In the next step, we evaluated the impact of the vectors on ADSC proliferation rate. Considering that the formazan-based assays such as MTT, MTS, and WST-1 possess potential for side reactions and ambiguities [40], we only eliminated the vectors from the pool that had more than a 25% negative impact on cell proliferation rate. We set this level of tolerance for screening purposes as well as to narrow down the field for more in-depth toxicity studies as will be described later. The cell proliferation rate study showed that only H4G (0.4 and 0.5 µg pEGFP) and Vanta-H4G (0.4µg pEGFP) had more than 25% efficiencies and acceptable negative impacts on ADSC proliferation rate (i.e., <25%). To confirm our cell proliferation rate observations for high performing H4G and Vanta-H4G vectors, the negative impact on MSC viability was also evaluated by a secondary method; i.e., by flow cytometry. Overall, the results showed an agreement between the two methods (Supporting Figure 7). GeneIn™, carrying 0.2µg of pEGFP, appeared to be the only viable vector that met our strict efficiency/toxicity guideline for transfecting ADSCs (Figures 5D and E). The adenoviral vector, rather than showing a negative impact on cell proliferation rates at high MOIs, actually induced cell proliferation (Figure 5F). This could be explained by the fact that toxic substances in low concentrations occasionally stimulate cellular metabolic activity. In order to protect themselves from such toxicities, cells upregulate their enzymatic activities at the initial stages. Cells will start to die when the concentration of toxic substances, in this case Ad-GFP, exceeds their level of tolerance.
We further characterized the screened and selected vectors from the studies mentioned above in terms of their impact on the cell membrane integrity during transfection. Considering the associated errors with the method and the ability of cells to recover from the assault, again we set our level of tolerance at 25% negative impact on cell membrane integrity for screening purposes. Given that the non-targeted, positively charged H4G and GeneIn™ vectors enter the cells through binding and temporarily disrupting the cell membranes, it is important to investigate whether the cellular entry process results in significant damage to the membrane integrity. Here, we performed an LDH release assay which showed both H4G and Vanta-H4G having minimal impact on the ADSCs membrane integrity (Figure 5G). This minimal disturbance could be attributed to the low surface positive charge associated with nanoparticles formed through complexation of pEGFP with either H4G or Vanta-H4G. The substantial release of LDH enzyme after transfection of the cells with GeneIn™ was somewhat expected as it bears a significantly high surface positive charge (see Figure 1F).
At this stage, we also carefully examined the morphology of the ADSCs by a light microscope to ensure that the selected vectors did not induce significant changes to the cells’ morphology. The observed pictures clearly show the deleterious effects of certain vector concentrations on the ADSCs, resulting in shrinkage and lysis of the cells. The cell morphology study also confirmed that our selected vectors did not alter the morphology of ADSCs as witnessed by the maintenance of their spindle-like shapes (Supporting Figures 8 and 9).
Evaluation of vectors’ impact on micronuclei formation (genotoxicity) and gene dysregulation
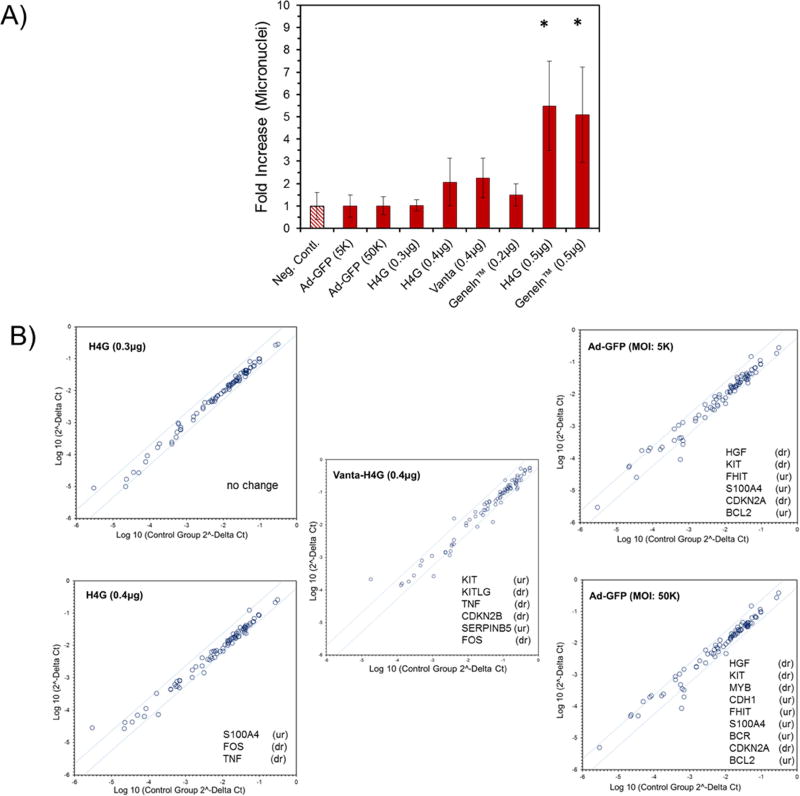
In addition to the tests that evaluate the somatic damages to stem cells during and post transfection such as LDH release and cell proliferation assays, it is also critically important to investigate the potential aberrations to the genome of the stem cells. In recent years, the need for evaluation of genotoxicity of gene delivery systems has been highlighted in several published articles [41–43]. Furthermore, the US Food and Drug Administration and International Conference on Harmonization in a published online record (https://www.fda.gov/downloads/drugs/guidances/ucm074931.pdf), recommend researchers and industries to report a genosafety profile of pharmaceutical formulation ingredients including nanocarriers [44]. Characterizing micronuclei formation requires an in vitro assay that uses the generation of nuclear blebs and micronuclei in the cytoplasm of interphase cells as an approximation of the cell’s genetic instability upon exposure to the reagents. Here, we adapted a flow cytometry-based method that could help quantitatively measure the micronuclei formation in transfected cells. From the efficiency/toxicity studies explained above, we identified that the H4G (0.4 and 0.5 µg pDNA) and Vanta-H4G (0.4 µg pDNA) are the most suitable vectors for ADSC transfection. To examine their genotoxicity, ADSCs were transfected with these vectors and the percentages of micronuclei formation were determined. For the negative control, we used the H4G vector carrying 0.3µg pDNA and as the positive control, we used GeneIn™ carrying 0.5µg of pDNA. Ad-GFP (MOI: 5K and 50K), which bears a negative surface charge and transfects ADSCs via CAR, was also used as a negative control. The selection of the vector controls was based on the data presented in Figure 5, which shows high toxicity for GeneIn™ (0.5 µg pDNA) and low toxicity for H4G (0.3µg pDNA) and Ad-GFP. Bryce et al. (2007), previously established that a genotoxic substance would increase the percentage of micronuclei by at least three folds higher than the untreated control group [24]. Based on this guideline, the results of this study showed that H4G (0.5µg pDNA) and GeneIn™ (0.5µg pDNA) produced significantly higher numbers of micronuclei in transfected ADSCs. Therefore, both vectors were considered genotoxic (*t-test, p<0.05), while all other vectors were non-genotoxic (p>0.05) (Figure 6A). The result of this study helped us eliminate H4G (0.5 µg) from the selected vectors despite the fact that the LDH release assay, WST-1 assay, and cell morphology studies had shown that it was acceptable. It was also very interesting to observe that the GeneIn™ carrying 0.2 µg pDNA did not show any significant genotoxicity despite the previous observations showing that it had some somatic toxicity.
Figure 6.
A) Evaluation of the impact of vectors on the formation of micronuclei in transfected ADSCs. The percentage of micronuclei in untransfected cells is normalized to a one-fold increase and is considered as the negative control. B) PCR microarray analysis of the dysregulated genes in cells transfected with H4G (0.3 and 0.4 µg pEGFP), Vanta-H4G (0.4 µg pEGFP) and Ad-GFP (MOI: 5K and 50K). Only the upregulated (ur) and downregulated (dr) genes are mentioned in each panel.
We further examined the effect of DBVs on up/down regulation of genes in ADSCs. Ideally, it is preferred not to observe any significant gene dysregulation. At post transfection, we sorted out the strong GFP-positive cells, reseeded them, and performed a PCR microarray assay to examine the extent of gene dysregulation in the transfected cells. As shown in Figure 6B, we did not observe any significant change in the genetic pathways of the cells that were transfected with the H4G vector carrying 0.3 µg of pDNA. ADSCs that were transfected with H4G (0.4 µg pDNA) showed dysregulation in three genes out of the 84 tested. Interestingly, in this group, the S100A4 tumor suppressor gene was upregulated, whereas the FOS/TNF gene pathway was downregulated. The FOS gene is a transcription factor, whose expression is most often positively correlated with TNF expression. It has been reported that the up-regulation of the FOS/TNF pathway could increase the probability of MSC transformation toward malignancy [45]. Considering that there is no negative report on downregulation of FOS/TNF pathway, it may be safe to conclude that such downregulation may reduce the probability of malignant transformation. Similar to the cells in the H4G-treated group, cells that were transfected with Vanta-H4G (0.4 µg pDNA) also exhibited downregulation of the FOS/TNF pathway. Additionally, it was observed that another signaling pathway; i.e., KITLG/KIT, was upregulated in this group. Both VEGF/VEGFR and KITLG/KIT signaling pathways play an essential role in stem cell hematopoiesis and new blood vessel formation [46, 47]. Reports also indicate that these pathways share multiple gene cross-talks during signal transduction [47, 48]. Because the Vanta-H4G complex competes with VEGF in the media for VEGFR-1 binding, the ADSC turns on the alternative KITLG/KIT pathway to adapt to the change. The upregulation of the KITLG/KIT pathway is a genetic level evidence supporting the cellular entry of the Vanta-H4G/pDNA nanoparticles through VEGFR binding. The PCR microarray data also showed that few genes were dysregulated within the adenovirus-transduced groups and there is a direct correlation between the MOI and number of dysregulated genes. Furthermore, it was noticeable that in addition to the upregulated BCL2 gene (anti-apoptotic), the genes that promote cell division and growth were downregulated (HGF, KIT, and MYB). The combination of these changes points to the potential toxicity of Ad-GFP to stem cells at such high MOIs. This observation provides genetic level evidence in support to our discussion of Figure 5F. Overall, the results of the genotoxicity assay and microarray analysis show that none of the selected vectors through the screening process had a significant detrimental effect on the genome of the transfected stem cells, validating our approach. For the complete list of dysregulated genes, please see (Supporting Table 1).
Evaluation of vectors’ impact on stem cell differentiation and surface biomarker expression
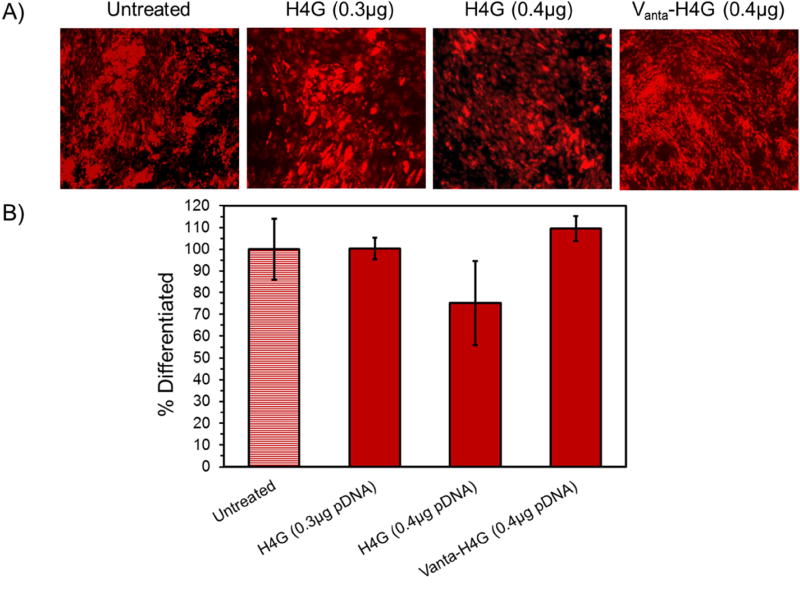
After verifying that H4G (0.3 and 0.4 µg pDNA) and Vanta-H4G (0.4 µg pDNA) groups were not genotoxic, we examined whether they, by any means, negatively affected the ADSCs potential for differentiation. This is important because the objective of most stem cell engineering studies is to ultimately differentiate them into a tissue. For this purpose, we first transfected the ADSCs with the above mentioned vectors using pEGFP, sorted out the ADSCs that were strongly positive in GFP expression, and then reseeded them for differentiation. Here, we sorted the strong GFP-positive cells because these cells received the maximum number of vector/pDNA nanocomplexes; thereby, demonstrating a higher probability of negative effects. The results of this study showed that none of the vectors negatively affected the ADSCs and the transfected cells could differentiate into adipocytes similar to that of the untreated cells (t-test, p>0.05) (Figure 7). Because Vanta-H4G (0.4 µg) appeared to be the most efficient and non-genotoxic construct, we transfected the ADSCs with this vector and examined their ability to differentiate into osteocytes as well. As expected, the transfected ADSCs could differentiate into osteocytes similar to the untransfected ADSCs (Supporting Figure 10).
Figure 7.
ADSC differentiation into adipocyte. A) Fluorescent microscopy images of the differentiated ADSCs. B) Bar chart showing the percentages of differentiated cells in each treated and untreated group using flow cytometry.
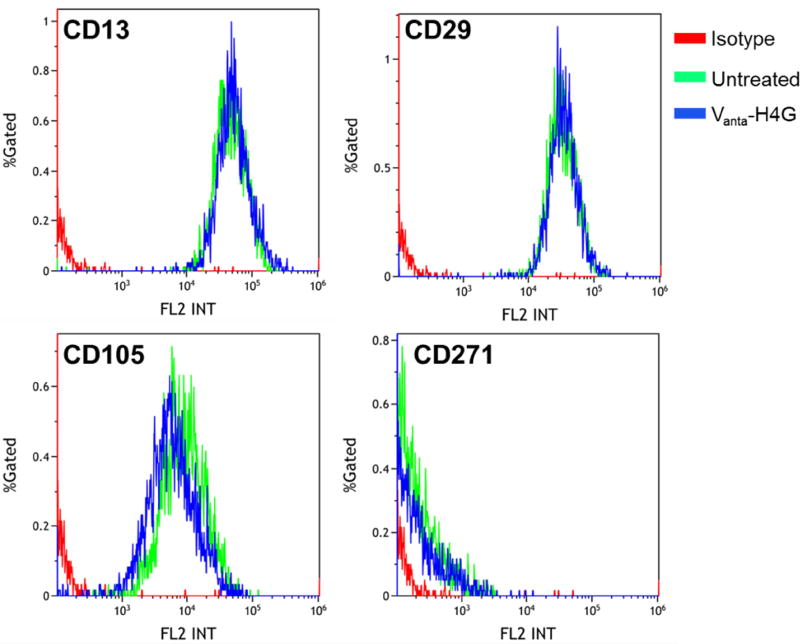
Furthermore, we evaluated the expression levels of a few typical and important ADSC surface biomarkers (i.e., CD13, CD29, and CD105) before and after transfection with Vanta-H4G 0.4 µg). This was to examine whether the vector had any negative impact on their expression levels. The results illustrated that the vector did not significantly alter the expression levels of the tested CD makers (Figure 8). In addition, we evaluated the expression of CD271 surface marker, which is not commonly present on the surface of ADSCs but is shown to upregulate in response to DNA damage [49]. The insignificant upregulation of CD271 in transfected ADSCs is another supporting data, which confirms that Vanta-H4G (0.4 µg) did not have a significant genotoxic effect. These observations demonstrate that the developed DBVs could indeed be used for efficient and safe genetic modification of ADSCs without any negative effect on their differentiation into the desired tissue.
Figure 8.
Expression of surface markers CD13, CD29, CD105 and CD271 before and after transfection of ADSCs with Vanta-H4G (0.4 µg).
Conclusions
The goal of this research was to develop a vector that is not only efficient in stem cell transfection, but also has the ability to maintain such efficiencies without inducing somatic or genetic toxicity. Overall, the efficiency and toxicity data show that among the developed DBVs, the VEGFR-1 targeted Vanta-H4G is not only the most efficient vector for ADSC transfection, but also one without any significant negative impact on physical integrity, metabolic activity, genetic composition, or cell differentiation. Considering that the adenoviral vector, which is also a targeted vector, could efficiently transfect stem cells with minimal acute toxicity in ADSCs, it may be safe to conclude that the best approach toward transfecting stem cells efficiently and safely is via receptor targeting rather than entry through the cellular membrane. In comparison to the tested commercially available non-viral and adenoviral vectors, the developed DBV appears to be the most efficient vector that meets the strict standards of safety for MSC engineering.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Biomedical Imaging and Bioengineering (R21EB016792) and National Cancer Institute (R01CA175318) to A. Hatefi. This research was also supported by the Shared Resources of the Rutgers Cancer Institute of New Jersey (P30CA072720).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
None
Author contributions
X.C. contributed to manuscript writing, vector production, ADSC characterization (cell cycle and VEGFR expression), cell transfection, transfection efficiency and toxicity measurements by WST-1 and LDH release assay, MOI measurements, CD biomarker expression and PCR microarray analysis.
A.N. contributed to manuscript writing, nanoparticle formulation, method development for quantitative genotoxicity assay and analysis and ADSC differentiation study.
N.P. contributed to vector production and purification, cell toxicity study by WST-1 assay, cell transfection and cell targeting study.
F.S.N. contributed to nanoparticle characterization in terms of size and charge.
A.H. (PI) contributed to the idea development, experimental design, data organization and analysis, manuscript writing and editing, project management and manuscript submission.
References
- 1.Shah K. Mesenchymal stem cells engineered for cancer therapy. Adv Drug Deliv Rev. 2012;64(8):739–48. doi: 10.1016/j.addr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamada H, Kobune M, Nakamura K, Kawano Y, Kato K, Honmou O, Houkin K, Matsunaga T, Niitsu Y. Mesenchymal stem cells (MSC) as therapeutic cytoreagents for gene therapy. Cancer Sci. 2005;96(3):149–56. doi: 10.1111/j.1349-7006.2005.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhl M, Weiler M, Wick W, Jacobs AH, Weller M, Herrlinger U. Migratory neural stem cells for improved thymidine kinase-based gene therapy of malignant gliomas. Biochem Biophys Res Commun. 2005;328(1):125–9. doi: 10.1016/j.bbrc.2004.12.164. [DOI] [PubMed] [Google Scholar]
- 4.Kucerova L, Altanerova V, Matuskova M, Tyciakova S, Altaner C. Adipose tissue-derived human mesenchymal stem cells mediated prodrug cancer gene therapy. Cancer Res. 2007;67(13):6304–13. doi: 10.1158/0008-5472.CAN-06-4024. [DOI] [PubMed] [Google Scholar]
- 5.Cavarretta IT, Altanerova V, Matuskova M, Kucerova L, Culig Z, Altaner C. Adipose tissue-derived mesenchymal stem cells expressing prodrug-converting enzyme inhibit human prostate tumor growth. Mol Ther. 2010;18(1):223–31. doi: 10.1038/mt.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noyan F, Diez IA, Hapke M, Klein C, Dewey RA. Induced transgene expression for the treatment of solid tumors by hematopoietic stem cell-based gene therapy. Cancer Gene Ther. 2012;19(5):352–7. doi: 10.1038/cgt.2012.8. [DOI] [PubMed] [Google Scholar]
- 7.Nouri FS, Wang X, Hatefi A. Genetically engineered theranostic mesenchymal stem cells for the evaluation of the anticancer efficacy of enzyme/prodrug systems. J Control Release. 2015;200:179–87. doi: 10.1016/j.jconrel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aboody KS, Najbauer J, Danks MK. Stem and progenitor cell-mediated tumor selective gene therapy. Gene Ther. 2008;15(10):739–52. doi: 10.1038/gt.2008.41. [DOI] [PubMed] [Google Scholar]
- 9.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4(5):346–58. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 10.Kim JA, Cho K, Shin MS, Lee WG, Jung N, Chung C, Chang JK. A novel electroporation method using a capillary and wire-type electrode. Biosens Bioelectron. 2008;23(9):1353–60. doi: 10.1016/j.bios.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Hubbs AF, Mercer RR, Benkovic SA, Harkema J, Sriram K, Schwegler-Berry D, Goravanahally MP, Nurkiewicz TR, Castranova V, Sargent LM. Nanotoxicology--a pathologist's perspective. Toxicol Pathol. 2011;39(2):301–24. doi: 10.1177/0192623310390705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norppa H, Catalan J, Falck G, Hannukainen K, Siivola K, Savolainen K. Nano-specific genotoxic effects. J Biomed Nanotechnol. 2011;7(1):19. doi: 10.1166/jbn.2011.1179. [DOI] [PubMed] [Google Scholar]
- 13.Canine BF, Wang Y, Hatefi A. Biosynthesis and characterization of a novel genetically engineered polymer for targeted gene transfer to cancer cells. J Control Release. 2009;138(3):188–96. doi: 10.1016/j.jconrel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangipudi SS, Canine BF, Wang Y, Hatefi A. Development of a genetically engineered biomimetic vector for targeted gene transfer to breast cancer cells. Mol Pharm. 2009;6(4):1100–9. doi: 10.1021/mp800251x. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Mangipudi SS, Canine BF, Hatefi A. A designer biomimetic vector with a chimeric architecture for targeted gene transfer. J Control Release. 2009;137(1):46–53. doi: 10.1016/j.jconrel.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deshayes S, Morris MC, Divita G, Heitz F. Interactions of amphipathic carrier peptides with membrane components in relation with their ability to deliver therapeutics. J Pept Sci. 2006;12(12):758–65. doi: 10.1002/psc.810. [DOI] [PubMed] [Google Scholar]
- 17.Deshayes S, Gerbal-Chaloin S, Morris MC, Aldrian-Herrada G, Charnet P, Divita G, Heitz F. On the mechanism of non-endosomial peptide-mediated cellular delivery of nucleic acids. Biochim Biophys Acta. 2004;1667(2):141–7. doi: 10.1016/j.bbamem.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 18.An JJ, Lee YP, Kim SY, Lee SH, Lee MJ, Jeong MS, Kim DW, Jang SH, Yoo KY, Won MH, Kang TC, Kwon OS, Cho SW, Lee KS, Park J, Eum WS, Choi SY. Transduced human PEP-1-heat shock protein 27 efficiently protects against brain ischemic insult. FEBS J. 2008;275(6):1296–308. doi: 10.1111/j.1742-4658.2008.06291.x. [DOI] [PubMed] [Google Scholar]
- 19.Muller J, Triebus J, Kretzschmar I, Volkmer R, Boisguerin P. The agony of choice: how to find a suitable CPP for cargo delivery. J Pept Sci. 2012;18(5):293–301. doi: 10.1002/psc.2396. [DOI] [PubMed] [Google Scholar]
- 20.Milletti F. Cell-penetrating peptides: classes, origin, and current landscape. Drug Discov Today. 2012;17(15–16):850–60. doi: 10.1016/j.drudis.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Karjoo Z, McCarthy HO, Patel P, Nouri FS, Hatefi A. Systematic engineering of uniform, highly efficient, targeted and shielded viral-mimetic nanoparticles. Small. 2013;9(16):2774–83. doi: 10.1002/smll.201300077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nouri FS, Wang X, Dorrani M, Karjoo Z, Hatefi A. A recombinant biopolymeric platform for reliable evaluation of the activity of pH-responsive amphiphile fusogenic peptides. Biomacromolecules. 2013;14(6):2033–40. doi: 10.1021/bm400380s. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Nomani A, Patel N, Hatefi A. Production of low-expressing recombinant cationic biopolymers with high purity. Protein Expr Purif. 2017;134:11–17. doi: 10.1016/j.pep.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryce SM, Bemis JC, Avlasevich SL, Dertinger SD. In vitro micronucleus assay scored by flow cytometry provides a comprehensive evaluation of cytogenetic damage and cytotoxicity. Mutat Res. 2007;630(1–2):78–91. doi: 10.1016/j.mrgentox.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul RW, Weisser KE, Loomis A, Sloane DL, LaFoe D, Atkinson EM, Overell RW. Gene transfer using a novel fusion protein, GAL4/invasin. Hum Gene Ther. 1997;8(10):1253–62. doi: 10.1089/hum.1997.8.10-1253. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy HO, Wang Y, Mangipudi SS, Hatefi A. Advances with the use of bio-inspired vectors towards creation of artificial viruses. Expert Opin Drug Deliv. 2010;7(4):497–512. doi: 10.1517/17425240903579989. [DOI] [PubMed] [Google Scholar]
- 27.Canine BF, Wang Y, Ouyang W, Hatefi A. Development of targeted recombinant polymers that can deliver siRNA to the cytoplasm and plasmid DNA to the cell nucleus. J Control Release. 2011;151(1):95–101. doi: 10.1016/j.jconrel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Canine BF, Hatefi A. HSV-TK/GCV cancer suicide gene therapy by a designed recombinant multifunctional vector. Nanomedicine. 2011;7(2):193–200. doi: 10.1016/j.nano.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Mangipudi SS, Canine BF, Hatefi A. A designer biomimetic vector with a chimeric architecture for targeted gene transfer. J Control Release. 2009;137:46–53. doi: 10.1016/j.jconrel.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balicki D, Putnam CD, Scaria PV, Beutler E. Structure and function correlation in histone H2A peptide-mediated gene transfer. Proc Natl Acad Sci U S A. 2002;99(11):7467–71. doi: 10.1073/pnas.102168299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Andrea LD, Iaccarino G, Fattorusso R, Sorriento D, Carannante C, Capasso D, Trimarco B, Pedone C. Targeting angiogenesis: structural characterization and biological properties of a de novo engineered VEGF mimicking peptide. Proc Natl Acad Sci U S A. 2005;102(40):14215–20. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Mousawi M, Tchistiakova L, Yurchenko L, Pietrzynski G, Moreno M, Stanimirovic D, Ahmad D, Alakhov V. A vascular endothelial growth factor high affinity receptor 1-specific peptide with antiangiogenic activity identified using a phage display peptide library. J Biol Chem. 2003;278(47):46681–91. doi: 10.1074/jbc.M308681200. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolanos-Garcia VM, Davies OR. Structural analysis and classification of native proteins from E. coli commonly co-purified by immobilised metal affinity chromatography. Biochim Biophys Acta. 2006;1760(9):1304–13. doi: 10.1016/j.bbagen.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 35.Nouri FS, Wang X, Chen X, Hatefi A. Reducing the Visibility of the Vector/DNA Nanocomplexes to the Immune System by Elastin-Like Peptides. Pharm Res. 2015;32(9):3018–28. doi: 10.1007/s11095-015-1683-5. [DOI] [PubMed] [Google Scholar]
- 36.Shah V, Taratula O, Garbuzenko OB, Patil ML, Savla R, Zhang M, Minko T. Genotoxicity of Different Nanocarriers: Possible Modifications for the Delivery of Nucleic Acids. Curr Drug Discov Technol. 2012 [PMC free article] [PubMed] [Google Scholar]
- 37.Hamma-Kourbali Y, Starzec A, Vassy R, Martin A, Kraemer M, Perret G, Crepin M. Carboxymethyl benzylamide dextran inhibits angiogenesis and growth of VEGF-overexpressing human epidermoid carcinoma xenograft in nude mice. Br J Cancer. 2003;89(1):215–21. doi: 10.1038/sj.bjc.6601029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mesti T, Savarin P, Triba MN, Le Moyec L, Ocvirk J, Banissi C, Carpentier AF. Metabolic impact of anti-angiogenic agents on U87 glioma cells. PLoS One. 2014;9(6):e99198. doi: 10.1371/journal.pone.0099198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lim SJ, Paeng JC, Kim SJ, Kim SY, Lee H, Moon DH. Enhanced expression of adenovirus-mediated sodium iodide symporter gene in MCF-7 breast cancer cells with retinoic acid treatment. J Nucl Med. 2007;48(3):398–404. [PubMed] [Google Scholar]
- 40.Jones CF, Grainger DW. In vitro assessments of nanomaterial toxicity. Adv Drug Deliv Rev. 2009;61(6):438–56. doi: 10.1016/j.addr.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parhamifar L, Andersen H, Wu L, Hall A, Hudzech D, Moghimi SM. Polycation-mediated integrated cell death processes. Adv Genet. 2014;88:353–98. doi: 10.1016/B978-0-12-800148-6.00012-2. [DOI] [PubMed] [Google Scholar]
- 42.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–5. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Adv Drug Deliv Rev. 2006;58(14):1523–31. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 44.Warheit DB, Donner EM. Rationale of genotoxicity testing of nanomaterials: regulatory requirements and appropriateness of available OECD test guidelines. Nanotoxicology. 2010;4:409–13. doi: 10.3109/17435390.2010.485704. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, Zhao Y, Liu Y, Akiyama K, Chen C, Qu C, Jin Y, Shi S. IFN-gamma and TNF-alpha synergistically induce mesenchymal stem cell impairment and tumorigenesis via NFkappaB signaling. Stem Cells. 2013;31(7):1383–95. doi: 10.1002/stem.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Broudy VC. Stem cell factor and hematopoiesis. Blood. 1997;90(4):1345–64. [PubMed] [Google Scholar]
- 47.Matsui J, Wakabayashi T, Asada M, Yoshimatsu K, Okada M. Stem cell factor/c-kit signaling promotes the survival, migration, and capillary tube formation of human umbilical vein endothelial cells. J Biol Chem. 2004;279(18):18600–7. doi: 10.1074/jbc.M311643200. [DOI] [PubMed] [Google Scholar]
- 48.Litz J, Krystal GW. Imatinib inhibits c-Kit-induced hypoxia-inducible factor-1alpha activity and vascular endothelial growth factor expression in small cell lung cancer cells. Mol Cancer Ther. 2006;5(6):1415–22. doi: 10.1158/1535-7163.MCT-05-0503. [DOI] [PubMed] [Google Scholar]
- 49.Redmer T, Walz I, Klinger B, Khouja S, Welte Y, Schafer R, Regenbrecht C. The role of the cancer stem cell marker CD271 in DNA damage response and drug resistance of melanoma cells. Oncogenesis. 2017;6(1):e291. doi: 10.1038/oncsis.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.